Genome-Wide Association Studies (GWAS) and related whole genome variant analyses have provided very compelling insights into the pathobiology of many diseases as well as the complexities surrounding general phenotypic expression. Unfortunately, they have not led to insights resulting in substantive improvements in health care. The paper by Khera et al. suggests, however, that we may be reaching a time when individual whole genome variant profiling can be used to assess individual disease risks in an impactful way and as part of routine health care. The results of the study by Khera et al. raise a number of very important questions about the delivery of genetic risk information in health care settings, however, and, along with the results of a number of other recent papers, are very likely to motivate additional studies.
Essentially, Khera et al. consider the use of disease risk scores computed from individual whole genome genetic variant profiles to identify subgroups of individuals that have a clinically significant risk of developing 5 diseases: coronary artery disease (CAD), atrial fibrillation (AFIB), type 2 diabetes (T2D), inflammatory bowel disease (IBD), and breast cancer (BC). They leveraged massive data sets and sophisticated analytical methods to construct ‘genome-wide polygenic risk scores’ or ‘GPS’ for each individual for each disease. The accuracy of these GPS in differentiating diseased and non-diseased individuals was assessed with both training and independent validation samples to ensure their reliability. They found that a large fraction of the individuals assessed with the GPS for each disease could be assigned a risk of developing that disease that was as high as the risk associated with individual genetic variants known to causally influence rare, monogenic forms of the disease that are often routinely considered in clinical diagnostic settings.
Khera et al. also suggested that preventive interventions that have proven to be effective for most of the conditions they studied could be deployed or suggested to high risk individuals based on their GPS. For example, statins and lifestyle change recommendations could be provided to individuals at elevated risk for CAD. In addition, they found that ~20% of all the individuals they studied had a 3-fold or greater risk for at least one of the five diseases, and that the number of individuals deemed at high risk from GPS analyses is, for at least CAD, 20-fold greater than those deemed at high risk via monogenic disease variant analyses. They concluded that there is little reason to question the potential utility of GPS and other whole genome variant analyses in health care settings.
There are some scientific and social impediments to the adoption of GPS in routine health care settings, however. There are also some very exciting directions and opportunities involving the use of GPS in health management and maintenance initiatives. Scientifically, as acknowledged by Khera et al., the construction of GPS must be pursued in individuals exhibiting greater geo-ethnic and ancestral diversity in order to generalize their results to large urban communities.(1) In addition, to truly assess risk, longitudinal studies of individuals exhibiting variation in GPS values must be pursued in order to establish age-specific disease incidence rates (and standard errors on those rates as well) for different GPS levels.(2) Age-specific risks will not only inform health maintenance practices, but also put in perspective individual GPS risks; e.g., if an individual has a high type 1 diabetes (T1D) GPS, but is 70 years old and has not yet manifested T1D, then that individual’s risk of developing T1D going forward is likely to be minimal given that the peak age-of-onset of of T1D is 14 years of age. Finally, GPS should be integrated with other factors that may be predictive of risk to improve risk assessment calculations, although, as Khera et al. point out, many traditional risk factors correlate with GPS and hence may provide redundant information. Genetic risk information, however, which reflects lifetime risk or the ‘trait’ health status of an individual, is truly complementary to many biomarkers or assays to identify the presence of signs of specific disease pathologies (such as cholesterol level or image-based evaluations of a tissue or organ), as these biomarkers reflect an individual’s ‘state’ health status. Thus, evaluating early signs of a condition using biomarkers reflecting an individual’s health state at a time for conditions for which the individual has a high lifetime or trait risk simply makes sense. It also reinforces the notion that genetic assessment is simply a part of a much greater preventive medicine practice whole for any given patient.(3, 4)
Social impediments to the use of GPS in routine health care settings are also complex. For example, physicians need to be able to interpret and understand relevant GPS reports, and patients would have to acknowledge GPS-derived information and reporting as having utility without suffering from often unwarranted fatalistic fears about their health or fears of discrimination at their workplace or by health insurers based on their GPS profiles.(5) In addition, payer/payee exchange and pricing standards would have to be established within the currently very complicated and burdensome health insurance climate, which may not be trivial without additional proof that use of GPS will save the system money. Regulatory agencies will have to introduce quality control standards for computing GPS, which is not necessarily trivial for available monogenic disease, individual variant diagnostics, let alone evaluations involving hundreds of thousands, if not millions, of variants. Standards for vetting various GPS products before approving them will have to be established, although the use of ‘real world’ data may have great utility in this context.(6) Finally, integration into the care stream via appropriate clinical and information workflows, reporting and follow-up procedures, will need to occur, and this can be costly and disruptive to legacy ways of processing risk-related information in clinical care streams.
Consideration of GPS in clinical contexts also opens up new ways of thinking about health care. Since GPS are derived by interrogating variants across the genome for their contribution to disease susceptibility, one could evaluate an individual’s risk for any number of diseases by computing the appropriate GPS. Thus, in some sense, individual whole genome analysis is a gift to lifetime disease risk assessment that can keep on giving. Being able to assess risk for many diseases and conditions simultaneously is of crucial importance in managing or seeking to maintain a patient’s health over a long period of time, if not a lifetime. This is in distinction to procedures used for making a diagnosis during a health crisis or only looking for symptoms and not necessarily evidence for future susceptibility to disease, during a routine health exam. Thus, treating a patient for one condition, knowing that he or she is susceptible to another, may inform needed treatments and prevention decisions.
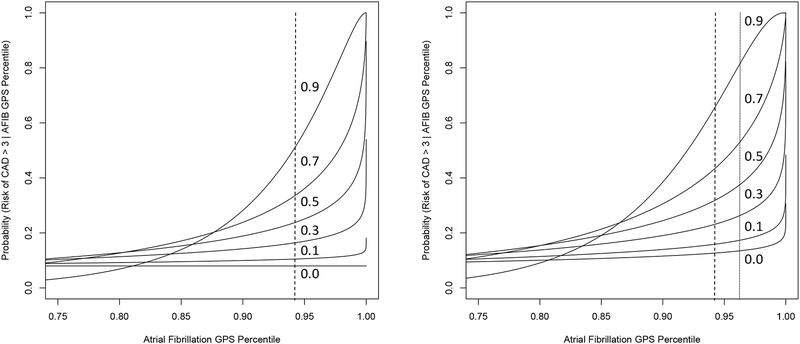
In addition, it is well known that many diseases exhibit genetic correlations; that is, the sets of genes and genetic variants associated with susceptibility to different diseases overlap, often substantially.(7–9) This has important implications. For example, two diseases with different prevalence rates but with a strong genetic correlation suggests that there may be mitigating factors for the disease with lower prevalence but not the other, despite the correlation, which could help identify both very specific and more general interventions for them. In addition, it may be that aspects of the pathologies associated with each disease could be governed or influenced by the genetic variants that do not overlap in the GPS for each, which could potentially lead to intervention targets. Finally, if diseases exhibit variation in their genetic correlations with most having non-zero genetic correlations, then insight into multiple disease risks are inevitable. This is exemplified in Figure 1, which uses risk percentiles from their CAD, AFIB and T2D GPS information from Khera et al as well as assumptions about their genetic correlations. Sensitivity to the implications of correlations among GPS for different diseases is of crucial importance as health care systems move away from merely dealing with a patient’s disease at the time towards preventing it in the first place. In addition, managing an individual as whole instead of as a set of isolated tissues and organs waiting to fail will require comprehensive health risk evaluations of the type envisioned.
Figure 1.
Conditional probability that an individual possesses a risk of coronary artery disease (CAD) at least 3 times as high as the average person (y-axis) given that the individual possesses a GPS score for atrial fibrillation (AFIB) in the percentiles reflected on the x axis. Left panel: the inset numbers give the strength of the genetic correlation between CAD and AFIB. The vertical dashed line is the percentile of the AFIB GPS distribution associated with a 3-fold a greater risk for AFIB. Right panel: same as the left panel except a third disease, type II diabetes (T2D), is included in the calculations in which it is assumed that T2D has genetic correlations of 0.2 and 0.1 with CAD and AFIB, respectively, and that the GPS percentile for T2D for the individual is the same for AFIB (as given on the x-axis). Note that the shift in the curves in right panel simply reflects the information about CAD risk provided by information about risk of diabetes even though it has weak assumed genetic correlations with CAD and AFIB. All calculations assumed multivariate normality of GPS values. The R package ‘condMVNorm’ was used to compute relevant conditional probabilities.(11)
There are other potential uses of GPS outside of routine clinical care, such as using GPS to enrich preventive clinical trials for individuals susceptible to a disease and thereby increase the power of the trials,(10) or coupling GPS to other genetic variant analyses, such as drug metabolizing enzyme profiling or nutrient use deficiency analyses, in order to craft even more individualized, and perhaps optimized, preventive and health maintenance procedures. Given the enthusiasm contemporary researchers have shown for genetics, there is little reason to doubt that the paper by Khera et al. will be the first of many that will expose the utility of genetic findings not only in drug target and clinical development, but also in routine health care settings as, e.g., simple prognostic tools – two activities that have been decades and hundreds of millions, if not billions, of dollars in the making.
References
- 1.Marigorta UM, Rodriguez JA, Gibson G, Navarro A. Replicability and Prediction: Lessons and Challenges from GWAS. Trends Genet. 2018;34(7):504–17. doi: 10.1016/j.tig.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, Flanders WD, Moonesinghe R, Ioannidis JP, Guessous I, Khoury MJ. Using lifetime risk estimates in personal genomic profiles: estimation of uncertainty. Am J Hum Genet. 2009;85(6):786–800. doi: 10.1016/j.ajhg.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel CJ, Sivadas A, Tabassum R, Preeprem T, Zhao J, Arafat D, Chen R, Morgan AA, Martin GS, Brigham KL, Butte AJ, Gibson G. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5(6):58. doi: 10.1186/gm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schork NJ. Genetic parts to a preventive medicine whole. Genome Med. 2013;5(6):54. doi: 10.1186/gm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364(6):524–34. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen TN, Trocio J, Kowal S, Ferrufino CP, Munakata J, South D. Leveraging Real-World Evidence in Disease-Management Decision-Making with a Total Cost of Care Estimator. Am Health Drug Benefits. 2016;9(9):475–85. [PMC free article] [PubMed] [Google Scholar]
- 7.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, Escott-Price V, Falcone GJ, Gormley P, Malik R, Patsopoulos NA, Ripke S, Wei Z, Yu D, Lee PH, Turley P, Grenier-Boley B, Chouraki V, Kamatani Y, Berr C, Letenneur L, Hannequin D, Amouyel P, Boland A, Deleuze JF, Duron E, Vardarajan BN, Reitz C, Goate AM, Huentelman MJ, Kamboh MI, Larson EB, Rogaeva E, St George-Hyslop P, Hakonarson H, Kukull WA, Farrer LA, Barnes LL, Beach TG, Demirci FY, Head E, Hulette CM, Jicha GA, Kauwe JSK, Kaye JA, Leverenz JB, Levey AI, Lieberman AP, Pankratz VS, Poon WW, Quinn JF, Saykin AJ, Schneider LS, Smith AG, Sonnen JA, Stern RA, Van Deerlin VM, Van Eldik LJ, Harold D, Russo G, Rubinsztein DC, Bayer A, Tsolaki M, Proitsi P, Fox NC, Hampel H, Owen MJ, Mead S, Passmore P, Morgan K, Nothen MM, Rossor M, Lupton MK, Hoffmann P, Kornhuber J, Lawlor B, McQuillin A, Al-Chalabi A, Bis JC, Ruiz A, Boada M, Seshadri S, Beiser A, Rice K, van der Lee SJ, De Jager PL, Geschwind DH, Riemenschneider M, Riedel-Heller S, Rotter JI, Ransmayr G, Hyman BT, Cruchaga C, Alegret M, Winsvold B, Palta P, Farh KH, Cuenca-Leon E, Furlotte N, Kurth T, Ligthart L, Terwindt GM, Freilinger T, Ran C, Gordon SD, Borck G, Adams HHH, Lehtimaki T, Wedenoja J, Buring JE, Schurks M, Hrafnsdottir M, Hottenga JJ, Penninx B, Artto V, Kaunisto M, Vepsalainen S, Martin NG, Montgomery GW, Kurki MI, Hamalainen E, Huang H, Huang J, Sandor C, Webber C, Muller-Myhsok B, Schreiber S, Salomaa V, Loehrer E, Gobel H, Macaya A, Pozo-Rosich P, Hansen T, Werge T, Kaprio J, Metspalu A, Kubisch C, Ferrari MD, Belin AC, van den Maagdenberg A, Zwart JA, Boomsma D, Eriksson N, Olesen J, Chasman DI, Nyholt DR, Avbersek A, Baum L, Berkovic S, Bradfield J, Buono R, Catarino CB, Cossette P, De Jonghe P, Depondt C, Dlugos D, Ferraro TN, French J, Hjalgrim H, Jamnadas-Khoda J, Kalviainen R, Kunz WS, Lerche H, Leu C, Lindhout D, Lo W, Lowenstein D, McCormack M, Moller RS, Molloy A, Ng PW, Oliver K, Privitera M, Radtke R, Ruppert AK, Sander T, Schachter S, Schankin C, Scheffer I, Schoch S, Sisodiya SM, Smith P, Sperling M, Striano P, Surges R, Thomas GN, Visscher F, Whelan CD, Zara F, Heinzen EL, Marson A, Becker F, Stroink H, Zimprich F, Gasser T, Gibbs R, Heutink P, Martinez M, Morris HR, Sharma M, Ryten M, Mok KY, Pulit S, Bevan S, Holliday E, Attia J, Battey T, Boncoraglio G, Thijs V, Chen WM, Mitchell B, Rothwell P, Sharma P, Sudlow C, Vicente A, Markus H, Kourkoulis C, Pera J, Raffeld M, Silliman S, Boraska Perica V, Thornton LM, Huckins LM, William Rayner N, Lewis CM, Gratacos M, Rybakowski F, Keski-Rahkonen A, Raevuori A, Hudson JI, Reichborn-Kjennerud T, Monteleone P, Karwautz A, Mannik K, Baker JH, O’Toole JK, Trace SE, Davis OSP, Helder SG, Ehrlich S, Herpertz-Dahlmann B, Danner UN, van Elburg AA, Clementi M, Forzan M, Docampo E, Lissowska J, Hauser J, Tortorella A, Maj M, Gonidakis F, Tziouvas K, Papezova H, Yilmaz Z, Wagner G, Cohen-Woods S, Herms S, Julia A, Rabionet R, Dick DM, Ripatti S, Andreassen OA, Espeseth T, Lundervold AJ, Steen VM, Pinto D, Scherer SW, Aschauer H, Schosser A, Alfredsson L, Padyukov L, Halmi KA, Mitchell J, Strober M, Bergen AW, Kaye W, Szatkiewicz JP, Cormand B, Ramos-Quiroga JA, Sanchez-Mora C, Ribases M, Casas M, Hervas A, Arranz MJ, Haavik J, Zayats T, Johansson S, Williams N, Dempfle A, Rothenberger A, Kuntsi J, Oades RD, Banaschewski T, Franke B, Buitelaar JK, Arias Vasquez A, Doyle AE, Reif A, Lesch KP, Freitag C, Rivero O, Palmason H, Romanos M, Langley K, Rietschel M, Witt SH, Dalsgaard S, Borglum AD, Waldman I, Wilmot B, Molly N, Bau CHD, Crosbie J, Schachar R, Loo SK, McGough JJ, Grevet EH, Medland SE, Robinson E, Weiss LA, Bacchelli E, Bailey A, Bal V, Battaglia A, Betancur C, Bolton P, Cantor R, Celestino-Soper P, Dawson G, De Rubeis S, Duque F, Green A, Klauck SM, Leboyer M, Levitt P, Maestrini E, Mane S, De-Luca DM, Parr J, Regan R, Reichenberg A, Sandin S, Vorstman J, Wassink T, Wijsman E, Cook E, Santangelo S, Delorme R, Roge B, Magalhaes T, Arking D, Schulze TG, Thompson RC, Strohmaier J, Matthews K, Melle I, Morris D, Blackwood D, McIntosh A, Bergen SE, Schalling M, Jamain S, Maaser A, Fischer SB, Reinbold CS, Fullerton JM, Guzman-Parra J, Mayoral F, Schofield PR, Cichon S, Muhleisen TW, Degenhardt F, Schumacher J, Bauer M, Mitchell PB, Gershon ES, Rice J, Potash JB, Zandi PP, Craddock N, Ferrier IN, Alda M, Rouleau GA, Turecki G, Ophoff R, Pato C, Anjorin A, Stahl E, Leber M, Czerski PM, Cruceanu C, Jones IR, Posthuma D, Andlauer TFM, Forstner AJ, Streit F, Baune BT, Air T, Sinnamon G, Wray NR, MacIntyre DJ, Porteous D, Homuth G, Rivera M, Grove J, Middeldorp CM, Hickie I, Pergadia M, Mehta D, Smit JH, Jansen R, de Geus E, Dunn E, Li QS, Nauck M, Schoevers RA, Beekman AT, Knowles JA, Viktorin A, Arnold P, Barr CL, Bedoya-Berrio G, Bienvenu OJ, Brentani H, Burton C, Camarena B, Cappi C, Cath D, Cavallini M, Cusi D, Darrow S, Denys D, Derks EM, Dietrich A, Fernandez T, Figee M, Freimer N, Gerber G, Grados M, Greenberg E, Hanna GL, Hartmann A, Hirschtritt ME, Hoekstra PJ, Huang A, Huyser C, Illmann C, Jenike M, Kuperman S, Leventhal B, Lochner C, Lyon GJ, Macciardi F, Madruga-Garrido M, Malaty IA, Maras A, McGrath L, Miguel EC, Mir P, Nestadt G, Nicolini H, Okun MS, Pakstis A, Paschou P, Piacentini J, Pittenger C, Plessen K, Ramensky V, Ramos EM, Reus V, Richter MA, Riddle MA, Robertson MM, Roessner V, Rosario M, Samuels JF, Sandor P, Stein DJ, Tsetsos F, Van Nieuwerburgh F, Weatherall S, Wendland JR, Wolanczyk T, Worbe Y, Zai G, Goes FS, McLaughlin N, Nestadt PS, Grabe HJ, Depienne C, Konkashbaev A, Lanzagorta N, Valencia-Duarte A, Bramon E, Buccola N, Cahn W, Cairns M, Chong SA, Cohen D, Crespo-Facorro B, Crowley J, Davidson M, DeLisi L, Dinan T, Donohoe G, Drapeau E, Duan J, Haan L, Hougaard D, Karachanak-Yankova S, Khrunin A, Klovins J, Kucinskas V, Lee Chee Keong J, Limborska S, Loughland C, Lonnqvist J, Maher B, Mattheisen M, McDonald C, Murphy KC, Nenadic I, van Os J, Pantelis C, Pato M, Petryshen T, Quested D, Roussos P, Sanders AR, Schall U, Schwab SG, Sim K, So HC, Stogmann E, Subramaniam M, Toncheva D, Waddington J, Walters J, Weiser M, Cheng W, Cloninger R, Curtis D, Gejman PV, Henskens F, Mattingsdal M, Oh SY, Scott R, Webb B, Breen G, Churchhouse C, Bulik CM, Daly M, Dichgans M, Faraone SV, Guerreiro R, Holmans P, Kendler KS, Koeleman B, Mathews CA, Price A, Scharf J, Sklar P, Williams J, Wood NW, Cotsapas C, Palotie A, Smoller JW, Sullivan P, Rosand J, Corvin A, Neale BM. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395). doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Bos SD, Harbo HF, Thompson WK, Schork AJ, Bettella F, Witoelar A, Lie BA, Li W, McEvoy LK, Djurovic S, Desikan RS, Dale AM, Andreassen OA. Genetic overlap between multiple sclerosis and several cardiovascular disease risk factors. Mult Scler. 2016;22(14):1783–93. doi: 10.1177/1352458516635873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama JS, Wang Y, Schork AJ, Thompson WK, Karch CM, Cruchaga C, McEvoy LK, Witoelar A, Chen CH, Holland D, Brewer JB, Franke A, Dillon WP, Wilson DM, Mukherjee P, Hess CP, Miller Z, Bonham LW, Shen J, Rabinovici GD, Rosen HJ, Miller BL, Hyman BT, Schellenberg GD, Karlsen TH, Andreassen OA, Dale AM, Desikan RS, Alzheimer’s Disease Neuroimaging I. Association Between Genetic Traits for Immune-Mediated Diseases and Alzheimer Disease. JAMA Neurol. 2016;73(6):691–7. doi: 10.1001/jamaneurol.2016.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schork NJ, Topol EJ. Genotype-based risk and pharmacogenetic sampling in clinical trials. J Biopharm Stat. 2010;20(2):315–33. doi: 10.1080/10543400903572779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Team RD. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2006. [Google Scholar]