Abstract
Genome-wide association studies have been successful in elucidating the genetic basis of colorectal cancer, but there remains unexplained variability in genetic risk. To identify new risk variants and to confirm reported associations, we conducted a genome-wide association study in 1,701 colorectal cancer cases and 14,082 cancer-free controls from the Finnish population. A total of 9,068,015 genetic variants were imputed and tested, and 30 promising variants were studied in additional 11,647 cases and 12,356 controls of European ancestry. The previously reported association between the single-nucleotide polymorphism rs992157 (2q35) and colorectal cancer was independently replicated (p=2.08×10−4; OR, 1.14; 95% CI, 1.06–1.23), and it was genome-wide significant in combined analysis (p=1.50×10−9; OR, 1.12; 95% CI, 1.08–1.16). Variants at 2q35, 6p21.2, 8q23.3, 8q24.21, 10q22.3, 10q24.2, 11q13.4, 11q23.1, 14q22.2, 15q13.3, 18q21.1, 20p12.3, and 20q13.33 were associated with colorectal cancer in the Finnish population (false discovery rate <0.1), but new risk loci were not found. These results replicate the effects of multiple loci on the risk of colorectal cancer and identify shared risk alleles between the Finnish population isolate and outbred populations.
Keywords: colorectal cancer, genetic predisposition to disease, genome-wide association study, single-nucleotide polymorphism
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and accounts for approximately 10% of global cancer incidence and mortality (http://globocan.iarc.fr/). Numerous genetic loci have been associated with CRC in genome-wide association studies (GWASs; https://www.ebi.ac.uk/gwas/), but much of its heritability remains unexplained, which limits personalized risk assessment and biological understanding of the disease.1,2 Discovery of new loci and replication of previously reported associations is thus important, and recent studies have continued to reveal novel CRC risk variants.3–7 The genetic architecture of CRC varies between populations, and studies in isolated founder populations can offer valuable insights into disease susceptibility.8
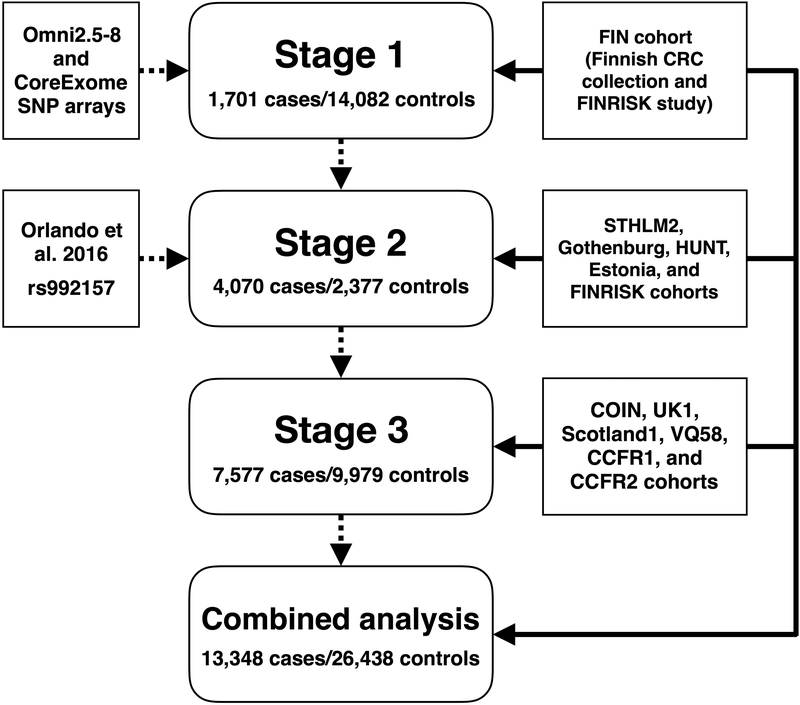
We conducted a GWAS of CRC in the Finnish population (the FIN cohort) using a large publicly available reference panel to impute genotypes and thus increase the odds of identifying disease-associated alleles across a wide range of allele frequencies.9 Thirty promising variants were investigated further in 11 European-ancestry studies (STHLM2, Gothenburg, HUNT, Estonia, FINRISK, COIN, UK1, Scotland1, VQ58, CCFR1, and CCFR2), adding to a total of 13,348 CRC cases and 26,438 controls.
In a recent meta-analysis of GWASs, the single-nucleotide polymorphism (SNP) rs992157 at 2q35, intronic to PNKD and TMBIM1, was found to be associated with CRC (p=3.15×10−8; odds ratio (OR), 1.10; 95% confidence interval (CI), 1.06–1.13).6 To replicate this finding, we genotyped and analyzed rs992157 in 4,439 CRC cases and 15,847 controls from five Northern European cohorts (STHLM2, Gothenburg, HUNT, Estonia, and a subset of the FIN cohort) that had not been previously studied for the association between rs992157 and CRC.
Materials and methods
This study was conducted in accordance with the Declaration of Helsinki and approved by the Finnish National Supervisory Authority for Welfare and Health, National Institute for Health and Welfare (THL/151/5.05.00/2017), and the Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS/408/13/03/03/09). We derived 1,627 cases with colorectal adenocarcinoma from the ongoing Finnish CRC collection and genotyped normal tissues (colorectal tissue or blood) with Illumina (San Diego, CA) HumanOmni2.5–8 SNP arrays.10,11 Illumina HumanCoreExome SNP array data for additional 91 CRC patients and 14,187 Finnish cancer-free controls were obtained from the National FINRISK Study (https://www.thl.fi/fi/web/thlfi-en/research-and-expertwork/population-studies/the-national-finrisk-study). Data on diagnosed cancers in the FINRISK study participants were collected from the Finnish Cancer Registry. PLINK v.1.90b3i (www.cog-genomics.org/plink/1.9/) was used for quality control.12 A total of 122 samples (17 genotyped with the HumanOmni2.5–8 array and 105 genotyped with HumanCoreExome array) were excluded on the basis of close relatedness (identity-by-descent coefficient >0.2), duplication, discordant sex information, or low genotyping rate. The FIN cohort consisted of the remaining 1,701 CRC cases and 14,082 cancer-free controls. By design, the HumanOmni2.5–8 SNP array contained 2,315,673 autosomal sites, 273,074 of which overlapped with the HumanCoreExome SNP array (https://support.illumina.com/downloads.html). Exclusion criteria for SNPs were genotyping rate <95%, excess homozygosity (frequency of rare homozygotes exceeding the frequency of heterozygotes, or any rare homozygous genotype with minor allele frequency (MAF) <2%), deviation from the Hardy-Weinberg equilibrium (p<1×10−8), differential missingness between genotyping batches (p<1×10−8), differential patterns of linkage disequilibrium (LD) in cases versus controls, and LD-based strand inconsistency. After quality control, 214,705 SNPs were pre-phased with SHAPEIT v2 (r790), and genotypes were imputed with a publicly available reference panel (https://imputation.sanger.ac.uk/; http://www.haplotype-reference-consortium.org/).9 Variants with low allele frequency (<0.4%) or low IMPUTE2 info score (<0.4) were excluded prior to association analysis. In stage 1, disease associations were tested with a linear mixed model (BOLT-LMM-inf; https://data.broadinstitute.org/alkesgroup/BOLT-LMM/), adjusting for log-transformed age and sex.13 A linear mixed model was used because it can control for population structure and cryptic relatedness.14 The age covariate was defined as age at CRC diagnosis in cases and age at right censoring (end of follow-up or death) in controls. An additive genetic model was assumed. The genomic inflation factor was estimated by dividing the observed median of the BOLT-LMM-inf test statistic by the median of the chi-squared distribution with one degree of freedom. The Benjamini-Hochberg method was used to adjust for false discovery rate.
In stage 2, the MassARRAY System by Agena Bioscience (San Diego, CA) was utilized at the Institute for Molecular Medicine Finland (FIMM) to genotype single-nucleotide variants in Nordic cohorts (STHLM2, 544 cases/541 controls; Gothenburg, 1,903 cases/258 controls; HUNT, 1,168 cases/1,147 controls; Estonia, 257 cases/259 controls; and FINRISK, 198 cases/172 controls), as well as 1,038 individuals from the FIN cohort who had also been genotyped with SNP arrays (925 with the HumanOmni2.5–8 array and 113 with the HumanCoreExome array). The STHLM2 cohort consisted of men who had been referred to prostate-specific antigen screening in Stockholm County, Sweden between 2010 and 2012; DNA samples were provided by the Karolinska Institute Biobank (http://ki.se/forskning/ki-biobank). The Gothenburg cohort was formed from CRC patients who had been operated at the Sahlgrenska University Hospital, Gothenburg, Sweden; DNA samples from cases and controls were provided by the Sahlgrenska Biobank (https://www.gothiaforum.com/sab). DNA samples from the HUNT cohort were provided by the Norwegian Nord-Trøndelag Health Study (HUNT) and Biobank (https://www.ntnu.edu/hunt). The Estonia cohort was derived from the sample collections of the Estonian Genome Center (www.geenivaramu.ee/en). The FINRISK cohort consisted of participants of the National FINRISK Study (198 CRC cases and 172 cancer-free controls) who had not been included in the FIN cohort due to unavailable SNP array data; DNA samples were provided by the THL Biobank, Finland (https://www.thl.fi/fi/web/thlfi-en/topics/information-packages/thl-biobank). When possible, cancer-free controls were matched to CRC cases on year of birth and sex. To assess imputation accuracy, squared Pearson correlation coefficients (r2) between IMPUTE2 genotype dosage and MassARRAY genotype were calculated.
To enable standard meta-analysis, data from the FIN cohort were reanalyzed by unconditional logistic regression under an additive genetic model, adjusting for sex, log-transformed age, and 10 principal components (SNPTEST v.2.5.2). In the MassARRAY-genotyped Nordic cohorts, unconditional logistic regression was applied using R v.3.3.3, provided that at least 10 minor alleles were observed. Details of the previously published GWASs (COIN, UK1, Scotland1, VQ58, CCFR1, and CCFR2) can be found in Reference 15.15 Genomic control was applied by multiplying the standard errors of regression coefficients by the square root of the inflation factor of the respective study. PLINK v.1.90b3i was used for LD-based SNP pruning and principal component analysis (PCA). PCA was performed using 13,012 LD-pruned SNPs with allele frequency >5% and IMPUTE2 info score >0.9. R v.3.3.3 was used for meta-analysis. Estimated log ORs and standard errors were combined to obtain summary p-values, ORs, and 95% CIs under inverse-variance weighted random-effects and fixed-effect models (function “rma.uni” in the metafor package v.1.9–9). All reported p-values are two-sided. The type I error rate (α) was 0.05, corresponding to a genome-wide significance threshold of 5×10−8.
Results
In stage 1, we used a linear mixed model (BOLT-LMM-inf)13 to test 9,068,015 single-nucleotide variants for association with CRC in the FIN cohort, which comprised 1,701 Finnish CRC cases and 14,082 population-matched, cancer-free controls. The median of the BOLT-LMM-inf test statistic was 0.512, corresponding to an inflation factor of 1.12, which was used for genomic control. A quantile-quantile (Q-Q) plot is shown in Supplementary Figure 1, PCA plots in Supplementary Figures 2 and 3, and a Manhattan plot in Supplementary Figure 4. A low-frequency variant at 12q14.3 (rs73121704; MAF, 0.860%) displayed the smallest p-value in stage 1 (p=4.07×10−9). Among the highest-ranking SNPs were the CRC-associated variants rs10505477 (p=5.29×10−8), rs6589219 (p=4.34×10−7; r2 with rs3802842, 0.942 in 1,000 Genomes Phase 3 European populations), and rs6983267 (p=1.38×10−6).16–18 Thirty-eight previously published CRC risk SNPs were tested for association with CRC in the FIN cohort, and 14 of the 38 SNPs showed associations with false discovery rate <0.1. Directions of effects were consistent with earlier publications for each of the 14 SNPs, which were located at 11q23.1 (rs3802842, q=1.77×10−5), 8q24.21 (rs6983267, q= 1.77×10−5; rs7014346, q=1.77×10−5), 20p12.3 (rs961253, q=6.92×10−5), 15q13.3 (rs4779584, q=1.29×10−3), 10q22.3 (rs704017, q=1.91×10−3), 18q21.1 (rs4939827, q=7.96×10−3), 2q35 (rs992157, q=7.96×10−3), 8q23.3 (rs16892766, q=0.0113), 14q22.2 (rs4444235, q=0.0231), 6p21.2 (rs1321311, q=0.0231), 20q13.33 (rs4925386, q=0.0501), 10q24.2 (rs1035209, q=0.0536), and 11q13.4 (rs3824999, q=0.0604). Stage 1 results and LocusZoom plots (http://locuszoom.org/) are shown in Supplementary Tables 1 and 2 and in Supplementary Figures 35 to 102, respectively.
From 20 loci that were ranked highest in stage 1, we selected 40 variants for MassARRAY genotyping in five Nordic cohorts (STHLM2, Gothenburg, HUNT, Estonia, and FINRISK; stage 2). Two variants were selected from each locus. rs992157 (2q35) was also selected for stage 2 because it had been recently reported as a CRC risk factor. We were unable to design genotyping assays for seven variants because of sequence context, and four variants failed genotyping. Consequently, 30 variants representing 20 loci were successfully genotyped in a total of 4,070 Nordic CRC cases and 2,377 controls. The MAF of 6:73457627G>C was low in all five Nordic cohorts, ranging from 0.000923 to 0.00954 (allele count, 2–7). To evaluate imputation accuracy, 1,038 individuals from the FIN cohort were directly genotyped with the MassARRAY platform. Squared Pearson correlation coefficients (r2) between IMPUTE2 genotype dosage and MassARRAY genotype for the 30 variants ranged from 0.816 to 1.00 (median, 0.978).
In stage 3, we obtained summary statistics from previously published GWASs that comprised 7,577 CRC cases and 9,979 controls of European ancestry.15 Summary-level data were available for 27 of the 30 variants that were genotyped in stage 2 (data for rs150509351, rs186867472, and 6:73457627G>C were missing).
To increase statistical power, datasets from stages 1 to 3 were combined (Figure 1), totaling 13,348 CRC cases and 26,438 controls.19 The FIN cohort was reanalyzed by logistic regression to obtain log ORs and corresponding standard errors; the inflation factor was 1.11. The post-imputation inflation factors for the COIN, UK1, Scotland1, VQ58, CCFR1 and CCFR2 studies were 1.10, 1.03, 1.04, 1.04, 1.03, and 1.08, respectively.15 Genomic control was applied for each of these studies. Inflation factors for the STHLM2, Gothenburg, HUNT, or Estonia studies were not estimated because of the small number of genotyped markers. Fixed-effect meta-analysis was performed, but because of possible study heterogeneity, we considered the random-effects model (Supplementary Table 3). Under the random-effects model, rs10505477 (8q24.21), rs6983267 (8q24.21), and rs992157 (2q35) were genome-wide significant (for rs10505477, p=7.63×10−14, phet=0.144, I2=34.4%; for rs6983267, p=7.45×10−13, phet=0.0985, I2=37.7%; for rs992157, p=1.50×10−9, phet=0.777, I2=0%), and rs6589219 (11q23.1) displayed suggestive evidence of association (p=9.14×10−6, phet=0.153, I2=36.5%). Combined effect size estimates and directions of effects for these four SNPs were consistent with prior studies.6,16–18
Figure 1.
Study scheme. Sources of genetic markers are shown on the left, analytic stages in the center, and sources of samples on the right.
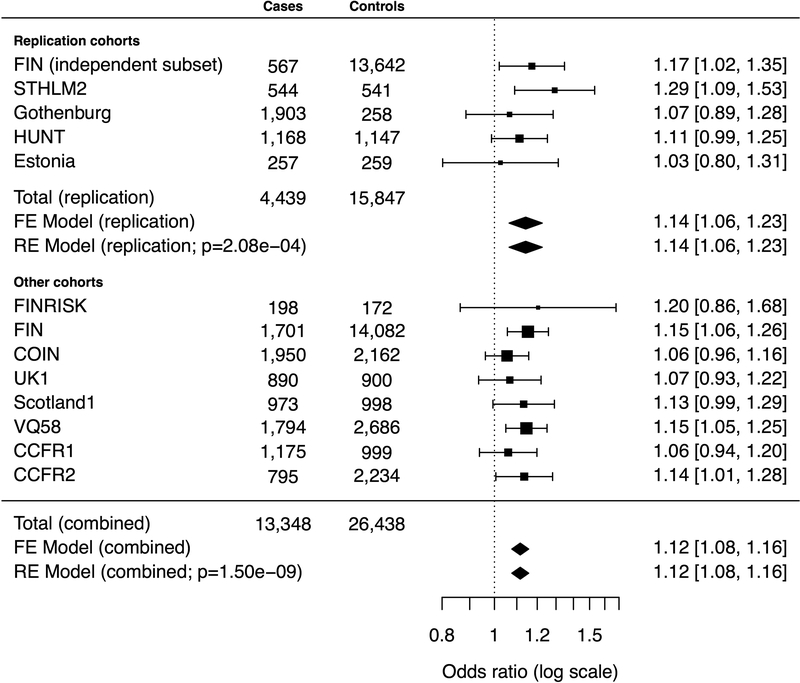
Next, we studied rs992157 (2q35) in a replication dataset comprising 4,439 CRC cases and 15,847 controls (STHLM2, Gothenburg, HUNT, Estonia, and a subset of the FIN cohort) who had not been previously studied for the association between rs992157 and CRC (Figure 2). In the FIN cohort, rs992157 had been directly genotyped with SNP arrays in both cases and controls, and the other Nordic cohorts were genotyped with the MassARRAY platform. Logistic regression models were fit within each cohort. In the independent subset of the FIN cohort (567 CRC cases and 13,642 cancer-free controls), the inflation factor was 1.11, and genomic control was applied accordingly. Estimated log ORs were combined under random-effects and fixed-effect models, the results of which were highly similar without notable study heterogeneity (phet=0.462, I2=0%). Applying Bonferroni correction for the 30 variants that were genotyped in the MassARRAY experiment (α=0.05/30≈0.00167), rs992157 was significantly associated with CRC with an OR of 1.14 (95% CI, 1.06–1.23; p=2.08×10−4). Consistent with prior results, the alternative allele (A) conferred a higher risk of CRC than the reference allele (G). For rs992157, r2 between IMPUTE2 genotype dosage and MassARRAY genotype was 1.00 in the FIN cohort.
Figure 2.
Study cohorts, sample sizes, and estimated odds ratios for rs992157. The vertical line corresponds to the null hypothesis (odds ratio=1). The horizontal lines and square brackets indicate 95% confidence intervals. Areas of the boxes are proportional to the weight of the study. Diamonds represent combined estimates. FE, fixed-effect. RE, random-effects.
Discussion
The identification of CRC susceptibility alleles and quantification of their effects is biologically and clinically meaningful. The genome-wide statistical analysis of tag SNPs has highlighted new genes and regulatory mechanisms in the pathogenesis of CRC while concurrently allowing more accurate estimation of the personalized risk of colorectal neoplasms.20,21 We conducted a GWAS of CRC in the Finnish population (stage 1), genotyped 30 promising variants in five Nordic cohorts (stage 2), and analyzed corresponding summary statistics from previously published GWASs (stage 3). A total of 39,786 individuals (13,348 CRC cases and 26,438 controls) were analyzed in stages 1 to 3. New genotype data generated in this study were used to analyze the recently reported effect of rs992157 (2q35) on CRC risk.
The association between rs992157 and CRC was independently replicated (p=2.08×10−4), and its effect size was approximately 1.1 (OR, 1.14; 95% CI, 1.06–1.23). In the combined analysis of 13,348 CRC cases and 26,438 controls, the p-value and OR for rs992157 were 1.50×10−9 and 1.12 (95% CI, 1.08–1.16), respectively, with no indication of study heterogeneity (phet=0.777, I2=0%). In addition to CRC, rs992157 has shown pleiotropic effects on adult human height and inflammatory bowel disease.6,22
In stage 1, we found evidence supporting multiple previously published SNPs as risk factors for CRC in the Finnish population with false discovery rate <0.1. The corresponding chromosomal regions and nearby genes were 2q35 (PNKD and TMBIM1), 6p21.2 (TRNAI25), 8q23.3 (LINC00536 and EIF3H), 8q24.21 (CCAT2 and LOC101930033), 10q22.3 (ZMIZ1-AS1), 10q24.2 (NKX2–3 and SLC25A28), 11q13.4 (POLD3), 11q23.1 (COLCA1 and COLCA2), 14q22.2 (RPS3AP46 and MIR5580), 15q13.3 (SCG5 and GREM1), 18q21.1 (SMAD7), 20p12.3 (FGFR3P3 and CASC20), and 20q13.33 (LAMA5).
We did not find Finnish population-specific CRC risk variants, which may reflect limitations in replicating them in other populations, their rarity, or small contributions to inherited risk. A low-frequency variant at 12q14.3 (rs73121704; MAF, 0.860%) displayed a notable association in stage 1 (p=4.07×10−9), but the finding was not supported by meta-analysis (random-effects p=0.466, fixed-effect p=0.122). Bias due to genotype imputation or population stratification remains a concern, and further data is needed.
A limitation of the study is that the number of variants selected for stages 2 and 3 was relatively small, and disease-associated variants may have been omitted from further investigation because of low rank in the primary analysis. It is also difficult to assess whether there was residual confounding due to population stratification or different genotyping platforms. For rs992157, r2 between IMPUTE2 genotype dosage and MassARRAY genotype was 1.00, making technical bias unlikely. Genomic control was applied for all primary GWASs to avoid type I error.
In conclusion, we replicated the association between rs992157 (2q35) and CRC in Northern European studies and found it to be genome-wide significant in a meta-analysis of 12 European-ancestry studies. SNPs at 2q35, 6p21.2, 8q23.3, 8q24.21, 10q22.3, 10q24.2, 11q13.4, 11q23.1, 14q22.2, 15q13.3, 18q21.1, 20p12.3, and 20q13.33 were associated with CRC in the Finnish population, which validates findings from previous studies and reveals shared genetic architecture of CRC between the Finnish population isolate and outbred populations.
Supplementary Material
Novelty & impact statements.
This study provides strong evidence for the association between rs992157 (2q35) and colorectal cancer by independent replication in 4,439 cases and 15,847 controls, as well as meta-analysis of 39,786 European-ancestry individuals. Previously published SNPs at 2q35, 6p21.2, 8q23.3, 8q24.21, 10q22.3, 10q24.2, 11q13.4, 11q23.1, 14q22.2, 15q13.3, 18q21.1, 20p12.3, and 20q13.33 were associated with colorectal cancer in the Finnish population, but new risk loci were not identified.
Acknowledgments
We are thankful to Sini Nieminen, Sirpa Soisalo, Marjo Rajalaakso, Inga-Lill Svedberg, Iina Vuoristo, Alison Ollikainen, and Heikki Metsola for their technical support. The study was supported by grants from Academy of Finland (Finnish Center of Excellence Program 2012–2017, Project No. 1250345), Cancer Society of Finland, Sigrid Juselius Foundation, Jane and Aatos Erkko Foundation, SYSCOL (an EU FP7 Collaborative Project No. 258236), Nordic Information for Action eScience Center (NIASC), Nordic Center of Excellence financed by NordForsk (Project No. 62721), NordForsk Colorectal Cancer Pilot Project (07 BM 11/424), Cancer Research UK (C348/A18927 for Edinburgh Colon Cancer Genetics Group (CCGG)), and UK Medical Research Council (MR/K018647/1 for Edinburgh CCGG), Estonian RC (IUT20–60 and PUT736), and European Regional Development Fund (Project No. 2014–2020.4.01.15–0012). Niko Välimäki received grant No. 287665 from the Academy of Finland. The Colon Cancer Family Registry (CCFR) was supported by grant UM1 CA167551 from the National Cancer Institute, USA, and through cooperative agreements with the following centres: Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01/U24 CA074800), Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783), Seattle Colorectal Cancer Family Registry (U01/U24 CA074794), University of Hawaii Colorectal Cancer Family Registry (U01/U24 CA074806), and USC Consortium Colorectal Cancer Family Registry U01/U24 CA074799). The CCFR GWASs were supported by grants U01 CA122839, R01 CA143237, and U19 CA148107 from National Cancer Institute, USA. We acknowledge Karolinska Institute Biobank, Sahlgrenska Biobank, HUNT Biobank, THL Biobank, and Estonian Genome Center for providing DNA samples from Nordic CRC cases and controls. The Nord-Trøndelag Health Study (HUNT Study) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Health Authority, and Norwegian Institute of Public Health.
Abbreviations
- CI
Confidence interval
- CRC
Colorectal cancer
- GWAS
Genome-wide association study
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- PCA
Principal component analysis
- Q-Q-plot
Quantile-quantile plot
- SNP
Single-nucleotide polymorphism
Footnotes
Conflict of interest statement
We have no conflicts of interest to declare.
References
- 1.Graff RE, Möller S, Passarelli MN, Witte JS, Skytthe A, Christensen K, Tan Q, Adami H-O, Czene K, Harris JR, Pukkala E, Kaprio J, et al. Familial Risk and Heritability of Colorectal Cancer in the Nordic Twin Study of Cancer. Clin Gastroenterol Hepatol 2017;15:1256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frampton MJE, Law P, Litchfield K, Morris EJ, Kerr D, Turnbull C, Tomlinson IP, Houlston RS. Implications of polygenic risk for personalised colorectal cancer screening. Ann Oncol 2016;27:429–34. [DOI] [PubMed] [Google Scholar]
- 3.Zeng C, Matsuda K, Jia W-H, Chang J, Kweon S-S, Xiang Y-B, Shin A, Jee SH, Kim D-H, Zhang B, Cai Q, Guo X, et al. Identification of Susceptibility Loci and Genes for Colorectal Cancer Risk. Gastroenterology 2016;150:1633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Gu D, Du M, Xu Z, Zhang S, Zhu L, Lu J, Zhang R, Xing J, Miao X, Chu H, Hu Z, et al. Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat Commun 2016;7:11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Schmit SL, Haiman CA, Keku TO, Kato I, Palmer JR, van den Berg D, Wilkens LR, Burnett T, Conti DV, Schumacher FR, Signorello LB, et al. Novel colon cancer susceptibility variants identified from a genome-wide association study in African Americans. Int J Cancer 2017;140:2728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlando G, Law PJ, Palin K, Tuupanen S, Gylfe A, Hänninen UA, Cajuso T, Tanskanen T, Kondelin J, Kaasinen E, Sarin A-P, Kaprio J, et al. Variation at 2q35 (PNKD and TMBIM1) influences colorectal cancer risk and identifies a pleiotropic effect with inflammatory bowel disease. Hum Mol Genet 2016;25:2349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher FR, Schmit SL, Jiao S, Edlund CK, Wang H, Zhang B, Hsu L, Huang S-C, Fischer CP, Harju JF, Idos GE, Lejbkowicz F, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun 2015;6:7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nyström-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Järvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med 1995;1:1203–6. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salovaara R, Loukola A, Kristo P, Kääriäinen H, Ahtola H, Eskelinen M, Härkönen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 2000;18:2193–200. [DOI] [PubMed] [Google Scholar]
- 11.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 1998;338:1481–7. [DOI] [PubMed] [Google Scholar]
- 12.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience [Internet] 2015;4 Available from: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh RP-, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, Patterson N, Price AL. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015;47:284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirinen M, Donnelly P, Spencer CCA.Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann Appl Stat 2013;7:369–90. [Google Scholar]
- 15.Al-Tassan NA, Whiffin N, Hosking FJ, Palles C, Farrington SM, Dobbins SE, Harris R, Gorman M, Tenesa A, Meyer BF, Wakil SM, Kinnersley B, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep 2015;5:10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanke BW, Greenwood CMT, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007;39:989–94. [DOI] [PubMed] [Google Scholar]
- 17.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007;39:984–8. [DOI] [PubMed] [Google Scholar]
- 18.Tenesa A, Farrington SM, Prendergast JGD, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 2008;40:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–13. [DOI] [PubMed] [Google Scholar]
- 20.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi L-Y, Domingo E, Smillie C, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet 2012;44:770–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I, Mecklin J-P, Järvinen H, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 2009;41:885–90. [DOI] [PubMed] [Google Scholar]
- 22.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J‘an, Kutalik Z, Amin N, Buchkovich ML, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46:1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.