Abstract
Forkhead box O (FOXO) transcription factors are central regulators of cellular homeostasis. FOXOs respond to a wide range of external stimuli, including growth factor signaling, oxidative stress, genotoxic stress, and nutrient deprivation. These signaling inputs regulate FOXOs through a number of posttranslational modifications, including phosphorylation, acetylation, ubiquitination, and methylation. Covalent modifications can affect localization, DNA binding, and interactions with other cofactors in the cell. FOXOs integrate the various modifications to regulate cell type-specific gene expression programs that are essential for metabolic homeostasis, redox balance, and the stress response. Together, these functions are critical for coordinating a response to environmental fluctuations in order to maintain cellular homeostasis during development and to support healthy aging.
1. INTRODUCTION: THE FOXO FAMILY
FOXO transcription factors are conserved regulators of cellular homeostasis, the stress response, and longevity. To carry out these functions, FOXOs regulate a variety of cellular processes, including cell cycle regulation, redox balance, proteostasis, apoptosis, metabolism, and repair of DNA damage (Brunet et al., 1999; Dijkers, Medema, Lammers, Koenderman, & Coffer, 2000; Kops et al., 2002; Lee, Kennedy, Tolonen, & Ruvkun, 2003; Mammucari et al., 2007; Medema, Kops, Bos, & Burgering, 2000; Nemoto & Finkel, 2002; Tran et al., 2002). In humans, the FOXO family comprises FOXO1, FOXO3, FOXO4, and FOXO6. Drosophila and C. elegans each contain a single FOXO gene, known as dFOXO and daf-16, respectively. All FOXO family members contain a forkhead box DNA binding domain that is highly conserved across species and nearly identical among mammalian FOXOs. FOXOs usually function as transcriptional activators, although repression of transcription has been observed in some cases (Ramaswamy, Nakamura, Sansal, Bergeron, & Sellers, 2002; Webb et al., 2013). In all species, FOXO activity is tightly regulated at the posttranslational level. FOXOs receive inputs from various signaling pathways in the form of covalent modifications and protein–protein interactions. Thus, FOXOs are influenced by a combination of external stimuli and intrinsic factors, which together direct transcriptional responses. In this review, we discuss our current understanding of how FOXOs are regulated, with a focus on mammalian systems.
1.1. FOXO Domain Structure and Posttranslational Modifications
FOXO transcription factors are characterized by a “forkhead” DNA binding domain of approximately 110 amino acids that recognizes the consensus sequence TGTTTAC (Furuyama, Nakazawa, Nakano, & Mori, 2000; Xuan & Zhang, 2005). In addition to the DNA binding domain, FOXOs contain short conserved sequences near their N- and C-termini, as well as conserved nuclear localization and export sequences (NLS and NES, respectively; Fig. 1) (Obsil & Obsilova, 2008). In all FOXO family members the NLS overlaps with the C-terminal end of the DNA binding domain and can be modified posttranslationally. Intriguingly, outside of these conserved domains, FOXOs contain long stretches of putative disordered domains. The intrinsically disordered domains of FOXO make up approximately 75% of the total protein and include the predicted transactivation domains (Wang, Marshall, & Ikura, 2015). The function of these domains is not fully understood, but they are likely to be more flexible regions involved in protein–protein interactions and chromatin remodeling, or serve as linkers to facilitate intra- and intermolecular interactions.
Fig. 1.
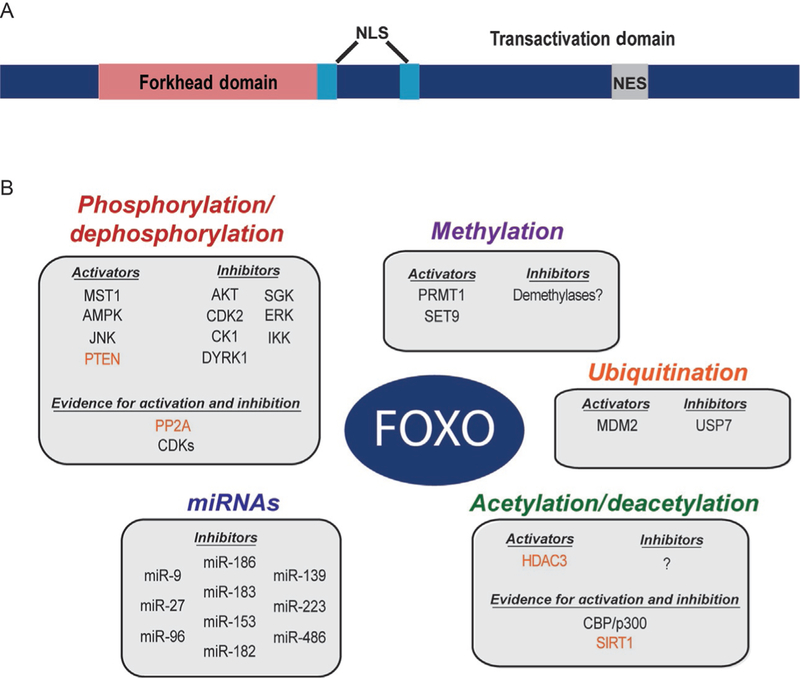
FOXO regulation through posttranslational modifications. (A) General domain structure of the FOXO family of transcription factors. The conserved DNA binding domain is termed the Forkhead box. NES, nuclear export signal; NLS, nuclear localization signal. (B) FOXOs are posttranslationally modified by a number of different enzymes that affect their transcriptional activity. FOXOs are also regulated at the posttranscriptional level by several different miRNAs. Proteins and miRNAs that have been identified as direct regulators of FOXOs in mammalian cells are listed. Note that proteins in red perform the opposite function than the categorized covalent modification (i.e., PTEN and PP2A are phosphatases and HDAC3 and SIRT1 are deacetylases).
FOXO transcription factors are extensively posttranslationally modified. Covalent modifications to FOXOs include phosphorylation, acetylation, methylation, and ubiquitination (Fig. 1). These modifications activate or inhibit FOXO activity by affecting localization, stability, DNA binding, or protein–protein interactions. For example, phosphorylation of FOXO transcription factors has been observed at multiple sites throughout the protein, and a number of kinases have been implicated in their regulation. The first evidence of direct regulation by a protein kinase was phosphorylation at three sites by the serine/threonine kinase AKT (also known as PKB) (Brunet et al., 1999; Kops et al., 1999). Additional kinases including SGK, AMPK, JNK, ERK, and MST1 have been observed to modify FOXOs (Asada et al., 2007; Brunet et al., 2001; Essers et al., 2004; Greer et al., 2007; Lehtinen et al., 2006). Activity of these kinases can inhibit (e.g., AKT, SGK) or augment (e.g., JNK, MST1) FOXO transcriptional activity. FOXOs also contain a number of lysine residues that are targeted by protein acetyltransferases, deacetylases, ubiquitin ligases, and methyltransferases (Brunet et al., 2004; Calnan et al., 2012; Fukuoka et al., 2003; Motta et al., 2004; Yamagata et al., 2008). Intriguingly, a number of these lysine modifications occur in and around the DNA binding domain and nuclear localization sequence. Covalent modification at these sites alters the activity of FOXOs through the regulation of DNA binding, protein stability, and protein–protein interactions. Mono- and polyubiquitination of FOXO factors have been observed and linked to transcriptional activity and stability, respectively (van der Horst et al., 2006; Yang et al., 2008).
The large number of modifications to the FOXO family has led to a proposed “FOXO Code,” whereby modifications act singly or in combination to selectively recruit protein partners to direct specific programs of gene expression (Calnan & Brunet, 2008). Indeed, cell type-specific gene expression networks have been identified at the chromatin level, which are likely to be influenced by particular extrinsic cues and lineage-specific cofactors (Webb, Kundaje, & Brunet, 2016). Here, we discuss the upstream regulatory inputs that influence FOXO activity in various cell types to ensure appropriate downstream responses.
2. REGULATION BY INSULIN/IGF SIGNALING AND OTHER GROWTH FACTORS
The insulin/insulin-like growth factor (IGF) signaling pathway functions in an evolutionarily conserved manner to regulate FOXO activity (Brunet et al., 1999; Lin, Dorman, Rodan, & Kenyon, 1997; Ogg et al., 1997). Insulin and IGF proteins regulate a variety of processes across tissues, including metabolism, growth, cognition, and proteostasis. Insulin, IGF, and several other growth factors (e.g., EGF, NGF, PDGF) activate a common downstream signaling pathway, which includes activation of AKT and MAPK. This signaling pathway is conserved from worms to humans and is a major regulatory input for FOXOs.
2.1. Inhibition by AKT and Other Kinases
Insulin and IGF receptors function as membrane-bound receptors with intracellular tyrosine kinase domains (Siddle, 2011). Once stimulated, the receptors autophosphorylate and recruit the intracellular insulin receptor substrate proteins (IRS; IRS1–3 in mammals). IRS proteins function as adapters that organize downstream signaling events, in particular through recruitment of phosphoinositol-3-kinase (PI3K) (Shaw, 2011). PI3K is a lipid kinase that generates phosphatidylinositol triphosphate (PIP3) and subsequent activation of phosphoinositide protein kinase (PDK) and AKT (Fig. 2). The protein kinase AKT is a key direct regulator of FOXO activity. Mammals have three AKT genes, named AKT1, 2, and 3, which have tissue-specific functions (Gonzalez & McGraw, 2009). Full activation of AKT involves phosphorylation by PDK-1 and mTORC2 at Thr308 and Ser473, respectively (Alessi et al., 1996; Sarbassov, Guertin, Ali, & Sabatini, 2005). Once activated, AKT directly phosphorylates FOXOs 1, 3, and 4 at three sites, resulting in sequestration in the cytoplasm (Brunet et al., 1999; Kops et al., 1999). The same sites that are targeted by AKT can also be phosphorylated by serum and glucocorticoid-induced kinase (SGK) (Brunet et al., 2001). The first two phosphosites are binding sites for the 14–3-3 proteins, which are localized to both the cytoplasm and the nucleus (Brunet et al., 1999; Obsil, Ghirlando, Anderson, Hickman, & Dyda, 2003). 14–3-3 binding in the cytoplasm prevents the cytoplasmic pool of FOXO from entering the nucleus, whereas 14–3-3-FOXO interaction in the nucleus is thought to expose the nuclear export sequence and increase transport back to the cytoplasm (Brunet et al., 2002; Obsilova et al., 2005). Once they have been shuttled to the cytoplasm, FOXOs can be degraded by the proteasome. The multisubunit E3 ligase SKP1-CUL-F-box (SCF) has been shown to recognize phosphorylated Ser256 on FOXO1 and polyubiquitinate the N-terminal half of the protein, tagging it for proteasomal degradation (Huang et al., 2005).
Fig. 2.
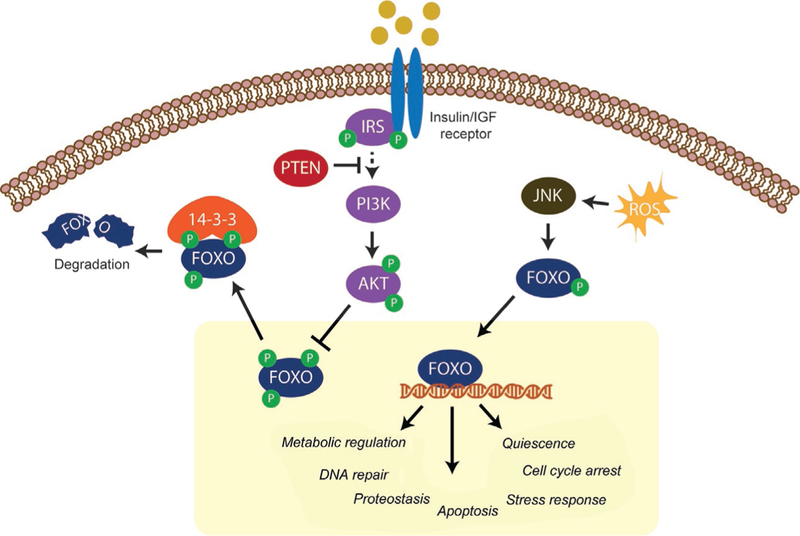
FOXO activity, stability, and localization are regulated by the insulin/IGF signaling pathway and reactive oxygen species. Insulin/insulin-like growth factors activate transmembrane receptors and in turn activate the insulin receptor substrate (IRS) proteins. IRS proteins then stimulate downstream signaling events, resulting in the phosphorylation and activation of AKT. Activated AKT phosphorylates FOXO proteins on three sites and triggers their inactivation and nuclear exclusion. FOXO phosphorylation by AKT enhances binding to 14–3-3 proteins and either cytoplasmic sequestration and/or degradation by the proteasome. Phosphatase and tensin homolog (PTEN) promotes FOXO activation and nuclear localization by inhibiting the activation of PI3K. In contrast to inhibition by insulin/IGF signaling, reactive oxygen species (ROS) promote nuclear localization and activation of FOXO by activating the c-Jun N-terminal kinase (JNK). JNK directly phosphorylates FOXOs, thereby stimulating activation of transcriptional regulation. Once activated, FOXOs regulate several cellular processes that promote cellular homeostasis.
14–3-3-independent regulation of FOXO localization has been observed in the case of FOXO1 (Rena, Prescott, Guo, Cohen, & Unterman, 2001). Although the precise mechanism has not been defined in all cases, additional protein kinases can potentiate the nuclear export of FOXOs. For example, cyclin-dependent kinase 2 (CDK2) phosphorylates FOXO1 at Ser249, which resides in the NLS (Huang, Regan, Lou, Chen, & Tindall, 2006). Casein kinase 1 (CK1) recognizes FOXO1 phosphorylated by AKT at Ser319, and subsequently phosphorylates Ser322 and Ser325 (Rena, Bain, Elliott, & Cohen, 2004; Rena et al., 2002). Modification of these sites enhances binding to the Ran-dependent export machinery, thereby reducing nuclear FOXO levels by accelerating nuclear export. Thus, tight regulation of the nuclear levels of FOXO proteins involves multiple mechanisms and phosphorylation events in addition to interaction with 14–3-3 proteins (Zhao et al., 2004).
AKT also phosphorylates and inhibits the transcriptional activity of FOXO6. However, FOXO6 stands apart from the other mammalian FOXOs because it is only phosphorylated by AKT at two sites, Thr26 and Ser184, and it does not appear to shuttle between the nucleus and the cytoplasm and instead is constitutively nuclear (van der Heide, Jacobs, Burbach, Hoekman, & Smidt, 2005).
2.2. Regulation by Phosphatases
Phosphatases function both upstream of FOXOs and directly on the FOXOs themselves to attenuate and fine-tune regulation by insulin/IGF signaling. The phosphatase and tensin homolog (PTEN) is a lipid/protein phosphatase that inhibits PI3 kinase signaling by dephosphorylating PIP3 to PI(4,5)P2 (Maehama & Dixon, 1998). PIP3 is critical for AKT activation; therefore, high PTEN activity reduces the active pool of AKT and increases FOXO activity. Genetic evidence supports an important role for PTEN upstream of FOXOs since genetic ablation of the PTEN and FOXO factors results in similar phenotypes in the mouse. For example, PTEN deficiency causes increased neural progenitor proliferation in vitro and larger brain size in vivo, similar to FOXO-deficient mice (Amiri et al., 2012; Groszer et al., 2006; Paik et al., 2009; Renault et al., 2009). Similarly, ablation of either PTEN or FOXOs in hematopoietic stem cells causes enhanced cell cycle entry and eventual exhaustion of the stem cell pool (Miyamoto et al., 2007; Tothova et al., 2007; Yilmaz et al., 2006; Zhang et al., 2006). PTEN is a tumor suppressor commonly mutated in cancers, including prostate, brain, and breast cancer (Hollander, Blumenthal, & Dennis, 2011). PTEN-negative prostate and renal carcinoma cells have elevated cytoplasmic FOXOs (Modur, Nagarajan, Evers, & Milbrandt, 2002; Nakamura et al., 2000), and expression of constitutively active FOXO in these cells can decrease tumorigenesis (Ramaswamy et al., 2002). However, the degree of FOXO dysregulation in most PTEN-associated cancers is not known, and the extent to which reduced FOXO activity in the absence of PTEN contributes to tumorigenicity will be an important question in the future. Nevertheless, PTEN is a critical upstream activator of FOXO transcription factors in healthy mammalian cells, and reduced FOXO activity in PTEN-negative tumors enhances tumorigenesis.
In addition to regulation by PTEN, FOXOs are regulated both directly and indirectly by protein phosphatase 2A (PP2A). Direct interactions between both FOXO1 and FOXO3 and PP2A have been observed, and pharmacological inhibition or ablation of PP2A enhances phosphorylation at the AKT phosphosites on FOXO1 (Yan et al., 2008). Dephosphorylation of the AKT phosphosites T32 and S253 by PP2A releases FOXOs from 14–3-3 binding and increases nuclear entry (Singh et al., 2010). The model that has emerged from these observations is one in which inhibition by AKT or SGK is tightly regulated in mammalian cells, whereas dephosphorylation by PP2A is constitutive and nonspecific. Under this model, the kinases are the important inhibitory nodes, and in their absence FOXOs are dephosphorylated and activated by default. However, upstream of FOXOs, AKT is targeted by specific PP2A regulatory subunits. In C. elegans, a key regulatory subunit of the PP2A holoenzyme, B56β/pptr-1, is actively regulated to balance FOXO stability with degradation (Padmanabhan et al., 2009). The B56β subunit specifically targets PP2A to dephosphorylate AKT-1, but not AKT-2 or AKT-3. Modulation of B56β/pptr-1 activity in the worm influences FOXO/DAF-16 localization and affects FOXO/ DAF-16-regulated phenotypes. Similarly, the Drosophila ortholog of B56β/pptr-1, widerborst, was identified as a regulator of the AKT–FOXO pathway in flies (Vereshchagina, Ramel, Bitoun, & Wilson, 2008). To what extent particular phosphatases target specific AKTs or FOXOs in mammalian cells remains largely unknown. Knockdown of B56β, but not the related factor B56α, in cultured adipocyte cell lines resulted in increased AKT-1 phosphorylation, suggesting that this interaction is conserved from worms to humans (Padmanabhan et al., 2009). However, whether this alters FOXO activity is not known. The B55α PP2A regulatory subunit has also been found to specifically target AKT (Kuo et al., 2008), but factors specific for FOXOs remain unknown. Thus, future studies will be required to dissect the context-specific regulation of FOXO activity by additional phosphatases.
3. NUTRIENT SIGNALING AND METABOLIC STRESS
FOXOs are activated by upstream signals under conditions of low nutrient availability or starvation. For example, in the liver, low nutrient status and thus low levels of insulin signaling activate FOXOs to restore glucose levels via glycogenolysis and gluconeogenesis. In this context, FOXOs regulate a number of target genes involved in energy metabolism, including the gluconeogenic enzymes glucose-6-phosphatase (G6Pase), phosphoenolpyruvate carboxykinase (Pepck), and (PGC1a) (Altomonte et al., 2003; Matsumoto, Pocai, Rossetti, Depinho, & Accili, 2007; Puigserver et al., 2003). FOXOs also maintain metabolic balance through regulation of autophagy genes, including Atg5, Atg12, and Becn1 (Webb & Brunet, 2014). Restoration of metabolic balance in response to starvation or limited nutrients is also a key function of FOXOs in beta cells of the pancreas and skeletal muscle. In addition to regulation by insulin/IGF/AKT signaling, FOXOs are also regulated by AMP-activated protein kinase (AMPK), a central intracellular regulator of energy homeostasis.
3.1. Regulation by AMPK
AMPK is an energy-sensing kinase that is activated in response to high AMP/ATP ratios as a result of ATP depletion (Hardie & Carling, 1997). AMPK functions as a trimeric complex that is allosterically activated by AMP binding. When cellular ATP levels are low, AMP-bound AMPK is directly phosphorylated at Thr172 by either LKB1 or CAMKK2 (Hawley et al., 2005; Shaw et al., 2004; Woods et al., 2005). Phospho-AMPK phosphorylates a number of enzymes and transcriptional regulators involved in metabolic regulation, including lipid metabolism, protein synthesis, and glucose uptake, in order to replenish the ATP supply. AMPK directly phosphorylates FOXO3 at six serine/threonine residues that are distinct from the AKT phosphosites (Thr179, Ser399, Ser413, Ser555, Ser588, and Ser626) (Greer et al., 2007). Unlike regulation by other kinases (e.g., AKT, SGK, MST1), modification by AMPK does not appear to alter the localization of FOXO3. Thus, the role of AMPK in the regulation of FOXOs may primarily be to promote interaction between cofactors and FOXO3 to affect specific target genes. Consistent with this notion, mutation of the six AMPK phosphorylation sites disrupted expression of novel targets involved in energy homoeostasis, including acetyl-CoA thioesterase (Acot12), ferroportin (Slc40a1), and the mitochondrial proton carrier uncoupling protein 2 (Ucp2). Alternatively, activation by AMPK may increase the overall efficiency of FOXO-mediated transcriptional activation. Indeed, AMPK phosphorylation of FOXO3 has been observed to enhance binding to CBP/p300 and transactivation in luciferase assays (Wang et al., 2012).
Some, but not all, of the AMPK sites are found in other mammalian and nonmammalian FOXO family members. Regulation of FOXOs by AMPK has been detected in C. elegans, and this pathway regulates life span extension in response to dietary restriction in worms (Greer et al., 2007). Thus, the AMPK–FOXO regulatory axis is conserved from worms to mammals.
Fasting conditions can activate Class IIa histone deacetylases (HDACs) to deacetylate and activate FOXOs in the liver. Specifically, in response to food deprivation, HDAC3 binds to target genes involved in gluconeogenesis and deacetylates FOXOs to stimulate FOXO-mediated gene activation (Mihaylova et al., 2011). This cellular context involves dual regulation by both phosphorylation and acetylation: low insulin levels drive FOXOs into the nucleus, which are then further activated by deacetylation by HDACs. Activation of G6pc and Pck1 (PEPCK) restores glucose homeostasis under these conditions, and the activation of FOXOs is reversed upon refeeding. In summary, FOXOs are critical for maintaining metabolic balance in conditions of nutrient deprivation. FOXOs integrate signals from insulin/IGF signaling, AMPK, HDACs, and possibly other inputs to respond to changing nutrient availability.
4. OXIDATIVE AND GENOTOXIC STRESS STIMULI
Regulation of the stress response is a key function of FOXO transcription factors. This important function was first observed in C. elegans where long-lived daf-2 mutants display resistance to oxidative stress (paraquat), a phenotype which is dependent on FOXO/DAF-16 (Honda & Honda, 1999). FOXOs are able to respond to stress signals independent of growth factor signaling since stress can induce nuclear entry in high growth factor conditions (Brunet et al., 2004; Frescas, Valenti, & Accili, 2005). Once they have been activated, FOXOs activate a number of downstream target genes encoding antioxidant enzymes that regulate redox homeostasis, including catalase, sestrins, and superoxide dismutase (SOD) (Klotz et al., 2015).
4.1. Regulation by the MAPK Family
Activation of FOXOs in response to stress is mediated in part by Jun-N-terminal kinase (JNK). JNK is activated by different cellular stresses, including UV irradiation and oxidative stress. In the presence of low-level oxidative stress JNK directly phosphorylates and activates FOXO4 at Thr447 and Thr451 (Fig. 2) (Essers et al., 2004). Modification of these sites disrupts the interaction with 14–3-3 proteins, allowing FOXO nuclear entry. JNK also directly phosphorylates 14–3-3, perturbing the interaction with FOXO and releasing it into the nucleus. Intriguingly, this regulatory axis is conserved across species and impacts life span in model organisms. JNK activation enhances dFOXO nuclear entry and extends life span in a FOXO-dependent manner in Drosophila (Wang, Bohmann, & Jasper, 2005). However, it remains unclear if FOXO is directly phosphorylated by JNK in this context, particularly because JNK has been found to phosphorylate other proteins in the insulin/IGF pathway, including IRS-1 (Hirosumi et al., 2002). Evidence from C. elegans suggests that JNK directly phosphorylates FOXO/DAF-16 and, similar to Drosophila, overexpression of JNK in worms extends life span in a FOXO/DAF-16-dependent manner (Oh et al., 2005). Together, these lines of evidence indicate that JNK can activate FOXOs via direct and indirect mechanisms. In Drosophila, activation of the JNK– FOXO axis culminates in expression of a program of genes that function in protection against oxidative stress (Wang, Bohmann, & Jasper, 2003). Similar targets are likely to reside downstream of JNK-stimulated FOXO in mammalian cells, but they have yet to be identified. In addition to JNK, p38 MAPK activates FOXO3 in response to genotoxic and environmental stress. p38 phosphorylates FOXO3 at Ser294 and Ser425, which are JNK target sites, as well as at a site distinct from JNK (Ser7) (Ho et al., 2012). Whether activation by p38 induces a response that completely overlaps with the JNK-induced response or is distinct from JNK-mediated stimulation remains unknown. However, mutation of Ser7 to Ala was sufficient to impair nuclear localization in response to genotoxic stress (doxorubicin treatment), suggesting that phosphorylation by p38 is a key input for full activation of FOXO in this context.
In contrast to the stimulation by JNK and p38, the MAPK family member ERK is a potent inhibitor of FOXO activity. In tumor cells, stimulation with growth factors activates ERK, which directly phosphorylates FOXO3 at three sites: Ser294, Ser344, and Ser425 (Asada et al., 2007; Yang et al., 2008). Phosphorylation at these sites promotes cytoplasmic retention, ubiquitination by the E3-ligase MDM2, and subsequent degradation by the proteasome. Mutation of the three ERK target sites causes resistance to MDM2-mediated degradation and can block tumor growth (Yang et al., 2008). This finding suggests parallel regulation of FOXOs downstream of growth factor signaling since the same stimuli simultaneously inhibit PI3K/AKT signaling and ERK activity.
4.2. Activation by Ste20-Like Kinase MST1
In addition to the MAPK family, the Ste20-like kinase (MST1) has been shown to phosphorylate and activate FOXOs in response to oxidative stress (Lehtinen et al., 2006; Yuan et al., 2009). MST1-mediated activation of FOXOs was first discovered in primary neurons, where treatment with hydrogen peroxide induced phosphorylation of FOXOs, followed by FOXO-dependent apoptosis. Similar to other activating modifications, the MST1 phosphosite was found to be within the forkhead domain (Ser212 and Ser207 in FOXO1 and FOXO3, respectively), and phosphorylation at this site disrupted 14–3-3 binding. This regulatory axis has been observed in other mammalian cell types and is evolutionarily conserved in C. elegans (Lehtinen et al., 2006). For example, Mst1—/— T cells are highly sensitive to oxidative stress, which correlates with reduced FOXO1 and FOXO3 stability and activity (Choi et al., 2009; Du et al., 2014). However, in contrast to what has been observed in neurons, MST1 does not regulate FOXO1 or FOXO3 localization in Treg cells. The MST1–FOXO interaction may have implications for FOXO’s tumor suppressor activity since treatment of lymphoma cell lines with the anticancer drug α-tocopheryl succinate, which causes accumulation of ROS, also induced apoptosis via the MST1–FOXO1 axis (Valis et al., 2011). The downstream effects of activation of the MST1–FOXO pathway are not fully understood, but appear to be cell type and species specific. In neurons, FOXO3 induces cell death in response to activation of MST1, and the proapoptotic gene BIM was identified as the key factor mediating this response (Lehtinen et al., 2006). In contrast, FOXOs have been proposed to activate ROS-detoxifying genes (e.g., MnSOD and catalase) and cell survival (e.g., Il7r) in naïve T cells (Choi et al., 2009). Activation of FOXOs by MST1 in Treg cells occurs during normal development, independent of exogenously induced stress (Du et al., 2014). In this situation, FOXOs are critical for regulating expression of Foxp3, which functions as a master regulator of Treg identity. Moreover, overexpression of the worm MST1 (cst-1) can extend life span in nematodes in a FOXO/DAF-16-dependent manner. Thus, the cellular and organismal context of FOXO activation is likely to direct the appropriate downstream response in each case.
The extent to which activation of FOXOs in this context differs from activation by JNK has not been determined. It is possible that JNK and MST1 function as redundant activators of the same gene programs that mediate the stress response. Alternatively, modification by each kinase may provide a binding site for particular binding partners that guide activation of specific gene networks. Additional work will be required to determine the precise downstream readouts of FOXO in each case and the extent to which they function as separate and overlapping networks.
4.3. Ubiquitination/Deubiquitination in Response to Stress
Monoubiquitination has also been observed to regulate FOXO activity in response to oxidative stress (peroxide treatment) (van der Horst et al., 2006). In this context, MDM2 has been observed to monoubiquitinate FOXO4 and increase its nuclear localization and transcriptional activity (Brenkman, de Keizer, van den Broek, Jochemsen, & Burgering, 2008). The deubiquitinating enzyme herpes virus-associated ubiquitin-specific protease (USP7) binds monoubiquitinated FOXO4 to attenuate the response, suggesting that, in this context, MDM2 alters the localization but not the stability of FOXOs. Interestingly, the authors observe that low levels of MDM2 potentiate FOXO4 activity, whereas high levels of MDM2 overexpression have a repressive effect. This result contrasts the observation that MDM2 induces FOXO degradation upon growth factor stimulation (Yang et al., 2008), and suggests that the effects of ubiquitination are context specific, depending either on the levels and/or nature of the stimulus.
4.4. Acetylation and Deacetylation Modulates FOXO Activity
Early studies from C. elegans identified an interaction between the nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase Sir2 and DAF-16/FOXO. Studies in mammalian cells confirmed that FOXOs are acetylated by p300 and CBP (CREB binding protein) in response to oxidative stress. The same conditions induce a physical association between various FOXO family members and the mammalian ortholog of Sir2, SIRT1. Knockout of SIRT1 enhances FOXO acetylation in fibroblasts and embryonic stem cells. SIRT1 is only present in the nucleus, and only associates with and deacetylates nuclear FOXOs in the presence of oxidative stress. Acetylation of FOXOs has been observed to both increase and decrease its transcriptional activity, depending on the target.
Motta et al. reported that acetylation of FOXO3 by p300 (E1A binding protein 300) enhanced transcriptional activity, and the effect could be reversed by the SIRT1 deacetylase (Motta et al., 2004). The authors extended their findings to other FOXOs and found that FOXOs 1 and 4 could also be deacetylated and inhibited by SIRT1. Moreover, knockout of SIRT1 correlated with increased transcription of two previously identified FOXO targets, Pepck (phosphoenolpyruvate carboxylase) and Igfpb1 (insulin-like growth factor binding protein 1). In contrast, others have observed that acetylation of FOXOs by CBP (CREB binding protein) inhibits transcriptional activity in the context of oxidative stress, and SIRT1 can relieve this inhibition (Daitoku et al., 2004; Fukuoka et al., 2003; van der Horst et al., 2004). In this case, overexpression of SIRT1 promoted FOXO-mediated cell cycle arrest and induction of p27/Cdkn1b and MnSOD. Brunet et al. observed that the effect of SIRT1–FOXO3 association was target dependent: SIRT1 enhanced induction of p27/CDKN1B and GADD45, but inhibited BIM activation (Brunet et al., 2004). This observation suggests that the acetyltransferases and deacetylases targeting FOXOs are modulators of FOXO activity, instead of simple on/off switches such as kinases (e.g., AKT and MAPK). Subsequent studies investigating the nature of the FOXO–acetyltransferase interaction supports this notion. Interestingly, the FOXO4–CBP interaction was found to be stabilized by the formation of a cysteine disulfide bridge that forms under conditions of high oxidative stress. In this case, the cysteine residues were observed to mediate the inhibition of FOXO4-induced cell cycle exit by p300, while promoting FOXO4-induced apoptosis (Dansen et al., 2009). Altogether, these findings suggest that acetylation of FOXOs has dual effects on modulating the response to oxidative stress: acetylation enhances apoptosis while reducing stress resistance. The precise mechanism by which acetylation tips the balance toward apoptosis is unknown, but the location of the acetylation sites, which primarily reside in the DNA binding domain, suggests that acetylation may affect binding specificity at different targets. Studies have demonstrated that acetylation of FOXOs can disrupt their ability to bind DNA (Brent, Anand, & Marmorstein, 2008; Matsuzaki et al., 2005; Tsai et al., 2007), but how this modification might alter target selection remains unclear. It must also be considered that histone acetylation and deacetylation, which activate and repress transcription, respectively, are performed by the same set of enzymes. Thus, it remains a challenge to unravel transcriptional effects due to alterations in histone acetylation vs FOXO activity in some experiments. Nevertheless, acetylation is a key covalent modification in the regulation of FOXO activity in mammalian systems.
4.5. Regulation by Cyclin-Dependent Kinases Under Genotoxic Stress
FOXOs function in the response to genotoxic stress, by integrating with the repair machinery and activating DNA damage response genes (Tran et al., 2002; Tsai, Chung, Takahashi, Xu, & Hu, 2008). CDK2 can directly phosphorylate FOXO1 in response to DNA damage. CDK2 plays an important role in the G1/S DNA damage checkpoint. CDK2 normally complexes with cyclin E to promote the G1/S transition. Under genotoxic stress, p21/CDKN1A inhibits CDK2, thereby inducing cell cycle arrest and preventing replication of damaged DNA (Satyanarayana & Kaldis, 2009). Huang et al. observed that CDK2 phosphorylates FOXO1 on Ser249, causing nuclear export and inhibition of FOXO1. DNA damage induced by a topoisomerase I inhibitor (camptothecin) inhibited CDK2 phosphorylation of FOXO1 and resulted in FOXO-induced apoptosis (Huang et al., 2006). CDK1, which regulates entry into mitosis, has been observed to phosphorylate the same site on FOXO1 (Ser249) in neurons. In this case, phosphorylation was observed to induce nuclear localization and activate FOXO1 by disrupting FOXO1–14-3–3 binding. CDK1 can induce apoptosis in neurons, and does so through the activation of FOXO1 in this context (Yuan et al., 2008). How phosphorylation at the same site can inhibit FOXO activity in one context (DNA damage/CDK2 activation) and enhance activity in another (postmitotic neurons/CDK1 activation) remains unclear. It is unlikely to be a difference between proliferating and terminally differentiated cells since Yuan et al. also observed activation of FOXO1 by CDK1 in dividing cells. An interesting possibility is that the same phosphosite is recognized by different cellular machinery in different phases of the cell cycle, resulting in differential regulation of FOXO1. Future work will be necessary to define the precise mechanism underlying this difference.
5. REGULATION OF FOXOS IN CANCER AND THE IMMUNE RESPONSE
FOXOs have emerged as critical regulators of the immune response. FOXOs generally function to restrain the immune response and inhibit tumor growth through the activation of genes that inhibit the cell cycle (e.g., Cdkn1b/p27), induce apoptosis (e.g., Bim), and limit proinflammatory cytokine production (e.g., TNFα). Mitogenic signals alter FOXO activity through both AKT-dependent and -independent signaling pathways, which we review below.
5.1. AKT-Independent Regulation of FOXO Localization by IkB Kinase in Tumors
In the tumor setting, regulation of FOXO activity can be entirely independent of AKT activity. In this context FOXOs can instead be inhibited by the IkB kinase/NK-kB signaling pathway. IkB kinase (IKK) is a trimeric complex that is activated by proinflammatory cytokines and viral infections. Once activated, IKK phosphorylates IkB proteins and targets them for degradation. IkB proteins bind and inhibit the NF-kB family of transcriptional regulators. Thus, high levels of cytokines and IKK signaling induce an NF-kB-mediated transcriptional program, promoting cell survival and the immune response (Luo, Kamata, & Karin, 2005). Hu et al. observed that some breast cancer tumors that are negative for activated (phosphorylated) AKT can have high levels of cytoplasmic FOXO protein, indicating an AKT-independent inhibition of FOXO activity in tumors (Hu et al., 2004). In such tumors, the authors found elevated IKK activity, indicating reciprocal activation states for FOXOs and IKK signaling. Interestingly, tumors with high nuclear FOXO levels and low IKK corresponded to improved survival rates. The authors went on to explore the mechanism underlying this correlation and discovered that the kinase subunits of IKK, IKKα, and IKKβ could inhibit the transcriptional activity of FOXO3 and induce export to the cytoplasm for degradation by the proteasome. The attenuation of FOXO3 activity was also observed using a version of FOXO3 with mutated AKT phosphosites, suggesting that inhibition by IKK signaling is independent of AKT. IKK was found to directly bind FOXO3 and phosphorylate it at Ser644, which is not present on FOXO1 or FOXO4, and direct regulation of these FOXOs by IKK has not been reported.
Regulation of FOXO3 by IKK has been observed in the context of leukemic cells as well. Similar to the observations in breast cancer cells, inhibition of IKK activity in acute myeloid leukemia blast cells induced FOXO3-mediated cell cycle arrest and apoptosis (Chapuis et al., 2010). Notably, AKT can also phosphorylate and stimulate IKK directly (Dan et al., 2008). Thus, regulation of FOXO3 by IKK can be independent of AKT in the case of activation by cytokines (TNF-α), but it may also work through AKT-induced IKK activity.
5.2. Regulation by T Cell Receptors, B Cell Receptors, and Cytokine Signaling
FOXO transcription factors play an important role in maintenance of the immune system, autoimmunity, and in the response to pathogens. Immune cell-specific ablation of FOXOs causes immune dysfunction phenotypes, depending on the particular FOXO and cell type that is targeted. For example, knockout of FOXO3 in mouse T cells causes hyperactivation of T cells and inflammatory disease.
T cells are stimulated in response to contact with antigen-presenting cells (APCs) and exposure to IL-2. IL-2 binds a trimeric cell surface receptor (IL-2R) and promotes T cell survival and expansion by activating the PI3K–AKT pathway (Minami, Kono, Miyazaki, & Taniguchi, 1993). Similar to the effect downstream of insulin/IGF signaling, signaling through AKT in this context inhibits FOXO activity (Stahl et al., 2002). Likewise, T cell receptor (TCR/CD28) stimulation through APC contact activates the PI3K–AKT pathway, which in turn drives T cell proliferation required for the immune response. Antigen-induced activation of PI3K in T cells inhibits FOXO1 activity through AKT-mediated phosphorylation and nuclear exclusion (Fabre et al., 2005). Moreover, introduction of a mutant form of FOXO1 that cannot be phosphorylated by AKT was constitutively nuclear and inhibited T cell proliferation even in the presence of APC contact. In the context of T cell stimulation, the Vav family of guanine exchange factors (GEFs) are activated by TCR/CD28 stimulation and regulate PI3K/ AKT activity upstream of FOXO1 (Charvet et al., 2006).
Cytokine withdrawal can also induce FOXO activation in primary B cells. Similar to T cells, cytokines stimulate PI3K/AKT to promote B cell survival, and cytokine deprivation induces FOXO-mediated apoptosis via activation of Bim and loss of mitochondrial integrity (Dijkers et al., 2002). In addition to inducing B cell death, FOXO1 has also emerged as a critical regulator of B cell development through the regulation of a target network that guides differentiation and homeostasis in the periphery (Szydlowski, Jablonska, & Juszczynski, 2014). In this context, FOXO1 and FOXO3 regulate B cell specialization through the regulation of Rag1 and Rag2 transcription, which induce genomic editing of the B cell receptor. Thus, in immature B cells PI3K/AKT signaling inhibits FOXO activity. Signaling through the B cell receptor involving the SLP-65 adapter protein inhibits PI3K/AKT, thereby stabilizing FOXOs and inducing genomic rearrangements that drive B cell maturation (Amin & Schlissel, 2008; Herzog et al., 2008).
In summary, FOXOs are important regulators of immune function and are under tight regulation by PI3K/AKT in T cells, B cells, and other immune cell types. This signaling network functions downstream of a variety of stimuli, including cytokines and antigen presentation through T cell and B cell receptors. FOXOs in turn regulate apoptosis, differentiation, cell cycle exit, and maturation to promote healthy immune system function and development.
6. OTHER INPUTS
6.1. Regulation by Methyltransferases
Methylation of FOXO1 and FOXO3 has been reported to alter their stability. Two arginine residues on FOXO1 (Arg248 and 250) can be methylated by protein arginine methyltransferase 1 (PRMT1). Methylation at these sites reduced phosphorylation by AKT at Ser253, thereby promoting FOXO1 nuclear entry and reducing degradation by the proteasome (Yamagata et al., 2008). Similarly, the Set9 methyltransferase can methylate Lys271 on FOXO3 to alter stability and transcriptional activity (Calnan et al., 2012). Similarly, Lys270 on FOXO3 was found to be methylated by Set9 in a different cell type (neurons) in response to oxidative stress (Xie et al., 2012). Interestingly, the Lys271 is a conserved residue among the FOXO family, and was previously identified as an acetylation site that is targeted by the SIRT1 deacetylase (Brunet et al., 2004). Although the precise interplay between these modifications has yet to be determined, these findings suggest that competition between different modifications may help balance the response to stress. Alternatively, the same residues may be modified differently in different cell types used to modulate FOXO activity.
6.2. Regulation of Feeding Behavior by DYRK1A
Dual-specificity tyrosine-phosphorylated and regulated kinase 1A (DYRK1A) is a nuclear-localized protein kinase that functions in the nervous system (Smith et al., 1997). Woods et al. demonstrated that DYRK1A binds and phosphorylates FOXO1 on Ser329, a site that is conserved in FOXO3, FOXO4, and DAF-16 (Woods et al., 2001). Phosphorylation by DYRK1 reduces the nuclear pool of FOXO1 and inhibits its transcriptional activity. Regulation of FOXOs by DYRK1 appears to be independent of insulin/IGF signaling, at least in 293T cells, because stimulation of the PI3K/AKT pathway did not affect phosphorylation at Ser329. More recent work in flies and mice established an in vivo function for FOXO regulation by DYRK1. In Drosophila, Dyrk1a can activate Sirt1, which in turn deacetylates and activates dFOXO to regulate food intake. This circuitry appears to be intact in the mouse hypothalamus since Dyrk1 transgenic mice displayed decreased FOXO acetylation and increased NPY transcription (Hong et al., 2012). Importantly, the initial study identifying a DYRK1–FOXO1 interaction reported direct phosphorylation and inhibition of FOXO1, whereas the more recent study reported that DYRK1 increases FOXO1 transcriptional indirectly through the activation of SIRT1. The reason for this difference is not clear, but it suggests that neurons regulating food intake may have specialized signaling networks to regulate FOXO activity to control neuropeptide expression and feeding behavior.
6.3. Modification of FOXOs by Additional Protein Kinases
A number of additional protein kinases have been reported to phosphorylate FOXO transcription factors and modulate their activity, including nemo-like kinase (NLK), glycogen synthase kinase 3 (GSK3), PERK, and protein kinase A (PKA) (Huo et al., 2014; Kim, Kim, Lee, & Chung, 2010; Zhang et al., 2013). Interestingly, PKA can phosphorylate FOXO1 at the three AKT target sites (Thr24, Ser256, and Ser319) and inhibit its transcriptional activity in endothelial cells (Lee, Chen, Pullikotil, & Quon, 2011). The PKA consensus site is nearly identical to the AKT consensus sequence ((R/K)XX (S/T) and (R/K)XRXX(S/T), respectively) (Huang, Tsai, Chen, Wu, & Chen, 2007; Kane et al., 2002). Thus, it remains possible that PKA targets FOXOs in a variety of cellular contexts. However, the extent to which PKA targets these sites in vivo remains unclear.
6.4. Regulation by miRNAs
Although most studies have focused on posttranslational regulation of FOXO factors, several groups have reported posttranscriptional regulation by miRNAs. For example, the FOXO1 30 untranslated region is targeted by miR-27a, miR-96, and miR-182 in breast cancer cells (MCF7 cells). Expression of these microRNAs maintains FOXO1 protein at a low level in breast cancer cells compared to normal breast tissue, thereby promoting an oncogenic state (Guttilla & White, 2009). Similarly, FOXO1 is maintained at a low level in endometrial cancer cells (HEC-1B and Ishikawa cells) through direct regulation by miR-9, miR-27, miR-96, miR-153, miR-182, miR-183, and miR-186 (Myatt et al., 2010). Other examples of miRNAs that target FOXOs include miR-139, miR-223, and miR-486 (Hasseine et al., 2009; Small et al., 2010; Wu et al., 2012). Thus, inhibition of FOXOs by miRNAs adds an additional layer of regulation, which is frequently used in the cancer cells to promote tumorigenesis.
7. CONCLUDING REMARKS
The FOXO family of transcription factors are key integrators of environmental cues and stimuli that maintain cellular homeostasis. Downstream functions of FOXOs are conserved across species, and include regulation of the stress response, nutrient signaling, DNA repair, and cell cycle status. Since the initial discovery that AKT is a potent inhibitor of FOXO nuclear entry and transcriptional activity, a wide range of stimuli and enzymes have been reported to impinge on FOXO activity. However, a number of important questions remain to be answered. For example, precisely how different enzymes and modifications are integrated to affect FOXO localization and fine-tune its transcriptional activity remains unclear. In addition, little is known about how different modifications or stimuli affect target selection at the chromatin level. It is likely that FOXOs bind and activate different target genes depending on the particular environmental cues involved, but this has yet to be explored. FOXOs are likely to interact with specific cofactors depending on their modification state, but to date the number of cofactors interacting with FOXOs remains limited. This will be important to clarify since binding partners are likely to be integral to binding site selection, and to help direct cell type-specific transcriptional responses. These studies would provide important insight into how FOXOs guide development, maintain cellular homeostasis, and promote healthy aging.
ACKNOWLEDGMENTS
Work in our laboratory is supported by NIH R01 AG053268, the American Federation for Aging Research, the Glenn Foundation for Research on the Biology of Aging, and a subproject award from the NIH/NIA U19 AG023122.
REFERENCES
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. (1996). Mechanism of activation of protein kinase B by insulin and IGF-1. The EMBO Journal, 15(23), 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Altomonte J, Richter A, Harbaran S, Suriawinata J, Nakae J, Thung SN, et al. (2003). Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. American Journal of Physiology. Endocrinology and Metabolism, 285(4), E718–728. 10.1152/ajpendo.00156.2003. [DOI] [PubMed] [Google Scholar]
- Amin RH, & Schlissel MS (2008). Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nature Immunology, 9(6), 613–622. 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Cho W, Zhou J, Birnbaum SG, Sinton CM, McKay RM, et al. (2012). Pten deletion in adult hippocampal neural stem/progenitor cells causes cellular abnormalities and alters neurogenesis. The Journal of Neuroscience, 32(17), 5880–5890. 10.1523/JNEUROSCI.5462-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada S, Daitoku H, Matsuzaki H, Saito T, Sudo T, Mukai H, et al. (2007). Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cellular Signalling, 19(3), 519–527. 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Brenkman AB, de Keizer PL, van den Broek NJ, Jochemsen AG, & Burgering BM (2008). Mdm2 induces mono-ubiquitination of FOXO4. PLoS One, 3(7), e2819 10.1371/journal.pone.0002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MM, Anand R, & Marmorstein R (2008). Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure, 16(9), 1407–1416. 10.1016/j.str.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96(6), 857–868. [DOI] [PubMed] [Google Scholar]
- Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, et al. (2002). 14–3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. The Journal of Cell Biology, 156(5), 817–828. 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, & Greenberg ME (2001). Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Molecular and Cellular Biology, 21(3), 952–965. 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science, 303(5666), 2011–2015. 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Calnan DR, & Brunet A (2008). The FoxO code. Oncogene, 27(16), 2276–2288. 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Calnan DR, Webb AE, White JL, Stowe TR, Goswami T, Shi X, et al. (2012). Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY), 4(7), 462–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis N, Park S, Leotoing L, Tamburini J, Verdier F, Bardet V, et al. (2010). IkappaB kinase overcomes PI3K/Akt and ERK/MAPK to control FOXO3a activity in acute myeloid leukemia. Blood, 116(20), 4240–4250. 10.1182/blood-2009-12-260711. [DOI] [PubMed] [Google Scholar]
- Charvet C, Canonigo AJ, Becart S, Maurer U, Miletic AV, Swat W, et al. (2006). Vav1 promotes T cell cycle progression by linking TCR/CD28 costimulation to FOXO1 and p27kip1 expression. Journal of Immunology, 177(8), 5024–5031. [DOI] [PubMed] [Google Scholar]
- Choi J, Oh S, Lee D, Oh HJ, Park JY, Lee SB, et al. (2009). Mst1-FoxO signaling protects naive T lymphocytes from cellular oxidative stress in mice. PLoS One, 4(11), e8011 10.1371/journal.pone.0008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, et al. (2004). Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America, 101(27), 10042–10047. 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, & Baldwin AS (2008). Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes & Development, 22(11), 1490–1500. 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, et al. (2009). Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nature Chemical Biology, 5(9), 664–672. 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Birkenkamp KU, Lam EW, Thomas NS, Lammers JW, Koenderman L, et al. (2002). FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: Protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. The Journal of Cell Biology, 156(3), 531–542. 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, & Coffer PJ (2000). Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Current Biology, 10(19), 1201–1204. [DOI] [PubMed] [Google Scholar]
- Du X, Shi H, Li J, Dong Y, Liang J, Ye J, et al. (2014). Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. Journal of Immunology, 192(4), 1525–1535. 10.4049/jimmunol.1301060. [DOI] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, et al. (2004). FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. The EMBO Journal, 23(24), 4802–4812. 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre S, Lang V, Harriague J, Jobart A, Unterman TG, Trautmann A, et al. (2005). Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. Journal of Immunology, 174(7), 4161–4171. [DOI] [PubMed] [Google Scholar]
- Frescas D, Valenti L, & Accili D (2005). Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. The Journal of Biological Chemistry, 280(21), 20589–20595. 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, & Fukamizu A (2003). Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. International Journal of Molecular Medicine, 12(4), 503–508. [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, & Mori N (2000). Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. The Biochemical Journal, 349(Pt. 2), 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, & McGraw TE (2009). The Akt kinases: Isoform specificity in metabolism and cancer. Cell Cycle, 8(16), 2502–2508. 10.4161/cc.8.16.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, et al. (2007). An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Current Biology, 17(19), 1646–1656. 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, et al. (2007). The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. The Journal of Biological Chemistry, 282(41), 30107–30119. 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, Zack JA, et al. (2006). PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proceedings of the National Academy of Sciences of the United States of America, 103(1), 111–116. 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, & White BA (2009). Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. The Journal of Biological Chemistry, 284(35), 23204–23216. 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, & Carling D (1997). The AMP-activated protein kinase—Fuel gauge of the mammalian cell? European Journal of Biochemistry, 246(2), 259–273. [DOI] [PubMed] [Google Scholar]
- Hasseine LK, Hinault C, Lebrun P, Gautier N, Paul-Bellon R, & Van Obberghen E (2009). miR-139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochemical and Biophysical Research Communications, 390(4), 1278–1282. 10.1016/j.bbrc.2009.10.135. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, et al. (2005). Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metabolism, 2(1), 9–19. 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Herzog S, Hug E, Meixlsperger S, Paik JH, DePinho RA, Reth M, et al. (2008). SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nature Immunology, 9(6), 623–631. 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, et al. (2002). A central role for JNK in obesity and insulin resistance. Nature, 420(6913), 333–336. 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Ho KK, McGuire VA, Koo CY, Muir KW, de Olano N, Maifoshie E, et al. (2012). Phosphorylation of FOXO3a on Ser-7 by p38 promotes its nuclear localization in response to doxorubicin. The Journal of Biological Chemistry, 287(2), 1545–1555. 10.1074/jbc.M111.284224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, & Dennis PA (2011). PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature Reviews Cancer, 11(4), 289–301. 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, & Honda S (1999). The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. The FASEB Journal, 13(11), 1385–1393. [PubMed] [Google Scholar]
- Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, et al. (2012). Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genetics, 8(8), e1002857 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. (2004). IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell, 117(2), 225–237. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, & Tindall DJ (2006). CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science, 314(5797), 294–297. 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, et al. (2005). Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proceedings of the National Academy of Sciences of the United States of America, 102(5), 1649–1654. 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Tsai ML, Chen GY, Wu CJ, & Chen SH (2007). A systematic MS-based approach for identifying in vitro substrates of PKA and PKG in rat uteri. Journal of Proteome Research, 6(7), 2674–2684. 10.1021/pr070134c. [DOI] [PubMed] [Google Scholar]
- Huo X, Liu S, Shao T, Hua H, Kong Q, Wang J, et al. (2014). GSK3 protein positively regulates type I insulin-like growth factor receptor through forkhead transcription factors FOXO1/3/4. The Journal of Biological Chemistry, 289(36), 24759–24770. 10.1074/jbc.M114.580738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S, Sano H, Liu SC, Asara JM, Lane WS, Garner CC, et al. (2002). A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. The Journal of Biological Chemistry, 277(25), 22115–22118. 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim Y, Lee J, & Chung J (2010). Regulation of FOXO1 by TAK1-Nemo-like kinase pathway. The Journal of Biological Chemistry, 285(11), 8122–8129. 10.1074/jbc.M110.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz LO, Sanchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, & Monsalve M (2015). Redox regulation of FoxO transcription factors. Redox Biology, 6, 51–72. 10.1016/j.redox.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. (2002). Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature, 419(6904), 316–321. 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, & Burgering BM (1999). Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature, 398(6728), 630–634. 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, & Chiang CW (2008). Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. The Journal of Biological Chemistry, 283(4), 1882–1892. 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- Lee JW, Chen H, Pullikotil P, & Quon MJ (2011). Protein kinase A-alpha directly phosphorylates FoxO1 in vascular endothelial cells to regulate expression of vascular cellular adhesion molecule-1 mRNA. The Journal of Biological Chemistry, 286(8), 6423–6432. 10.1074/jbc.M110.180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, & Ruvkun G (2003). DAF-16 target genes that control C. elegans life-span and metabolism. Science, 300(5619), 644–647. 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, et al. (2006). A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell, 125(5), 987–1001. https://doi.org/10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, & Kenyon C (1997). daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science, 278(5341), 1319–1322. [DOI] [PubMed] [Google Scholar]
- Luo JL, Kamata H, & Karin M (2005). IKK/NF-kappaB signaling: Balancing life and death—A new approach to cancer therapy. The Journal of Clinical Investigation, 115(10), 2625–2632. 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, & Dixon JE (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of Biological Chemistry, 273(22), 13375–13378. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metabolism, 6(6), 458–471. 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, & Accili D (2007). Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metabolism, 6(3), 208–216. 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, & Fukamizu A (2005). Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proceedings of the National Academy of Sciences of the United States of America, 102(32), 11278–11283. 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, & Burgering BM (2000). AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature, 404(6779), 782–787. 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, et al. (2011). Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell, 145(4), 607–621. 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Kono T, Miyazaki T, & Taniguchi T (1993). The IL-2 receptor complex: Its structure, function, and target genes. Annual Review of Immunology, 11, 245–268. 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, et al. (2007). Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell, 1(1), 101–112. 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Modur V, Nagarajan R, Evers BM, & Milbrandt J (2002). FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. The Journal of Biological Chemistry, 277(49), 47928–47937. 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell, 116(4), 551–563. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, et al. (2010). Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Research, 70(1), 367–377. 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, & Sellers WR (2000). Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Molecular and Cellular Biology, 20(23), 8969–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, & Finkel T (2002). Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science, 295(5564), 2450–2452. 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- Obsil T, Ghirlando R, Anderson DE, Hickman AB, & Dyda F (2003). Two 14–3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14–3-3 proteins and inhibition of DNA binding. Biochemistry, 42(51), 15264–15272. 10.1021/bi0352724. [DOI] [PubMed] [Google Scholar]
- Obsil T, & Obsilova V (2008). Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene, 27(16), 2263–2275. 10.1038/onc.2008.20/. [DOI] [PubMed] [Google Scholar]
- Obsilova V, Vecer J, Herman P, Pabianova A, Sulc M, Teisinger J, et al. (2005). 14–3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry, 44(34), 11608–11617. 10.1021/bi050618r. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature, 389(6654), 994–999. 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, & Tissenbaum HA (2005). JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proceedings of the National Academy of Sciences of the United States of America, 102(12), 4494–4499. 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, & Tissenbaum HA (2009). A PP2A regulatory subunit regulates C. elegans insulin/ IGF-1 signaling by modulating AKT-1 phosphorylation. Cell, 136(5), 939–951. 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, et al. (2009). FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell, 5(5), 540–553. 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, et al. (2003). Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature, 423(6939), 550–555. 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, & Sellers WR (2002). A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell, 2(1), 81–91. [DOI] [PubMed] [Google Scholar]
- Rena G, Bain J, Elliott M, & Cohen P (2004). D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Reports, 5(1), 60–65. 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Prescott AR, Guo S, Cohen P, & Unterman TG (2001). Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14–3-3 binding, transactivation and nuclear targetting. The Biochemical Journal, 354(Pt. 3), 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Woods YL, Prescott AR, Peggie M, Unterman TG, Williams MR, et al. (2002). Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. The EMBO Journal, 21(9), 2263–2271. 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell, 5(5), 527–539. 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, & Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science, 307(5712), 1098–1101. 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, & Kaldis P (2009). A dual role of Cdk2 in DNA damage response. Cell Division, 4, 9 10.1186/1747-1028-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM (2011). The insulin receptor substrate (IRS) proteins: At the intersection of metabolism and cancer. Cell Cycle, 10(11), 1750–1756. 10.4161/cc.10.11.15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. (2004). The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proceedings of the National Academy of Sciences of the United States of America, 101(10), 3329–3335. 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle K (2011). Signalling by insulin and IGF receptors: Supporting acts and new players. Journal of Molecular Endocrinology, 47(1), R1–10. 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- Singh A, Ye M, Bucur O, Zhu S, Tanya Santos M, Rabinovitz I, et al. (2010). Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14–3-3 and AKT. Molecular Biology of the Cell, 21(6), 1140–1152. 10.1091/mbc.E09-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proceedings of the National Academy of Sciences of the United States of America, 107(9), 4218–4223. 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Stevens ME, Sudanagunta SP, Bronson RT, Makhinson M, Watabe AM, et al. (1997). Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nature Genetics, 16(1), 28–36. 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, et al. (2002). The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. Journal of Immunology, 168(10), 5024–5031. [DOI] [PubMed] [Google Scholar]
- Szydlowski M, Jablonska E, & Juszczynski P (2014). FOXO1 transcription factor: A critical effector of the PI3K-AKT axis in B-cell development. International Reviews of Immunology, 33(2), 146–157. 10.3109/08830185.2014.885022. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. (2007). FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell, 128(2), 325–339. 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr., DiStefano PS, et al. (2002). DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science, 296(5567), 530–534. 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- Tsai WB, Chung YM, Takahashi Y, Xu Z, & Hu MC (2008). Functional inter-action between FOXO3a and ATM regulates DNA damage response. Nature Cell Biology, 10(4), 460–467. 10.1038/ncb1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, & Hsiao CD (2007). Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Research, 35(20), 6984–6994. 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valis K, Prochazka L, Boura E, Chladova J, Obsil T, Rohlena J, et al. (2011). Hippo/ Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Research, 71(3), 946–954. 10.1158/0008-5472.CAN-10-2203. [DOI] [PubMed] [Google Scholar]
- van der Heide LP, Jacobs FM, Burbach JP, Hoekman MF, & Smidt MP (2005). FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleocytoplasmic shuttling. The Biochemical Journal, 391(Pt. 3), 623–629. 10.1042/BJ20050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. (2006). FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biology, 8(10), 1064–1073. 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, & Burgering BM (2004). FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). The Journal of Biological Chemistry, 279(28), 28873–28879. 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Vereshchagina N, Ramel MC, Bitoun E, & Wilson C (2008). The protein phosphatase PP2A-B’ subunit Widerborst is a negative regulator of cytoplasmic activated Akt and lipid metabolism in Drosophila. Journal of Cell Science, 121(Pt. 20), 3383–3392. 10.1242/jcs.035220. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, & Jasper H (2003). JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Developmental Cell, 5(5), 811–816. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, & Jasper H (2005). JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell, 121(1), 115–125. 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wang F, Marshall CB, & Ikura M (2015). Forkhead followed by disordered tail: The intrinsically disordered regions of FOXO3a. Intrinsically Disordered Proteins, 3(1), e1056906 10.1080/21690707.2015.1056906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Marshall CB, Yamamoto K, Li GY, Gasmi-Seabrook GM, Okada H, et al. (2012). Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment. Proceedings of the National Academy of Sciences of the United States of America, 109(16), 6078–6083. 10.1073/pnas.1119073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, & Brunet A (2014). FOXO transcription factors: Key regulators of cellular quality control. Trends in Biochemical Sciences, 39(4), 159–169. 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Kundaje A, & Brunet A (2016). Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell, 15(4), 673–685. 10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Pollina EA, Vierbuchen T, Urban N, Ucar D, Leeman DS, et al. (2013). FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Reports, 4(3), 477–491. 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, et al. (2005). Ca2 +/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metabolism, 2(1), 21–33. 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Woods YL, Rena G, Morrice N, Barthel A, Becker W, Guo S, et al. (2001). The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. The Biochemical Journal, 355(Pt. 3), 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M, et al. (2012). MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Letters, 586(7), 1038–1043. 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- Xie Q, Hao Y, Tao L, Peng S, Rao C, Chen H, et al. (2012). Lysine methylation of FOXO3 regulates oxidative stress-induced neuronal cell death. EMBO Reports, 13(4), 371–377. 10.1038/embor.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Z, & Zhang MQ (2005). From worm to human: Bioinformatics approaches to identify FOXO target genes. Mechanisms of Ageing and Development, 126(1), 209–215. 10.1016/j.mad.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, et al. (2008). Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Molecular Cell, 32(2), 221–231. 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, & Yang E (2008). PP2A regulates the pro-apoptotic activity of FOXO1. The Journal of Biological Chemistry, 283(12), 7411–7420. 10.1074/jbc.M708083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. (2008). ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nature Cell Biology, 10(2), 138–148. 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, et al. (2006). Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature, 441(7092), 475–482. 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, et al. (2008). Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science, 319(5870), 1665–1668. 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Lehtinen MK, Merlo P, Villen J, Gygi S, & Bonni A (2009). Regulation of neuronal cell death by MST1-FOXO1 signaling. The Journal of Biological Chemistry, 284(17), 11285–11292. 10.1074/jbc.M900461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, et al. (2006). PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature, 441(7092), 518–522. 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hietakangas V, Wee S, Lim SC, Gunaratne J, & Cohen SM (2013). ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes & Development, 27(4), 441–449. 10.1101/gad.201731.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Gan L, Pan H, Kan D, Majeski M, Adam SA, et al. (2004). Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: Characterization of phosphorylation- and 14–3-3-dependent and -independent mechanisms. The Biochemical Journal, 378(Pt. 3), 839–849. 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]


