Abstract
Purpose
Liquid biopsies that noninvasively detect molecular correlates of aggressive prostate cancer (PCa) could be used to triage patients, reducing the burdens of unnecessary invasive prostate biopsy and enabling early detection of high-risk disease. DNA hypermethylation is among the earliest and most frequent aberrations in PCa. We investigated the accuracy of a six-gene DNA methylation panel (Epigenetic Cancer of the Prostate Test in Urine [epiCaPture]) at detecting PCa, high-grade (Gleason score greater than or equal to 8) and high-risk (D’Amico and Cancer of the Prostate Risk Assessment] PCa from urine.
Patients and Methods
Prognostic utility of epiCaPture genes was first validated in two independent prostate tissue cohorts. epiCaPture was assessed in a multicenter prospective study of 463 men undergoing prostate biopsy. epiCaPture was performed by quantitative methylation-specific polymerase chain reaction in DNA isolated from prebiopsy urine sediments and evaluated by receiver operating characteristic and decision curves (clinical benefit). The epiCaPture score was developed and validated on a two thirds training set to one third test set.
Results
Higher methylation of epiCaPture genes was significantly associated with increasing aggressiveness in PCa tissues. In urine, area under the receiver operating characteristic curve was 0.64, 0.86, and 0.83 for detecting PCa, high-grade PCa, and high-risk PCa, respectively. Decision curves revealed a net benefit across relevant threshold probabilities. Independent analysis of two epiCaPture genes in the same clinical cohort provided analytical validation. Parallel epiCaPture analysis in urine and matched biopsy cores showed added value of a liquid biopsy.
Conclusion
epiCaPture is a urine DNA methylation test for high-risk PCa. Its tumor specificity out-performs that of prostate-specific antigen (greater than 3 ng/mL). Used as an adjunct to prostate-specific antigen, epiCaPture could aid patient stratification to determine need for biopsy.
INTRODUCTION
Prostate cancer (PCa) is the most common noncutaneous malignancy in men and the third leading cause of cancer-related deaths in men in the Western world. In 2012, there were an estimated 1.1 million men diagnosed with this disease. With an aging population, spread of Westernized diet, and increasing use of prostate-specific antigen (PSA) testing, this figure is predicted to double by 2035.1
PCa demonstrates extreme clinical heterogeneity. Overall, 10-year survival rates are close to 100%2; thus, there is much interest in strategies to reduce overtreatment of low/intermediate-risk tumors.3 However, more than 300,000 men die as a result of PCa each year. This stark figure, together with the knowledge that overtreatment comes with the high price of prevalent life-changing side effects, illustrates the need for tools to selectively identify high-risk PCa (those tumors with a high propensity to metastasize) at an early stage, while the disease is potentially curable.
Research is ongoing into methods to address this need and better risk-stratify patients. Developments include algorithms around PSA isoforms (ie, the four-kallikrein and Prostate Health Index tests),4 and multifactorial risk-calculator nomograms, such as the Prostate Cancer Prevention Trial and European Randomized Study of Screening for Prostate Cancer.5 Urine in particular is an attractive noninvasive route, with the potential to reduce the number of patients subjected to prostate biopsy. Urinary analysis of molecular correlates of aggressive disease could be used to triage patients and identify those who actually require an invasive prostate biopsy to histologically diagnose and grade their disease.
In 2012, Movember launched four collaborative Global Action Plan (GAP) initiatives, focusing on PCa biomarkers in urine, plasma, serum, and exosomes. The aim of the GAP1 urine project was to develop multiparametric urine biomarkers to facilitate early detection of aggressive PCa. Underpinning this aim was the cooperative establishment of prebiopsy post–digital rectal examination (DRE) urine biorepositories at each institution, with a confederated database housing accompanying clinical, pathologic, and lifestyle data. The GAP1 urine project united investigators from seven different countries studying proteomic, transcriptomic, and epigenetic analytes.
Herein, we present the results of one of two DNA methylation panels investigated in GAP1 urine, adhering to the REMARK (Reporting Recommendations for Tumor Markers) guidelines on correct reporting of prognostic biomarker research.6 The Epigenetic Cancer of the Prostate Test in Urine (epiCaPture) is a multibiomarker panel that quantitatively measures DNA hypermethylation at the 5′-regulatory regions of six genes (GSTP1, SFRP2, IGFBP3, IGFBP7, APC, and PTGS2), all of which have previously been reported in PCa.7-10 Cumulative evidence supports promoter hypermethylation and concomitant gene silencing in PCa initiation7,11 and as a strong prognostic indicator.12,13 The purpose of this study was to determine the accuracy of epiCaPture for noninvasive detection of PCa and aggressive PCa, as defined by Gleason score (greater than or equal to 8) on biopsy, as well as two widely used risk-stratification systems, namely, D’Amico14 and Cancer of the Prostate Risk Assessment (CAPRA).15
PATIENTS AND METHODS
Patient Population
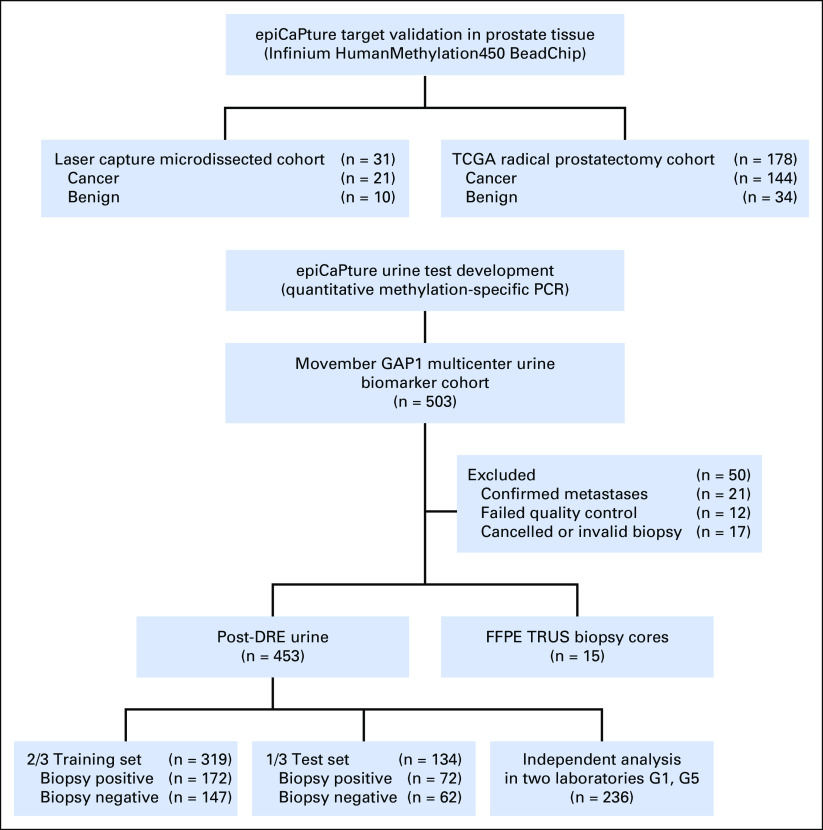
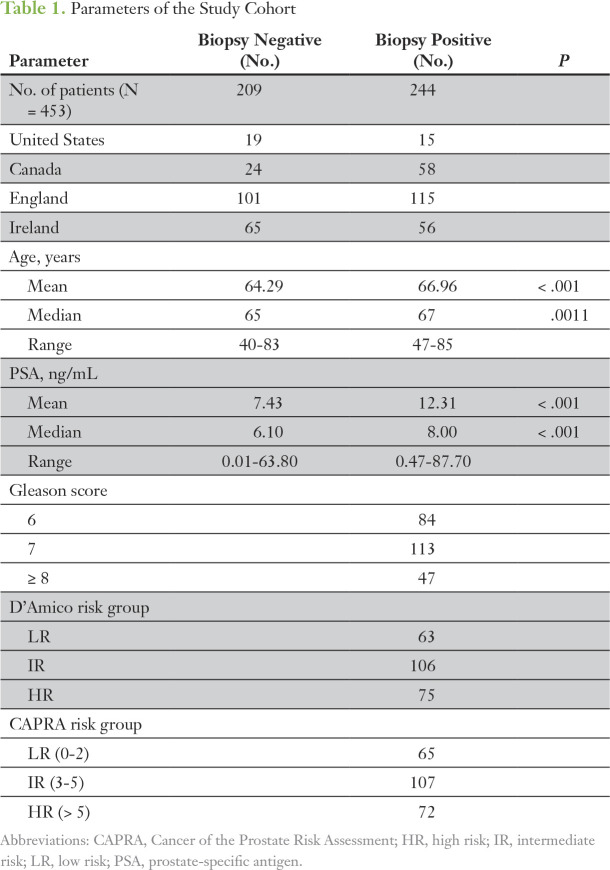
This institutional review board–approved, multicenter prospective study enrolled men who were scheduled for transrectal ultrasound-guided (TRUS) biopsy because of an elevated and/or increasing PSA and/or a suspicious DRE. Six centers from the United States (Emory Healthcare, Atlanta, GA), Canada (University Health Network and Lunenfeld-Tanenbaum Research Institute, Sinai Health Systems, Toronto), and Europe (St James’s Hospital, Mater Misericordiae University Hospital, and Tallaght Hospital, all in Ireland; Norfolk and Norwich University Hospital, England), collected prebiopsy, post-DRE urine (≤ 50 mL) between January 2012 and October 2015. Exclusion criteria included previous diagnosis of PCa, active urinary tract infection, recent prostate biopsy or transurethral resection of the prostate (less than 6 wk), and confirmed presence of metastatic disease (by pelvic magnetic resonance imaging or bone scan). Histopathologic evaluation of TRUS biopsy cores was carried out at each site as per standard procedure by local consultant histopathologists. In total, urine samples were collected from 503 men, with matched formalin-fixed paraffin-embedded TRUS biopsy cores acquired, where available (Fig 1). Signed informed consent was obtained from all patients. The demographic, clinical, and pathologic data for the whole cohort are listed in Table 1. In addition to this prebiopsy population, a retrospective analysis of the epiCaPture gene loci was performed on two independent cohorts of prostate tissues (Fig A2; Data Supplement).
Fig 1.
Flow diagram of specimens used in the study. DRE, digital rectal examination; epiCaPture, Epigenetic Cancer of the Prostate Test in Urine; FFPE, formalin-fixed paraffin-embedded; GAP, Global Action Plan; PCR, polymerase chain reaction; TCGA, The Cancer Genome Atlas; TRUS, transrectal ultrasound.
Table 1.
Parameters of the Study Cohort
Sample Collection, Processing, and epiCaPture Analysis
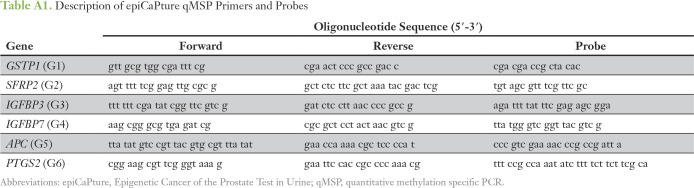
Urine samples were stored on ice and processed within 4 hours of collection, following a Movember GAP1 standard operating procedure. Details of nucleic acid isolation and quantification and epiCaPture analysis are provided in Appendix Table A1 and the Data Supplement.
Statistical Analysis
Statistical analyses were performed with R version 3.4.1 and GraphPad Prism version 6. Two-tailed tests were used, and P < .05 was considered significant. The ability of epiCaPture genes (or the combination thereof), when applied to urine, to predict cancer (v no cancer detected on biopsy), high-grade cancer (Gleason score greater than or equal to 8 v all else), and high-risk cancer (v all else) as classified by D’Amico and CAPRA was assessed by logistic regression models, built in a two thirds training set (n = 319) and validated in a one third test set (n = 134). Discriminatory ability of the models was assessed via sensitivity, specificity, positive predictive value, and negative predictive value (NPV) using the Youden index. Diagnostic potential was quantified using the area under the receiver operating characteristic curve (AUC). Clinical utility was assessed by decision curve analysis (DCA).16
RESULTS
epiCaPture Genes Demonstrate Prognostic Utility in Prostate Tissues
Mindful that most potential biomarkers fail to translate to the clinic,17 we first set out to verify the prognostic utility of each epiCaPture constituent gene by measuring its methylation in two independent cohorts of radical prostatectomy samples. There was strong evidence for differences in mean methylation β-values between four different groups (benign, low-risk, aggressive, and metastatic PCa) in a small cohort of prostate tissues (subjected to epithelial cell enrichment) measured using the Infinium HumanMethylation450 BeadChip (Fig 2A). Post hoc analyses revealed significantly higher methylation of each gene in aggressive and/or metastatic lesions compared with benign tissue. Extracting freely available data from the same analytical platform for a larger The Cancer Genome Atlas cohort yielded similar results; each epiCaPture gene demonstrated significantly higher levels of DNA methylation in clinically significant and high-risk tumors, relative to histologically benign tissue (Fig 2B). Although there was some evidence of methylation in low-risk disease, only G1 (GSTP1) was methylated at significantly higher levels than benign tissue in both cohorts.
Fig 2.
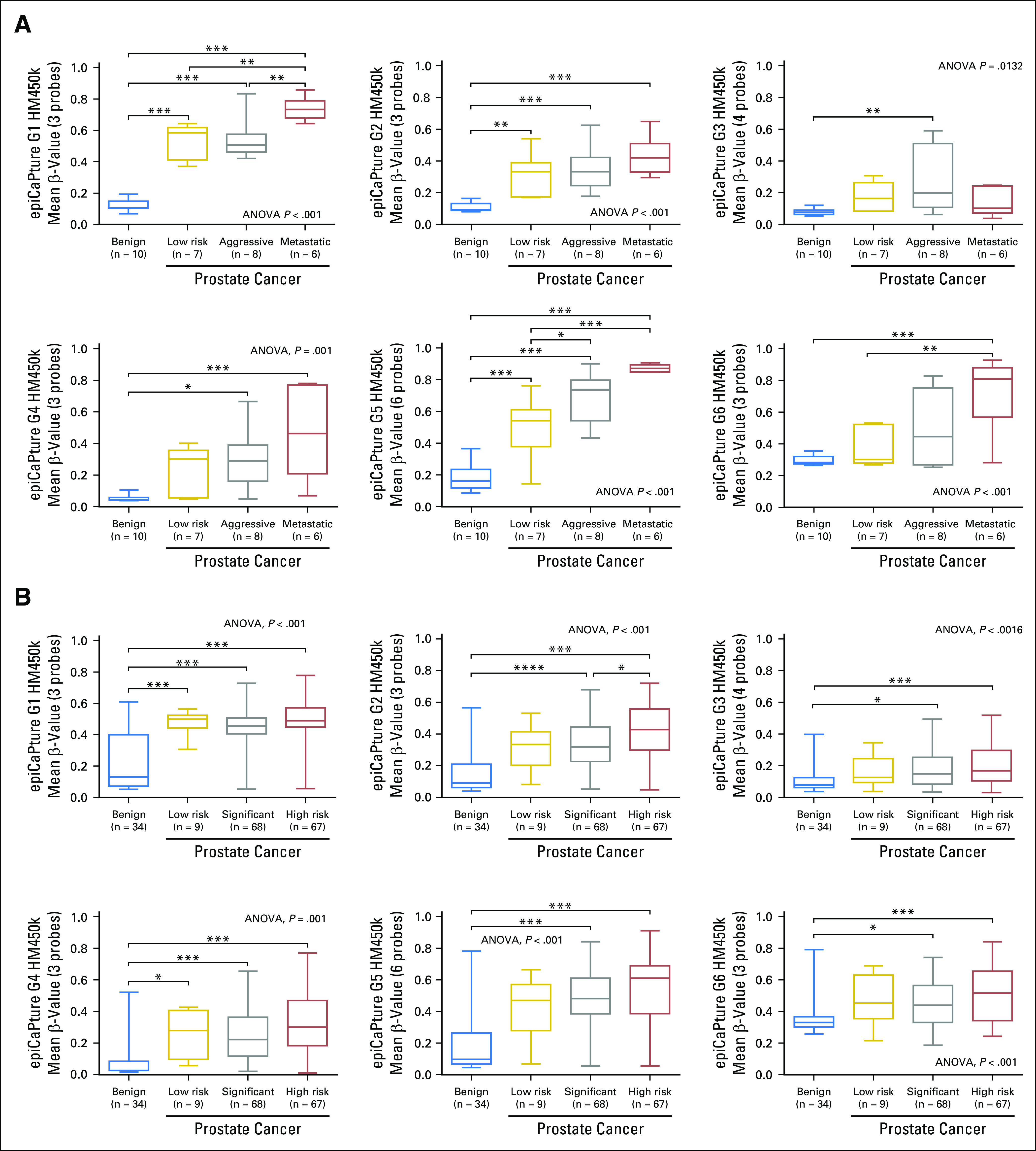
Epigenetic Cancer of the Prostate Test in Urine (epiCaPture) gene validation in prostate tissue specimens. DNA methylation was measured using the Infinium HumanMethylation450 BeadChip (HM450k) in two independent cohorts of patients undergoing radical prostatectomy. (A) Laser capture microdissected enriched prostate epithelial cells from benign prostate tissue (procured from men undergoing radical cystoprostatectomy for bladder cancer, with no clinical or histologic evidence of prostate cancer [PCa]) and low-risk, aggressive, and metastatic PCa. (B) Data extracted from the TCGA repository for matched-benign prostate tissue and low-risk, significant, and high-risk PCa, all procured from men undergoing radical prostatectomy. A full description of the two cohorts is provided in the Data Supplement. Each data set was analyzed by one-way analysis of variance (ANOVA), followed by post hoc analyses using the Tukey-Kramer multiple comparisons test. Boxes indicate the 25th and 75th percentiles, the line represents the median, and the whiskers show the minimum and maximum value in each cohort of samples. Significance is indicated by *P < .05, **P < .01, and *** P < .001, and **** P < .0001
epiCaPture Performance Characteristics in Urine
In total, epiCaPture was performed on 453 urine samples (Fig 3). Logistic regression models were developed using a 319-specimen training set (corresponding to 70% of the total cohort) and then evaluated for their ability to correctly predict cancer/high-grade cancer/high-risk cancer in the remaining test set. Analyzing all six genes together (epiCaPture score) consistently performed better than individual genes or combinations thereof. epiCaPture showed a comparable ability to predict cancer on biopsy as PSA (AUC for both, 0.64; Fig 3A), with a superior tumor specificity (0.98 v 0.81), considering both as continuous variables (Appendix Table A2). The prognostic value of epiCaPture was next studied. AUCs for detecting high-grade, high-risk D’Amico and CAPRA were 0.86, 0.83, and 0.80, respectively (Figs 3B-3D), with sensitivities of 0.85, 0.70, and 0.67 and NPV for all three prognostic end points greater than 0.94 (Appendix Table A2). Combining epiCaPture and PSA correctly predicted all high-grade cancers on biopsy (AUC, 0.95). Not forgetting the clinical significance of Gleason 7 disease, we also applied the revised five-grade grouping system,18 with a view to detecting group 3 (Gleason 4 and 3) and above. The combination of epiCaPture and PSA gave the best model (AUC, 0.82; sensitivity, 0.73; specificity, 0.76; Appendix Fig A1; Appendix Table A2).
Fig 3.
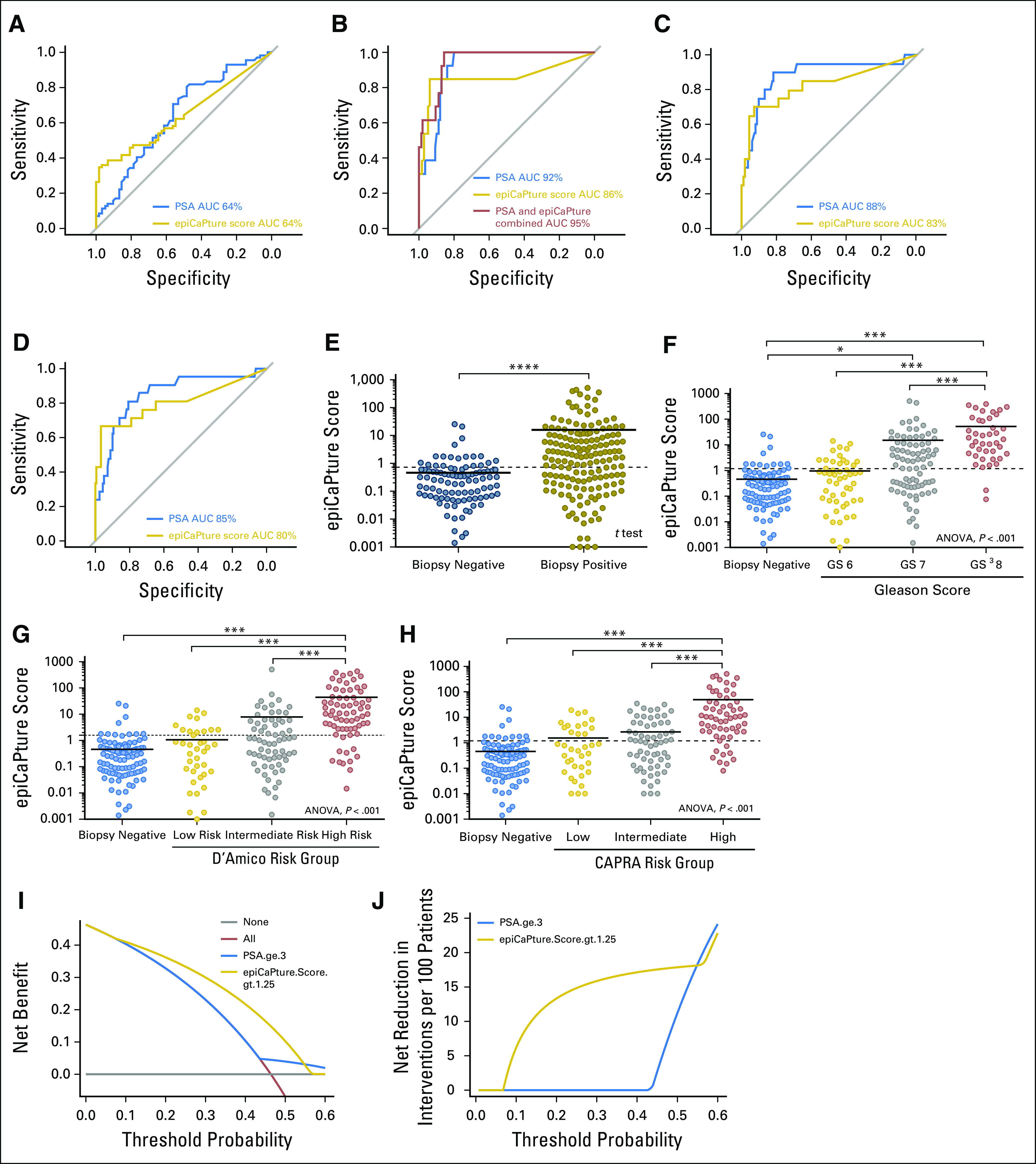
Epigenetic Cancer of the Prostate Test in Urine (epiCaPture) performance at noninvasive detection of prostate cancer in urine. The performance of epiCaPture compared with/in conjunction with prostate-specific antigen (PSA; treated as a continuous variable) in the test set (n = 134) for predicting each end point was assessed by receiver operating characteristic curves: (A) cancer (v no cancer detected on transrectal ultrasound biopsy), (B) high-grade cancer (Gleason score greater than or equal to 8 v all else), (C) D’Amico high-risk cancer (v all else), and (D) CAPRA (Cancer of the Prostate Risk Assesment) high-risk cancer (v all else). The distribution of epiCaPture scores in the whole study population (n = 453), viewed by (E) biopsy outcome, (F) Gleason grade, (G) risk categorization by D’Amico risk-classification systems, and (H) risk categorization by CAPRA risk-classification system. The epiCaPture score was calculated for each sample by summing the normalized index of methylation (Data Supplement) for G1 to G6. epiCaPture score distributions were analyzed by t test or one-way analysis of variance (ANOVA), followed by post hoc analyses using the Tukey-Kramer multiple comparisons test. For ease of interpretation, epiCaPture scores are plotted on a logarithmic axis, and samples that were negative (epiCaPture score = 0) are not shown. Solid black line represents the mean of each group. Dashed lines indicate the epiCaPture score thresholds derived in the training set to maximize both sensitivity and specificity. Significance is indicated by *P < .05, **P < .01, ***P < .001, and ****P < .0001. (I) Decision curve analysis demonstrated net clinical benefit of performing biopsy on patients in the test set on the basis of an epiCaPture score greater than 1.25 versus PSA greater than or equal to 3 ng/mL across a range of clinically relevant threshold probabilities (the risk of cancer, such that a patient would choose to undergo biopsy by weighing the relative harms of false-positive and false-negative predictions). The horizontal gray line represents the decision curve for the performing biopsy on no patients and the red line represents the decision curve for performing biopsy on all patients. (J) This translates to a net reduction in biopsies of approximately 15%, as compared with PSA, across a range of threshold probabilities. AUC, area under the receiver operating characteristic curve.
We next evaluated the training set data to select an epiCaPture score cutoff that could best inform the decision on whether to perform a biopsy. Applying a threshold greater than 0.73 detected 38% of all cancers in the test set. Increasing this cutoff to greater than 1.25 detected 85% of high-grade disease, with a tumor specificity of 86% (Figs 3E-3H). Compared with the clinically accepted PSA threshold of 3 ng/mL, this represents a 75% improvement in tumor specificity. Within the total study cohort, 245 men fell within the prognostically challenging PSA 4 to 10 ng/mL bracket. Of these 245, 35 (14.29%) men had high-grade cancer. At the greater than 1.25 threshold, 14 of 35 (40%) of these men with high-grade disease tested positive for epiCaPture. Finally, DCA showed that at this threshold, epiCaPture demonstrated a net benefit over PSA in the decision on whether to perform a biopsy, enabling a 15% reduction in prostate biopsies (Figs 3I and 3J).
epiCaPture Genes Show High Reproducibility by Independent Analysis
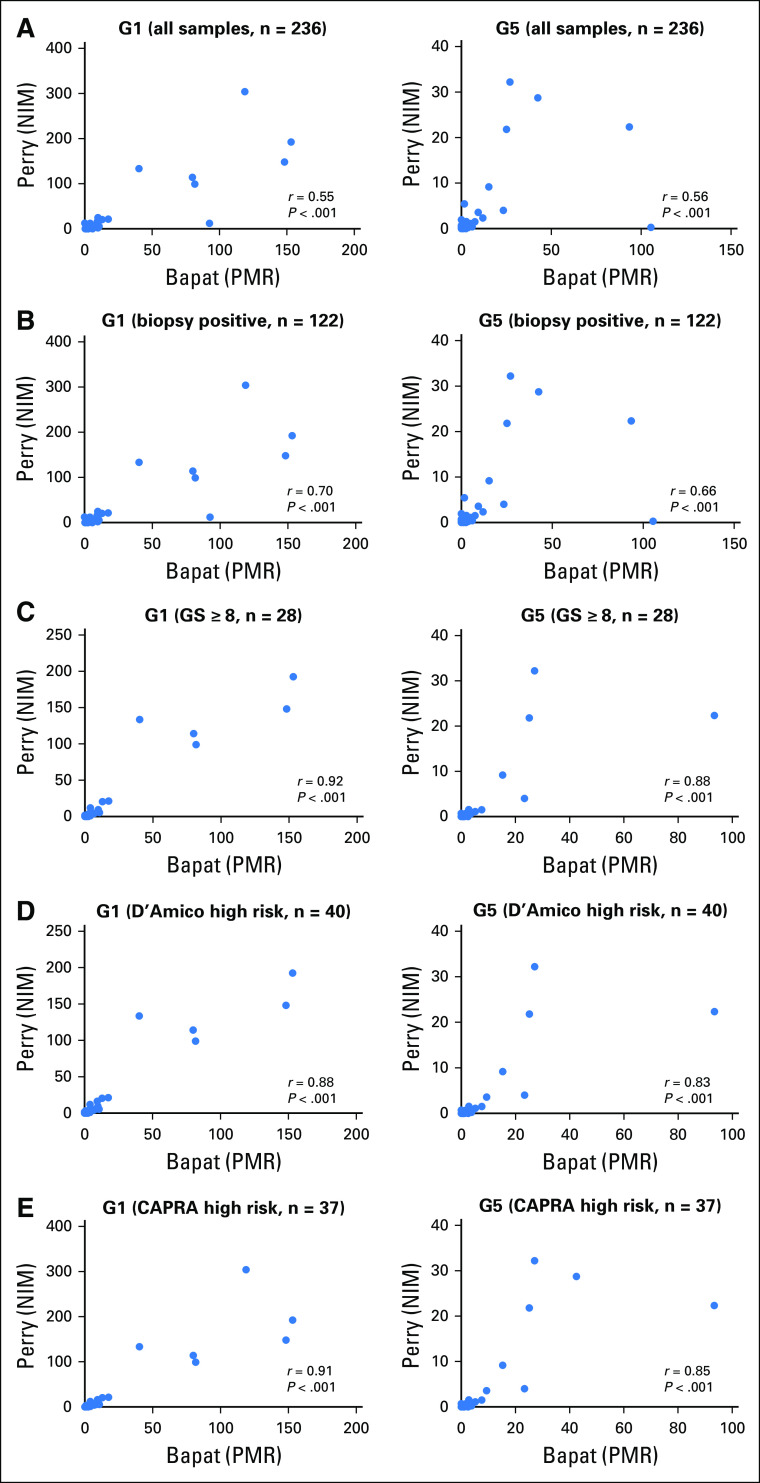
To examine the robustness of epiCaPture, a matched quantitative methylation specific PCR analysis of two of the panel (G1: GSTP1, and G5: APC) was carried out in two independent laboratories on 236 of the urine samples. Although the genes were the same, some differences existed in the target and control amplicon sequences and post-PCR analytical formulae (Data Supplement). Considering the sample set as a whole, irrespective of grade or other factors, both genes demonstrated a moderate correlation in methylation values between the two laboratories (r = 0.55, r = 0.56; P < .001; Fig 4A). However, the correlation increased for both genes, when considering only the biopsy-positive samples (n = 122; r = 0.70, r = 0.66; P < .001; Fig 4B), the high-grade samples (r = 0.92, r = 0.88; P < .001; Fig 4C) or high-risk samples by applying either D’Amico (r = 0.88, r = 0.83; P < .001; Fig 4D), or CAPRA (r = 0.91, r = 0.85; P < .001; Fig 4E) risk stratification. Furthermore, matched data were available for 18 of 30 high-risk/grade samples that were epiCaPture negative, of which 15 (83.33%) and 16 (88.89%) were also negative for G1 and G5, respectively, in the independent analysis.
Fig 4.
Robustness of Epigenetic Cancer of the Prostate Test in Urine (epiCaPture) assays by independent verification. (A) Two of the six epiCaPture assays (G1: GSTP1 and G5: APC) were analyzed independently in laboratories in Ireland (Perry) and Canada (Bapat) by quantitative methylation specific PCR in urine sediments from 236 of 453 men in the study. Relevant subsets were then considered by applying the study end points: (B) men with biopsy-positive disease, (C) men with high-grade (Gleason score greater than or equal to 8) prostate cancer, and men with high-risk prostate cancer defined by (D) D’Amico, and (E) CAPRA (Cancer of the Prostate Risk Assessment) classification. Data from the two laboratories are differentiated by the labels Perry and Bapat to indicate the principle investigator at each site. In each data set, the normalized index of methylation (NIM) and percent methylated ratio (PMR) represent the normalized data value for each sample. Correlation was calculated using Spearman rank correlation.
Parallel epiCaPture Analysis in Urine and Matched Biopsy Cores Highlights Value of Liquid Biopsy
PCa is renowned for its multifocal nature. A concern for any liquid biopsy is therefore how well it captures molecular aberrations occurring within discrete tumor foci. Matched formalin-fixed paraffin-embedded biopsy cores were available for a proportion of patients, enabling parallel epiCaPture analysis on multiple tumor foci and urine from individual men (n = 15). G1 to G6 methylation was higher in biopsy cores than in urine (Appendix Fig A2A), indicative of the higher tumor content in tissue over liquid biopsies. Similarly, individuals demonstrated higher epiCaPture scores in tissues than in urine (Appendix Fig A2B). The most interesting insights came from the matched analysis of urine with multiple biopsy cores, of which two notable cases are highlighted.
Patient SJH149 was diagnosed with a minute focus of Gleason score 6 adenocarcinoma. His urine epiCaPture score was strongly positive. A repeat biopsy revealed high-grade cancer in 40% of cores on the right side (Fig 5A). epiCaPture analysis of the original biopsy cores revealed DNA methylation, albeit at a magnitude lower than detected in urine. Notably, the epiCaPture fingerprint (ie, the relative contribution of each gene to the epiCaPture score) differed between tissue cores and urine. This finding could suggest that the source of the tumor DNA analyzed in urine could have been the high-grade tumor, which was not sampled on the original biopsy.
Fig 5.
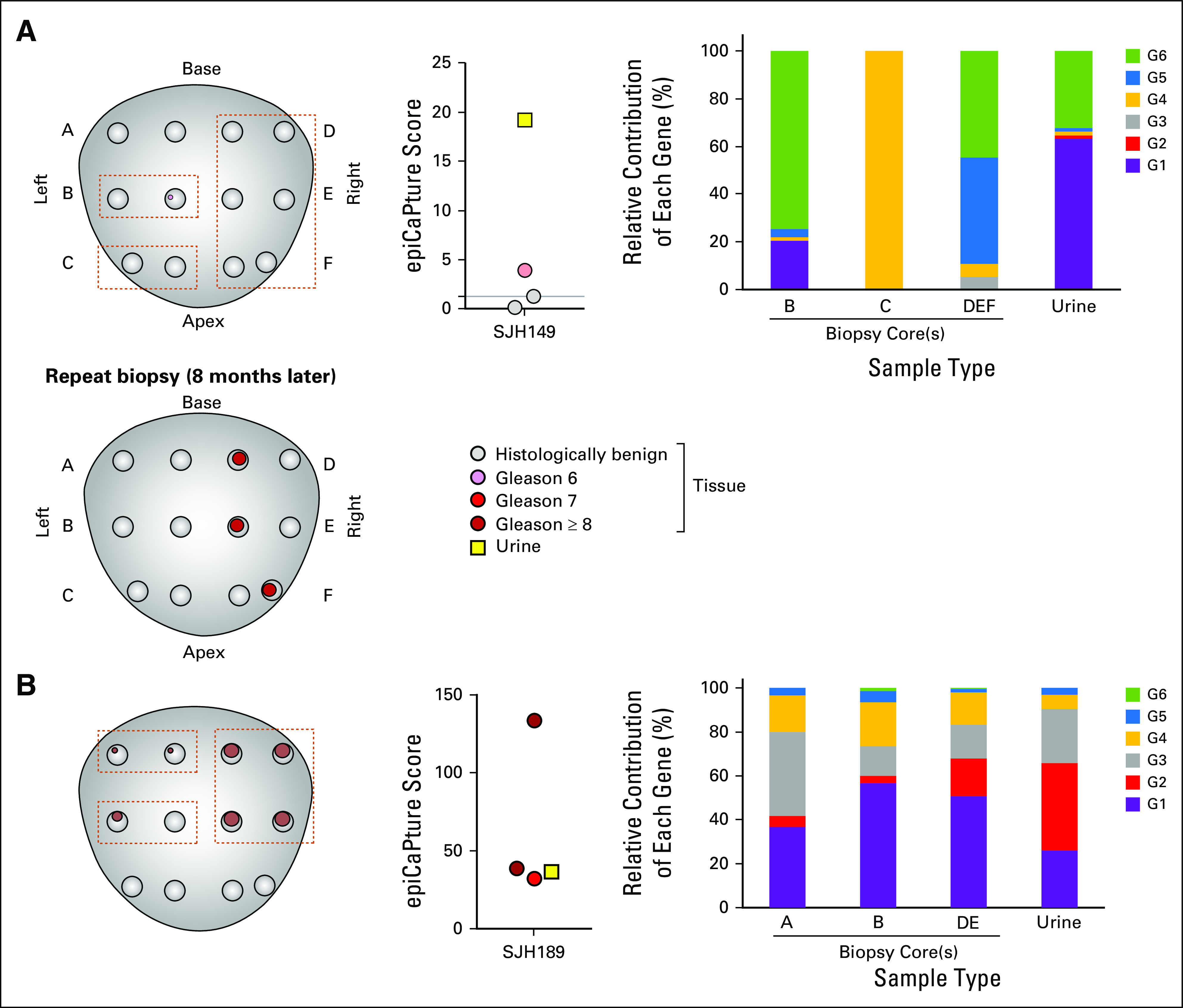
Parallel Epigenetic Cancer of the Prostate Test in Urine (epiCaPture) analysis in liquid and tissue biopsies. Prostate maps are for indicative purposes only and are not drawn to scale. A standard 12-core biopsy is illustrated. In some cases, more than 12 cores were taken. Tumor presence is denoted by pink (Gleason score 6), red (Gleason score 7), or brown (Gleason score greater than or equal to 8) circles, with size indicative of percentage tumor in each core. Dashed brown lines indicate cores that were used/combined for DNA extraction. epiCaPture analysis was performed in parallel on matched biopsy cores (circles) and prebiopsy urine (yellow square). The threshold for positivity (epiCaPture score greater than 1.25) is indicated on the graph. epiCaPture fingerprints indicate the relative contribution of each of the six genes to the epiCaPture score calculated for matched tissue cores and urine. (A) Low-risk prostate cancer SJH149 (62 years of age; prostate-specific antigen, 3.08 ng/mL) demonstrated that urine as a liquid biopsy is a viable alternative to sampling bias inherent to tissue needle biopsies. Original biopsy: tumor occupying less than 1% total volume in one of two cores on the left-hand side. DNA was extracted from cores B, C, and D + E + F. Repeat biopsy: tumor was present in the medial, anterior, and posterior of the right-hand side of the gland. (B) High-risk prostate cancer SJH189 (80 years of age; prostate-specific antigen, 6.63 ng/mL) demonstrated similar epiCaPture fingerprint across different tumor areas sampled on biopsy and in urine.
In contrast, patient SJH189 was diagnosed with a high-percentage, high-grade (intermixed patterns of grade 4 and 5) PCa on both sides of his prostate. His urine epiCaPture score was strongly positive, as were his tissue cores (Fig 5B). His epiCaPture fingerprint was comparable across urine and tissue cores, with a strong contribution from G1 to G4, smaller contribution from G5, and G6 negligible. So, despite pathologic heterogeneity within the gland, the three tumor areas seemed to be epigenetically homogeneous.
DISCUSSION
In this urine PCa biomarker study, we evaluated the performance of epiCaPture, a six-gene DNA methylation panel, for noninvasive detection of PCa and more specifically high-grade or high-risk PCa in urine from patients referred for prostate biopsy on the basis of elevated PSA and/or abnormal DRE. Complying with REMARK guidelines, we describe a new DNA methylation-based urine test for early detection of aggressive PCa.
Many prostate tumors display a protracted disease trajectory, almost indolent in behavior.19 The widespread use of the serum PSA test in many countries has significantly increased the incidence and thus overtreatment of these low-risk cancers with little likelihood of clinical manifestation.20-23 An important first consideration in developing epiCaPture was to therefore critically appraise the evidence for prognostic utility of each constituent gene. Using two independent cohorts of prostate tissues, we showed that the DNA methylation of epiCaPture genes increases with aggressiveness. Conversely, however, it is estimated that approximately two thirds of men undergoing invasive prostate biopsy have no tumor diagnosed, because of the high false-positive rate of serum PSA.23,24 As such, millions of unnecessary biopsies are performed worldwide each year, at great cost to our health care systems and causing significant anxiety, trauma, and comorbidities for patients.25,26 PSA accuracy has been improved by the development of algorithms incorporating different PSA isoforms, namely the four-kallikrein and Prostate Health Index tests, with AUC values in the region of 0.7 (all PCa) and 0.71 to 0.79 (high-grade PCa, Gleason score greater than or equal to 7).4,27,28 However, PSA is a single biomarker; used alone it does not address PCa heterogeneity or inform on disease biology.
The multifocal nature of PCa together with small sampling (less than 5%) of the gland by TRUS biopsy is problematic; approximately 20% of tumors are upgraded on radical prostatectomy because of sampling error.29 Although sophisticated imaging technologies, such as magnetic resonance imaging, may improve the accuracy of prostate biopsy,30 their widespread application is hampered by high cost, access to specialist equipment and personnel, interobserver subjectivity, and requirement for anesthesia. Alternatively, transperineal biopsy enables a more complete sampling of the gland, including the anterior prostate, but also requires anesthesia and is thus unsuitable as a population-based detection method for such a prevalent disease.
The dearth of blood-based prognostic biomarkers as a screening tool for aggressive PCa in conjunction with limitations of the prostate biopsy in its various forms has led to excitement around urine biomarkers. Physical manipulation of the prostate by DRE, followed by a first-void urinalysis, could provide a simple, holistic insight into molecular alterations occurring in the prostate gland. Indeed, many studies have demonstrated significant value in detecting and noninvasively monitoring several genitourinary malignancies through urine.31-34 Some other prognostic urine indicators of aggressive PCa have already been described. For example, the Mi Prostate Score (MiPS), which incorporates the prostate-specific TMPRSS2-ERG fusion and PCA3 transcripts in urine, in conjunction with serum PSA, delivers AUCs of 0.75 and 0.78 for detecting all PCa and high-grade PCa, respectively.35 However, the prognostic value of this combination has been questioned by others.36,37 Combining expression of three genes (HOXC6, TRD1, and DLX1) functionally implicated in PCa with serum PSA detected clinically significant disease (Gleason score greater than or equal to 7 on biopsy), with an AUC of 0.81.38
Epigenetic alterations represent a particularly attractive source of biomarkers: promoter hypermethylation of many regulatory genes occurs early during prostate cancer initiation, it is often prognostic, and assessment of DNA is a more robust analyte than RNA. Early studies demonstrated detection of GSTP1 methylation in urine from patients with PCa, although sensitivity was poor (less than 30%).39 Follow-up work expanded the repertoire of markers (GSTP1, APC, and RARβ2), which improved sensitivity to approximately 60%.40 The epiCaPture panel represents five commonly perturbed pathways in PCa: intracellular detoxification, Wnt and IGF axes, cell cycle, and prostaglandin biosynthesis. Assessing its diagnostic performance in terms of ability to predict cancer (irrespective of grade) on biopsy, epiCaPture was equivalent to PSA, with AUCs of 0.64. However, the reality of clinical practice is that PSA is not used as a continuous variable, with a threshold applied to guide decisions on whether to biopsy. Taking the widely used cut-off of 3 ng/mL, the performance of PSA decreased to AUC of 0.55, in line with previous reports. In contrast, applying an epiCaPture threshold (greater than 0.73; developed on training set) did not markedly alter its diagnostic performance (AUC, 0.68) and delivered a 79% improvement over the tumor specificity of PSA.
In an effort to address the prognostic utility of epiCaPture within the context of the clinically heterogeneous Gleason 7 disease, we applied the revised five-grade grouping system,18 in addition to two widely used risk-classification systems. It should be noted all men with aggressive cancers in this study had an elevated PSA; however, its specificity was poor. The epiCaPture performance characteristics represent a potential improvement on existing methods for patient stratification and for determining the need to perform biopsy, detecting 85% of high-grade tumors, with an NPV of 98%. Furthermore, the overlap of GSTP1 and APC in the two GAP1 urine DNA methylation biomarker panels revealed excellent correlation in detection of aggressive PCa, illustrating the robustness of these biomarkers. In the context of the diagnostic gray zone, PSA 4 to 10 ng/mL, epiCaPture demonstrated potential for further classifying patients, positive in 40% of men with high-grade PCa. Applying DCA with epiCaPture showed a net reduction of approximately 15% in prostate biopsies. Given the high prevalence of this disease, this has the potential to dramatically reduce the number of biopsies being performed. Although these data are promising, a large prospective clinical trial is needed to assess whether epiCaPture offers a survival advantage.
A limitation of our study is that the cohort is young; analysis was only possible using biopsy as the end point. We cannot rule out the possibility that a proportion of our epiCaPture-positive urine samples in men with low-grade or benign tissues were indeed tumor cases missed on biopsy. Longer-term follow-up is thus needed to more accurately assess the false-positive rate of epiCaPture (calculated at 5% in this study, but presumed lower because of sampling error of TRUS biopsy). Evidence actually shows that DNA methylation detected in histologically benign tissue (due to an epigenetic field effect from the cancer), can be used as a tool to rule out the possibility that the biopsy missed the tumor. For example, the Confirm MDx test, which measures DNA methylation at three genes (GSTP1, APC, and RASSF1) in histologically benign biopsy tissue cores, is used to guide the decision to repeat a biopsy.41
Conversely, 14 of 47 (29.79%) high-grade tumors were epiCaPture-negative. Independent analysis revealed concordant negativity in more than 80% of these men, and lowering the epiCaPture threshold did alter this result; they were negative across all six markers. Possible explanations such as low tumor volume were excluded. We believe that the most likely reason is interoperator differences in DRE; these high-risk/grade cancers were epiCaPture negative because they had insufficient/undetectable prostate cells present in their urine. Future work will focus on incorporating expression of a prostate-specific marker (ie, KLK2 or KLK3) into epiCaPture, to enable confirmation of the presence of prostate cells in the urine, validating its suitability for epiCaPture analysis and ultimately improving the performance of the test. Future work will also investigate the intra-individual reproducibility of epiCaPture by repeated measures in men.
Finally, to the best of our knowledge, this is the first PCa biomarker study to perform a matched analysis of tissue and liquid biopsies. A more extensive project is now warranted to fully assess the effects of the multifocal nature of PCa on signatures derived from liquid biopsies.
epiCaPture could be used in conjunction with existing tools (ie, PSA and risk calculators) to help guide the decision to biopsy. On further validation and cost-benefit analysis, epiCaPture may have utility as a population-based stratification tool to identify those men who require a biopsy, to alleviate patient harms and hospital burdens of over-performing biopsies on patients.
ACKNOWLEDGMENT
We thank the urologists and their teams at participating hospitals and the Health Research Board–funded Dublin Centre for Clinical Research Network, affiliated research nurses, and technical staff, in particular John McCourt. We thank investigators Drs Robert Vessella, and Colm Morrissey, and the patients and families who participated in the prostate cancer donor program at the University of Washington Medical Center. This article is dedicated to the late Professor John Fitzpatrick, MD, a wonderful mentor.
Movember GAP1 Urine Biomarker Collaborative Members:
Colin Cooper, Jeremy Clark, Schools of Medicine and Biological Sciences, University of East Anglia, Norwich, Norfolk.
Bharati Bapat, Lunenfeld-Tanenbaum Research Institute, Sinai Health Systems, University of Toronto, Toronto, Ontario, Canada.
Robert G. Bristow, Director, Manchester Cancer Research Centre, Chief Academic Officer and Honorary Consultant, Christie NHS Trust, University Professor of Cancer Studies, University of Manchester, Manchester Cancer Research Centre, University of Manchester, Manchester, UK.
Chris Parker, Royal Marsden Hospital, Sutton, UK.
Ian Mills, Queen’s University Belfast, Centre for Cancer Research and Cell Biology, Movember/Prostate Cancer UK Centre of Excellence, School of Medicine, Dentistry, and Biomedical Sciences. Institute for Health Sciences, Belfast, UK; Centre for Molecular Medicine Norway, Oslo University and Oslo University Hospitals, Forskningsparken, Oslo, Norway.
Hardev Pandha, University of Surrey, Guildford, Surrey, UK.
Hayley Whitaker, Molecular Diagnostics and Therapeutics Group, Cruciform Building, University College London, London, UK.
David Neal, Cancer Research UK Cambridge Research Institute, Cambridge, UK.
Mireia Olivan, Andreas Doll, Vall d’Hebron Research Institute and Hospital and Autonomous University of Barcelona, Barcelona, Spain.
Hing Leung, The Beatson Institute for Cancer Research, University of Glasgow, Glasgow, UK.
Antoinette Perry, Cancer Biology and Therapeutics Laboratory, School of Biomolecular and Biomedical Science, Conway Institute, University College Dublin, Dublin, Ireland.
Martin Sanda, Urology, Emory University and Emory Healthcare, Atlanta, GA.
Jack Schalken, Nijmegen Medical Centre, Radboud University, Nijmegen, The Netherlands.
Appendix
Fig A1.
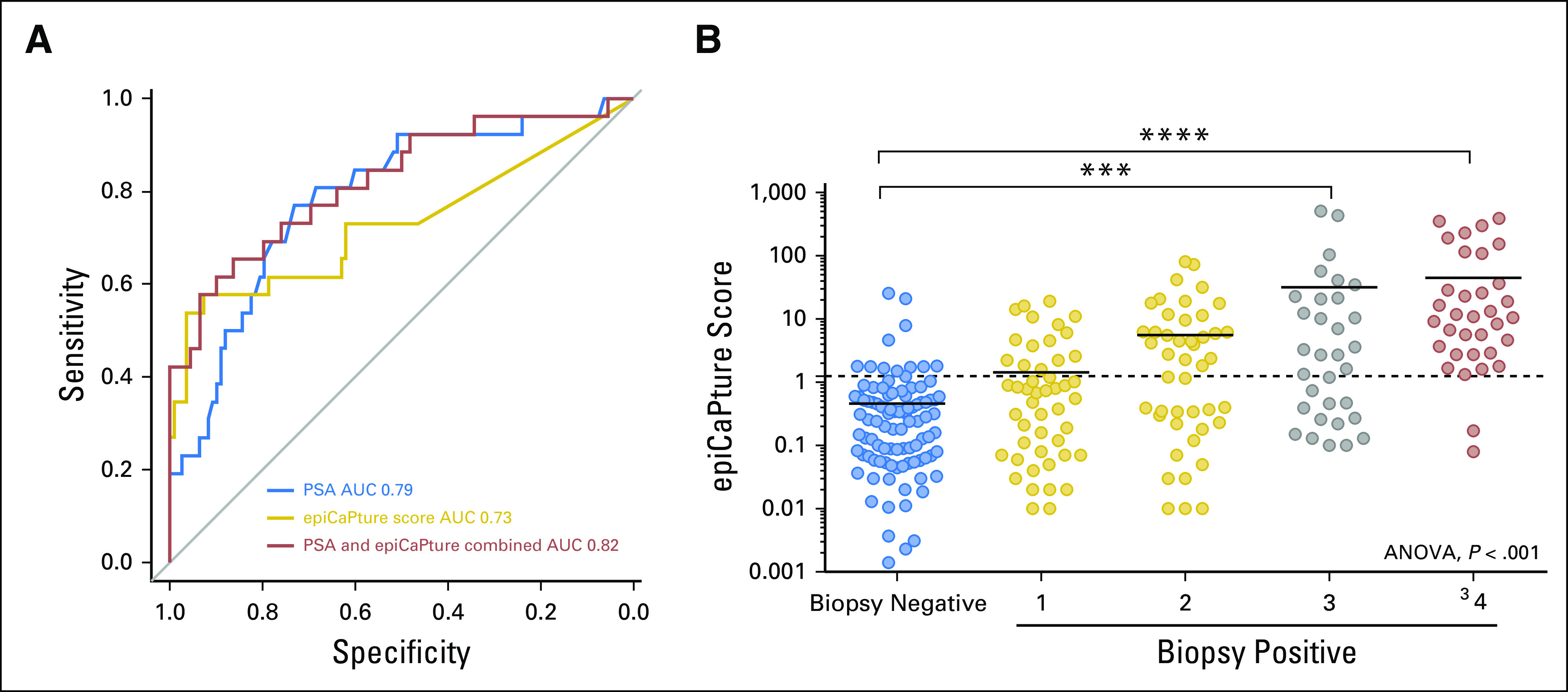
(A) The performance of epiCaPture compared to and in conjunction with prostate-specific antigen (PSA; treated as a continuous variable) in the test set (n = 134) for predicting ISUP group 3, 4 and 5 PCa on biopsy was assessed by receiver operator characteristic curves. (B) The distribution of epiCaPture scores in biopsy-negative (no cancer detected) v biopsy-positive patients, categorized by ISUP group, shows increasing methylation by group. Significance is indicated by ***P < .001 and ****P < .0001. ANOVA, analysis of variance; AUC, area under the receiver operating curve.
Fig A2.
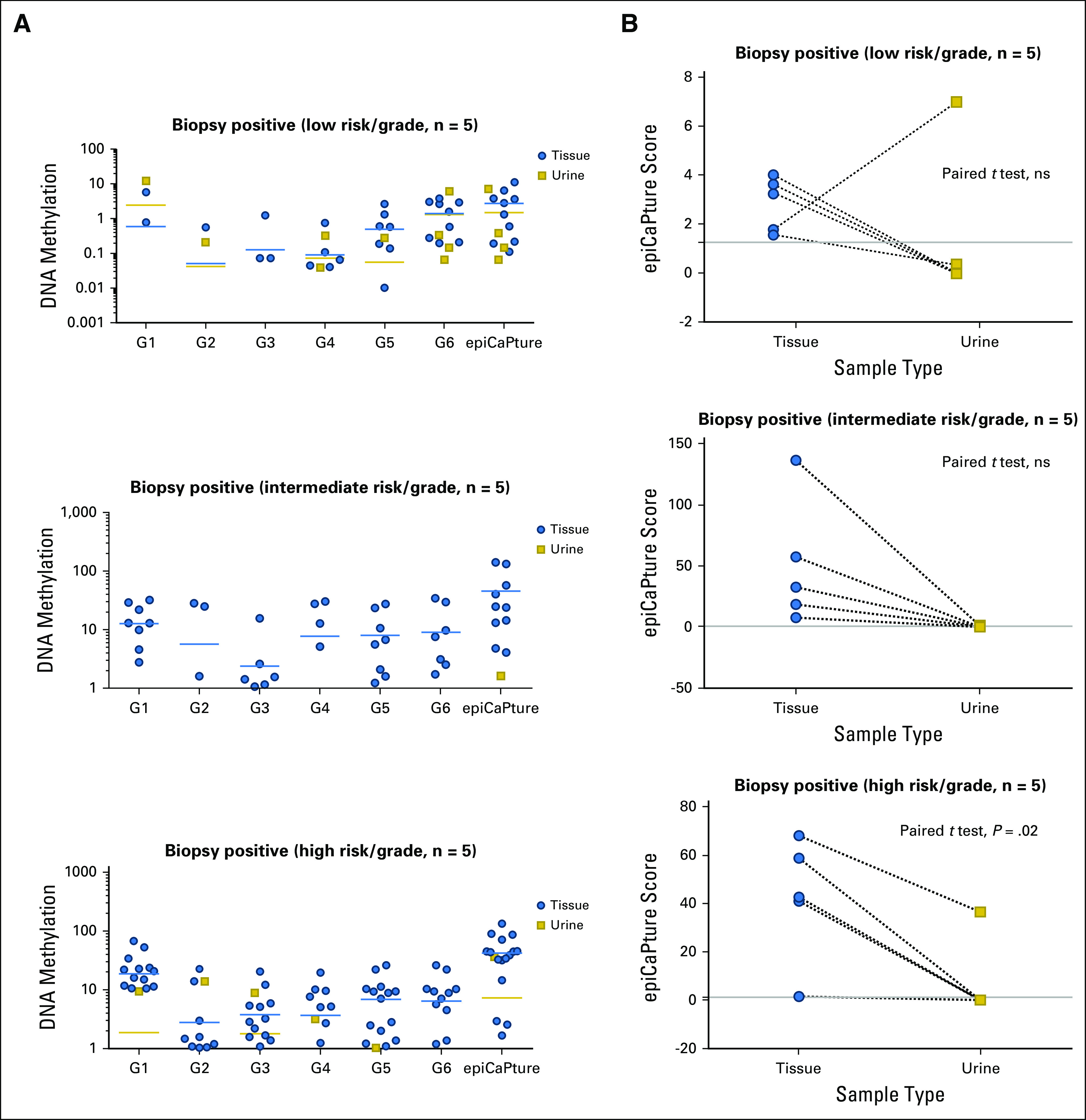
Parallel epiCaPture analysis in urine and matched biopsy cores. epiCaPture was performed in urine and matched formalin-fixed paraffin embedded biopsy cores from randomly selected biopsy-positive (n = 15: low-risk [CAPRA and D’Amico, Gleason score 6; intermediate risk, Gleason score 7; and high risk, Gleason score ≥ 8]) patients. (A) Parallel analysis of epiCaPture genes (G1-G6) in biopsy tissue cores and urine. DNA methylation of each gene is indicated by the NIM (normalised index of methylation), and for the collective panel, by the epiCaPture score (NIM sum G1-G6). Each point represents a single sample (biopsy core or urine), with the mean for each group indicated by a horizontal line (yellow, urine; blue, tissue). For ease of interpretation, DNA methylation is plotted on a logarithmic axis, and samples with no methylation are not shown. (B) Paired T tests on matched biopsy cores and urine reveal higher epiCaPture scores in tissues than in urine. For each patient, a single tissue epiCaPture score was calculated using the average across all cores studied (1-5). The epiCaPture threshold for urine positivity (1.25) is indicated by a gray line. ns, not significant.
Table A1.
Description of epiCaPture qMSP Primers and Probes
Table A2.
epiCaPture Performance Characteristics in Test Set
Footnotes
Supported by grant funding from Movember (GAP1 Urine Biomarker Award), US Prostate Cancer Foundation (Young Investigator Award, A.S.P.), Science Foundation Ireland Grants No. 14/TIDA/2348 (A.S.P.) and 15/IA/3104 (W.M.G.), Royal Irish Academy (cost-sharing award, A.S.P. and J.C.), Irish Cancer Society Grant No. CRF09PER (A.S.P.), Bob Champion Cancer Trust, the Masonic Charitable Foundation, the King family (C.S.C.), and the National Institutes of Health Grant No. U01-113913 (M.G.S.).
AUTHOR CONTRIBUTIONS
Conception and design: Colin Cooper, Bharati Bapat, Martin G. Sanda, Jeremy Clark, Antoinette S. Perry
Provision of study material or patients: Neil Fleshner, Robert Mills, Marcelino Y. Hanna, Elizabeth McEvoy, Rustom P. Manecksha, Colin Cooper, Bharati Bapat, Martin G. Sanda, Jeremy Clark
Collection and assembly of data: Eve O’Reilly, Alexandra V. Tuzova, Anna L. Walsh, Odharnaith O’Brien, Sarah Kelly, Odharna Ni Dhomhnallain, Liam DeBarra, Connie M. Dale, Rick Brugman, Gavin Clarke, Olivia Schmidt, Dattatraya Patil, Kathryn L. Pellegrini, Neil Fleshner, Julia Garcia, Fang Zhao, Stephen Finn, Robert Mills, Marcelino Y. Hanna, Rachel Hurst, Elizabeth McEvoy, Colin S. Cooper, Daniel S. Brewer, Bharati Bapat, Antoinette S. Perry
Data analysis and interpretation: Eve O'Reilly, Alexandra V. Tuzova, Niamh M. Russell, Sarah Kelly, Rick Brugman, Gavin Clarke, Olivia Schmidt, Shane O’Meachair, Dattatraya Patil, William M. Gallagher, Rustom P. Manecksha, Daniel S. Brewer, Jeremy Clark, Antoinette S. Perry
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Eve O'Reilly
No relationship to disclose
Alexandra V. Tuzova
No relationship to disclose
Anna L. Walsh
No relationship to disclose
Niamh M. Russell
No relationship to disclose
Odharnaith O'Brien
No relationship to disclose
Sarah Kelly
No relationship to disclose
Odharna Ni Dhomhnallain
No relationship to disclose
Liam DeBarra
No relationship to disclose
Connie M. Dale
No relationship to disclose
Rick Brugman
No relationship to disclose
Gavin Clarke
No relationship to disclose
Olivia Schmidt
No relationship to disclose
Shane O'Meachair
No relationship to disclose
Dattatraya Patil
No relationship to disclose
Kathryn L. Pellegrini
No relationship to disclose
Neil Fleshner
Honoraria: Amgen, Janssen Oncology, Bayer, Sanofi, AbbVie, Ferring Pharmaceuticals, Astellas Medivation
Consulting or Advisory Role: Hybridyne Health
Research Funding: Ferring (Inst), Astellas Pharma (Inst), Janssen Oncology (Inst), Amgen (Inst), Nucleix (Inst), Progenix (Inst), Spectracure AB (Inst)
Julia Garcia
No relationship to disclose
Fang Zhao
No relationship to disclose
Stephen Finn
Honoraria: Roche
Research Funding: Amgen (Inst)
Travel, Accommodations, Expenses: Pfizer
Robert Mills
No relationship to disclose
Marcelino Y. Hanna
No relationship to disclose
Rachel Hurst
No relationship to disclose
Elizabeth McEvoy
No relationship to disclose
William M. Gallagher
Employment: OncoMark
Leadership: OncoMark
Stock and Other Ownership Interests: OncoMark
Consulting or Advisory Role: Carrick Therapeutics
Research Funding: Carrick Therapeutics
Patents, Royalties, Other Intellectual Property: Two patents which have been licensed to OncoMark
Travel, Accommodations, Expenses: OncoMark
Rustom P. Manecksha
Honoraria: Janssen, Boston Scientific
Travel, Accommodations, Expenses: Ferring Pharmaceuticals, Astellas Pharma
Colin S. Cooper
Patents, Royalties, Other Intellectual Property: I have two patents for tests for aggressive prostate cancer filed in the past 2 years. These patents are not related to the current work. (Inst)
Daniel S. Brewer
Patents, Royalties, Other Intellectual Property: Patents held in drug discovery with Novartis (I), patents pending concerning cancer subtype detection and biomarkers with University of East Anglia
Bharati Bapat
No relationship to disclose
Martin G. Sanda
No relationship to disclose
Jeremy Clark
No relationship to disclose
Antoinette S. Perry
Patents, Royalties, Other Intellectual Property: University College Dublin holds a patent that relates to this work
REFERENCES
- 1.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer; http://globocan.iarc.fr [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Wilt TJ, Jones KM, Barry MJ, et al. Follow-up of prostatectomy versus observation for early prostate cancer. N Engl J Med. 2017;377:132–142. doi: 10.1056/NEJMoa1615869. [DOI] [PubMed] [Google Scholar]
- 4.Nordström T, Vickers A, Assel M, et al. Comparison between the four-kallikrein panel and prostate health index for predicting prostate cancer. Eur Urol. 2015;68:139–146. doi: 10.1016/j.eururo.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roobol MJ, Vedder MM, Nieboer D, et al. Comparison of two prostate cancer risk calculators that include the prostate health index. Eur Urol Focus. 2015;1:185–190. doi: 10.1016/j.euf.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Altman DG, McShane LM, Sauerbrei W, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry AS, Loftus B, Moroose R, et al. In silico mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer. 2007;96:1587–1594. doi: 10.1038/sj.bjc.6603767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry AS, O’Hurley G, Raheem OA, et al. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int J Cancer. 2013;132:1771–1780. doi: 10.1002/ijc.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan L, Murphy TM, Barrett C, et al. IGFBP7 promoter methylation and gene expression analysis in prostate cancer. J Urol. 2012;188:1354–1360. doi: 10.1016/j.juro.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Bastian PJ, Ellinger J, Heukamp LC, et al. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur Urol. 2007;51:665–674. doi: 10.1016/j.eururo.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Kang GH, Lee S, Lee HJ, et al. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 12.Richiardi L, Fiano V, Vizzini L, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27:3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 13.Mahapatra S, Klee EW, Young CY, et al. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18:2882–2895. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- 14.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 15.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes DF, Allen J, Compton C, et al. Breaking a vicious cycle. Sci Transl Med. 2013;5:196cm6. doi: 10.1126/scitranslmed.3005950. [DOI] [PubMed] [Google Scholar]
- 18.Epstein JI, Zelefsky MJ, Sjoberg DD, et al. A contemporary prostate cancer grading system: A validated alternative to the Gleason score. Eur Urol. 2016;69:428–435. doi: 10.1016/j.eururo.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson JE, Andrén O, Andersson SO, et al. Natural history of early, localized prostate cancer. JAMA. 2004;291:2713–2719. doi: 10.1001/jama.291.22.2713. [DOI] [PubMed] [Google Scholar]
- 20.Hugosson J, Carlsson S, Aus G, et al. Mortality results from the Göteborg randomised population-based prostate-cancer screening trial. Lancet Oncol. 2010;11:725–732. doi: 10.1016/S1470-2045(10)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 22.Andriole GL, Crawford ED, Grubb RL, III, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heijnsdijk EA, Wever EM, Auvinen A, et al. Quality-of-life effects of prostate-specific antigen screening. N Engl J Med. 2012;367:595–605. doi: 10.1056/NEJMoa1201637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andriole GL. Pathology: The lottery of conventional prostate biopsy. Nat Rev Urol. 2009;6:188–189. doi: 10.1038/nrurol.2009.46. [DOI] [PubMed] [Google Scholar]
- 25.Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol. 2013;64:876–892. doi: 10.1016/j.eururo.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 26.Lundström KJ, Drevin L, Carlsson S, et al. Nationwide population based study of infections after transrectal ultrasound guided prostate biopsy. J Urol. 2014;192:1116–1122. doi: 10.1016/j.juro.2014.04.098. [DOI] [PubMed] [Google Scholar]
- 27.Loeb S, Sanda MG, Broyles DL, et al. The prostate health index selectively identifies clinically significant prostate cancer. J Urol. 2015;193:1163–1169. doi: 10.1016/j.juro.2014.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vickers AJ, Cronin AM, Roobol MJ, et al. A four-kallikrein panel predicts prostate cancer in men with recent screening: Data from the European Randomized Study of Screening for Prostate Cancer, Rotterdam. Clin Cancer Res. 2010;16:3232–3239. doi: 10.1158/1078-0432.CCR-10-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haffner MC, De Marzo AM, Yegnasubramanian S, et al. Diagnostic challenges of clonal heterogeneity in prostate cancer. J Clin Oncol. 2015;33:e38–e40. doi: 10.1200/JCO.2013.50.3540. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–397. doi: 10.1001/jama.2014.17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa VL, Henrique R, Danielsen SA, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16:5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 32.Torres-Ferreira J, Ramalho-Carvalho J, Gomez A, et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol Cancer. 2017;16:26. doi: 10.1186/s12943-017-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junker K, Heinzelmann J, Beckham C, et al. Extracellular vesicles and their role in urologic malignancies. Eur Urol. 2016;70:323–331. doi: 10.1016/j.eururo.2016.02.046. [DOI] [PubMed] [Google Scholar]
- 34.Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol. 2014;32:4066–4072. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70:45–53. doi: 10.1016/j.eururo.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leyten GH, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65:534–542. doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leyten GH, Hessels D, Smit FP, et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin Cancer Res. 2015;21:3061–3070. doi: 10.1158/1078-0432.CCR-14-3334. [DOI] [PubMed] [Google Scholar]
- 39.Cairns P, Esteller M, Herman JG, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–2730. [PubMed] [Google Scholar]
- 40.Baden J, Adams S, Astacio T, et al. Predicting prostate biopsy result in men with prostate specific antigen 2.0 to 10.0 ng/ml using an investigational prostate cancer methylation assay. J Urol. 2011;186:2101–2106. doi: 10.1016/j.juro.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 41.Van Neste L, Partin AW, Stewart GD, et al. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate. 2016;76:1078–1087. doi: 10.1002/pros.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]