Abstract
Bacteria are under constant attack from bacteriophages (phages), bacterial parasites that are the most abundant biological entity on earth. To resist phage infection, bacteria have evolved an impressive arsenal of anti-phage systems. Recent advances have significantly broadened and deepened our understanding of how bacteria battle phages, spearheaded by new systems like CRISPR-Cas. This review aims to summarise bacterial anti-phage mechanisms, with an emphasis on the most recent developments in the field.
Introduction
Throughout evolution, bacteria have been preyed upon by parasitic bacteriophages (phages). Everywhere bacteria are found, they coexist with their respective phages, undergoing continuous cycles of infection. As a consequence, in order to survive and thrive, bacteria have developed an arsenal of anti-phage mechanisms. Due to the immense evolutionary pressure imposed by phages, the diversity and sophistication of bacteria’s anti-phage mechanisms are astounding, and we are only now beginning to appreciate the complexity of the interactions between bacteria and these parasites. In addition, the study of anti-phage mechanisms has resulted in invaluable tools, such as restriction enzymes and CRISPR-based gene editing techniques. This review summarises the mechanisms employed by bacteria to resist their phages, with an emphasis of novel developments in the field.
Bacteriophage-host interactions
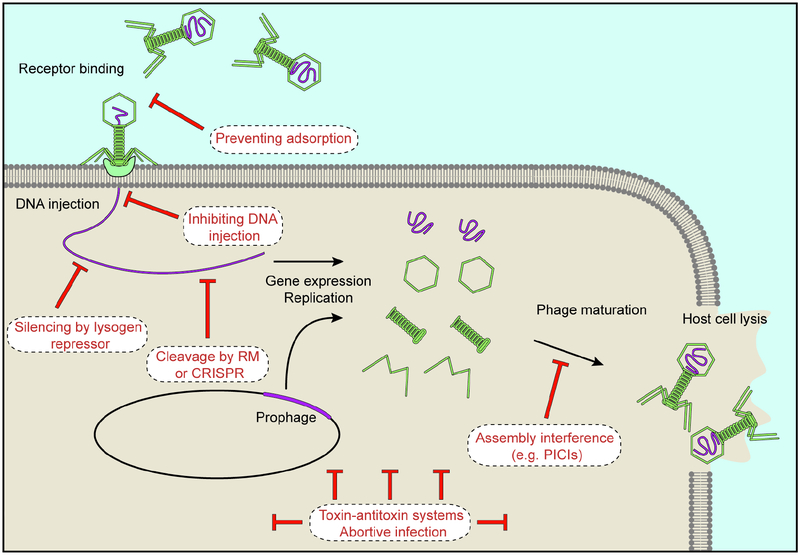
The red queen hypothesis states that an organism must constantly evolve to maintain their relative fitness in the face of a predator (McLaughlin and Malik, 2017). In the context of the bacteria-phage relationship, this means that bacteria continuously evolve and update anti-phage mechanisms, while phages adapt to overcome these mechanisms. Competitive bacteria-phage coevolution, often referred to as an “evolutionary arms race”, has produced a multitude of bacterial defence mechanisms that act to inhibit every stage of the phage life cycle (Figure 1). Although not discussed extensively in this review, phages have developed as many means to circumvent these defence strategies. For example, a metagenomic study in this issue (Uribe et al., 2019) identified phylogenetically widespread phage-encoded anti-CRISPR genes, encoding proteins that neutralise CRISPR immunity.
Figure 1. Stages of a phage’s life cycle that can be targeted by different anti-phage mechanisms.
Upon recognising a surface receptor, a phage injects its DNA into the host cell. Either after injection or after prophage induction, the viral genome is subject to several rounds of replication and gene expression that leads to the assembly and accumulation of new viral particles, which are released upon lysis of the host cell. As indicated, anti-phage mechanisms can interfere with any part of this process.
As a result of this arms race, bacteria and phages coevolve, and seem to exist in stable equilibria without dramatic fluctuations or extinction events in natural environments (Fernandez et al., 2018). Key to this arms race is the propensity of bacterial defence systems to spread through horizontal gene transfer (Stern and Sorek, 2011). Whereas this in principle could lead to an extensive proliferation of defence mechanisms to provide more protection to the host population, bacteria only tend to have a subset of the available diversity of anti-phage mechanisms. This in part due to fitness costs associated with carrying defence systems. Therefore bacteria, even in the context of a race for survival against their parasites, must tune the trade-off between the cost of carrying anti-phage systems and the benefit of resisting phage infection (van Houte et al., 2016a).
Preventing phage entry
Infection begins with the binding to specific surface proteins or cell wall components of the host cell, an event that is followed by the injection of the phage’s genome. Consequently, bacteria use both broad and phage-specific mechanisms to prevent phage adsorption and injection (Figure 2).
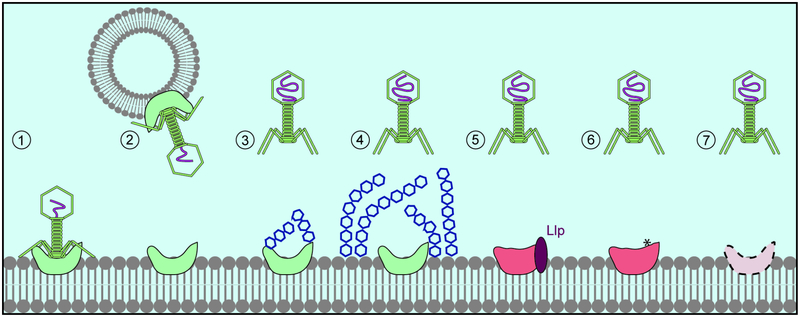
Figure 2. Prevention of phage adsorption.
1. Successful binding of the phage to its receptor (green). 2. Sequestration of phage particles by OMVs containing the phage receptor. 3. Prevention of phage adsorption due to receptor post-translational modifications (glycosylation). 4. Prevention of phage adsorption due to receptor occlusion by surface structures (glycan capsule). 5. Receptor modification through interaction with another protein. 6. Receptor mutations that abolish phage binding. 7. Regulation of receptor expression.
Preventing phage adsorption
Many bacteria spend much of their life cycle embedded in biofilms, an extracellular matrix made up of polymers where bacteria live in close proximity, often on surfaces. Biofilms protect bacteria in various ways, but how these structures affect phage-bacteria interactions remains incompletely understood. Computational modelling suggests that biofilms can conditionally survive and grow in the presence of phage (Simmons et al., 2018). This was also shown experimentally in Escherichia coli with a virulent mutant of phage P1 where, depending on nutrient availability and phage infectivity, the bacterial colony size reaches an equilibrium (Eriksen et al., 2018). In this scenario, cells inside the colony divide, and are shielded by peripheral cells that get infected. Another study showed that while early biofilms were quickly eradicated, mature E. coli biofilms avoided clearance by T7 phage (Vidakovic et al., 2018). Fluorescent labelling of cells and phages revealed that the biofilm structure prevents phage access to the biofilm interior. This depended on the presence of the host protein curli, which forms amyloid fibres that promote the formation of an extracellular matrix and a dense cell packing.
In addition to the protective shield provided by biofilms, Gram-negative bacteria can secrete outer membrane vesicles (OMVs), spherical structures made up of outer membrane components and periplasmic cargo which pinch off the cell (Schwechheimer and Kuehn, 2015). Since they contain exposed outer membrane proteins that can act as phage receptors, OMVs can act as decoys, sequestering extracellular phage. One report showed that pre-incubation with OMVs reduced T4 infectivity in E. coli, and phage-bound OMV complexes could be visualized with electron microscopy (Manning and Kuehn, 2011). Similarly, it was shown recently that Vibrio cholerae OMVs could bind three different phages, an interaction that was dependent on the presence of phage receptor in the OMVs (Reyes-Robles et al., 2018).
Bacteria can also prevent adsorption by hiding or masking surface receptors. For example, in Pseudomonas aeruginosa, type IV pili can be glycosylated to prevent the binding of several pilus-specific phages (Harvey et al., 2018). Receptors can also be blocked by polysaccharide capsules, which shield the whole bacterial surface. The polysialic acid capsule of E. coli K1 prevents phage T7 attachment to its receptor, lipopolysaccharide (LPS), thereby reducing infectivity (Scholl et al., 2005). In response, phages can have enzymes in their tails that degrade various capsules, giving rise to an evolutionary arms race that results in the extreme diversification of capsule synthesis and hydrolysing enzyme genes of the host and phage, respectively (Fernandes and Sao-Jose, 2018). Finally, surface proteins can also hide phage receptors. E. coli lytic phage T5 uses the outer membrane iron uptake protein FhuA as its receptor and expresses the lipoprotein Llp to mask it. This prevents additional T5 particles, and possibly other phages that use FhuA as receptor, such as T1 and phi80, from entering and disturbing T5’s infection cycle (Pedruzzi et al., 1998). This phenomenon is an example of superinfection exclusion (Sie), a process where intracellular phages, including prophages, block the infection of the same (homotypic Sie) or a different (heterotypic Sie) phage.
Another mechanism to prevent adsorption is the introduction of mutations within receptor genes that affect the protein or its expression. This is a common mode of resistance that is perhaps best exemplified by the identification of mutations in LamB, the phage lambda receptor, in E. coli resistant cells (Clement et al., 1983). More recently it was found that receptor expression can be modulated by lysogenic phages via Sie. The P. aeruginosa prophage D3112 expresses the protein Tip, which interacts with the ATPase PilB to prevent type IV pili extension. D3112, as well as other phages that use these pili as receptors, are therefore unable to infect D3112 lysogens (Chung et al., 2014). Indeed, a systematic screen of P. aeruginosa Sie mechanisms identified many prophages interfering with either type IV pilus function, or with the O-antigen, another typical P. aeruginosa phage receptor in the surface polysaccharide (Bondy-Denomy et al., 2016).
Preventing DNA injection
Blocking the entry of phage DNA into the cytoplasm is another mechanism of preventing phage infections. The E. coli prophage HK97 confers both homotypic as well as heterotypic (against the closely related phage HK75) Sie thorough the expression of gp15 (Cumby et al., 2012). This is an inner membrane (transmembrane) protein that interacts with the host glucose transporter PtsG, and most likely disrupts its association with phage components required for translocating the viral genome across the inner membrane, thereby preventing the transfer of DNA into the cytoplasm (Cumby et al., 2015). Another recent example of a heterotypic Sie mechanism preventing DNA injection is found in the mycobacteriophage Fruitloop. During the lytic cycle, Fruitloop gp52 inactivates Wag31, an essential mycobacterial protein involved in cell wall synthesis at the cell poles (Ko and Hatfull, 2018). This prevents DNA injection by an unrelated group of mycobacteriophages that rely on Wag31, including the phages Hedgerow and Rosebush.
Targeting bacteriophage nucleic acids
Once the phage genome is injected into the host cell, it will initiate the lytic cycle, a genetic programme to achieve viral propagation. Temperate phages have the choice of entering the lysogenic cycle, which requires the repression of the lytic genes by a transcription factor. Possibly the most widespread homotypic Sie mechanism that targets the phage nucleic acid is the use of this repressor, expressed by prophages to maintain lysogeny, to abort the lytic programme of other closely related phages that infect the lysogen (Johnson et al., 1981). However, the most direct mechanism to target the viral nucleic acids is the employment of nucleases that degrade the injected genome.
Restriction-modification systems
Restriction-modification (RM) systems (see (Tock and Dryden, 2005) for a more detailed review) are an ubiquitous and extremely diverse mode of anti-phage defence. They are normally made up of two activities; a restriction endonuclease and a methyltransferase (the modification component). The restriction endonuclease recognises short DNA motifs, usually 4–8 base-pairs long, and cuts the phage DNA. These DNA motifs exist in both the bacterial host and invading phage, but the host protects its genome by using the methyltransferase to modify its own DNA to avoid recognition by the restriction enzyme. An invading phage is usually not methylated, and will therefore be cut upon injection. RM systems are classified into four major types, based their mechanism of action and subunit composition (Tock and Dryden, 2005). Both type I and III systems translocate along DNA and cleave away from the recognition sites. Type II, known for their use in molecular cloning, cleave within or near the recognition site. Type IV systems lack a methylase and only contain a restriction endonuclease which only cleaves modified DNA. Finally, there are examples of “inverted” RM systems that do not belong to any of these types. The phage ϕC31 can propagate in Streptomyces coelicolor A2(3) harbouring the four-gene “phage growth limiting” (pgl) locus, but only mounts one cycle of infection. The released phages are unable to reinfect Pgl+ hosts (Chinenova et al., 1982), presumably due to the action of the methyltransferase pglX (Sumby and Smith, 2002), which modifies new phage DNA to make it susceptible for restriction in the next Pgl+ host by an unknown mechanism.
RM systems and DNA modfications exemplify an elaborate “arms race” between E. coli and phage T4. T4 contains hydroxymethylcytosine (HMC) instead of cytosine in its DNA, inhibiting all type I-III RM systems that recognise sites containing cytosine. To counter this, E. coli uses McrBC, a type IV system specific for HMC-containing DNA (Raleigh and Wilson, 1986). In response, T4 can glycosylate its DNA, which impairs McrBC activity. Against this, E. coli has evolved an additional type IV system, the GmrSGmrD system, that can cleave glycosylated DNA (Bair and Black, 2007).
CRISPR-Cas systems
One of the most significant scientific advances in the last decades was the discovery of CRISPR-Cas bacterial immune systems [for a recent review, see (Hille et al., 2018)]. These systems are present in approximately 50% of sequenced bacteria and 90% of sequenced archaea (Makarova et al., 2015), and provide resistance against invading phages (Barrangou et al., 2007) and plasmids (Marraffini and Sontheimer, 2008). Uniquely, CRISPR systems are adaptive rather than innate immune systems, where exposure to a previous infection is memorised. The molecular basis for immunological memory are short (30–40 base pairs) “spacer” sequences acquired from invader genomes, flanked by similarly short semi-palindromic repeats. Repeats and spacers are transcribed and processed into small CRISPR RNA (crRNA) guides. CRISPR associated (cas) genes, usually adjacent to the CRISPR locus, encode the protein machinery required for the acquisition of new spacer sequences upon infection (the adaptation phase) and for the sequence-specific elimination of the invader (the targeting phase). During the latter, RNA-guided Cas nucleases use the crRNAs to recognise and cleave the invader’s nucleic acids via complementary base pairing. Based on the composition of cas genes, CRISPR systems can be classified into two classes, six types, and multiple subtypes (Koonin et al., 2017), with diverse mechanisms of action (Figure 3).
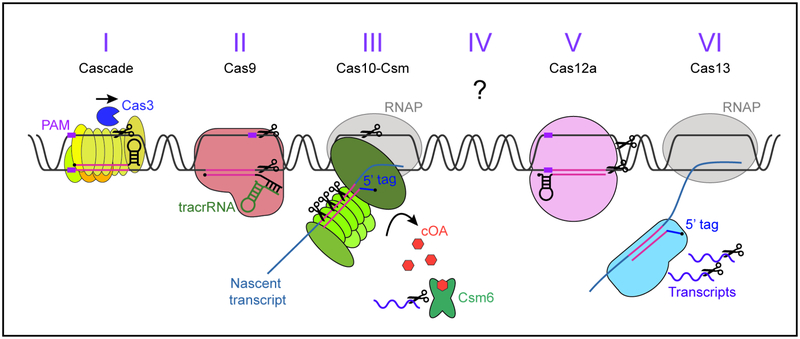
Figure 3. Different CRISPR targeting mechanisms.
See text for details. Purple box: PAM, black circles: crRNAs 5’ end, pink: spacer/protospacer sequences, blue: 5’ crRNA tags inhibiting type III/VI autoimmunity. For types I, II, and V, the DNA double helix is unwound by the main effector complex in a PAM-dependent manner, and DNA is cut by Cas3 (type I) or Cas9/Cas12a (types II/V). Type III and VI recognise the protospacer within a nascent transcript in a PAM-independent manner, this is followed by the cleavage of DNA and/or RNA from the invader. Type III also produces cyclic oligoadenylates (orange hexagons) which allosterically activate the accessory RNase Csm6 (Csx1 for III-B).
CRISPR targeting – destroying the invader
Types I, II, and V use the crRNA guide to recognise the complementary target sequence in the DNA of the invader, known as the protospacer. In addition to this complementarity, cleavage requires the presence of a conserved protospacer adjacent motif (PAM) in one flank of the target (Gasiunas et al., 2012; Jinek et al., 2012; Sashital et al., 2012; Zetsche et al., 2015). As a consequence of this targeting requirements, phages harbouring mutations that eliminate the PAM or the complementarity between the protospacer and the crRNA, can escape targeting (Deveau et al., 2008; Semenova et al., 2011). Whereas type II and V systems employ a single-subunit RNA-guided nuclease (Cas9 and Cas12, respectively) (Sapranauskas et al., 2011; Zetsche et al., 2015), type I systems use a multi-subunit crRNA Cas complex known as Cascade to locate the protospacer and a second nuclease, Cas3, recruited by Cascade to the target, to cleave its DNA (Brouns et al., 2008; Sinkunas et al., 2013).
Type III CRISPR systems are composed of the main effector complex (Csm or Cmr for III-A or III-B, respectively), and of an accessory RNase (Csm6 or Csx1 for III-A or III-B, respectively). Transcription across the target is an absolute requirement for immunity (Goldberg et al., 2014) since the effector complex binds the protospacer within the nascent RNA through complementary RNA-RNA base-pairing (rather than recognising target DNA). This binding unleashes the non-specific single-stranded DNase activity of Cas10 (the main subunit of both the Csm and Cmr complexes), which cuts the non-template strand of the transcribed viral DNA (Kazlauskiene et al., 2016; Samai et al., 2015). In addition, target recognition results in the synthesis of cyclic oligo-adenylates (cOA) by another domain of Cas10, a ligand that activates the Csm6/Csx1 non-specific RNase (Kazlauskiene et al., 2017; Niewoehner et al., 2017). This accessory RNase is required for targets that are poorly recognised by the crRNA guide and thus provide inefficient activation of the Cas10 ssDNase, such as weakly transcribed (Rostøl and Marraffini, 2019) or mutated targets (Jiang et al., 2016). Finally, the Csm3/Cmr4 subunit of the effector complex cleaves the protospacer RNA (Hale et al., 2009; Tamulaitis et al., 2014), an event that neutralizes both activities of the Cas10 subunit. Type III-A immunity does not require the recognition of a PAM on the RNA target and can tolerate between 6–8 mismatches between the protospacer and the crRNA (Pyenson et al., 2017). Consequently, type III-A CRISPR systems offer more robust defence against rapidly mutating phage invaders than a type II (Pyenson et al., 2017) and Type I (Silas et al., 2017) systems.
Type VI CRISPR systems are characterised by the effector protein Cas13 which, uniquely, only targets RNA (Abudayyeh et al., 2016). Like in type III CRISPR systems, transcription across the target is required for interference, and there is no PAM sequence requirement (Meeske and Marraffini, 2018). Binding to a target RNA results in the cleavage of both target and non-target transcripts, the latter causing a growth delay observed in cells undergoing type VI CRISPR immunity (Meeske and Marraffini, 2018; Abudayyeh et al., 2016). Although Leptotrichia shahii Cas13a with a reprogrammed spacer was able to confer protection against the single-stranded RNA virus MS2 in an E. coli heterologous host, none of the spacers found so far (neither on type VI nor in any other CRISPR loci) match RNA viral genomes, and therefore the role of Cas13 in anti-phage immunity still remains unclear.
CRISPR adaptation – remembering the invader
Adaptation is the process where a short sequence from an invader is incorporated into the CRISPR array as a new spacer, offering protection against future invaders containing the same or very similar protospacer sequences [for a recent review, see (McGinn and Marraffini, 2018)]. Because different bacteria in the population acquire different new spacers from the invader, the result of CRISPR adaptation is the extreme diversification of spacer repertoire of the population, which in turn is fundamental to prevent the emergence of escape phages with mutated target sequences (van Houte et al., 2016b). CRISPR adaptation has two steps; the selection of functional sequences from the invader’s genome (known as prespacers) and their insertion into the leader end of the CRISPR array.
A functional spacer must (i) not target the host chromosome (i.e. avoid autoimmunity), and (ii) be flanked by the correct PAM in type I, II or V systems, or produce a crRNA complementary to a transcript in type III and VI systems. It has been found that both type I and II systems prefer the acquisition from DNA molecules with free DNA ends such as DNA breaks or phage cos sites (the phage DNA end that is injected first) (Levy et al., 2015; Modell et al., 2017). These observations suggest a model where phages (and plasmids), which have relatively small genomes and therefore replicate more frequently than the host chromosome, are more prone to breaks and stalls at replication forks, increasing the probability of prespacer acquisition from their genomes. In addition, DNA injection during phage infection or plasmid conjugation offers a free DNA end for the CRISPR acquisition machinery. The selection of invader sequences that are flanked by a functional PAM can be performed by either the nuclease that recognizes the motif during targeting, Cas9, for type II systems (Heler et al., 2015), or by Cas proteins dedicated to this task, such as Cas4 for some type I systems (Lee et al., 2018; Kieper et al., 2018; Shiimori et al., 2018). Interestingly, a study published in this issue of Cell Host & Microbe suggests that the spacer repertoire of the surviving population, at least in type II systems, is determined by the rate of acquisition of each sequence, not so much by the potency of the DNA cleavage it mediates (Heler et al., 2019). Very little is known about spacer acquisition in type III, V and VI systems. Adaptation has only been observed in the Marinomonas mediterranea III-B system, which harbours a relatively rare reverse transcriptase-Cas1 (RT-Cas1) fusion. In this marine bacterium, integration of DNA spacers derived from cellular RNA is achieved via a reverse transcription reaction mediated by this fusion protein (Silas et al., 2016). This is thus an elegant way for the transcription-dependent type III systems to sample from well-transcribed regions and ensure functional spacers.
Once selected, functional prespacers are integrated into the CRISPR array by the Cas1-Cas2 complex via a reaction mechanism similar to that of retroviral integrases and transposases (Nunez et al., 2015). Spacers are added to the leader end of the CRISPR array, and in type II systems this is achieved by the recognition of a ‘leader-anchoring sequence’ directly upstream of the first repeat (McGinn and Marraffini, 2016; Wright and Doudna, 2016). Significantly, leader-proximal spacers provide better immunity, and integrating the spacers from the most recent infection in this polarized manner ensures protection against the most pressing viral threat (McGinn and Marraffini, 2016).
A unique aspect of type I adaptation is the presence of priming, where pre-existing spacers against an invader enhances further adaptation against the same threat (Datsenko et al., 2012). This allows immunity to keep up with rapidly mutating phages that might have altered their target sequence and thus escaped CRISPR targeting. Primed adaptation requires the binding by Cascade to mutated protospacers, harbouring a non-functional PAM or seed sequence mismatch with the crRNA. Moreover, a study in this issue (Jackson et al., 2019) revealed that “slipped” (imprecisely acquired) spacers, although providing less efficient interference, enhance primed acquisition. After binding the DNA, Cascade recruits Cas1-Cas2 along with Cas3 (Redding et al., 2015) to capture prespacers from the target.
Prokaryotic Argonautes
Prokaryotic argonautes (pAgos) represent a recently discovered bacterial innate defence mechanism found in approximately 9% of bacterial genomes and 32% of archaeal genomes [for a recent review, see (Hegge et al., 2018)]. They are often encoded within defence islands, regions enriched for phage resistance systems, and have undergone extensive horizontal gene transfer (Makarova et al., 2009), two factors which suggest a defensive role. So far, several mechanisms have been demonstrated, including DNA-guided DNA silencing and RNA-guided DNA silencing. For the former mechanism, it was shown in two systems that the apo form of pAgo can first degrade invader DNA sequence non-specifically. Degradation products from this DNA are used as guide DNAs, which allows sequence-specific interference against the same target (Swarts et al., 2017; Zander et al., 2017). Some pAgos are also predicted to be catalytically inactive, but are encoded near other nuclease genes that might be guided by pAgo to the invader. So far, however, only in vivo immunity against invasive plasmids has been shown; the role of pAgo in defence against phages, if any, remains elusive.
Abortive infection and toxin-antitoxin systems
Abortive infection (Abi) and toxin-antitoxin (TA) systems are widespread, albeit poorly understood, stress systems. They can work as antiviral systems by stressing the infecting cells to disturb the phage life cycle and prevent virion release. Abi systems are phenotypically, rather than genetically, defined, and are always involved in disrupting phage infection. TAs, on the other hand, usually comprise a genetically well-defined TA gene pair, with the toxin causing the stress and the antitoxin inhibiting the toxin’s catalytic activity. The line between Abis and TAs is blurred and they share some overlap, with some previously identified Abi mechanisms employing TA genes to create the antiviral cell stress, for example the lactococcal Abi systems AbiQ (Samson et al., 2013) and AbiE (Dy et al., 2014). A central outstanding question is to what extent activation of these systems upon infection leads to permanent cell death as opposed to temporary growth arrest and dormancy.
Abortive infection
Abi is a process by which cells prevent release of functional phage virions at the expense of host cell survival/fitness. It is considered an altruistic action; a “programmed cell death” that prevents the spread of the phage to the surrounding clonal bacterial population. This is achieved through the perturbation of essential cellular processes such as translation, transcription, and replication, or by inducing membrane leakage. How phage infection is recognised to trigger the Abi response is often unknown.
Although Abi systems are widespread, most have been characterised in E. coli and Lactococcus lactis, a Gram-positive bacterium used in dairy production. The Lit and PrrC systems of E. coli are activated by phage T4 and disrupt translation. The Lit protease of E. coli K12 is activated by the Gol peptide of the T4 major capsid protein, a gene that is transcribed late in the phage infection cycle (Bingham et al., 2000) and cleaves the ribosomal elongation factor EF-Tu, thereby arresting translation for both the phage and its host. PrrC of E. coli CT196 cleaves the tRNALys in the anticodon loop; this depletes the tRNALys pool and inhibits global translation (Kaufmann, 2000). In L. lactis, more than 20 Abi systems have been identified. The single-gene abiK system is able to reduce infectivity of many prevalent lactococcal phage groups 106-fold. AbiK possesses polymerase activity, synthesising long DNA molecules with random sequences in vitro (Wang et al., 2011). Since phage mutants that escape AbiK have mutations in phage-encoded recombinases, it was hypothesized that AbiK-synthesised DNA interferes with phage recombination, preventing phage replication and maturation. How this activity is also detrimental to the host, and what triggers AbiK during infection, remains unclear. Another lactococcal Abi system, AbiZ, reduces the burst size of phage Φ31 by 100-fold. AbiZ seems to act cooperatively with the phage pore-forming protein holin to induce premature lysis and the release of immature, non-infectious phage particles (Durmaz and Klaenhammer, 2007).
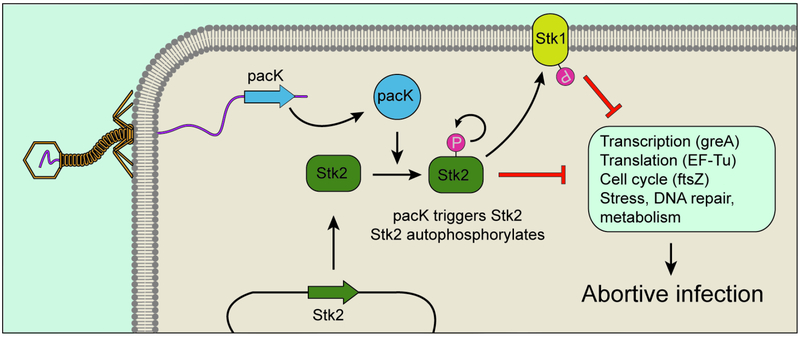
Recently, a kinase-mediated Abi mechanism protecting against Siphoviridae phages was uncovered in the common skin bacterium Staphylococcus epidermidis (Depardieu et al., 2016) (Figure 4). Upon infection, the eukaryotic-like Serine/Threonine kinase Stk2 was found to phosphorylate a range of targets in diverse cellular pathways, including transcription, translation, replication, and metabolism. This widespread phosphorylation presumably disrupts these pathways to result in host death. Phages able to escape Stk2 activation carry mutations in the pacK gene, suggesting that PacK induces Stk2 auto-phosphorylation to initiate this defence pathway.
Figure 4. The staphylococcal Stk2 Abi system.
The phage protein pacK triggers autophosphorylation (and activation) of the S. epidermidis Stk2 kinase. Activated Stk2 phosphorylates Stk1 and miscellaneous cellular factors, eventually leading to the abortion of the viral infectious cycle and cell death.
Toxin-antitoxin systems
A fundamental feature of TA systems is the instability of the antitoxin: it is labile and must be continuously expressed to remain at appropriate stoichiometric ratios with and neutralize the toxin [for a recent review, see (Harms et al., 2018)]. Toxins can possess various catalytic activities, including DNase and RNase, or can inhibit DNA replication, ATP synthesis, or the cell division machinery. There are at least six TA types, categorised based on the nature of the toxin and antitoxin (protein or RNA), and the mechanism of toxin neutralisation, with many bacteria harbouring dozens of TA gene pairs (E. coli K-12 has more than 35 TA pairs (Harms et al., 2018)). This high genetic diversity reflects the many functions found for TA systems: in addition to phage defence, they have been implicated in stress responses, plasmid maintenance, and persister cell formation.
Some TA systems can directly inhibit the phage life cycle. The plant pathogen Pectobacterium atrosepticum possesses the ToxN/ToxI TA pair, where the endoribonuclease ToxN is sequestered by binding the noncoding RNA antitoxin ToxI (Fineran et al., 2009). Upon phage infection, the RNase activity of ToxN is unleashed to destroy both host and phage transcripts (Blower et al., 2011), arresting the infection. Similarly, the MazF/MazE TA system of E. coli can suppress phage T4 infection by activating MazF’s ribonuclease activity (Alawneh et al., 2016). To counter this, phage T4 carries the ADP-ribosyltransferase Alt, which modifies and inhibits MazF (Alawneh et al., 2016). The finding that phages have evolved mechanisms that inactivate TA systems strongly indicates that they are part of the host-phage arms race. For example, in addition to Alt, the T4 dmd gene directly binds and suppresses two other toxins, LsoA and RnlA (Otsuka and Yonesaki, 2012).
As mentioned above, it is not clear whether TA induction causes altruistic suicide of the infected cell or only a temporary growth arrest. The activation of toxins can in some cases be reversible. With the MazF/MazE and ToxN/ToxI systems, cells where the toxin MazF or ToxN was expressed could be rescued and were viable upon induction of the expression of the antitoxin (MazE or ToxI, respectively) after a delay (Fineran et al., 2009; Pedersen et al., 2002). Recently, a comprehensive analysis of MazF cleavage sites revealed that most mRNAs, as well as rRNA precursors, are cleaved at multiple sites (Culviner and Laub, 2018). Presumably, upon toxin neutralization, cells can replenish their RNA pool and resume growth. Given the extensive diversity of TA systems, it is probable that they can work both as dormancy induction and programmed cell death (Abi) systems, and the outcome will depend on a range of factors including the toxin mechanism of action, the duration of the toxin’s activity, and the life cycle of the phage.
Bacteriophage assembly interference
Phage-Inducible Chromosomal Islands (PICIs) form a group of genetic elements that parasitise phages for replication and transmission [for a recent review, see (Penades and Christie, 2015)]. PICIs are integrated into a bacterial chromosome and excise in the presence of a specific “helper phage” (by infection or lysogen induction). Although the main role of PICIs seems to be the dissemination of the genetic material they harbour (in many cases bacterial virulence determinants), they interfere with the phage life cycle, and can therefore be classified as an anti-phage mechanism.
PICI genomes are often small (~15 kb), encoding genes required for excision and integration, factors that promote PICI packaging and dissemination, and a repressor that inhibits their expression in the absence of the helper phage. PICIs are best characterized in Staphylococcus aureus, where they are named “SaPIs” (S. aureus Pathogenicity Islands). SaPIs are induced when their repressor, Stl, is sequestered away by an anti-repressor expressed early during the helper phage lytic cycle (e.g. helper phage 80α) (Tormo-Mas et al., 2010) (Figure 5). Derepression induces the expression of SaPI genes and couples the SaPI’s propagation cycle with that of the helper phage. Specialized SaPI structural proteins modulate the assembly of the helper phage’s capsid to produce a capsid that can only be packaged with the smaller SaPI genome. As a result, the host bacterium lyses, but primarily releases SaPI virions that infect neighbouring cells to disseminate the SaPI genomes and the virulence factors they encode. Recently, PICIs were found to also be widespread in gram-negative bacteria (Fillol-Salom et al., 2018). Instead of being regulated by a repressor, Gram-negative PICIs are induced by a PICI-encoded activator whose expression requires the helper phage.
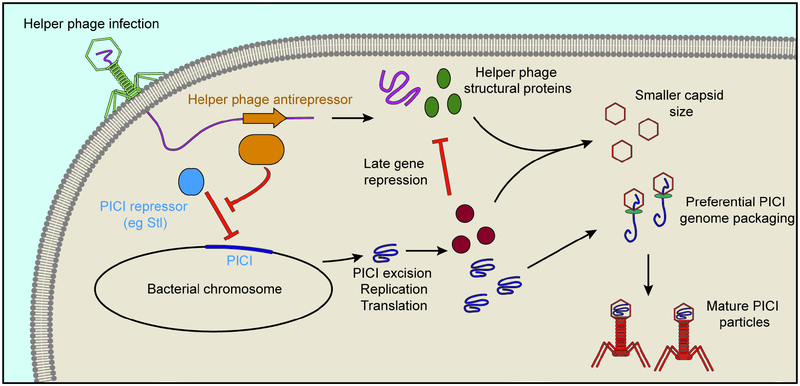
Figure 5. PICI-mediated interference of phage assembly.
In Gram-positive organisms, PICI expression is inhibited by a transcription repressor. Helper phages produce an antirepressor, leading to the excision of the PICI from the host chromosome. The PICI genome replicates, and expresses proteins that repress late helper phage genes and alter the phage capsid size to be more appropriate for the PICI genome size. This in turn leads to both the preferential packaging of PICI genomes and the prevention of the formation of helper phage virions.
To fight phage, V. cholerae encodes PICI-like elements (PLEs) (Seed et al., 2013), which are similar to other gram-negative PICIs though with somewhat different gene content. Upon infection of the host bacterium by the phage ICP1, the PLE-encoded recombinase Int excises PLE from the V. cholerae chromosome (McKitterick and Seed, 2018). PLE then replicates to high levels and inhibits ICP1 phage replication by unknown mechanisms. Interestingly, several ICP1 phage isolates encoded their own CRISPR system capable of neutralising PLEs (Seed et al., 2013), allowing ICP1 propagation in PLE+ V. cholerae strains.
Recently discovered anti-phage mechanisms
Given the astronomical number and diversity of bacteriophages in our planet, it is likely that we only know a minority of the anti-phage mechanisms present in prokaryotes. For example, a recent extensive screen of mycobacterial phages revealed varied mechanisms of phage-encoded Sie, which included a (p)ppGpp synthetase and a single-subunit RM system, as well as classical Sie modes like promoter repression and inhibition of entry (Dedrick et al., 2017). Sie, being as much a phage-phage interaction as a bacterium-phage interaction, is likely a more important and diverse resistance mechanism than previously appreciated.
Interestingly, bacterial defence systems often cluster in defence islands (Makarova et al., 2011). This has allowed a “guilt-by-association” approach to uncover new anti-phage mechanisms. The BREX (BacteRiophage EXclusion) system, which was discovered in this way, mediates methylation of a non-palindromic, six-nucleotide motif, most likely to achieve self/non-self discrimination (Goldfarb et al., 2015; Gordeeva et al., 2018). Phage DNA is inactivated after injection prior to DNA replication by an unknown mechanism that is not thought to involve cleavage. The BREX systems characterised recently share two genes (pglX and pglZ) with the Pgl system of S. coelicolor (see above) (Goldfarb et al., 2015), with the Pgl system being denoted type 2 BREX (of six types in total). Another recently discovered system is DISARM (Ofir et al., 2018), which provides broad anti-phage immunity through a novel RM-like mechanism that includes a methyltransferase modifying a five-nucleotide motif and a multi-component restriction element that probably cleaves unmodified phage DNA early in the phage life cycle.
Also using an approach based on identifying defence gene neighbourhoods, a recent study identified 26 candidate systems that were heterologously expressed in E. coli or Bacillus subtilis and assayed for anti-phage activity (Doron et al., 2018). Of these, nine provided robust protection against at least one type of phage (and one against plasmids), with one, named “Zorya”, most likely being an Abi system that may cause membrane depolarization of the host using a proton channel. Finally, the guilt-by-association approach was used to identify genes enriched near CRISPR loci (Shah et al., 2018; Shmakov et al., 2018). A diverse range of CRISPR-associated accessory candidate genes were identified, which likely complement or expand the functions of the core cas gene machinery.
Putting it all together
The phage resistance mechanisms discussed so far have mostly been studied in the lab individually, though this is rarely how a bacterium’s arsenal is applied against phage in nature. Bacteria employ several complementary lines of defence, none of which are mutually exclusive, and a phage has to overcome each system to allow successful infection. On the other hand, contrary to experimental settings, the environment typically contains a heterogeneous mix of phages.
Synergistic effects between anti-phage mechanisms are starting to be recognised. RM and CRISPR systems often co-exist, and spacer acquisition by the CRISPR system is enhanced in the presence of an RM system (Hynes et al., 2014). Cooperation between different CRISPR types has also been detected: M. mediterranea contains two CRISPR systems, and spacers incorporated from phage into the type I-B array can be used by the III-B machinery against phage (Silas et al., 2017). The cross-talk between the type I and III systems makes protection more robust since it is harder for phages to escape type III targeting through protospacer mutations (Pyenson et al., 2017). Although not experimentally demonstrated, it is tempting to speculate that there can also be synergy between dormancy-inducing components (TA systems) and effector components (i.e. CRISPR and RM systems), which are often clustered in genomic defence islands (Makarova et al., 2013). The rationale is that TA or Abi systems could “buy time” for the cell, inducing short-term dormancy while CRISPR or RM mechanisms eliminate the phage. This concept is illustrated in the type III-A CRISPR-Cas response, where the non-specific RNase Csm6 causes a transient growth arrest until the DNase activity of the Cas10-Csm complex has eliminated the plasmid invader (Rostøl and Marraffini, 2019). Induction of dormancy could also afford time for the acquisition of new spacers during CRISPR adaptation. Alternatively, if the invader is not cleared and the toxin remains active, the infected cell would die and prevent further spread of the phage.
Outlook and future directions
Recently, novel technologies and experimental approaches, as well as renewed interest in bacteriophages, have dramatically boosted our knowledge of how bacteria resist their parasites. Scientists have probed both broader (new systems) and deeper (expanding repertoire of known systems). Future studies will surely continue this trend, which will most likely translate into clinical outcomes and technological innovations. Still, there are hurdles to overcome. The study of more integrative models where it is possible to appreciate how different immune mechanisms interact and complement each other, as well as more ecological approaches, where the fluxes of multiple and different bacteria and phages can be observed, will undoubtedly expand our understanding of prokaryotic immunity. The extent to which phages interfere with the life cycle of other phages is also underappreciated. Last but not least, given the importance of phages as mediators of horizontal gene transfer, the study of defence mechanisms will help us understand prokaryotic evolution. Considering the millions of years bacteria and phages have coevolved, there surely remains a cornucopia of unknown unknowns for us to discover.
Bacteria are preyed upon by parasitic bacteriophages, which undergo continuous cycles of infection. In order to survive, bacteria have developed a complex arsenal of anti-phage mechanisms. Rostøl and Marraffini review the mechanisms employed by bacteria to resist their phages, with an emphasis on recent developments in the field.
Acknowledgements.
We would like to thank members of the Marraffini laboratory, and David Ding, for helpful discussions. J.T.R. was supported by a Boehringer Ingelheim Fonds PhD fellowship. L.A.M. is supported by a Burroughs Wellcome Fund PATH Award, and an NIH Director’s Pioneer Award (DP1GM128184).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests. L.A.M. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics and a cofounder of Eligo Biosciences.
References
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawneh AM, Qi D, Yonesaki T, and Otsuka Y (2016). An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol Microbiol 99, 188–198. [DOI] [PubMed] [Google Scholar]
- Bair CL, and Black LW (2007). A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol 366, 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bingham R, Ekunwe SI, Falk S, Snyder L, and Kleanthous C (2000). The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J Biol Chem 275, 23219–23226. [DOI] [PubMed] [Google Scholar]
- Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, and Salmond GP (2011). A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol 18, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, and Maxwell KL (2016). Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10, 2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenova TA, Mkrtumian NM, and Lomovskaia ND (1982). [Genetic characteristics of a new phage resistance trait in Streptomyces coelicolor A3(2)]. Genetika 18, 1945–1952. [PubMed] [Google Scholar]
- Chung IY, Jang HJ, Bae HW, and Cho YH (2014). A phage protein that inhibits the bacterial ATPase required for type IV pilus assembly. Proc Natl Acad Sci U S A 111, 11503–11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement JM, Lepouce E, Marchal C, and Hofnung M (1983). Genetic study of a membrane protein: DNA sequence alterations due to 17 lamB point mutations affecting adsorption of phage lambda. EMBO J 2, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culviner PH, and Laub MT (2018). Global Analysis of the E. coli Toxin MazF Reveals Widespread Cleavage of mRNA and the Inhibition of rRNA Maturation and Ribosome Biogenesis. Mol Cell 70, 868–880 e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Edwards AM, Davidson AR, and Maxwell KL (2012). The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol 194, 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, and Maxwell KL (2015). The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol Microbiol 96, 437–447. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, and Semenova E (2012). Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3, 945. [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, Reyes JC, Russell DA, Adair T, Alvey R, et al. (2017). Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol 2, 16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depardieu F, Didier JP, Bernheim A, Sherlock A, Molina H, Duclos B, and Bikard D (2016). A Eukaryotic-like Serine/Threonine Kinase Protects Staphylococci against Phages. Cell Host Microbe 20, 471–481. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, and Moineau S (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, and Sorek R (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz E, and Klaenhammer TR (2007). Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J Bacteriol 189, 1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy RL, Przybilski R, Semeijn K, Salmond GP, and Fineran PC (2014). A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res 42, 4590–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen RS, Svenningsen SL, Sneppen K, and Mitarai N (2018). A growing microcolony can survive and support persistent propagation of virulent phages. Proc Natl Acad Sci U S A 115, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, and Sao-Jose C (2018). Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Rodriguez A, and Garcia P (2018). Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J 12, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillol-Salom A, Martinez-Rubio R, Abdulrahman RF, Chen J, Davies R, and Penades JR (2018). Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J 12, 2114–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, and Salmond GP (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106, 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, and Siksnys V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109, E2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, and Marraffini LA (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, and Sorek R (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeeva J, Morozova N, Sierro N, Isaev A, Sinkunas T, Tsvetkova K, Matlashov M, Truncaite L, Morgan RD, Ivanov NV, et al. (2018). BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, and Terns MP (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Brodersen DE, Mitarai N, and Gerdes K (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol Cell 70, 768–784. [DOI] [PubMed] [Google Scholar]
- Harvey H, Bondy-Denomy J, Marquis H, Sztanko KM, Davidson AR, and Burrows LL (2018). Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat Microbiol 3, 47–52. [DOI] [PubMed] [Google Scholar]
- Hegge JW, Swarts DC, and van der Oost J (2018). Prokaryotic Argonaute proteins: novel genome-editing tools? Nat Rev Microbiol 16, 5–11. [DOI] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, and Marraffini LA (2015). Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, Bratovic M, Ressel S, and Charpentier E (2018). The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259. [DOI] [PubMed] [Google Scholar]
- Hynes AP, Villion M, and Moineau S (2014). Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat Commun 5, 4399. [DOI] [PubMed] [Google Scholar]
- Jiang W, Samai P, and Marraffini LA (2016). Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164, 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, and Ptashne M (1981). lambda Repressor and cro--components of an efficient molecular switch. Nature 294, 217–223. [DOI] [PubMed] [Google Scholar]
- Kaufmann G (2000). Anticodon nucleases. Trends Biochem Sci 25, 70–74. [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, and Siksnys V (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C, and Siksnys V (2016). Spatiotemporal Control of Type III-A CRISPR-Cas Immunity: Coupling DNA Degradation with the Target RNA Recognition. Mol Cell 62, 295–306. [DOI] [PubMed] [Google Scholar]
- Kieper SN, Almendros C, Behler J, McKenzie RE, Nobrega FL, Haagsma AC, Vink JNA, Hess WR, and Brouns SJJ (2018). Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Rep 22, 3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CC, and Hatfull GF (2018). Mycobacteriophage Fruitloop gp52 inactivates Wag31 (DivIVA) to prevent heterotypic superinfection. Mol Microbiol 108, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, and Zhang F (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhou Y, Taylor DW, and Sashital DG (2018). Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Mol Cell 70, 48–59 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, and Sorek R (2015). CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, and Koonin EV (2013). Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41, 4360–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Snir S, and Koonin EV (2011). Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol 193, 6039–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, van der Oost J, and Koonin EV (2009). Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, and Kuehn MJ (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, and Sontheimer EJ (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, and Marraffini LA (2016). CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol Cell 64, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, and Marraffini LA (2018). Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol. [DOI] [PubMed] [Google Scholar]
- McKitterick AC, and Seed KD (2018). Anti-phage islands force their target phage to directly mediate island excision and spread. Nat Commun 9, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN Jr., and Malik HS (2017). Genetic conflicts: the usual suspects and beyond. J Exp Biol 220, 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, and Marraffini LA (2018). RNA Guide Complementarity Prevents Self-Targeting in Type VI CRISPR Systems. Mol Cell 71, 791–801 e793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Jiang W, and Marraffini LA (2017). CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostol JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, and Jinek M (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, and Doudna JA (2015). Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, Yaakov G, Doron S, and Sorek R (2018). DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, and Yonesaki T (2012). Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol 83, 669–681. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, and Gerdes K (2002). Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45, 501–510. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Rosenbusch JP, and Locher KP (1998). Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5-encoded lipoprotein. FEMS Microbiol Lett 168, 119–125. [DOI] [PubMed] [Google Scholar]
- Penades JR, and Christie GE (2015). The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu Rev Virol 2, 181–201. [DOI] [PubMed] [Google Scholar]
- Pyenson NC, Gayvert K, Varble A, Elemento O, and Marraffini LA (2017). Broad Targeting Specificity during Bacterial Type III CRISPR-Cas Immunity Constrains Viral Escape. Cell Host Microbe 22, 343–353 e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh EA, and Wilson G (1986). Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A 83, 9070–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, and Greene EC (2015). Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell 163, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Robles T, Dillard RS, Cairns LS, Silva-Valenzuela CA, Housman M, Ali A, Wright ER, and Camilli A (2018). Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostøl JT, and Marraffini LA (2019). Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat Microbiol In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, and Marraffini LA (2015). Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Spinelli S, Cambillau C, and Moineau S (2013). Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol Microbiol 87, 756–768. [DOI] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, and Siksnys V (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39, 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, and Doudna JA (2012). Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46, 606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Adhya S, and Merril C (2005). Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl Environ Microbiol 71, 4872–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, and Kuehn MJ (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13, 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Lazinski DW, Calderwood SB, and Camilli A (2013). A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, and Severinov K (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 108, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Alkhnbashi OS, Behler J, Han W, She Q, Hess WR, Garrett RA, and Backofen R (2018). Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiimori M, Garrett SC, Graveley BR, and Terns MP (2018). Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci. Mol Cell 70, 814–824 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov SA, Makarova KS, Wolf YI, Severinov KV, and Koonin EV (2018). Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci U S A 115, E5307–E5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silas S, Lucas-Elio P, Jackson SA, Aroca-Crevillen A, Hansen LL, Fineran PC, Fire AZ, and Sanchez-Amat A (2017). Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silas S, Mohr G, Sidote DJ, Markham LM, Sanchez-Amat A, Bhaya D, Lambowitz AM, and Fire AZ (2016). Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M, Drescher K, Nadell CD, and Bucci V (2018). Phage mobility is a core determinant of phage-bacteria coexistence in biofilms. ISME J 12, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, and Siksnys V (2013). In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, and Sorek R (2011). The phage-host arms race: shaping the evolution of microbes. Bioessays 33, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, and Smith MC (2002). Genetics of the phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2). Mol Microbiol 44, 489–500. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Szczepaniak M, Sheng G, Chandradoss SD, Zhu Y, Timmers EM, Zhang Y, Zhao H, Lou J, Wang Y, et al. (2017). Autonomous Generation and Loading of DNA Guides by Bacterial Argonaute. Mol Cell 65, 985–998 e986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, and Siksnys V (2014). Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol Cell 56, 506–517. [DOI] [PubMed] [Google Scholar]
- Tock MR, and Dryden DT (2005). The biology of restriction and anti-restriction. Curr Opin Microbiol 8, 466–472. [DOI] [PubMed] [Google Scholar]
- Tormo-Mas MA, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa I, Barbe J, Novick RP, Christie GE, and Penades JR (2010). Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 465, 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Buckling A, and Westra ER (2016a). Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol Mol Biol Rev 80, 745–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, et al. (2016b). The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidakovic L, Singh PK, Hartmann R, Nadell CD, and Drescher K (2018). Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol 3, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Villion M, Semper C, Coros C, Moineau S, and Zimmerly S (2011). A reverse transcriptase-related protein mediates phage resistance and polymerizes untemplated DNA in vitro. Nucleic Acids Res 39, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, and Doudna JA (2016). Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol 23, 876–883. [DOI] [PubMed] [Google Scholar]
- Zander A, Willkomm S, Ofer S, van Wolferen M, Egert L, Buchmeier S, Stockl S, Tinnefeld P, Schneider S, Klingl A, et al. (2017). Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat Microbiol 2, 17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]