Abstract
Purpose
Krüppel-like factor 4 (KLF4) promotes corneal epithelial (CE) cell fate while suppressing mesenchymal properties. TGF-β plays a crucial role in cell differentiation and development, and if dysregulated, it induces epithelial-mesenchymal transition (EMT). As KLF4 and TGF-β regulate each other in a context-dependent manner, we evaluated the role of the crosstalk between KLF4 and TGF-β-signaling in CE homeostasis.
Methods
We used spatiotemporally regulated ablation of Klf4 within the adult mouse CE in ternary transgenic Klf4Δ/ΔCE (Klf4LoxP/LoxP/ Krt12rtTA/rtTA/ Tet-O-Cre) mice and short hairpin RNA (shRNA)-mediated knockdown or lentiviral vector-mediated overexpression of KLF4 in human corneal limbal epithelial (HCLE) cells to evaluate the crosstalk between KLF4 and TGF-β-signaling components. Expression of TGF-β signaling components and cyclin-dependent kinase (CDK) inhibitors was quantified by quantitative PCR, immunoblots, and/or immunofluorescent staining.
Results
CE-specific ablation of Klf4 resulted in (1) upregulation of TGF-β1, -β2, -βR1, and -βR2; (2) downregulation of inhibitory Smad7; (3) hyperphosphorylation of Smad2/3; (4) elevated nuclear localization of phospho-Smad2/3 and Smad4; and (5) downregulation of CDK inhibitors p16 and p27. Consistently, shRNA-mediated knockdown of KLF4 in HCLE cells resulted in upregulation of TGF-β1 and -β2, hyperphosphorylation and nuclear localization of SMAD2/3, downregulation of SMAD7, and elevated SMAD4 nuclear localization. Furthermore, overexpression of KLF4 in HCLE cells resulted in downregulation of TGF-β1, -βR1, and -βR2 and upregulation of SMAD7, p16, and p27.
Conclusions
Collectively, these results demonstrate that KLF4 regulates CE cell cycle progression by suppressing canonical TGF-β signaling and overcomes the undesirable concomitant decrease in TGF-β–dependent CDK inhibitors p16 and p27 expression by directly upregulating them.
Keywords: KLF4, TGF-β, SMAD, EMT, corneal epithelium, squamous neoplasia
The stratified squamous corneal epithelium (CE) plays an important role in protecting the rest of the eye from external threats and ensuring a clear and refractile optical path for the incident light to reach the photoreceptors in the retina.1 A proper balance between proliferation in the basal cell layers and differentiation of the suprabasal cells is necessary for CE homeostasis.2,3 Disruption of this balance results in a spectrum of tumors collectively called ocular surface squamous neoplasia (OSSN).4 Solar ultraviolet (UV) radiation, human immunodeficiency virus (HIV), and human papilloma virus (HPV) infections are considered the main risk factors for OSSN.5 Despite their devastating effects on vision, the pathophysiology of OSSN remains understudied, and their molecular underpinnings are unexplored.4
Krüppel like factor-4 (KLF4) and KLF5, two of the most highly expressed transcription factors in the cornea,6 play important roles in CE homeostasis.7–13 KLF4 regulates its target gene expression by binding GC-rich sequences such as CACCC elements and promotes epithelial cell fate by suppressing epithelial-mesenchymal transition (EMT).14–19 The expression and function of KLF4 is regulated in a context-dependent manner at the level of transcription, mRNA- and protein-stability, and by post-translational modification.20–22 KLF4 is induced by serum starvation,23,24 oxidative stress,25 sodium butyrate,26 selenium,27 IFNγ,28 and cAMP29 but is suppressed by TGF-β.10 Within the CE, KLF4 is expressed at a low level in a mosaic pattern in the basal epithelial cells and in a uniformly robust pattern in the differentiated superficial cell layers consistent with its prodifferentiation role.7–11 Although deletion, mutation, or altered expression of KLF4 is associated with different tumors,19,30 its involvement in OSSN has not been investigated.
TGF-β signaling plays a crucial role in epithelial cell growth, proliferation, differentiation, and development, and if dysregulated, it induces epithelial-mesenchymal transition (EMT).31–36 TGF-β pathway is disrupted in different cancers including hepatocellular,37 colorectal,38 gastrointestinal,12 and head and neck squamous cell carcinomas.39 Different steps of tumor progression, including tumor initiation, stemness, invasion, metastasis, and resistance to therapy are associated with specific transitional states of EMT defined by unique transcriptional landscapes regulated by EMT transcription factors such as Zeb1, Zeb2, Snail, Slug, Twist1, and Twist2.40 Previously, we reported that CE-specific ablation of Klf4 results in upregulation of these EMT transcription factors and that KLF4 expression is downregulated in human corneal limbal epithelial (HCLE) cells undergoing TGF-β–induced EMT, suggesting a reciprocal relationship between KLF4 and TGF-β signaling within the CE.9,10
Both KLF4 and TGF-β are expressed in the cornea, where they regulate CE integrity and wound healing.6,10,41 KLF4 and TGF-β influence each other in a context-dependent manner.42,43 Similar to KLF4, TGF-β serves dual functions in tumors in a context-dependent manner, as it inhibits initial stage tumor development by acting as a cytostatic factor and promotes EMT and metastasis in late stage tumors.44 Although the individual roles of KLF4 and TGF-β have been studied within the CE,10,41 the precise connection between KLF4 and TGF-β is largely unexplored. Considering that (1) the CE-specific ablation of Klf4 resulted in dysregulated cell proliferation, loss of epithelial features, and gain of mesenchymal characteristics reminiscent of EMT,9,10 (2) the loss of KLF4 exacerbates oncogenic TGF-β signaling in hepatocellular carcinomas,37 and (3) TGF-β–induced EMT is accompanied by KLF4 downregulation in both HCLE cells10 and prostate tumors,10,45 here we tested the hypothesis that KLF4 promotes the antitumorigenic environment and contributes to CE homeostasis by suppressing TGF-β signaling and upregulating cell cycle inhibitors. Our results indicate that KLF4 promotes the CE phenotype by suppressing SMAD2/3-mediated TGF-β signaling and overcomes the undesirable concomitant decrease in TGF-β–dependent expression of p16 and p27 by directly upregulating them.
Methods
Mice
CE-specific ablation of Klf4 was achieved by feeding 8- to 10-week-old ternary transgenic Klf4Δ/ΔCE (Klf4LoxP/LoxP/Krt12rtTA/rtTA/ Tet-O-Cre/−) mice of a mixed genetic background with doxycycline chow as detailed previously.9 Age-matched littermates fed with regular chow served as wild-type (WT) controls. All procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee and conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
HCLE Cells
HCLE cells (from Dr. Ilene Gipson, Harvard University, Cambridge, MA, USA) were cultured as previously described46 in keratinocyte serum-free medium (Gibco-Invitrogen Corp., Rockville, MD, USA) supplemented with 25 μg/mL bovine pituitary extract, 0.2 ng/mL epidermal growth factor, and 0.3 M calcium chloride in a humidified incubator with 5% CO2 at 37°C.
Lentiviral Vector Construction and Transduction in HCLE Cells
A lentiviral vector containing cytomegalovirus (CMV) promoter-directed green fluorescent protein (GFP) and KLF4 (pLenti-CMV-GFP-T2A-KLF4) was constructed by cloning PCR-amplified KLF4 and GFP cDNAs using primers that inserted the ribosome skipping motif peptide T2A (see Supplementary Table S1 for primer sequences) in the plasmid pLenti-CMV-GFP-Puro (Addgene, Cambridge, MA, USA).47 This resulted in the addition of a single N-terminal amino acid (proline) on KLF4. Lentiviral packaging was performed in Lenti-X 293T cells with Lenti-X Packaging Single Shots (TaKaRa, Kusatsu, Japan). HCLE cells at 70% confluence were infected with pLenti-CMV-GFP or pLenti-CMV-GFP-T2A-KLF4 lentivirus at a multiplicity of infection of 3 in the presence of 6 μg/mL polybrene. After 5 days of transduction, GFP-positive cells were sorted via flow cytometry and propagated to generate HCLE-WT or HCLE-KLF4 cells, respectively.
Short hairpin RNA-Mediated KLF4 Knockdown
HCLE cells at 70% to 80% confluency were transfected with plasmids encoding short hairpin RNA targeting KLF4 (shRNA1-4), or a scrambled negative control shRNA (shRNA-5; Qiagen, Germantown, MD, USA). The transfected cells were selected using media containing 1 mg/mL G418 for 2 weeks. Mixed pools of the shRNA-transfected KLF4 knocked down HCLE cells (HCLE-KD) were tested for KLF4 expression by quantitative PCR (qPCR), immunoblots, and immunofluorescent staining and used in further experiments.
Antibodies
All antibodies used are presented in Supplementary Table S2 and were previously tested and confirmed to cross-react with corresponding mouse antigens.
Total RNA Isolation and qPCR
Total RNA from mouse corneas or HCLE cells was isolated using EZ-10 spin column total RNA mini-prep kit (Bio Basic, Inc., Amherst, NY, USA) and used for cDNA synthesis with mouse Maloney leukemia virus reverse transcriptase (Promega, Madison, WI, USA). qPCR was performed using SYBR Green gene expression assays in triplicate using an ABI StepOne Plus thermocycler with appropriate endogenous controls (Applied Biosystems, Foster City, CA, USA). The sequence of primers used for qPCR is shown in Supplemental Table S1.
Immunoblots
Immunoblots were carried out as detailed previously.10 WT or Klf4Δ/ΔCE corneas were homogenized in urea buffer (8.0 M urea, 0.8 % Triton X-100, 3% β-mercaptoethanol, 0.2% SDS, and protease inhibitors). HCLE cells were lysed in RIPA buffer with protease inhibitors and phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA, USA). A total of 20 to 30 μg total protein in the supernatant was separated on 4% to 12% gradient polyacrylamide gels using 2-(N-morpholino)ethanesulfonic acid (MES) or 3-(N-morpholino)propanesulfonic acid (MOPS) buffer and transferred onto polyvinylidine fluoride (PVDF) membranes. The membranes were blocked, incubated overnight with primary antibody at 4°C, washed thrice with PBST, incubated with appropriate secondary antibody for 1 hour at 23°C, and washed thrice with PBST, followed by a wash with PBS to remove traces of Tween-20. Blots were scanned on Odyssey scanner (Li-Cor Biosciences, Lincoln, NE, USA) and quantified using Image-J software (National Institutes of Health, Bethesda, MD, USA). β-Actin was used as a loading control for normalizing the data.
Immunofluorescent Staining
HCLE cells grown on coverslips or 8-μm-thick sections from OCT-embedded eyeballs were fixed in freshly prepared buffered 4 % paraformaldehyde for 10 minutes, washed three times for 5 minutes each with PBS (pH 7.4), permeabilized (0.1 % Triton X-100 in PBS), and washed thrice with PBS for 5 minutes each. Samples were then treated with glycine for 20 minutes, washed with PBS, blocked with 10% goat or donkey serum in PBS for 1 hour at 23°C, washed twice with PBS, and incubated with the appropriate dilution of the primary antibody for 2 hours at 23°C or overnight at 4°C. After further washing in PBS, the slides were incubated with appropriate Alexafluor-conjugated secondary antibody (Molecular Probes, Carlsbad, CA, USA) for 1 hour at 23°C, washed, counterstained with 4,6-diamidino-2-phenylindole (DAPI), and mounted with Aqua-Poly/Mount (Polysciences, Warrington, PA, USA). Images were acquired using an Olympus IX81 microscope (Olympus America, Inc., Center Valley, PA, USA). Image processing was performed using FV10-ASW4.2 Viewer software. After converting the images to grayscale, their relative fluorescence intensity was quantified using Image J software (National Institutes of Health).
Statistical Analysis
The results presented here are representative of at least three independent experiments and shown as mean ± SEM. Statistical significance was tested by Student's t-test, with P ≤ 0.05 considered statistically significant.
Results
KLF4 Negatively Regulates the Expression of TGF-β1, -β2, and Their Receptors in the CE
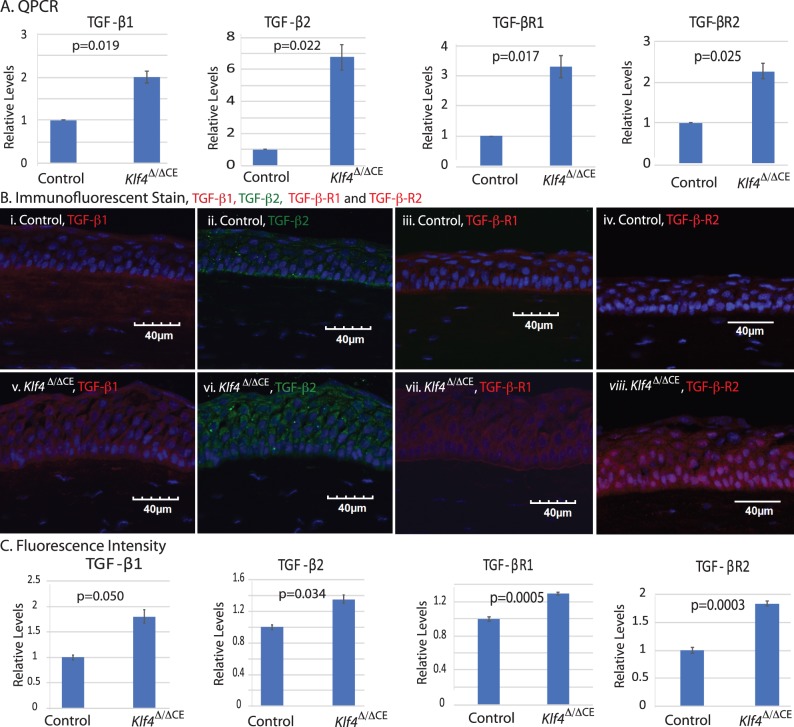
Three lines of evidence warranted a further examination of the relationship between KLF4 and TGF-β signaling within the CE: (1) KLF4 inhibits EMT by upregulating epithelial genes and suppressing mesenchymal genes9,10,48; (2) TGF-β induces EMT by suppressing KLF410; and (3) KLF4 and TGF-β regulate each other in a context-dependent manner.42,43,49 Toward this, we quantified TGF-β signaling components in Klf4Δ/ΔCE corneas. qPCR revealed statistically significant upregulation of TGF-β1, -β2, -βR1, and -βR2 in the Klf4Δ/ΔCE corneas compared with the control (Fig. 1A). Consistently, immunofluorescent stain revealed significant upregulation of TGF-β1 -β2, -βR1, and -βR2 expression in Klf4Δ/ΔCE corneas (Figs. 1B, 1C), indicating that KLF4 negatively regulates TGF-β1, -β2, and their receptors within the CE.
Figure 1.
Klf4Δ/ΔCE cells display increased expression of TGF-β1, -β2, -βR1, and -βR2. (A) qPCR reveals that the Klf4Δ/ΔCE corneas display significant upregulation of TGF-β1, -β2, -βR1, and -βR2 transcripts. (B) Immunofluorescent stain confirms the elevated CE expression and localization of TGF-β1, -β2, -βR1, and -βR2 in Klf4Δ/ΔCE (v–viii) compared with the control (i–iv) cornea. (C) Bar graph showing quantified relative fluorescence intensity. Significant increase in TGF-β1, -β2, -βR1, and -βR2 was observed in Klf4Δ/ΔCE corneas compared with the control. Data show results from three independent experiments performed in duplicate and reported as means ± SEM. Images were acquired at 40×; scale bar: 40 μm.
To further confirm the role of KLF4 in regulating TGF-β signaling pathway in CE, we overexpressed KLF4 in HCLE cells using lentiviral transduction. qPCR revealed a 67-fold increase in KLF4 transcripts in HCLE-KLF4 cells compared with the HCLE-WT control (Fig. 2A). Robust overexpression and predominantly nuclear accumulation of KLF4 in HCLE-KLF4 cells were confirmed by immunoblots and immunofluorescent stain, respectively (Figs. 2B, 2C). qPCR also revealed that KLF4 overexpression resulted in a significant decrease in TGF-β1 (0.26-fold), TGF-β2 (0.89-fold), TGF-βR1 (0.44-fold), and TGF-βR2 (0.29-fold) in HCLE-KLF4 compared with the HCLE-WT cells, concomitant with a significant 15-fold increase in SMAD7, a cellular inhibitor of TGF-β signaling (Fig. 2D). Immunofluorescent stain displayed a consistent decrease in TGF-β1 and -β2 expression in the HCLE-KLF4 compared with the HCLE-WT cells (Fig. 2E). Together with Fig. 1, these results suggest that KLF4 negatively regulates TGF-β signaling within the CE.
Figure 2.
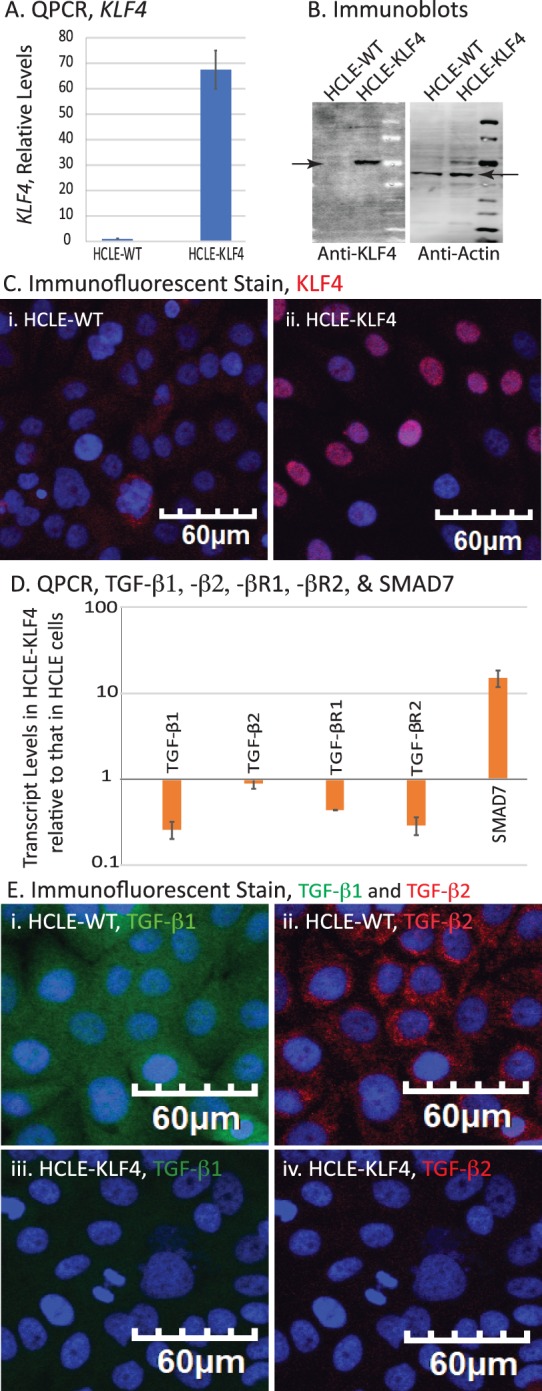
Overexpression of KLF4 in HCLE cells alters the expression levels of TGF-β signaling pathway components. (A) qPCR showing increased levels of KLF4 mRNA in HCLE-KLF4 compared with HCLE-WT. (B) Immunoblot showing overexpressed KLF4 in HCLE-KLF4 cells. (C) Immunofluorescent stain showing increased expression and nuclear localization of KLF4 in HCLE-KLF4. (D) HCLE-KLF4 cells show decreased levels of TGF-β1, -β2, -βR1, and -βR2 and increased expression of TGF-β signaling inhibitor SMAD7. Data are presented in logarithmic scale to accommodate large range in values. Data show results from three independent experiments performed in triplicates and reported as means ± SEM. (E) Immunofluorescent stain confirming the increased expression of TGF-β1and TGF-β2 in HCLE-KLF4 (iii, iv) compared with HCLE-WT cells (i, ii). Images were acquired at 20×; scale bar: 60 μm.
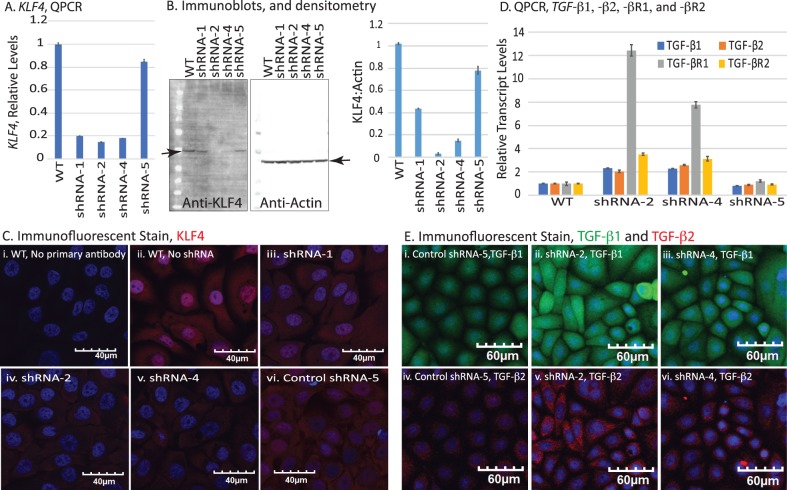
In a reciprocal approach, we knocked down the endogenous KLF4 expression levels in HCLE cells using plasmids expressing different anti-KLF4 shRNAs. qPCR revealed efficient knockdown of KLF4 in HCLE cells transfected with anti–KLF4-shRNA-1 (by 80%), –shRNA-2 (by 85%), and –shRNA-4 (by 82%), but not in those transfected with the control scrambled shRNA-5 (by 15%) compared with the untransfected cells (Fig. 3A). Immunoblots confirmed efficient knockdown of KLF4 protein in HCLE cells transfected with shRNA-2 (by 97%) and -4 (by 85%), but only a moderate decrease in those transfected with shRNA-1 (by 60%) and a marginal decrease in those transfected with control shRNA-5 (by 20%; Fig. 3B), which was further corroborated by immunofluorescent stain (Fig. 3C). qPCR also revealed a significant increase in TGF-β1, -β2, -βR1, and -βR2 transcripts in shRNA-2– and -4–transfected cells compared with shRNA-5 or control HCLE cells (Fig. 3D), which was further confirmed by immunofluorescent stain (Fig. 3E). Taken together, these results are consistent with a strong inverse relationship between of KLF4 and TGF-β signaling within the CE cells.
Figure 3.
Confirmation of shRNA-mediated KLF4 knockdown in HCLE (HCLE-KD) cells. (A) qPCR showing decreased KLF4 transcripts in HCLE cells transfected with anti-KLF4 shRNA-1, -2, and -4. shRNA-5 serves as a scrambled control. (B) Immunoblot confirms KLF4 knockdown. Bar graph shows densitometric quantification of the immunoblots. (C) Immunofluorescent stain showing the decreased expression and nuclear localization of KLF4 in shRNA-2– and -4–transfected cells. Images acquired at 40×; scale bar, 40 μm. (D) qPCR showing increased levels of TGF-β genes in HCLE-KD cells transfected with shRNA-2 and -4, relative to shRNA-5–transfected controls. Results from three independent experiments are reported as mean ± SEM. (E) Immunofluorescent stain showing expression of TGF-β1 and -β2 in control and HCLE-KD cells. Images acquired at 20×; scale bars as shown.
Klf4 Ablation Activates SMAD2/3-Mediated Canonical TGF-β Signaling
TGF-β mediates its activity either by canonical SMAD signaling or via the noncanonical mitogen-activated protein kinase pathway.50 To determine which pathway is activated on Klf4 ablation within the CE, we next examined Smad2/3 expression and phosphorylation status in the control and Klf4Δ/ΔCE corneas and in shRNA-2–, shRNA-4–, or shRNA-5–transfected HCLE cells. Immunofluorescent stain revealed no significant change in Smad2/3 expression in the Klf4Δ/ΔCE corneas (Figs. 4Ai, Aii). In contrast, phospho-Smad2/3 (pSmad2/3) levels were significantly increased, with elevated nuclear translocation in the Klf4Δ/ΔCE compared with the control corneas (Figs. 4Aiii, 4Aiv, 4B). Although there was no significant change in the overall levels of Smad4, it was predominantly localized in the nucleus in the Klf4Δ/ΔCE corneas (Figs. 4Av, 4Avi). Consistent with these results, immunoblots and immunofluorescent stain revealed elevated SMAD2/3 phosphorylation and nuclear localization in shRNA-2– and shRNA-4–transfected HCLE cells compared with those transfected with scrambled shRNA-5 and the untransfected controls (Figs. 5A–5C). Furthermore, SMAD4 was predominantly localized in the nucleus in shRNA-2– and -4–transfected HCLE cells compared with the shRNA-5 control transfected cells (Fig. 5D). Together, these results indicate that KLF4 knockdown activates SMAD2/3-mediated canonical TGF-β signaling.
Figure 4.
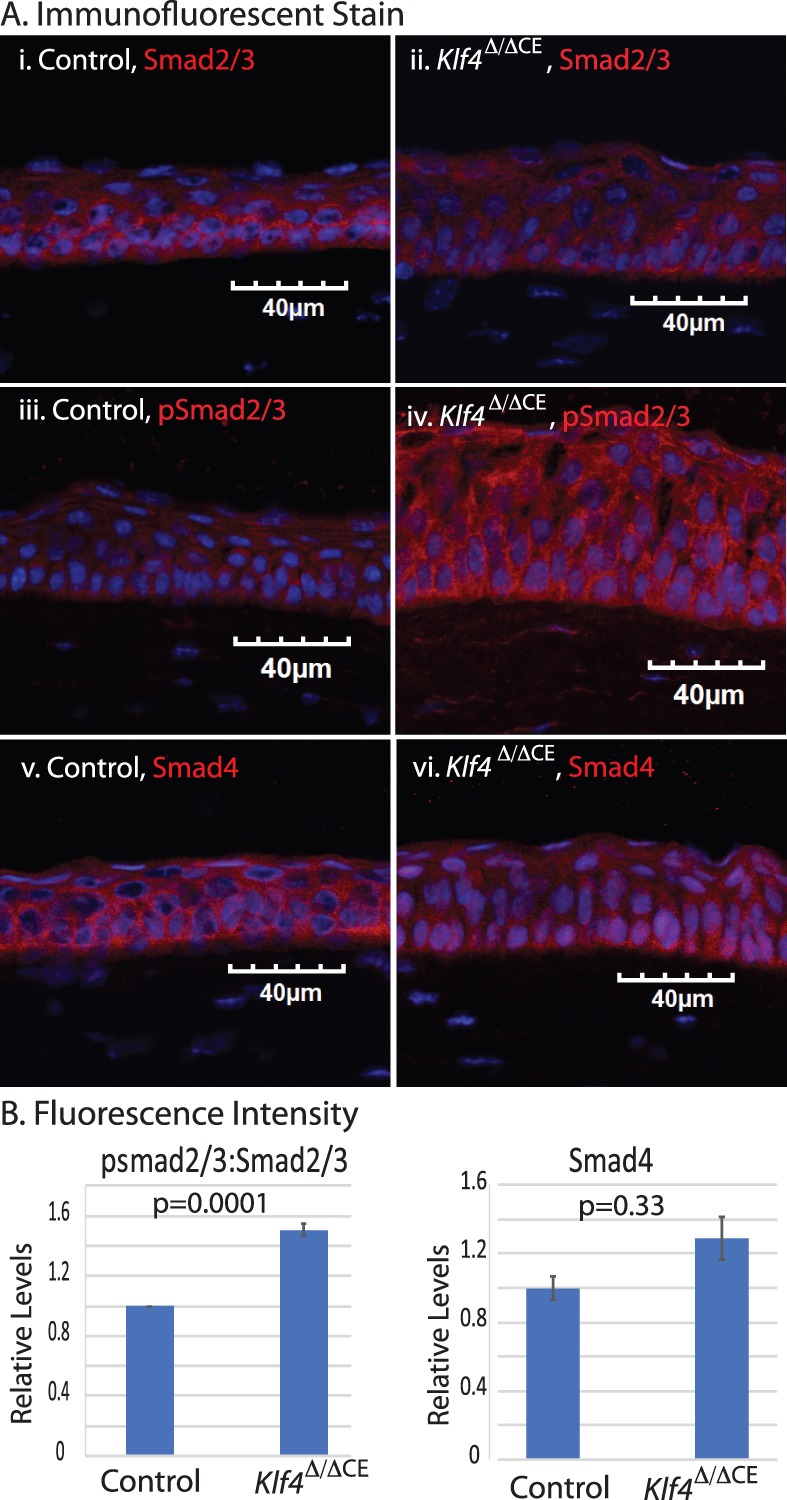
Canonical Smad2/3 signaling pathway is activated in Klf4Δ/ΔCE cells. (A) Expression of Smad2/3, pSmad2/3, and Smad4 in Klf4Δ/ΔCE and control CE visualized by immunofluorescent stain. (B) Bar graph shows fluorescence intensity, pSmad2/3:Smad2/3 ratio (left), and Smad4 (right). Data show results from three independent experiments reported as means ± SEM. Image acquired at 40×; scale bar: 40 μm.
Figure 5.
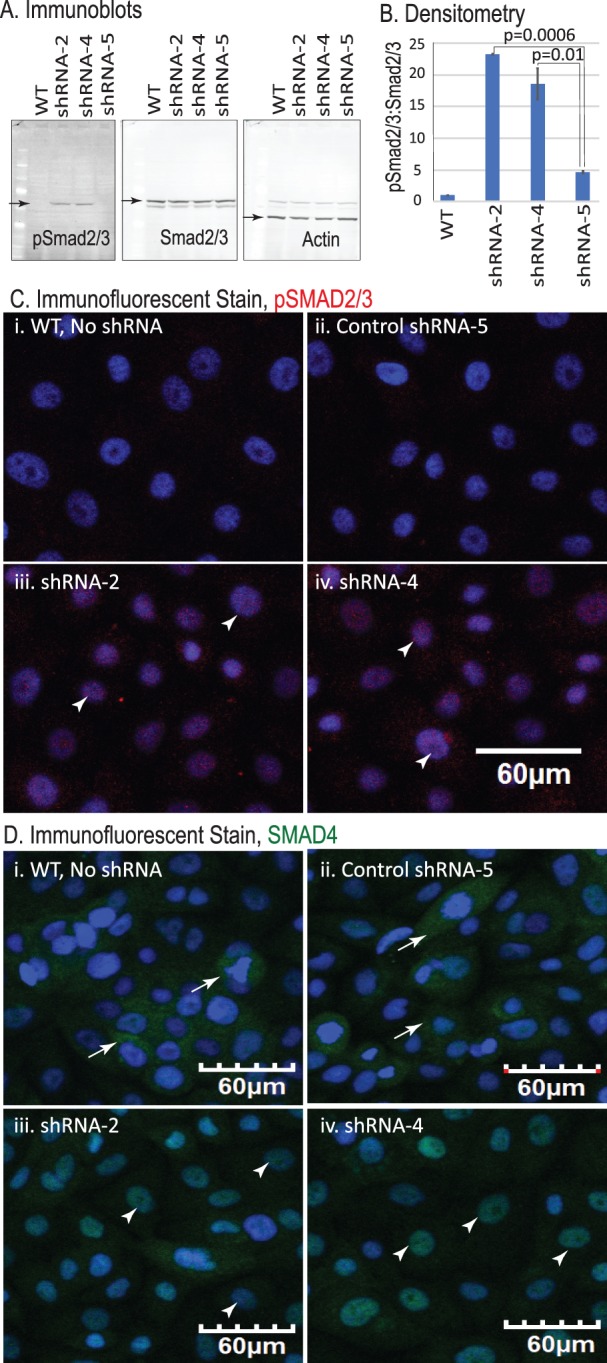
SMAD2/3 signaling pathway is activated in HCLE-KD cells. (A) Immunoblots showing pSMAD2/3 and total smad2/3 in control and HCLE-KD cells. β-Actin serves as loading control. (B) Bar diagram shows the quantified densitometric ratio of pSmad2/3:Smad2/3. Data show results from three independent experiments reported as mean ± SEM. (C) Immunofluorescent stain showing increased expression and localization of pSMAD2/3 in shRNA-2 and -4. No nuclear pSMAD2/3 was observed in controls. (D) Immunofluorescent stain showing increased nuclear translocation of SMAD4 in HCLE-KD cells compared with the control. Images were acquired at 20×; scale bar: 60 μm.
KLF4 Inhibits TGF-β Signaling by Upregulating SMAD7 Expression
As qPCR revealed a robust 15-fold increase in expression of TGF-β signaling inhibitor SMAD7 in HCLE-KLF4 cells (Fig. 2B), we further examined Smad7 in Klf4Δ/ΔCE and HCLE cells. Immunofluorescent stain revealed significant decrease in SMAD7 expression in Klf4Δ/ΔCE corneas and in shRNA-2 and -4 HCLE cells compared with the controls (Figs. 6A, 6B). Reciprocally, SMAD7 expression was significantly upregulated in HCLE-KLF4 cells (Fig. 6C). Collectively, these results indicate that KLF4 controls SMAD2/3-mediated TGF-β signaling in CE cells by upregulating SMAD7.
Figure 6.
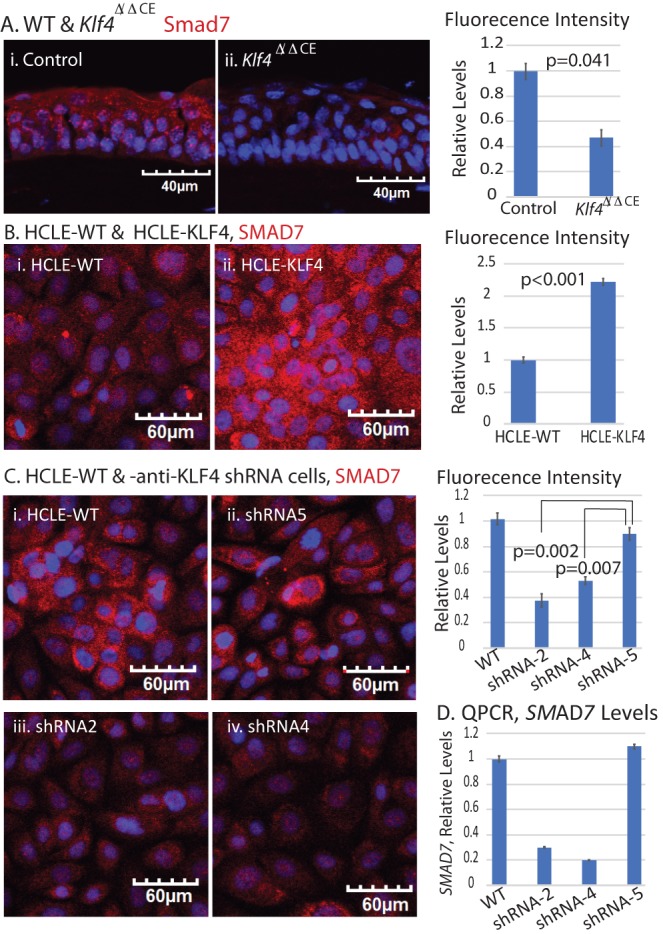
KLF4 regulates SMAD7 expression in CE cells. (A) Immunofluorescent stain and corresponding fluorescence intensity bar graph, showing significant decrease in Smad7 in Klf4Δ/ΔCE cells compared with the control. (B) Immunofluorescent stain and corresponding fluorescence intensity bar graph, showing significant increase in SMAD7 expression in HCLE-KLF4 cells. (C) Immunofluorescent stain and corresponding fluorescence intensity bar graphs showing significant decrease in SMAD7 expression in HCLE-KD compared with the control cells. Images were acquired at 20×; scale bar as shown. (D) qPCR confirming the downregulation of SMAD7 transcripts in HCLE-KD compared with the control cells. Data show results from two independent experiments performed in triplicate and reported as means ± SEM.
KLF4 Upregulates Cell Cycle Arrest Proteins p16 and p27
TGF-β signaling upregulates cyclin-dependent kinase (CDK) inhibitors p16 and p27 in different cell types.51–54 Considering that the results above suggest that TGF-β signaling is activated upon suppression of KLF4, it was not clear how Klf4 ablation results in increased proliferation in Klf4Δ/ΔCE cells.8,9,11 To clarify this, we evaluated p16 and p27 expression in Klf4Δ/ΔCE and HCLE-KLF4 cells. Klf4Δ/ΔCE corneas displayed a decrease in p16 and p27 expression compared with the control (Fig. 7A). Consistent with these results, HCLE-KLF4 cells expressed increased levels of p16 (2.5-fold) and p27 (1.5-fold) compared with the control HCLE-WT cells (Fig. 7B). Taken together, these results suggest that KLF4 overcomes the undesirable effect of unabated cell proliferation upon suppression of TGF-β signaling by promoting the expression of CDK inhibitors p16 and p27, which in turn inhibit cell cycle progression.
Figure 7.
KLF4 regulates p16 and p27. (A) Immunofluorescent stain for p16 (i, ii) and p27 (iii, iv) in control and Klf4Δ/ΔCE corneas. (B) Immunofluorescent stain in HCLE-KLF4 and HCLE-WT cells for p16 (i, ii) and p27 (iii, iv). Bar graphs on the right indicate significant decrease in p16 levels in Klf4Δ/ΔCE cells compared with the control or in HCLE-KLF4 compared with the HCLE-WT. Change in p27 levels was not significant. Data show results from three independent experiments reported as mean ± SEM. Images were acquired at 40×; scale bar: 40 μm for control and Klf4Δ/ΔCE cells. For HCLE-WT and HCLE-KLF4 cells, images were acquired at 20×; scale bar: 60 μm.
Discussion
Previously we reported that Klf4 ablation results in CE hyperplasia with compromised barrier function, increased proliferation, and migration, consistent with EMT commonly associated with squamous cell carcinomas.9,10 Here, we provide evidence that (1) activation of Smad2/3-mediated TGF-β signaling underlies the initiation of EMT upon downregulation of Klf4, and (2) decreased expression of CDK inhibitors p16 and p27 underlies the higher rate of proliferation in Klf4Δ/ΔCE corneas (Fig. 8). Collectively, these results suggest that KLF4, when present in normal levels, suppresses TGF-β signaling by promoting the expression of inhibitory SMAD7 and overcomes the undesirable concomitant decrease in TGF-β–dependent expression of p16 and p27 by directly upregulating them (Fig. 8).
Figure 8.
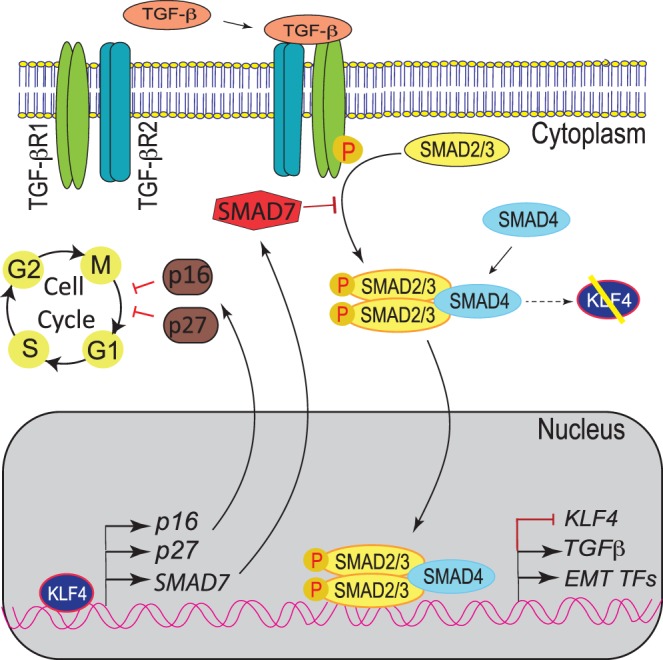
Crosstalk between KLF4 and TGF-β signaling plays an important role in CE homeostasis. In homeostatic condition, KLF4 controls cell proliferation by regulating proper cellular levels of SMAD7, p16, and p27. SMAD7 inhibits TGF-β signaling cascade suppressing EMT, whereas p16 and p27 regulate cell cycle progression. Decreased levels of KLF4 result in downregulation of SMAD7, which in turn leads to activation of TGF-β signaling and EMT, as well as downregulation of p16 and p27 that leads to dysregulated cell cycle progression through the G1/S phase.
The unique positioning of proliferating and differentiated cells in the complex architecture of stratified squamous corneal epithelium makes it a useful model to understand the important biological events essential for epithelial homeostasis and pathology. KLF4 and TGF-β both influence cell proliferation and serve as anti-or protumorigenic agents in a context-dependent manner. For example, TGF-β regulates KLF4 positively in vascular smooth muscle cells and negatively in mink lung epithelial cells.42,43 Although the individual roles of KLF4 and TGF-β are well known in the cornea, the crosstalk between them was not studied previously.10,55 Our observation of altered expression of canonical TGF-β signaling pathway components in Klf4Δ/ΔCE corneas and HCLE-KLF4 cells provides evidence that KLF4 helps maintain appropriate levels TGF-β signaling in CE cells. In corneas subjected to injuries, and/or pathologic conditions, elevated TGF-β coupled with lower KLF4 levels initiates EMT and tumorigenic signaling.10,56
TGF-β functions through the SMAD-dependent or SMAD-independent signaling cascade, depending on the cellular context.57–60 We observed increased SMAD2/3 phosphorylation in Klf4Δ/ΔCE corneas and upon shRNA-mediated knockdown of KLF4 in HCLE (HCLE-KD) cells, suggesting that canonical SMAD signaling is activated. The increased shuttling of Smad4 to the nucleus in Klf4Δ/ΔCE cornea, an essential step for nuclear entry of pSmad2/3, provides a favorable platform for transcription of TGF-β target genes. SMAD7 inhibits TGF-β signaling by interacting with TGF-β receptors, impeding with TGF-β receptor–SMAD2/3-SMAD4 complex formation.61,62 Upregulation of SMAD7 in HCLE-KLF4, and its downregulation in Klf4Δ/ΔCE corneas as well as HCLE-KD cells, suggests that KLF4 upregulates SMAD7, which in turn suppresses TGF-β signaling in CE cells (Fig. 8). Taken together, the lower levels of SMAD7 and increased pSMAD2/3 and nuclear SMAD4 in HCLE-KD cells and Klf4Δ/ΔCE corneas provide evidence that KLF4 regulates SMAD-mediated TGF-β signaling in CE cells.
The data presented here demonstrate that Klf4Δ/ΔCE corneas display elevated TGF-β signaling that induces EMT. EMT creates a pool of differentiated myofibroblast-like cells that secrete extracellular matrix proteins, leading to fibrosis.63,64 EMT also results in apoptosis in the presence of TGF-β in many cell types.65,66 The sustained presence of TGF-β is also known to carve out a small population of proliferative cells that can evade EMT-induced apoptosis.65 Our previous data and current observations indicate no signs of growth arrest or apoptosis in Klf4Δ/ΔCE corneas. Instead, we observed hyperproliferation of CE cells,9,10 likely due to the release of p16- and p27-mediated cell cycle arrest. KLF4 suppresses cyclin-D1 and promotes p16 and p27, resulting in cell cycle arrest at G1/S, which further results in cellular senescence and arrested growth.67 The loss of KLF4 creates a proliferative milieu by downregulating p16 and p27 and favors mesenchymal phenotype by elevating TGF-β signaling, creating a pathologic environment conducive for squamous neoplasia.
Corneal tumors are relatively rare despite frequent exposure of the cornea to adverse conditions such as UV radiation, chemical and physical insults, pathogens, and mutagens. Ocular surface squamous neoplasia (OSSN), a rare form of ocular tumor in elderly patients, occurs mainly due to UV exposure,4,5 p53 mutation, and hereditary deficiency of DNA damage.68 Although OSSN is visually debilitating and clinically relevant, the lack of appropriate study systems and low abundance of clinical samples makes it difficult to understand the molecular mechanisms that underlie OSSN. Considering that KLF4 is highly expressed in the CE cells, and its ablation results in EMT, it is likely that KLF4 serves as a strong tumor suppressor in CE cells, protecting them from frequent oncogenic challenges.
In summary, this report provides evidence that KLF4 regulates corneal epithelial cell cycle progression by suppressing canonical TGF-β signaling and upregulating CDK inhibitors P16 and P27. Interestingly, the related transcription factor KLF5 also serves as a tumor suppressor by downregulating EGF signaling and upregulating p15, indicating that multiple redundant mechanisms operate in concert to regulate cell proliferation.37,39,40 Although KLF4, KLF5, and TGF-β are highly expressed in the cornea, and are frequently altered in many cancers, their involvement in OSSN is not well established. It would be worthwhile examining human OSSN samples to further verify our findings, as better understanding of the crosstalk between KLF4, KLF5, and TGF-β in the CE would help us further unravel their involvement in epithelial tumor development and progression.
Supplementary Material
Acknowledgments
The authors thank Kate Davoli (Histology Core Facility), Kira Lathrop (Imaging Core Facility), and the Molecular Biology Core Facility for help during this study.
Supported by National Institutes of Health Grant RO1 EY026533 (SKS), National Eye Institute Core Grant P30 EY08098, and unrestricted grants from Research to Prevent Blindness Inc. and the Eye & Ear Foundation of Pittsburgh.
Disclosure: A. Tiwari, None; S. Swamynathan, None; N. Alexander, None; J. Gnalian, None; S. Tian, None; P.R. Kinchington, None; S.K. Swamynathan, None
References
- 1.Swamynathan SK. Ocular surface development and gene expression. J Ophthalmol. 2013;2013:103947. doi: 10.1155/2013/103947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhouailly D, Pearton DJ, Michon F. The vertebrate corneal epithelium: from early specification to constant renewal. Dev Dyn. 2014;243:1226–1241. doi: 10.1002/dvdy.24179. [DOI] [PubMed] [Google Scholar]
- 3.Thoft RA, Friend J. The X, Y, Z hypothesis of corneal epithelial maintenance. Invest Ophthalmol Vis Sci. 1983;24:1442–1443. [PubMed] [Google Scholar]
- 4.Gichuhi S, Ohnuma S, Sagoo MS, Burton MJ. Pathophysiology of ocular surface squamous neoplasia. Exp Eye Res. 2014;129:172–182. doi: 10.1016/j.exer.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carreira H, Coutinho F, Carrilho C, Lunet N. HIV and HPV infections and ocular surface squamous neoplasia: systematic review and meta-analysis. Br J Cancer. 2013;109:1981–1988. doi: 10.1038/bjc.2013.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norman B, Davis J, Piatigorsky J. Postnatal gene expression in the normal mouse cornea by SAGE. Invest Ophthalmol Vis Sci. 2004;45:429–440. doi: 10.1167/iovs.03-0449. [DOI] [PubMed] [Google Scholar]
- 7.Swamynathan S, Kenchegowda D, Piatigorsky J, Swamynathan S. Regulation of corneal epithelial barrier function by Kruppel-like transcription factor 4. Invest Ophthalmol Vis Sci. 2011;52:1762–1769. doi: 10.1167/iovs.10-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swamynathan SK, Davis J, Piatigorsky J. Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol Vis Sci. 2008;49:3360–3370. doi: 10.1167/iovs.08-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delp EE, Swamynathan S, Kao WW, Swamynathan SK. Spatiotemporally regulated ablation of Klf4 in adult mouse corneal epithelial cells results in altered epithelial cell identity and disrupted homeostasis. Invest Ophthalmol Vis Sci. 2015;56:3549–3558. doi: 10.1167/iovs.15-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiwari A, Loughner CL, Swamynathan S, Swamynathan SK. KLF4 plays an essential role in corneal epithelial homeostasis by promoting epithelial cell fate and suppressing epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci. 2017;58:2785–2795. doi: 10.1167/iovs.17-21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, Piatigorsky J. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Molec Cell Biol. 2007;27:182–194. doi: 10.1128/MCB.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, Chen XQ, Li P. The role of TGF-beta and its receptors in gastrointestinal cancers. Translational Oncol. 2018;12:475–484. doi: 10.1016/j.tranon.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loughner CL, Tiwari A, Kenchegowda D, Swamynathan S, Swamynathan SK. Spatiotemporally controlled ablation of Klf5 results in dysregulated epithelial homeostasis in adult mouse corneas. Invest Ophthalmol Vis Sci. 2017;58:4683–4693. doi: 10.1167/iovs.17-22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A. KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Molec Cell Biol. 2011;31:2513–2527. doi: 10.1128/MCB.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 19.Ghaleb AM, Yang VW. Kruppel-like factor 4 (KLF4): what we currently know. Gene. 2017;611:27–37. doi: 10.1016/j.gene.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q, Hong Y, Zhan Q, Shen Y, Liu Z. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009;69:8284–8292. doi: 10.1158/0008-5472.CAN-09-1345. [DOI] [PubMed] [Google Scholar]
- 21.Du JX, McConnell BB, Yang VW. A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Kruppel-like factor 4. J Biol Chem. 2010;285:28298–28308. doi: 10.1074/jbc.M110.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 23.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- 25.Cullingford TE, Butler MJ, Marshall AK, Tham el L, Sugden PH, Clerk A. Differential regulation of Kruppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta. 2008;1783:1229–1236. doi: 10.1016/j.bbamcr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shie JL, Chen ZY, O'Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointestinal Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Zhang H, Zhu L, Zhao L, Dong Y. Kruppel-like factor 4 is a novel mediator of selenium in growth inhibition. Molec Cancer Res. 2008;6:306–313. doi: 10.1158/1541-7786.MCR-07-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZY, Shie J, Tseng C. Up-regulation of gut-enriched kruppel-like factor by interferon-gamma in human colon carcinoma cells. FEBS Lett. 2000;477:67–72. doi: 10.1016/s0014-5793(00)01764-6. [DOI] [PubMed] [Google Scholar]
- 29.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M, Hao B, Zhan Y, Luo G. Kruppel-like factor 4 expression in solid tumor prognosis: a meta-analysis. Clin Chim Acta. 2018;485:50–59. doi: 10.1016/j.cca.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 31.Anumanthan G, Halder SK, Osada H, et al. Restoration of TGF-beta signalling reduces tumorigenicity in human lung cancer cells. Br J Cancer. 2005;93:1157–1167. doi: 10.1038/sj.bjc.6602831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis-Dusenbery BN, Chan MC, Reno KE, et al. Down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh MI, Chang Y, Jung JC. Temporal and spatial distribution of TGF-beta isoforms and signaling intermediates in corneal regenerative wound repair. Histol Histopathol. 2009;24:1405–1416. doi: 10.14670/HH-24.1405. [DOI] [PubMed] [Google Scholar]
- 34.Massague J, Cheifetz S, Laiho M, Ralph DA, Weis FM, Zentella A. Transforming growth factor-beta. Cancer Surv. 1992;12:81–103. [PubMed] [Google Scholar]
- 35.Saarma M. GDNF: a stranger in the TGF-beta superfamily? Eur J Biochem. 2000;267:6968–6971. doi: 10.1046/j.1432-1327.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 36.Wilson SE, Schultz GS, Chegini N, Weng J, He YG. Epidermal growth factor, transforming growth factor alpha, transforming growth factor beta, acidic fibroblast growth factor, basic fibroblast growth factor, and interleukin-1 proteins in the cornea. Exp Eye Res. 1994;59:63–71. doi: 10.1006/exer.1994.1081. [DOI] [PubMed] [Google Scholar]
- 37.Sun H, Peng Z, Tang H, et al. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-beta signaling in and promote progression of hepatocellular carcinoma. Oncogene. 2017;36:2957–2968. doi: 10.1038/onc.2016.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soleimani A, Pashirzad M, Avan A, Ferns GA, Khazaei M, Hassanian SM. Role of the transforming growth factor-beta signaling pathway in the pathogenesis of colorectal cancer. J Cell Biochem. doi: 10.1002/jcb.28331. [published online ahead of print December 16, 2018] [DOI] [PubMed]
- 39.Pang X, Tang YL, Liang XH. Transforming growth factor-beta signaling in head and neck squamous cell carcinoma: insights into cellular responses. Oncol Lett. 2018;16:4799–4806. doi: 10.3892/ol.2018.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. doi: 10.1016/j.tcb.2018.12.001. [published online ahead of print December 26, 2018] [DOI] [PubMed]
- 41.Tandon A, Tovey JC, Sharma A, Gupta R, Mohan RR. Role of transforming growth factor Beta in corneal function, biology and pathology. Curr Molec Med. 2010;10:565–578. doi: 10.2174/1566524011009060565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu D, Wan Y. Regulation of Kruppel-like factor 4 by the anaphase promoting complex pathway is involved in TGF-beta signaling. J Biol Chem. 2011;286:6890–6901. doi: 10.1074/jbc.M110.179952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li HX, Han M, Bernier M, et al. Kruppel-like factor 4 promotes differentiation by transforming growth factor-beta receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells. J Biol Chem. 2010;285:17846–17856. doi: 10.1074/jbc.M109.076992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. J Pathol. 2011;223:205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 45.Liu YN, Abou-Kheir W, Yin JJ, et al. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor beta-initiated prostate cancer epithelial-mesenchymal transition. Molec Cell Biol. 2012;32:941–953. doi: 10.1128/MCB.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argueso P, Tisdale A, Spurr-Michaud S, Sumiyoshi M, Gipson IK. Mucin characteristics of human corneal-limbal epithelial cells that exclude the rose bengal anionic dye. Invest Ophthalmol Vis Sci. 2006;47:113–119. doi: 10.1167/iovs.05-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campeau E, Ruhl VE, Rodier F, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksoy I, Giudice V, Delahaye E, et al. Klf4 and Klf5 differentially inhibit mesoderm and endoderm differentiation in embryonic stem cells. Nat Commun. 2014;5:3719. doi: 10.1038/ncomms4719. [DOI] [PubMed] [Google Scholar]
- 49.He M, Zheng B, Zhang Y, et al. KLF4 mediates the link between TGF-beta1-induced gene transcription and H3 acetylation in vascular smooth muscle cells. FASEB J. 2015;29:4059–4070. doi: 10.1096/fj.15-272658. [DOI] [PubMed] [Google Scholar]
- 50.Clark DA, Coker R. Transforming growth factor-beta (TGF-beta) Int J Biochem Cell Biol. 1998;30:293–298. doi: 10.1016/s1357-2725(97)00128-3. [DOI] [PubMed] [Google Scholar]
- 51.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robson CN, Gnanapragasam V, Byrne RL, Collins AT, Neal DE. Transforming growth factor-beta1 up-regulates p15, p21 and p27 and blocks cell cycling in G1 in human prostate epithelium. J Endocrinol. 1999;160:257–266. doi: 10.1677/joe.0.1600257. [DOI] [PubMed] [Google Scholar]
- 53.Pillaire MJ, Casagrande F, Malecaze F, Manenti S, Darbon JM. Regulation by transforming growth factor-beta 1 of G1 cyclin-dependent kinases in human retinal epithelial cells. Exp Eye Res. 1999;68:193–199. doi: 10.1006/exer.1998.0583. [DOI] [PubMed] [Google Scholar]
- 54.Kamesaki H, Nishizawa K, Michaud GY, Cossman J, Kiyono T. TGF-beta 1 induces the cyclin-dependent kinase inhibitor p27Kip1 mRNA and protein in murine B cells. J Immunol. 1998;160:770–777. [PubMed] [Google Scholar]
- 55.Mita T, Yamashita H, Kaji Y, et al. Effects of transforming growth factor beta on corneal epithelial and stromal cell function in a rat wound healing model after excimer laser keratectomy. Graefes Arch Clin Exp Ophthalmol. 1998;236:834–843. doi: 10.1007/s004170050168. [DOI] [PubMed] [Google Scholar]
- 56.Jester JV, Barry-Lane PA, Cavanagh HD, Petroll WM. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea. 1996;15:505–516. [PubMed] [Google Scholar]
- 57.Bazan HE, Varner L. A mitogen-activated protein kinase (MAP-kinase) cascade is stimulated by platelet activating factor (PAF) in corneal epithelium. Curr Eye Res. 1997;16:372–379. doi: 10.1076/ceyr.16.4.372.10699. [DOI] [PubMed] [Google Scholar]
- 58.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 59.Li DQ, Luo L, Chen Z, Kim HS, Song XJ, Pflugfelder SC. JNK and ERK MAP kinases mediate induction of IL-1beta, TNF-alpha and IL-8 following hyperosmolar stress in human limbal epithelial cells. Exp Eye Res. 2006;82:588–596. doi: 10.1016/j.exer.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saika S. TGF-beta signal transduction in corneal wound healing as a therapeutic target. Cornea. 2004;23:S25–S30. doi: 10.1097/01.ico.0000136668.41000.73. [DOI] [PubMed] [Google Scholar]
- 61.Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Fei T, Zhang L, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Molec Cell Biol. 2007;27:4488–4499. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiwari A, Kumar R, Ram J, Sharma M, Luthra-Guptasarma M. Control of fibrotic changes through the synergistic effects of anti-fibronectin antibody and an RGDS-tagged form of the same antibody. Sci Rep. 2016;6:30872. doi: 10.1038/srep30872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27:1218–1230. doi: 10.1038/sj.onc.1210741. [DOI] [PubMed] [Google Scholar]
- 66.Tiwari N, Meyer-Schaller N, Arnold P, et al. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8) PLoS One. 2013;8:e57329. doi: 10.1371/journal.pone.0057329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans PM, Liu C. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sinica. 2008;40:554–564. doi: 10.1111/j.1745-7270.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basti S, Macsai MS. Ocular surface squamous neoplasia: a review. Cornea. 2003;22:687–704. doi: 10.1097/00003226-200310000-00015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.