Abstract
Background
The bioactive metabolites of omega 3 and omega 6 polyunsaturated fatty acids (ω-3 and ω-6) are known as oxylipins and endocannabinoids (eCBs). These lipid metabolites are involved in prompting and resolving the inflammatory response that leads to the onset of inflammatory bowel disease (IBD). This study aims to quantify these bioactive lipids in the colonic mucosa and to evaluate the potential link to cytokine gene expression during inflammatory events in ulcerative colitis (UC).
Methods
Colon biopsies were taken from 15 treatment-naïve UC patients, 5 deep remission UC patients, and 10 healthy controls. Thirty-five oxylipins and 11 eCBs were quantified by means of ultra-high-performance liquid chromatography coupled with tandem mass spectrometry. Levels of mRNA for 10 cytokines were measured by reverse transcription polymerase chain reaction.
Results
Levels of ω-6-related oxylipins were significantly elevated in treatment-naïve patients with respect to controls, whereas the levels of ω-3 eCBs were lower. 15S-Hydroxy-eicosatrienoic acid (15S-HETrE) was significantly upregulated in UC deep remission patients compared with controls. All investigated cytokines had significantly higher mRNA levels in the inflamed mucosa of treatment-naïve UC patients. Cytokine gene expression was positively correlated with several ω-6 arachidonic acid–related oxylipins, whereas negative correlation was found with lipoxin, prostacyclin, and the eCBs.
Conclusions
Increased levels of ω-6-related oxylipins and decreased levels of ω-3-related eCBs are associated with the debut of UC. This highlights the altered balance between pro- and anti-inflammatory lipid mediators in IBD and suggests potential targets for intervention.
Keywords: EPEA, DHEA, IBD, PUFA, eicosanoids
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic, relapsing inflammatory disorder in the gastrointestinal tract that affects up to 0.5% of the population of the Western world.1 The 2 major forms of IBD, ulcerative colitis (UC) and Crohn’s disease (CD), are characterized by a dysregulated mucosal immune response triggered by intestinal commensal flora.2 The onset of IBD symptoms appears to be caused by an imbalance between pro- and anti-inflammatory molecules.3 However, several factors might be involved in the chronic inflammatory state observed in IBD. These include cytokines, interleukins (ILs), nitric oxide (NO), free radicals, activated Toll-like receptors, oxylipins, and microbiota.3 Furthermore, it has previously been shown that colitis is associated with a disruption in the lipid metabolism.4
Oxylipins are bioactive derivatives mainly from omega 3 and omega 6 polyunsaturated fatty acids (ω-3 and ω-6 PUFAs) such as ω-6 arachidonic acid (AA), ω-6 linoleic acid (LA), ω-3 eicosapentaenoic acid (EPA), and ω-3 docosahexaenoic acid (DHA).5 Oxylipins are synthetized through 3 main enzymatic pathways, namely cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP450), resulting in more than 100 active mediators. The AA-derived oxylipins, also known as eicosanoids, are involved in chemotaxis and promoting the recruitment of neutrophils to the site of inflammation. The role of oxylipins in IBD is very complex and not completely understood; for example, prostaglandin E2 (PGE2) induces epithelial proliferation in response to mucosal damage and suppresses the release of tumor necrosis factor (TNF) from macrophages.6 Leukotriene B4 (LTB4) has chemotactic effects by stimulating leucocyte activation and adhesion to the vascular endothelium and promotes the production of inflammatory cytokines.7 Furthermore, inflammation-resolving oxylipins termed resolvins, lipoxins, protectins, and maresins are produced from AA, EPA, and DHA.8
The endocannabinoids (eCBs) are a family of bioactive lipids that are biosynthesized from membrane glycerophospholipids and bind to cannabis receptors (CB), specifically, CB1 and CB2.9 The primary eCBs are arachidonoyl ethanolamine, known as anandamide (AEA), and 2-arachidonoylglycerol (2-AG). The secondary or “atypical” eCBs, such as docohexaenoic ethanolamine (DHEA) and eicosapentaenoyl ethanolamide (EPEA), play an important synergetic role to AEA.10 CB1 receptors are highly expressed in several brain regions that mediate the psychoactive effects of cannabinoids, whereas CB2 receptors are found in a number of immune cells and in a few neurons.11 It has been shown that eCBs regulate immune homeostasis in the gut–pancreas axis. For instance, eCBs inhibit the release of a wide class of pro-inflammatory mediators, including IL-1β, TNF, and NO.9, 12 Some studies have reported changes in endocannabinoid system expression during UC.13 However, a previous targeted analysis of eCBs in inflamed mucosa in IBD was inconclusive and was restricted to ω-6 AA derivatives.14–17
A quantitative analysis of all bioactive lipid metabolites in UC colon biopsies is needed to fully understand their involvement in promoting and resolving the inflammatory event in IBD. Therefore, in this study, we have quantified 35 nonesterified oxylipins and 11 eCB metabolites (Supplementary Table 1) simultaneously in colon biopsies taken from 3 different groups, namely treatment-naïve UC patients in the debut of the disease, deep remission UC patients, and healthy subjects. We have further analyzed the cytokine profile in colon biopsies from the same patients to evaluate a potential link between the lipid profile and the inflammatory events mediated by pro- and anti-inflammatory cytokines.
METHODS
Collection of Biopsies
Mucosal biopsies were collected from newly diagnosed treatment-naïve UC patients and UC patients in deep remission. UC diagnosis was established based on clinical, endoscopic, and histological criteria defined by European Crohn’s and Colitis Organization (ECCO) guidelines.18 Furthermore, the degree of inflammation was evaluated during colonoscopy using the scoring system of the Ulcerative Colitis Disease Activity Index (UCDAI).19 Moreover, TNF mRNA levels were measured by real-time reverse transcription polymerase chain reaction (RT-PCR) to assess the level of UC activity.20 Deep remission was defined by endoscopically healed mucosa (Mayo score = 0) and a normalized mucosal TNF gene expression level induced by anti-TNF treatment.21 Subjects admitted for a cancer screening and with normal colonoscopy histological findings served as healthy controls. None of the recruited subjects suffered from irritable bowel syndrome, and they were not taking nonsteroidal anti-inflammatory drugs (NSAIDs) before the colonoscopy. The patients in deep remission were on regular UC medications including 5-aminosalicylic acid (5-ASA), azathioprine, and anti-TNF. From each study participant, 2 adjacent biopsies were obtained from the inflamed mucosa, and 1 biopsy was immediately immersed in RNAlater (Qiagen, Hilden, Germany). The second biopsy was immediately frozen in a dry cryotube tube at –70°C. The biopsies from both UC patients and the UC remission group were obtained from the rectum or sigmoid colon, whereas biopsies from the control group were obtained from the rectum area only. The dry weight of the biopsies ranged from 2 to 8 mg. All biopsies were kept at –70°C until further analysis.
Chemical and Reagents
The eCB analytical standards, the oxylipin analytical standards, and 12- (cyclohexylamino)carbonyl[amino]-dodecanoic acid (CUDA) were purchased from Cayman Chemicals (Ann Arbor, MI, USA). Acetonitrile (ACN) and methanol (MeOH) were acquired from Merck (Darmstadt, Germany). Isopropanol was obtained from VWR PROLABO (Fontenay-sous-Bois, France). Acetic acid was purchased from Aldrich Chemical Company, Inc. (Milwaukee, WI, USA). All solvents were of HPLC grade or higher. Water was purified by a Milli-Q Gradient system (Millipore, Milford, MA, USA). Oasis HLB cartridges (3 cc, 60 mg) were obtained from Waters (Milford, MA, USA).
Endocannabinoid and Oxylipin Quantification
Analysis of eCBs and nonesterified oxylipins was performed by a previously published method.22 Briefly, 500 μL of methanol and a tungsten bead were added to each sample; the samples were then mixed in Qiagen plates (Qiagen, Valencia, CA, USA) for 3 minutes at a speed of 30 Hz. After removing the beads, the samples were centrifuged for 3 minutes at a speed of 14,000 rpm (2125 × g) and +4°C. The metabolites were extracted by a solid phase extraction (SPE) protocol described elsewhere.23 In brief, the samples were spiked with 10 μL of the following internal standard solution: 50 ng/mL 12,13-DiHOME-d4 and 12,13-EPOME-d4, 25 ng/mL 9-HODE-d4, PGE2-d4, 5-HETE-d8, 20-HETE-d6 and TXB2-d4, 800 ng/mL 2-AG-d8, 40 ng/mL PGF2α-EA-d4 and PGE2-EA-d4, 20 ng/mL AEA-d4, OEA-d4, and SEA-d3. Then, the samples were applied to the SPE columns and washed by a mix of 5% MeOH and 0.1% acetic acid, before eluting the metabolites with 3 mL of ACN and 2 mL of MeOH. Finally, the samples were dried using a vacuum concentrator (MIVac, SP, Warminster, PA, USA), reconstituted in 100 µL of MeOH, and spiked with 10 µL of the recovery standard CUDA (0.025 µg/mL). The analysis was conducted using an Agilent UPLC system (Infinity 1290) coupled with an electrospray ionization source (ESI) to an Agilent 6490 triple quadrupole system equipped with iFunnel Technology (Agilent Technologies, Santa Clara, CA, USA). Metabolite separation was performed using a Waters BEH C18 column (2.1 mm × 150 mm, 130 Å, 1.7-μm particle size). A flow rate of 300 μL/min and 10-μL injection volume were employed for each run. Separate injections were used for subsequent ionization in positive (eCB) and negative (oxylipin) mode. The mobile phase consisted of (1) 0.1% acetic acid in MilliQ water and (2) acetonitrile:isopropanol (90:10). The gradient and ESI applied conditions were optimized and have been described elsewhere.23 MassHunter Workstation software was used to control the instrument and to integrate all peaks manually.
An 8-point calibration curve was constructed using pure standards. Furthermore, the recovery of each internal standard was calculated by adding the recovery standard CUDA to each sample as quality control.
Quantification of Cytokine mRNA Using Real-time PCR
Total RNA was isolated from patient biopsies using the Allprep DNA/RNA Mini Kit (Qiagen, Hilden, Germany, Cat No: 80204) and the automated QIAcube instrument (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Quantity and purity of the extracted RNA were determined using the Qubit 3 Fluorometer (Cat No: Q33216; Invitrogen by Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription of the total RNA was performed using the QuantiTect Reverse Transcription Kit (Cat. No: 205314; Qiagen, Hilden, Germany). Levels of mRNA for IL-1β, IL-4, IL-5, IL-6, IL-10, IL-12, IL-17, IL-23, IFN-γ, TNF, and the housekeeping gene β-actin were quantified by a previously published method.24 The primers and probe sequences are shown in Supplementary Table 3. Cytokine mRNA expression was reported according to the ∆CT and ∆∆CT method described by Schmittgen, with fold change as 2-∆∆CT.25 For the TNF assay, we used an in-house absolute standard based on a serially diluted PCR product. By using this standard curve, we derived a copy number per μg of total RNA for each sample.
Statistical Analysis
The concentration of each metabolite was normalized by sample weight, and the results were reported as pg/mg of colon tissue. Statistical analysis was carried out using MetaboAnalyst 3.0, a web tool for metabolomics data analysis (http://www.metaboanalyst.ca/).26 Two samples had extremely low and high concentrations (below/higher than the mean plus/minus 3 standard deviations) of 60% of the metabolites and were consequently excluded. One percent of reported metabolites were below the level of detection. Therefore, they were replaced by a small value (half of the minimum positive value in the original data).
First, metabolite concentrations were autoscaled to reduce differences in magnitude.27 Second, the Shapiro-Wilk test for normality was run. The data were found to be non–normally distributed, and nonparametric univariate analysis (Mann-Whitney U test) was performed. Differences in the mean concentration of metabolites between the study groups were identified at a fold change (FC) of 2 and a false discovery rate (FDR; Benjamini Hochberg) cutoff of 0.1, as previously described.28 The 2-FC cutoff was chosen to minimize the effects of biological variation, whereas the FDR cutoff was set to 0.1 due to the exploratory nature of our study, and the low risk of reporting false positivity. Finally, significant variation in the metabolite concentrations among the 3 study groups was detected by Kruskal-Wallis nonparametric analysis of variance. For multiple testing correction, acquired P values were adjusted using the Benjamini and Hochberg method. Adjusted P values <0.05 were considered significant.
Frequency distribution analysis and tests of normality (Shapiro Wilk) were run on ΔCT values from RT-PCR analyses. The data were found to be normally distributed, and cytokine gene expression differences between the study groups were compared using a 2-tailed Student t test. To account for the multiple group testing, acquired P values were adjusted by the Dunett post hoc test. Adjusted P values <0.05 were considered significant.
Pair-wise Spearman’s rank correlation coefficients between metabolites, transcripts, and between metabolites and transcripts (autoscaled values) were computed and are presented in a heatmap. This was done using RStudio: Integrated Development Environment (version 1.0.143); and R package “corrplot”: Visualization of a Correlation Matrix (version 0.84; https://github.com/taiyun/corrplot).
Ethical Considerations
The Regional Committee of Medical Ethics of North Norway and the Norwegian Social Science Data Services approved the study and the storage of biological material under the number REK NORD 2012/1349. In addition, all enrolled subjects have signed an informed written consent.
RESULTS
Subjects Characteristic
In total, 15 newly diagnosed treatment-naïve UC patients with mild to severe disease activity, 5 UC patients in deep remission, and 10 healthy controls were enrolled in this study. The study group characteristics are described in Table 1. Ulcerative colitis patients’ disease activity ranged from mild to severe; a UCDAI score of 3 to 6 was defined as mild, 7 to 10 as moderate, and 11 to 12 as severe UC. Accordingly, 7 patients had mild UC, 4 patients had moderate UC, and 4 patients had severe UC.
TABLE 1:
Description of Study Group Characteristics
| Study Group | No. Subjects | Age, Mean (Range), y | Sex, Female/Male | UCDAI Score, Median (Range) | TNF-α, Mean (Range), Copies/μg of Total RNA |
|---|---|---|---|---|---|
| UC patients | 15 | 37 (14–69) | 6/9 | 9 (4–15) | 15,207 (4300–44,600) |
| Healthy controls | 10 | 68 (25–86) | 4/6 | — | 3430 (1100–7900) |
| UC remission | 5 | 46 (41–70) | 0/5 | <1 | 4083 (1400–8500) |
Mucosal eCB and Oxylipin Profiles in Treatment-Naïve UC Patients, UC Remission Patients, and Controls
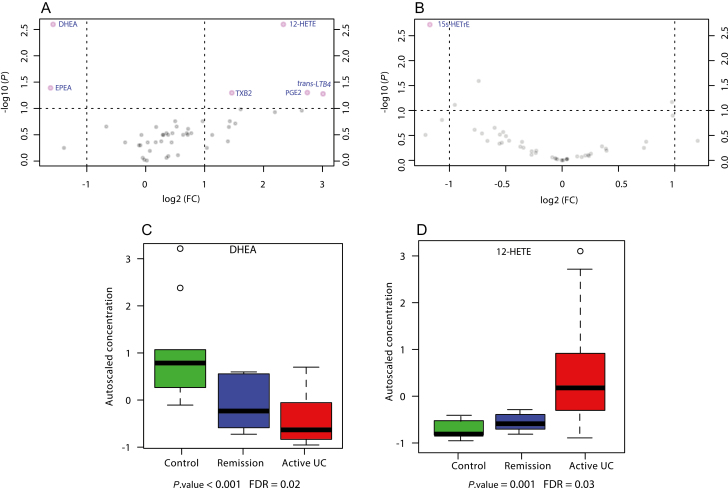
The concentrations of eCBs and oxylipins in colon biopsies from treatment-naïve UC and deep remission UC patients were compared with controls. As seen in Figure 1A, the volcano plot shows that the mucosal levels of PGE2, thromboxane (TXB2), trans- leukotriene (trans-LTB4), and 12-Hydroxy-eicosatetraenoic acid (12-HETE) were significantly upregulated (FDR ≤ 0.1) in colon biopsies taken from treatment-naïve patients. The mean concentrations of these oxylipins were increased by 7-, 3-, 8-, and 5-fold, respectively. In contrast, DHEA and EPEA were significantly downregulated. The mean concentration was decreased by 3-fold for both eCBs, with respect to the control group.
FIGURE 1.
Results from univariate analysis of oxylipin and eCB mean mucosal concentrations. A, B, Volcano plots of changes in mean mucosal concentrations of oxylipins and eCBs in treatment-naïve patients vs healthy controls (HCs), and UC deep remission patients vs HCs, respectively. The vertical lines correspond to 2.0-fold up- and downregulation, and the horizontal lines represent a P value of 0.05 (Mann-Whitney U test) at a cutoff FDR value of 0.1. The points in the plots represent metabolite mean concentrations. Metabolites in pink have passed the volcano plot filtering. C, D, Box plots of the autoscaled concentration of DHEA and 12-HETE, respectively. The mean concentrations of these metabolites were found to have significantly changed among the study groups according to Kruskal-Wallis analysis of variance.
The comparison between the mucosal concentration of the investigated metabolites in deep remission UC patients and healthy controls is demonstrated in Figure 1B. Only 15Ss-Hydroxy-eicosatrienoic acid (15s-HETrE) was significantly upregulated by 2-fold in deep remission UC patients compared with healthy controls.
Furthermore, the Kruskal-Wallis test was used to compare the metabolite mucosal profiles between all 3 groups, as shown in Figure 2A. The metabolites that showed significant variance between the study groups were 1 ω-3 eCB, specifically DHAE (Fig. 1D, C), and 1 ω-6 AA oxylipin, specifically HETE-12 (Fig. 1D). The mean concentration of DHAE decreased in a stepwise manner from UC-naïve treatment patients to UC remission patients and controls. In contrast, the concentration of HETE-12 was the highest in the treatment-naïve UC group.
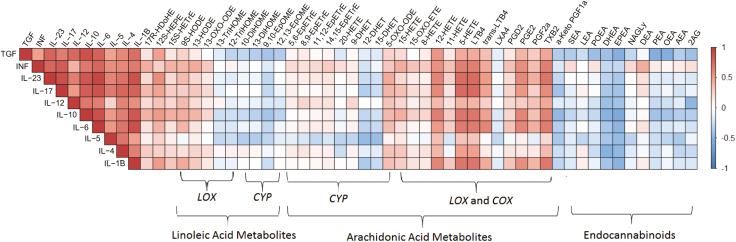
FIGURE 2.
Colored heatmap of the pair-wise Spearman’s rank correlation coefficients computed for cytokines vs cytokines, cytokines vs eCBs, and cytokines vs oxylipins. The colors refer to the correlation coefficient direction and magnitude, ranging from –1 (blue) to 1 (red). Each box in the heatmap is constructed from the metabolites-cytockines data of the 28 enrolled subjects. The metabolites are ordered according to the corresponding PUFA and the metabolic pathway. The correlation coefficients and the significance P values corresponding to all computed correlations are provided in the Supplementary Data.
Mucosal Cytokine Gene Expression in Treatment-Naïve UC Patients, UC Remission Patients, and Controls
Cytokine gene expression in colon biopsies was investigated by the quantification of mRNA using real-time PCR. Comparative analysis of mean differences in the cytokine gene expression levels between treatment-naïve UC patients, UC remission patients, and controls was done by Student t test. The results are shown in Table 2. All investigated cytokines had significantly higher mRNA levels in the inflamed mucosa of treatment-naïve UC patients compared with healthy controls. However, IL-5 did not differ significantly (P = 0.057). Furthermore, no significant differences were found in the gene expression of all investigated cytokines between the UC remission group and healthy controls (Table 2).
TABLE 2:
Comparison of Cytokine Gene Expressions Between the Study Groups
| Cytokine | Treatment-Naïve UC Patients, Cytokine Gene Expression Fold Changea | Deep Remission UC Patients, Cytokine Gene Expression Fold Changea | Treatment-Naïve UC Patients vs Controlsb | Deep Remission vs Controlsb |
|---|---|---|---|---|
| IL-1β | 7.88 | 1.33 | <0.001 | 0.88 |
| IL-6 | 15.98 | 1.35 | <0.001 | 0.88 |
| IL-12A | 3.35 | 0.79 | 0.0404 | 0.80 |
| IFN | 6.53 | 0.76 | 0.001 | 0.80 |
| IL-4 | 4.58 | 1.29 | 0.008 | 0.95 |
| IL-5 | 4.18 | 1.15 | 0.07 | 0.98 |
| IL-17A | 33.38 | 0.84 | <0.001 | 0.76 |
| IL-23A | 4.26 | 0.73 | 0.001 | 0.35 |
| IL-10 | 4.13 | 1.15 | <0.001 | 0.54 |
| TGF- β | 1.94 | 1.17 | 0.009 | 0.82 |
aMean fold change with respect to controls.
bAdjusted P value from the 2-tailed Student t test by Dunett post hoc.
Cytokine Gene Expression Correlation With Oxylipins and eCBs
To assess the association between cytokine gene expression and the investigated metabolites in the mucosal biopsies of the study groups, Spearman’s rank correlation between cytokines-cytokines and cytokines-metabolites was computed and is presented as an asymmetric heatmap (Fig. 2), where red represents a positive correlation and blue a negative correlation. All investigated cytokines were positively correlated with each other. Furthermore, the cytokines were found to be negatively correlated with several eCBs, mainly EPEA and DHEA (r ≈ –0.4). In contrast, the cytokines were positively correlated with nearly all AA metabolites, specifically PGE2, 12-HETE, 5-Hydroxy-eicosatetraenoic acid (5-HETE), TXB2, and LTB4 (r ≈ 0.5). However, there was a negative correlation with lipoxin (LXA4) and α-Keto prostaglandin F1 (α-keto PGF1), as shown in Figure 2. Furthermore, the correlation matrix revealed a negative correlation between cytokines and ω-6 AA–derived vicinal diols (DHETs) and a positive correlation with ω-6 AA–derived epoxides (EpETrEs).
Spearman’s rank correlation coefficients were computed for metabolites-metabolites, cytokines-cytokines, and cytokines-metabolites and are represented as a symmetric heatmap (Supplementary Fig. 1). In addition, the correlation coefficients and the significance P values corresponding to all computed correlations are provided in the Supplementary Data.
DISCUSSION
This study provides a unique, quantitative, and comprehensive analysis of a large number of oxylipins and eCBs in the colon mucosa of treatment-naïve newly diagnosed UC patients and deep remission UC patients. Previous studies were restricted to investigating oxylipins related to selected enzymatic pathways (COX-2 and 5-LOX) in UC.29–31 Moreover, in these studies, oxylipins were determined by liquid chromatography–tandem mass spectrometry (LC-MS/MS) untargeted analysis.32 For a more accurate quantification,33 we have quantified 35 oxylipin and 11 eCB metabolites using a fully validated targeted high-performance LC-MS/MS method. In addition, previously published studies were performed on a mix of treated and treatment-naïve UC patients,29–31 which might be a limitation. Therefore, in this study, only treatment-naïve patients were included in the active UC group (Table 1). Moreover, the deep remission patients were selected based on their endoscopy scores and TNF measurement results.
Our findings suggest that inflammation of the colonic mucosa in UC at debut is associated with a significant elevation in concentrations of ω-6 AA–related oxylipins, specifically, PGE2, TXB2, trans-LTB4, and 12-HETE, in addition to lower concentrations of ω-3 eCBs (DHEA and EPEA) (Fig. 1). The ω-6 AA–related oxylipins are potent immune response regulators. For example, PGE2, produced via COX within the AA cascade, has a pro-inflammatory effect via IL-6 production and dendritic cell activation and an anti-inflammatory effect via local Treg-cell accumulation and lipoxin induction.32, 34 Moreover, 12-HETE, produced via 12-LOX within the AA cascade, is a potent chemoattractant for neutrophils.35 In addition, LTB4 and its isomer trans-LTB4 stimulate the neutrophil chemotaxis in UC.36 Furthermore, TXB2 is the stable downstream metabolite of thromboxane A2 (TXA2), which is known for causing vasoconstriction, platelet aggregation, and T-cell activation.37 Studies on ω-6 and ω-3 PUFAs and their bioactive lipid metabolites in IBD patients have revealed an alteration in their mucosal levels.5, 32, 38 To our knowledge, our study is the first to report alterations in EPEA and DHAE levels in colonic mucosa in UC.
Interestingly, our data showed differences in the oxylipin profiles between deep remission patients and healthy controls. The ω-6-related oxylipin 15s-HETrE was significantly higher in the UC deep remission group in comparison with the control group, whereas the other investigated lipid metabolites did not differ significantly. Studies suggest that 15s-HETrE has an anti-inflammatory role via suppressing COX-2 overexpression39 and inhibiting platelet reactivity and thrombosis.40 The UC remission patients enrolled in this study, however, had completely resolved inflammation in the colonic mucosa. Higher levels of 15(s)-HETrE could indicate the importance of this anti-inflammatory oxylipin in maintaining the state of remission. However, due to the low number of patients, this finding was not conclusive, and a further confirmatory study is needed.
We also investigated cytokine gene expression to have an overview of the association between cytokine production at the transcriptomic level and the lipid mediators at the metabolomic level. This allows a deeper interpretation of the variation in the eCB and oxylipin profiles. As our study is purely descriptive, we were more interested in describing the direction and degree of correlation than the statistical significance.
The gene expression of all investigated cytokines was higher in debut patients compared with healthy controls. This finding is in agreement with previous studies.24, 41–43 Interestingly, cytokine gene expression was positively correlated with AA-related oxylipins, except for LXA4 and α-keto PGF1, where a negative correlation was found (Fig. 2). These 2 oxylipins play an important anti-inflammatory role. For instance, α-keto PGF1 is a stable metabolite of prostacyclin (PGI2),44 which inhibits platelet activation and reduces the intensity of the inflammatory response.45 LXA4 is a potent inflammation resolution oxylipin that promotes the clearance of apoptotic cells by macrophages and limits the infiltration of pro-inflammatory leukocytes.46 In fact, an LXA4 analog was found to inhibit TNF and IL-2 mucosal expression in induced colitis in mice.47 Accordingly, increasing the levels of LXA4 and PGI2 may represent promising targets for intervention. Our data also revealed imbalances in the CYP pathway (Fig. 2), namely between the anti-inflammatory EpETrEs and the pro-inflammatory DHETs. This has previously been studied in obesity-induced colonic inflammation48 but needs to be further explored in IBD.
In contrast to AA-related oxylipins, the correlation matrix revealed a negative correlation between the cytokine profiles and the eCB profile, in particular regarding EPEA and DHEA. Therefore, our findings suggest a potential role of ω-3-derived eCBs in the resolution of inflammation, and we propose novel therapeutic targets. Cannabinoid agonists and endocannabinoid degradation inhibitors in rodent models of IBD have identified a potential therapeutic role for eCBs.14, 49 Recently, a potential anti-inflammatory role for EPEA and DHEA was suggested.50 This is through the epoxide forms (EEQ-EA and EDP-EA), which inhibit the production of the pro-inflammatory cytokine IL-6 and promote the anti-inflammatory cytokine IL-10.50 However, studies on the effectiveness of ω-3 supplementation in the prevention and treatment of UC have been inconlusive and have failed to establish daily recommended intake.51, 52 In contrast, a trial study aiming to restore the lipid signaling balance in the intestinal tract by alkaline sphingomyelinase (Alk-SMase) rectal installation found significantly reduced inflammation and TNF expression.9
The small sample size in our study precludes subgroup analysis according to the severity of the disease in the UC treatment-naïve group. In addition, the healthy control group was considerably older than the 2 UC groups, which might affect our results. Furthermore, the small size of the UC remission group, which only consisted of males, is considered a weakness in this study. Due to the imbalanced distribution in the analyzed cohort, the effects of both sex and age were not included in our data analysis. Therefore, our findings are exploratory and need to be validated in a larger cohort, in which, preferably, only sex- and age-matched healthy controls are included. This approach might give normally distributed data, and thus allow for the use of parametric statistical tests, which have more statistical power.
CONCLUSIONS
We demonstrated for the first time that the onset of UC is associated with increased levels of ω-6-related oxylipins and decreased levels of ω-3-related eCBs. Furthermore, we have revealed an association between bioactive lipid mediators and pro- and anti- cytokine production. Our findings highlight the mucosal fingerprints of the metabolism of PUFAs, which may be involved in the progression of inflammation and may be considered as potential targets for intervention that need to be explored in more detail in a larger study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Renate Meyer for administrating the patient samples and Ingrid Christiansen for the technical help performing the TNF levels measurements.
Supported by: This project was funded by University of Tromsø–The Arctic University of Norway and Helse Nord RHF (SFP-1134-13).
REFERENCES
- 1. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–727. [DOI] [PubMed] [Google Scholar]
- 2. Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y). 2010;6:339–346. [PMC free article] [PubMed] [Google Scholar]
- 3. Das UN. Inflammatory bowel disease as a disorder of an imbalance between pro- and anti-inflammatory molecules and deficiency of resolution bioactive lipids. Lipids Health Dis. 2016;15:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sjöqvist U, Hertervig E, Nilsson A, et al. . Chronic colitis is associated with a reduction of mucosal alkaline sphingomyelinase activity. Inflamm Bowel Dis. 2002;8:258–263. [DOI] [PubMed] [Google Scholar]
- 5. Wolfer AM, Gaudin M, Taylor-Robinson SD, et al. . Development and validation of a high-throughput ultrahigh-performance liquid chromatography-mass spectrometry approach for screening of oxylipins and their precursors. Anal Chem. 2015;87:11721–11731. [DOI] [PubMed] [Google Scholar]
- 6. Wallace JL. Prostaglandin biology in inflammatory bowel disease. Gastroenterol Clin North Am. 2001;30:971–980. [DOI] [PubMed] [Google Scholar]
- 7. Martel-Pelletier J, Lajeunesse D, Reboul P, Pelletier JP. Therapeutic role of dual inhibitors of 5-LOX and COX, selective and non-selective non-steroidal anti-inflammatory drugs. Ann Rheum Dis. 2003;62:501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111:5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Acharya N, Penukonda S, Shcheglova T, et al. . Endocannabinoid system acts as a regulator of immune homeostasis in the gut. Proc Natl Acad Sci U S A. 2017;114:5005–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leinwand KL, Gerich ME, Hoffenberg EJ, Collins CB. Manipulation of the endocannabinoid system in colitis: a comprehensive review. Inflamm Bowel Dis. 2017;23:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mackie K. Cannabinoid receptors: where they are and what they do. J Neuroendocrinol. 2008;20(Suppl 1):10–14. [DOI] [PubMed] [Google Scholar]
- 12. Esposito G, Filippis DD, Cirillo C, et al. . Cannabidiol in inflammatory bowel diseases: a brief overview. Phytother Res. 2013;27:633–636. [DOI] [PubMed] [Google Scholar]
- 13. Marquéz L, Suárez J, Iglesias M, et al. . Ulcerative colitis induces changes on the expression of the endocannabinoid system in the human colonic tissue. PLoS One. 2009;4:e6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alhouayek M, Muccioli GG. The endocannabinoid system in inflammatory bowel diseases: from pathophysiology to therapeutic opportunity. Trends Mol Med. 2012;18:615–625. [DOI] [PubMed] [Google Scholar]
- 15. Di Sabatino A, Battista N, Biancheri P, et al. . The endogenous cannabinoid system in the gut of patients with inflammatory bowel disease. Mucosal Immunol. 2011;4:574–583. [DOI] [PubMed] [Google Scholar]
- 16. D’Argenio G, Valenti M, Scaglione G, et al. . Up-regulation of anandamide levels as an endogenous mechanism and a pharmacological strategy to limit colon inflammation. Faseb J. 2006;20:568–570. [DOI] [PubMed] [Google Scholar]
- 17. Darmani NA, Izzo AA, Degenhardt B, et al. . Involvement of the cannabimimetic compound, N-palmitoyl-ethanolamine, in inflammatory and neuropathic conditions: review of the available pre-clinical data, and first human studies. Neuropharmacology. 2005;48:1154–1163. [DOI] [PubMed] [Google Scholar]
- 18. Stange EF, Travis SP, Vermeire S, et al. ; European Crohn’s and Colitis Organisation (ECCO) European evidence-based consensus on the diagnosis and management of ulcerative colitis: definitions and diagnosis. J Crohns Colitis. 2008;2:1–23. [DOI] [PubMed] [Google Scholar]
- 19. Sutherland LR, Martin F, Greer S, et al. . 5-aminosalicylic acid enema in the treatment of distal ulcerative colitis, proctosigmoiditis, and proctitis. Gastroenterology. 1987;92:1894–1898. [DOI] [PubMed] [Google Scholar]
- 20. Olsen T, Goll R, Cui G, et al. . Tissue levels of tumor necrosis factor-alpha correlates with grade of inflammation in untreated ulcerative colitis. Scand J Gastroenterol. 2007;42:1312–1320. [DOI] [PubMed] [Google Scholar]
- 21. Johnsen KM, Goll R, Hansen V, et al. . Repeated intensified infliximab induction - results from an 11-year prospective study of ulcerative colitis using a novel treatment algorithm. Eur J Gastroenterol Hepatol. 2017;29:98–104. [DOI] [PubMed] [Google Scholar]
- 22. Wu J, Gouveia-Figueira S, Domellöf M, et al. . Oxylipins, endocannabinoids, and related compounds in human milk: levels and effects of storage conditions. Prostaglandins Other Lipid Mediat. 2016;122:28–36. [DOI] [PubMed] [Google Scholar]
- 23. Gouveia-Figueira S, Nording ML. Validation of a tandem mass spectrometry method using combined extraction of 37 oxylipins and 14 endocannabinoid-related compounds including prostamides from biological matrices. Prostaglandins Other Lipid Mediat. 2015;121:110–121. [DOI] [PubMed] [Google Scholar]
- 24. Rismo R, Olsen T, Cui G, et al. . Mucosal cytokine gene expression profiles as biomarkers of response to infliximab in ulcerative colitis. Scand J Gastroenterol. 2012;47:538–547. [DOI] [PubMed] [Google Scholar]
- 25. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 26. Xia J, Wishart DS. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. [DOI] [PubMed] [Google Scholar]
- 27. van den Berg RA, Hoefsloot HC, Westerhuis JA, et al. . Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vinaixa M, Samino S, Saez I, et al. . A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2:775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boughton-Smith NK, Hawkey CJ, Whittle BJ. Biosynthesis of lipoxygenase and cyclo-oxygenase products from [14C]-arachidonic acid by human colonic mucosa. Gut. 1983;24:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jupp J, Hillier K, Elliott DH, et al. . Colonic expression of leukotriene-pathway enzymes in inflammatory bowel diseases. Inflamm Bowel Dis. 2007;13:537–546. [DOI] [PubMed] [Google Scholar]
- 31. Carty E, De Brabander M, Feakins RM, Rampton DS. Measurement of in vivo rectal mucosal cytokine and eicosanoid production in ulcerative colitis using filter paper. Gut. 2000;46:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masoodi M, Pearl DS, Eiden M, et al. . Altered colonic mucosal polyunsaturated fatty acid (PUFA) derived lipid mediators in ulcerative colitis: new insight into relationship with disease activity and pathophysiology. PLoS One. 2013;8:e76532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem. 2016;88:524–545. [DOI] [PubMed] [Google Scholar]
- 34. Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szczuko M, Zapałowska-Chwyć M, Maciejewska D, et al. . Significant improvement selected mediators of inflammation in phenotypes of women with PCOS after reduction and low GI diet. Mediators Inflamm. 2017;2017:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wéra O, Lancellotti P, Oury C. The dual role of neutrophils in inflammatory bowel diseases. J Clin Med. 2016;5:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lone AM, Taskén K. Proinflammatory and immunoregulatory roles of eicosanoids in T cells. Front Immunol. 2013;4:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee Y, Choo J, Kim SJ, et al. . Analysis of endogenous lipids during intestinal wound healing. PLoS One. 2017;12:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pham H, Banerjee T, Ziboh VA. Suppression of cyclooxygenase-2 overexpression by 15S-hydroxyeicosatrienoic acid in androgen-dependent prostatic adenocarcinoma cells. Int J Cancer. 2004;111:192–197. [DOI] [PubMed] [Google Scholar]
- 40. Yeung J, Tourdot BE, Adili R, et al. . 12(S)-hetre, a 12-lipoxygenase oxylipin of dihomo-γ-linolenic acid, inhibits thrombosis via gαs signaling in platelets. Arterioscler Thromb Vasc Biol. 2016;36:2068–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nemeth ZH, Bogdanovski DA, Barratt-Stopper P, et al. . Crohn’s disease and ulcerative colitis show unique cytokine profiles. Cureus. 2017;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsuda R, Koide T, Tokoro C, et al. . Quantitive cytokine mrna expression profiles in the colonic mucosa of patients with steroid naïve ulcerative colitis during active and quiescent disease. Inflamm Bowel Dis. 2009;15:328–334. [DOI] [PubMed] [Google Scholar]
- 43. Korolkova OY, Myers JN, Pellom ST, et al. . Characterization of serum cytokine profile in predominantly colonic inflammatory bowel disease to delineate ulcerative and Crohn’s colitides. Clin Med Insights Gastroenterol. 2015;8:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olofsson J, Norjavaara E, Selstam G. Synthesis of prostaglandin F2 alpha, E2 and prostacyclin in isolated corpora lutea of adult pseudopregnant rats throughout the luteal life-span. Prostaglandins Leukot Essent Fatty Acids. 1992;46:151–161. [DOI] [PubMed] [Google Scholar]
- 45. Yoshida H, Granger DN. Inflammatory bowel disease: a paradigm for the link between coagulation and inflammation. Inflamm Bowel Dis. 2009;15:1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Biasi F, Leonarduzzi G, Oteiza PI, Poli G. Inflammatory bowel disease: mechanisms, redox considerations, and therapeutic targets. Antioxid Redox Signal. 2013;19:1711–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fiorucci S, Wallace JL, Mencarelli A, et al. . A beta-oxidation-resistant lipoxin A4 analog treats hapten-induced colitis by attenuating inflammation and immune dysfunction. Proc Natl Acad Sci U S A. 2004;101:15736–15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang W, Yang J, Zhang J, et al. . Lipidomic profiling reveals soluble epoxide hydrolase as a therapeutic target of obesity-induced colonic inflammation. Proc Natl Acad Sci U S A. 2018;115:5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee Y, Jo J, Chung HY, et al. . Endocannabinoids in the gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2016;311:G655–G666. [DOI] [PubMed] [Google Scholar]
- 50. McDougle DR, Watson JE, Abdeen AA, et al. . Anti-inflammatory ω-3 endocannabinoid epoxides. Proc Natl Acad Sci U S A. 2017;114:E6034–E6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scaioli E, Liverani E, Belluzzi A. The imbalance between n-6/n-3 polyunsaturated fatty acids and inflammatory bowel disease: a comprehensive review and future therapeutic perspectives. Int J Mol Sci. 2017;18:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ungaro F, Rubbino F, Danese S, D’Alessio S. Actors and factors in the resolution of intestinal inflammation: lipid mediators as a new approach to therapy in inflammatory bowel diseases. Front Immunol. 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.