Abstract
Small Ruminant Lentiviruses (SRLV) include at least 4 viral highly divergent genotypes. Genotypes A and B are widely distributed and genotypes C and E have been recognized in restricted geographic areas. New phylogroups have been identified targeting conserved regions. However, this approach suffers from the potential risk to misamplify highly divergent strains. Pathogenic strains are easily adapted to fibroblastic cells, but non-pathogenic strains isolation may require a different approach. We developed a fast and effective method for SRLV full genome characterization after cell culture isolation. Spleen samples were collected during regular slaughter from sheep and goats in northwestern Italy. Spleen-derived macrophage cultures were monitored for reverse transcriptase activity and RNA was extracted from the supernatant of positive cultures. Using Illumina MiSeq platform 22 new full genome sequences were obtained. The success of this approach is based on the following features: spleen is one of the main target for SRLV persistence; red pulp is a reserve of resident macrophages, the main target for SRLV replication in vivo; RTA is a sensitive assay for any replicating retrovirus; de novo sequencing do not require genetic knowledge in advance.
Introduction
Small Ruminant Lentiviruses (SRLV) include, to date, 4 highly divergent viral genotypes. The genetic differences among viral strains are related to antigenic and biological properties both in vitro and in vivo. Historically, Visna Maedi virus (MVV) and Caprine Arthritis Encephalitis virus (CAEV) were first isolated from sheep and goat respectively, and have been considered for long time to be strictly associated to specific clinical features and host. While those viruses are still considered prototypes of the widely distributed genotypes A and B, a number of sub-genotypes, within group A and B, and new genotypes (C and E) have been recognized. Consequently, SRLV are now considered host adapted but not strictly host associated. Moreover, the differences defined in the past [1] are becoming less clear due to the increasing number of available sequences.
Pathogenic strains are frequently isolated from specific or suggestive gross lesions by tissue explantation (i.e. lung, udder, synovial membrane) or co-cultured with permissive cells, typically fibroblast-like cells. Strains that achieve adaptation to fibroblastic cells, overgrow and show the typical cytopathic effect characterized by syncitia formation. In the early nineties, through the viral isolation method, a number of sheep and goat pathogenic strains were isolated and later characterized as B2 and B1 subtype respectively [2,3]. Unfortunately this approach may potentially fail in the detection of new genotypes (i.e. unsuccessful isolation or unclear cytopathic effect) due to the limited capacity of some low pathogenic strains to adapt to fibroblasts [4]. In order to overcome this problem, molecular approaches based on new PCR protocols were developed. These tools strongly support the SRLVs characterization, increasing the knowledge about their genetic heterogeneity [4]. This was the case of the genotype E: biological characterization in vitro and in vivo of the subtype E1, known as Roccaverano strain, opened new insights into putative non-pathogenic strains, being able to grow productively only in macrophage culture and including their role in mitigating the pathogenic potential of more virulent strains [5,6].
With the advent of the Next Generation Sequencing (NGS) technology, new opportunities became available to fully characterize SRLV isolates, even in the absence of previous knowledge of genetic and biological properties.
Keeping in mind that low pathogenic SRLVs may have a restricted cell tropism and may be difficult to isolate from standard tissue explantation, we developed a fast and effective method for SRLV full genome characterization after cell culture isolation. Spleen explant cultures were performed from goats and sheep sampled during slaughtering and NGS protocols from reverse transcriptase activity positive cultures were applied. By this approach, 22 full genomes were assembled representing two major genotypes. In addition, beside the pathogenic B1 and B2 subtypes, a large number of isolates belonging to the subtype A8 was found.
Material and methods
Sample collection and virus isolation
Blood and spleen paired samples were collected from adult ovine and caprine local breeds. Two slaughterhouses in the Piedmont Region, Northwestern Italy, were chosen in order to cover different geographical areas and to increase the variability of samples. Animals were collected randomly during the standard slaughtering activities. (see details in Table 1)
Table 1. Samples characterized in the present study.
| Isolate (Accession num) |
Host | Elisa Screening |
Elisa Genotying |
CPE | RT activity (passage) |
Gag subtype |
|---|---|---|---|---|---|---|
| To1_89 (MH374290) | goat | na | na | yes | 1 | B1 |
| Taccone (MH374289) | goat | na | na | yes | 1 | B1 |
| VdA (MH374291) | goat | Positive | A | no | 1 | A8 |
| It001.2017 (MG554402) | sheep | Positive | Indet | yes | 4 | B2 |
| It002.2017 (MG554403) | goat | Positive | Indet | no | 1 | A8 |
| It003.2017 (MG554404) | goat | Positive | A | no | 1 | A8 |
| It004.2017 (MG554405) | goat | Positive | A | no | 1 | A8 |
| It005.2017 (MG554406) | goat | Positive | Indet | no | 1 | A8 |
| It006.2017 (MG554407) | goat | Positive | E | yes | 1 | A8 |
| It007.2017 (MG554408) | goat | Positive | Indet | no | 1 | A8 |
| It009.2017 (MG554409) | goat | Positive | A | yes | 1 | A18 (A3-A4) |
| It010.2017 (MG554410) | goat | Positive | B | yes | 1 | B1 |
| It014.2017 (MG554411) | goat | Positive | Indet | yes | 1 | B1 |
| It016.2017 (MG554412) | goat | Positive | B | yes | 1 | B1 |
| It017.2017 (MG554413) | goat | Positive | Indet | yes | 1 | B1 |
| It020.2017 (MG554414) | goat | Positive | B | yes | 1 | B1 |
| It024.2017 (MH374283) | goat | Positive | A | yes | 3 | A8 |
| It025.2017 (MH374284) | goat | Positive | A | no | 1 | A8 |
| It026.2017 (MH374285) | goat | Positive | A | no | 1 | A8 |
| It032.2017 (MH374286) | goat | Negative | Negative | no | 3 | A8 |
| It038.2017 (MH374287) | sheep | Positive | A | yes | 3 | A19 (A9-11) |
| It042.2017 (MH374288) | sheep | Positive | Indet | yes | 3 | B2 |
Blood serum was used for antibody screening and genotyping using a commercially available ELISA kit (Eradikit—SRLV Starter kit, In3diagnostic, Italy); spleen was promptly delivered to the laboratory for tissue explantation. Briefly, the splenic capsule was disinfected with 70% ethanol and a 5 ml syringe with G21 needle was inserted into the splenic pulp. A negative pressure was applied by the syringe plunger while the needle was guided in different directions into the splenic tissue. When enough material was extracted into the nozzle, the syringe was removed and pulp material resuspended into 5ml of DMEM supplemented with L-glutamine 1mM and 2X antibiotic/antimycotic solution (Sigma Aldrich). During the second sampling period the described preliminary procedure was accomplished directly at the slaughterhouse. After 4h incubation at 37°C, medium was removed and tissue fragments were seeded on 25cm2 flasks in complete macrophage medium consisting in RPMI medium supplemented with L-glutamine 2mM, 1% non-essential amino acids, vitamins, sodium pyruvate 1mM, 2-mercaptoethanol 17μM, gentamicin 50μg/ml and FBS 10%. Cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2 and medium was partially replaced every 3–7 days.
Once a week, medium was collected and reverse transcriptase activity was determined using Lenti RT activity kit (Cavidi, Uppsala, Sweden). RT activity was tested and recorded for each culture passage, in order to evaluate its trend. In the case of RT activity positive outcome, all medium was collected and further processed for whole genome sequencing. The presence of cytopathic effect in fibroblastic cells was recorded at each culture passage. In the presence of fibroblastic overgrowth, trypsin sensitive cells were periodically removed from the original flask.
MiSeq run
RT positive cell culture supernatants were centrifuged for 20 minutes at 600 g to eliminate cell debris and were concentrated with Amicon-15 100 kDa centrifugal filter tubes (Millipore Merck KGaA, Darmstadt, Germany).
Viral RNA was extracted with QiAmp Viral RNA Kit (Qiagen, Hilden, Germany) and quantified using Nanodrop system (Thermo Fisher Scientific). Viral RNA was reverse transcribed into double stranded cDNA with Maxima H Minus Double–stranded cDNA Synthesis kit (Thermo Fisher Scientific) in accordance with manufacturer instructions and quantified with a fluorimetric method, Qubit dsDNA kit (Life Technologies). Samples were used for DNA library preparation using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocol. The quantity of DNA was assessed using Agilent DNA High Sensitivity chip assay (Agilent Technologies) and the Qubit dsDNA kit (Life Technologies). Paired-end libraries were sequenced using Illumina V2 chemistry and Illumina MiSeq platform.
Data analysis
Reads obtained by the MiSeq runs were checked for quality (FastQC) and trimmed (Trimmomatic ver. 0.32). Two parallels pipelines were followed. Reads were aligned to all known reference genomes in order to identify and confirm the viral genotype (Geneious ver. 11.1.2); the reads were further aligned to the consensus sequence obtained after the first step, in order to confirm the genome sequence. In parallel, the reads were used for de novo assembling (Velvet software ver. 1.2.10); the obtained contigs were compared to the consensus sequence derived from resequencing. Annotation of the main genes was performed manually, by comparing amino acidic sequences among new and reference genomes, as well as the LTR regions. Genotype was determined basing on the Gag gene sequence alignment as previously reported [1,6]. Gag gene and the complete genome sequence were used to depict phylogenetic relationships between the newly characterized and the reference strains using a Bayesian approach implemented in MrBayes package [7]. Basing on the phylogenetic tree topologies, the association between genetic sequence features and RT activity and CPE was calculated using the algorithms implemented in Bats software ver. 0.9 based on Bayesian Markov-Chain Monte Carlo approach to the investigation of phylogeny–trait correlations [8].
Results
A total of 42 paired samples (spleen and blood serum) were collected from 16 sheep and 26 goats. Thirty-three were antibody positive and 26 were serotyped according to the reactivity against an immunodominant linear epitope of the capsid antigen, able to discriminate among genotype A, B, and E [2,9,10].
Twenty-six RT activity positive cultures from spleen explants were obtained (25 from seropositive and 1 from a seronegative animal) and further processed for whole genome sequencing. Nineteen isolates were readily available after the first collection time (about 10 days post-culture) or after the first passage (after 17 days of culture), while additional 7 isolates showed RT activity after 3–4 weeks of culture. Interestingly, only half (13 out of 26) of the strains showed cytopathic effect (CPE) on overgrowing fibroblastic-like cells, characterized by typical cell fusion.
Considering a coverage cut-off of 100x, full genome sequence were obtained from 22 isolates out of 26, using the proposed method. Four samples which did not meet the coverage criteria were not taken into account. Sequence analysis revealed the presence of genotypes A and B. The heterogeneity within each genotype was quite high, confirming the circulation of subtypes A8, B1 and B2, based on the similarity of gag genes. Moreover, one sample from sheep (It038.2017) showed a large difference with known sequences (at least around the 24%) belonging to the same monophyletic clade together with A9 and A11 subtypes. In the same way, the isolate It009.2017 was genetically related to subtypes A1 and A4 but differences were within the 25%-15%. Following the criteria published before [1], these samples suggest the presence of new subtypes (A18 and A19) and confirm the very high heterogeneity of VMV-like viral strains.
As reported before, genotypes A and subtype B1 were identified in samples from goats and sheep, whereas B2 was only found in ovine samples. Sequences obtained from animals belonging to the same flock clustered together, suggesting a clonal origin of the viral strain.
No differences in terms of SRLV positivity or genotyping between the two slaughterhouses were recorded. A positive association was observed between the phylogroup A8 and the absence of CPE in culture: the observed Monophyletic clade value mean was 5.015 (p < 0.10) and indicated a significant correlation between in vitro features and phylogenetic relationship among A8 new strains. The relevant data are summarized in Table 1. All but one serum sample were correctly classified using the serotyping ELISA according to the paired strain sequence analysis, while a single sample (It006.2017) gave spurious results (E in serotyping vs A8 subtype in sequencing). Sequence analysis of the gag gene encompassing the immunodominant epitope revealed a single non-synonymous mutation P231Q, a specific signature of genotype E [4] (Table 2). On the other hand HV1 and HV2 regions along env gene sequence did not show motifs that can be clearly associated to a single subtype or to low pathogenic features (Table 3).
Table 2. Alignment of the immunodominant epitope within the gag gene.
Reference genotype are reported in bold. Dots indicate identical residue comparing to the reference K1514 (MVVlike A genotype).
| Isolate | Subtype | Capsid epitope |
|---|---|---|
| K1514 | A1 | QKELIQGKLNEEAERWVRQNPPGP--NVLTVDQ |
| It025.2917 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--. . . . . . . |
| It007.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--. . . . . . . |
| It005.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--. . . . . . . |
| ItVdA.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--. . . . . . . |
| It038.2017 | A18 | . . . . . . . . . . . . . . . . . . . . . . . .--. . . . . . . |
| It009.2017 | A19 | . . . . . . . . . . . . . . . .I. . . . . . .--. . . . . . . |
| It024.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--Q. . . . . . |
| It026.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . . .--Q. . . . . . |
| It004.2017 | A8 | . . . . . . . . . . . . . . . . . . . . . . .Q--. . . . . . . |
| It002.2017 | A8 | . . . . . . . . . . . . . . . .I. . . . . . .--. . . . . . . |
| It003.2017 | A8 | . . . . . . . . . . . . . . . .M. . . . . .Q--. . . . . . . |
| It006.2017 | A8 | . . . . . . . . . . . . . . . .M. . . .Q----.A. . . . . |
| Cork | B1 | . . . . . . . . . . . . . . . .R.N . . .P.AGGG. . . . . |
| It016.2017 | B1 | . . . . . . . . . . . . . . . .R.N . . .P.AGGG. . . . . |
| It010.2017 | B1 | . . . . . . . . . . . . . . . .R.N . . .P.QGGG. . . . . |
| It020.2017 | B1 | . . . . . . . . . . . . . . . .R.N . . .PQAGGG. . . . . |
| It017.2017 | B1 | . . . . . . . . . . . . . . . .R.N . . .PQGGGG. . . .L |
| It014.2017 | B1 | . . . . . . . . . . . . . .Q.R.N . . .PQAGGA. . . . . |
| It001.2017 | B2 | . . . . . . . . . . . . . . . .R.N . . .PQAGGG. . . . . |
| It042.2017 | B2 | . . . . . . . . . . . . . . . .R.N . . .PQAGGG. . . . . |
| EU010124 Roccaverano | E1 | V . . .V.D . . .K . . .T.M. . . .QP.--GG. . . . . |
Table 3. Variable regions within env gene among SRLV A genotype strains.
HV1 and HV2: hypervariable (HV) region; TM: transmembrane domain within env.
| ENVELOPE REGIONS | HV1 | HV2 | V5 | TM |
|---|---|---|---|---|
| VLVLV1A K1514 | VGNGTITGNCSVTNWDG | NKWTCAARRK--GSRRDSLYIAG-RD | QSYMEAQGENRRS | ELDCWHYQHYCVTS |
| VLVLV1B K1514 | . . . . . . . . . . . . . . . . . | . . . . . . . .TGRK. .Q. . . . . . . .-. . | . . . . . . . .K. . . . | . . . . . . . . . . . . . . |
| NC 001452 kv1772 | . . . . . . . . . . . . . . . . . | . . . . .------K.Q . . .-. . . .-. . | . . . . . . . . . .K. . | . . . . . . . . . . . . . . |
| VLVGAGA_kv1772 | . . . . . . . . . . . . . . . . . | . . . . .------K.Q . . .-. . . .-. . | . . . . . . . . . .K. . | . . . . . . . . . . . . . . |
| VLVCG_Visna/Maedi | . . . . . . . . . . . . . . . . . | . . . . .------K.Q . . .-. . . .-. . | . . . . .ER. . . . . . | . . . . . . . . . . . . . . |
| VLVCGA_LV1.1 | . . . . . . . . . . . . . . . . . | . . . . .------K.Q . . .-. . . .-. . | . . . . .ER. . . . . . | . . . . . . . . . . . . . . |
| It038.2017 | VGNGTITGNCSVTNWDG | . . . . . .P.WGKG. .--. . . . . . .G.Q | DQ.LKTNKRRK. . | . . . . . . .H.F. . . . |
| AF479638_P1OLV | . . . . .L. . . . . . .D . . . | RQ . . .S. .VG--.TT. . . . . . . .-.N | KA.S.KKKRQPQ- | . . . . . . . . . . . . . . |
| OLVCG_SAOMVV | . . . . . . . . . . . . .D.E. | . . . . . . . .NS--KKK. . . . . . . .-. . | KA.R.KNMR.K. . | . . . . . . . . . . . . . . |
| NC_001511_Ovinelentivirus | . . . . . . . . . . . . .D.E. | . . . . . . . .NS--KKK. . . . . . . .-. . | KA.R.KNMR.K. . | . . . . . . . . . . . . . . |
| It009.2017 | . . . . . . . . . . . . .D . . . | K. . . . . . .AN--DK. . . . . . . . .-.N | N. . . .KNRKKQKR | Q. . . . . . . . . . . . . |
| KT453988_g6221 | I. . . . . . . . . . . . . . . . | .Q. . . . . .TA--.KK. . . . . . . .-. . | . . . .QN.EK.K.A | . . . . . .H. .F. . . . |
| KT453990_s7631 | I. . . . . . . . . . . . . . . . | .Q. . . . . .TA--.E. . . . . . . . .-. . | . . . .QN.EK.K.A | . . . . . .H. .F. . . . |
| KT453989_s7385 | I. . . . . . . . . . . . . . . . | .Q. . . . . .TA--.KK. . . . . . . .-. . | . . . .QN.EK.K.A | . . . . . .H. .F. . . . |
| HQ848062_Ov697 | . . . . .V. . . . . . . . . . . | .T . . .S. .K.--.-. . . . . . . . .-. . | . . .I.S.EK.K. . | . . . . . .H. .F. . . . |
| KY358788_USMARC-199906011-2 | . . . . . . . . . . .A. . . . . | .Q . . .K . . .S--.NKT. . . . . . .-GE | . . . . .T.RRKK. . | . . . . . .H. .F. . . . |
| KY358787_USMARC-200303013-1 | . . . . . . . . . . . .K. . . . | .L. . . .P. .R--.NVT. . . . . . .-GK | K. .I.T.RRKK.A | . . . . . .H. .F. . . . |
| ItVdA.2017 | . . .D. . . . . . . . .D . . . | .E. . . . . .QR--NDK. . . . . . . .-.N | T. .VDQKRGKKKR | . . . . . . . . .F. . . . |
| It024.2017 | . . . . . . . . . . . . . . . . . | . . . . . . . .KEKGKGQQ. . . . . . .-. . | T. .V.QS.KSKNR | . . . . . . . . .F. . . . |
| It026.2017 | . . . . . . . . . . . . . . . . . | . . . . . . . .K.--KGQQ. . . . . . .-. . | T. .V.QS.K.KNR | . . . . . . . . .F. . . . |
| It007.2017 | . . . . . . . . . . . . .D . . . | .M. . . . . .QS--.E. . . . . . . . .-.N | TN.V.L.KRRQKR | . . . . . . . . .F. . . . |
| It006.2017 | . . . . . . . . . . . . . . . . . | G. . . . . . .Q.--RDKQ. . . . . . .-.N | T. .V.QN.KKKKR | . . . . . . . . .F. . . . |
| It005.2017 | . . .N. . . . . . . . .D . . . | . . . . . . . .KQ--EEQW. . . . . . .-.N | T. .I.Q.KGKKKR | . . . . . . . . .F. . . . |
| It002.2017 | K. . . . . . .A.N. . . . . . | .R. . . . . .Q.--DG. . . . . . . . .-.N | T. . . .QTRGKKKR | . . . . . . . . .F. . . . |
| It025.2017 | . . .N. . . . . . . . . . . . . | D. . . . . . .QN--NEAQ. . . . . . .-.N | TA.V.Q.-KRKKR | . . . . . . . . .F. . . . |
| It003.2017 | A. .N. . . . . . . . .D . . . | S. . . . .E.Q.--ENKT. .V. . . .-.E | T. .V.Q.TRKKKR | . . . . . . . . .F. . . . |
| It004.2017 | . . .D. . . . . . . . .D . . . | RH. . . .E.QR--.NKT. .V. . . .-.E | A. .V.Q.TRKK.R | . . . . . . . . .F. . . . |
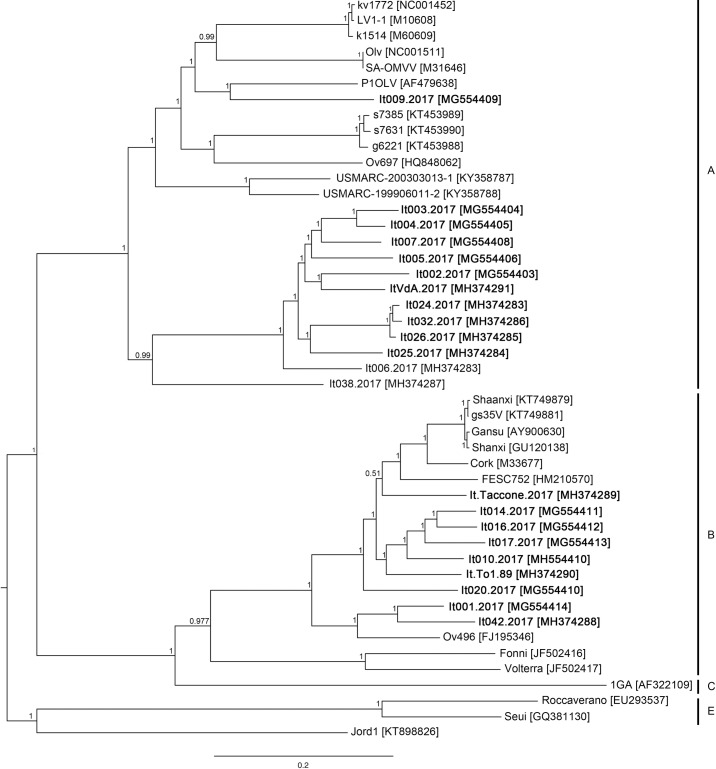
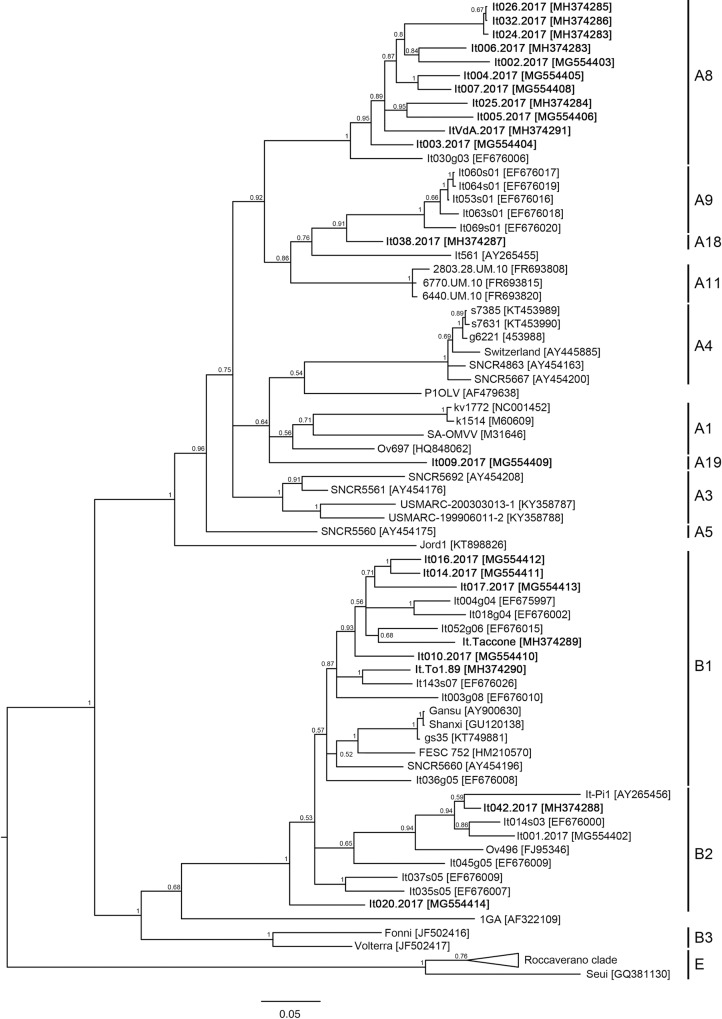
Phylogenetic trees clearly confirmed the genotype and the subtypes clustering of the new isolates considering both the complete genome (Fig 1) and partial gag gene sequence (Fig 2) alignments.
Fig 1. Bayesian tree based on the full genome sequences alignment.
Newly characterized isolates are reported in bold. SRLV genotypes are reported. Posterior probability of each node is showed above branches.
Fig 2. Bayesian tree based on the partial gag sequence alignment.
Newly characterized isolates are reported in bold. SRLV subtypes are reported. Posterior probability of each node is showed above branches.
Full genome sequences of newly assembled isolates are available on GenBank (MG554402-MG554414 and MH374283-MH374291).
Discussion
In this study, a new approach was developed addressing the full genetic characterization of SRLV isolates by NGS technologies. Since 1985, when the full genome sequence of the first isolate was published, a number of full genomes has been deposited, representing the prototype strains associated to specific diseases of relevant genotypes. These first genetic data represented a useful hallmark for SRLV diagnosis and control, opening new perspectives for recombinant antigen technologies and development of sensitive serological tests, covering the heterogeneity of lentiviruses in both sheep and goats. Moreover, genetic alignment has been progressively improved (in terms of quality and quantity) allowing the design of primer sets facilitating the amplification of at least different conserved genome regions. To date 17 subtypes of genotype A [11], 4 subtypes of genotype B [12], and two subtypes of genotypes C [13] and E [4,14] have been identified using standard end point partial amplification of the LTR and gag, pol and env genes followed by Sanger sequencing. This has led to a significant improvement of the knowledge on viral epidemiology, pathogenesis, diagnosis and control.
The method developed in this study allowed the characterization of 22 SRLV full genomes from fresh isolates, providing a fast and economically feasible tool for SRLV investigation. The availability of full genome, along with a viral isolate, is often necessary to evaluate antigenic and biological properties in detail. Moreover, the sequence coverage obtained with the described procedure supports the evidence of high heterogeneity among the isolates and within each genotype, as reported in previous studies [1,6].
According to de novo sequencing strategy potentially novel highly divergent genomes can be virtually detected, even in the presence of mixed infections, providing the genetic bases for the development of specific diagnostic tools. In this context, results presented in the study are quite intriguing and can be extrapolated to other known infected regions or to unexplored populations even if they are representative of a limited geographic area. This approach also allowed the genetic characterization of hypervariable regions such as HV3-5 within the env gene that may be difficult to amplify by conventional PCR due to strain specificity [15,16].
Keeping in mind the biological constraint of SRLV in the asymptomatic stage, the success of virus isolation highly depends on the viral load present in explant cultures from the main sites of viral persistence. According to previous experiences, spleen tissue gave the higher rate of success, followed by mammary gland and peripheral blood mononuclear cells (PBMC) co-culture [17]. Spleen ex vivo biopsy by needle aspiration produced enough red pulp fragments with the desired size that rapidly established a culture of terminally differentiated macrophages, possibly derived by red pulp resident cells. This explains the high proportion of viral strains obtained in the first passage. Contamination of fibroblastic-like culture is a normal feature in many tissue explants and spleen is not an exception. Terminally differentiated macrophages and dendritic cells are often replaced by overgrowing fibroblastic cells during cell culture propagation. In many instances, this results in adaptation of SRLV isolates able to infect fibroblasts and producing the characteristic cell fusion. However, this phenomenon is not always observed. The prototype strain Roccaverano was firstly isolated from spleen and mammary gland explants. Adaptation to standard foetal cells (synovial membrane, lung or choroid plexus) was unsuccessful [18] presumably due to restricted macrophage tropism [5]. About half of the strains isolated in the present study belonging to the genotype A did not show cell membrane fusion and a subset of them demonstrated impaired adaptation to overgrowing fibroblasts, as RT activity trended to decrease over time, as soon as infected macrophages died and were replaced by fibroblasts. This behavior is in agreement with a previous study on low pathogenic SRLV strains [19]. Since viral microevolution seems to be essential to drive tissue compartmentalization [16], we cannot exclude that viral isolation in asymptomatic animals may be restricted to canonical cell types compared with isolation from lentivirus specific gross lesions (i.e. lung, udder, synovial membrane).
However, in the present study all strains belonging to subtype B1 in goats and B2 in sheep were fusogenic in vitro, suggesting enhanced ability of the latter subtypes to efficiently replicate in spleen-derived fibroblasts. The first virological survey in the same area was carried out in the early nineties and later genetically characterized [2]. All strains were isolated from gross lesions and belonged to genotype B and now referred as B1 in goats and B2 in sheep [1]. More than 20 years later, the majority of goat isolates from asymptomatic animals belong to subtype A8; this finding strongly suggests that subtype A8 may represent a low pathogenic subtype that may have passed inadvertently causing persistent infections. Flock owners were not aware of any of the clinical signs attributable to SRLV before the French breeds entered the population in the early 1980s, further supporting this hypothesis. It should be noted that in the sampled area no control measures have been ever implemented and the great number of A8 isolates is unlikely to have emerged as a consequence of diagnostic escape, as happened for subtype A4 in Swiss goats [19,20]. In the latter experience, the poor performance of serological test in detecting SRLV A4 subtypes, most likely favored the spread of genotype A4 in goats, although the same test was quite effective in genotype B eradication campaigns (subtype B1).
Interestingly, the A4 subtype in Swiss goat is probably endemic in some Swiss regions but no SRLV-induced pathology has been recorded in Switzerland in the last 15 years [20]. A similar picture could be attributable to subtype A8 in northwest Italy.
It is noteworthy to consider the amino acid sequence spanning the hypervariable regions of the envelope protein (HV1 and HV2) which are believed to play a crucial role in cellular receptor binding. Interestingly, the A8 fusogenic strain contained an insertion of 2 residues within the HV2 motif and a greater proportion of basic amino acids, mimicking the prototype fusogenic strain A1 (Table 3). On the other hand, the sequence analysis of U3 region of the long terminal repeats was similar in A8 subgroup, being the transcription factor binding sites well conserved among isolates. These data taken together suggest that receptor binding, rather than transcription factors, may be associated to low pathogenic potential (Table 3) [21].
A certain degree of antigenic heterogeneity was observed in some isolates of subtype A8 and this may influence the sensitivity of some diagnostic tests. Sequence analysis of the immunodominant transmembrane epitope [22] revealed a non-synonymous mutation Y to F within the loop of disulfide bond (Table 2). In addition, the above mentioned P231Q mutation within the major capsid antigen was sufficient to drive the serotyping reactivity versus genotype E antigen leading to misclassification by standard ELISA tests. Since the genotype E derived antigen is usually not included in commercially available antigens, this A8 variant may potentially escape from standard serological test.
Only one goat showed an A8 virological positive outcome in the absence of serological reaction. The genome analysis of this isolate did not reveal any atypical epitope signature which may explain serological misdiagnosis. We cannot exclude a very recent infection in the window in which antibody response is not yet detectable or, instead, an impaired antibody production in late infection steps [23].
Among the sequences described in the present study, two possible new subtypes within genotype A were found (It009.2017 and It038.2017). Both were fusogenic in vitro (Table 1) and showed highest reactivity against genotype A-derived antigen in the genotyping ELISA test. In both cases, the new isolates did not show significant similarity with the available sequence data set (similarity values lower than the 85%). The former isolate is similar to A4 and A1 subtypes, belonging to the same monophyletic clade within the gag gene-based tree. The latter isolate is phylogenetically related to the sheep strain It-561 isolated in Tuscany in 1995 and to the A9 SRLV subtype (Fig 2). The structure of these clades suggests that the new isolates belong to different evolutionary lineages compared to the reference subtypes, but their position within the clade highlights the very high heterogeneity of SRLV, especially within the genotype A. This result shows how the SRLV population structure is complex and its evolutionary patterns are still largely unknown. The two different host species are characterized by different farming management and population sizes; this may influence the animal-pathogen interactions and consequently may drive the SRLV to evolve in different manners in different behaviors. Given the improvement of diagnostic and viral characterization tools, these results may help to understand the complexity of SRLV viral heterogeneity and should lead to consider an update in SRLV classification, considering both genetic and in vitro properties of the new isolates.
Moreover, the pathogenic potential of these less frequent subgroups (i.e. A18 and A19) will require additional studies since the serological tools are not fully able to differentiate the strains within each genotype.
In conclusion the proposed approach, involving virus purification from spleen biopsy followed by NGS, allowed the isolation and full genome characterization of 22 novel SRLV strains. The success of this method is based on the following features: i) spleen is one of the main target organs for SRLV persistence; ii) red pulp is a reserve of resident macrophages, the main target for SRLV replication in vivo; iii) RT activity is a sensitive and specific assay for revealing SRLV grown in cell culture; iv) de novo sequencing and assembling do not require previous genetic knowledge. Even if further studies are needed in order to validate the method and to asses its diagnostic performances we were able to detect both pathogenic and non pathogenic viral strains in goats and sheep, despite the limited sampling area, increasing the knowledge about SRLV genetic diversity.
Acknowledgments
This work was funded by “Ricerca Locale ex60% 2016” grant provided by the University of Torino. The authors acknowledge all the slaughterhouse owners for the collaboration during the sample collection. RR was supported by Ramón y Cajal contract.
Data Availability
All relevant data are within the manuscript. Full genome sequences of newly assembled isolates are available on GenBank (MG554402-MG554414 and MH374283-MH374291.
Funding Statement
This research was funded by the University of Torino (Ricerca Locale ex60% 2016 grant to SR). RR was supported by Ramón y Cajal contract. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shah C, Böni J, Huder JB, Vogt H-R, Mühlherr J, Zanoni R, et al. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: evidence for regular sheep-to-goat transmission and worldwide propagation through livestock trade. Virology. 2004;319: 12–26. 10.1016/j.virol.2003.09.047 [DOI] [PubMed] [Google Scholar]
- 2.Grego E, Profiti M, Giammarioli M, Giannino L, Rutili D, Woodall C, et al. Genetic Heterogeneity of Small Ruminant Lentiviruses Involves Immunodominant Epitope of Capsid Antigen and Affects Sensitivity of Single-Strain-Based Immunoassay. Clin Vaccine Immunol. 2002;9: 828–832. 10.1128/CDLI.9.4.828-832.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosati S, Kwang J, Keen JE. Genome Analysis of North American Small Ruminant Lentiviruses by Polymerase Chain Reaction and Restriction Enzyme Analysis. J Vet Diagnostic Investig. 1995;7: 437–443. 10.1177/104063879500700403 [DOI] [PubMed] [Google Scholar]
- 4.Grego E, Bertolotti L, Quasso A, Profiti M, Lacerenza D, Muz D, et al. Genetic characterization of small ruminant lentivirus in Italian mixed flocks: evidence for a novel genotype circulating in a local goat population. J Gen Virol. 2007;88: 3423–3427. 10.1099/vir.0.83292-0 [DOI] [PubMed] [Google Scholar]
- 5.Juganaru M, Reina R, Bertolotti L, Stella MC, Profiti M, Armentano M, et al. In vitro properties of small ruminant lentivirus genotype E. Virology. 2011;410 10.1016/j.virol.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 6.Bertolotti L, Mazzei M, Puggioni G, Carrozza ML, dei Giudici S, Muz D, et al. Characterization of new small ruminant lentivirus subtype B3 suggests animal trade within the mediterranean basin. J Gen Virol. 2011;92 10.1099/vir.0.032334-0 [DOI] [PubMed] [Google Scholar]
- 7.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8: 239–46. 10.1016/j.meegid.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 9.Reina R, Grego E, Profiti M, Glaria I, Robino P, Quasso A, et al. Development of specific diagnostic test for small ruminant lentivirus genotype E. Vet Microbiol. 2009;138: 251–257. 10.1016/j.vetmic.2009.04.005 [DOI] [PubMed] [Google Scholar]
- 10.de Andrés X, Ramírez H, Bertolotti L, San Román B, Glaria I, Crespo H, et al. An insight into a combination of ELISA strategies to diagnose small ruminant lentivirus infections. Vet Immunol Immunopathol. 2013;152 10.1016/j.vetimm.2012.12.017 [DOI] [PubMed] [Google Scholar]
- 11.Olech M, Valas S, Kuźmak J. Epidemiological survey in single-species flocks from Poland reveals expanded genetic and antigenic diversity of small ruminant lentiviruses. Roques P, editor. PLoS One. 2018;13: e0193892 10.1371/journal.pone.0193892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santry LA, de Jong J, Gold AC, Walsh SR, Menzies PI, Wootton SK. Genetic characterization of small ruminant lentiviruses circulating in naturally infected sheep and goats in Ontario, Canada. Virus Res. 2013;175: 30–44. 10.1016/j.virusres.2013.03.019 [DOI] [PubMed] [Google Scholar]
- 13.Gjerset B, Rimstad E, Teige J, Soetaert K, Jonassen CM. Impact of natural sheep–goat transmission on detection and control of small ruminant lentivirus group C infections. Vet Microbiol. 2009;135: 231–238. 10.1016/j.vetmic.2008.09.069 [DOI] [PubMed] [Google Scholar]
- 14.Reina R, Bertolotti L, Dei Giudici S, Puggioni G, Ponti N, Profiti M, et al. Small ruminant lentivirus genotype E is widespread in Sarda goat. Vet Microbiol. 2010;144: 24–31. 10.1016/j.vetmic.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 15.Mordasini F, Vogt H-R, Zahno M-L, Maeschli A, Nenci C, Zanoni R, et al. Analysis of the Antibody Response to an Immunodominant Epitope of the Envelope Glycoprotein of a Lentivirus and Its Diagnostic Potential. J Clin Microbiol. 2006;44: 981–991. 10.1128/JCM.44.3.981-991.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramírez H, Reina R, Bertolotti L, Cenoz A, Hernández M-M, San Román B, et al. Study of compartmentalization in the visna clinical form of small ruminant lentivirus infection in sheep. BMC Vet Res. 2012;8 10.1186/1746-6148-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolari F, Guarda F, Rosati S, Valenza F. Correlazione fra reperti anatomo-istologici ed isolamento virale nella Maedi degli ovini. Atti della Società Italiana delle Scienze Veterinarie. 1991. pp. 1083–1087. [Google Scholar]
- 18.Reina R, Grego E, Bertolotti L, De Meneghi D, Rosati S. Genome analysis of small-ruminant lentivirus genotype E: a caprine lentivirus with natural deletions of the dUTPase subunit, vpr-like accessory gene, and 70-base-pair repeat of the U3 region. J Virol. 2009;83: 1152–1155. 10.1128/JVI.01627-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardinaux L, Zahno M-L, Deubelbeiss M, Zanoni R, Vogt H-R, Bertoni G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet Microbiol. 2013;162: 572–581. 10.1016/j.vetmic.2012.11.017 [DOI] [PubMed] [Google Scholar]
- 20.Deubelbeiss M, Blatti-Cardinaux L, Zahno M-L, Zanoni R, Vogt H-R, Posthaus H, et al. Characterization of small ruminant lentivirus A4 subtype isolates and assessment of their pathogenic potential in naturally infected goats. Virol J. 2014;11: 65 10.1186/1743-422X-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juganaru M, Reina R, Grego E, Profiti M, Rosati S. LTR promoter activity of SRLV genotype E, strain Roccaverano. Vet Res Commun. 2010;34: 47–51. 10.1007/s11259-010-9390-5 [DOI] [PubMed] [Google Scholar]
- 22.Bertoni G, Zahno ML, Zanoni R, Vogt HR, Peterhans E, Ruff G, et al. Antibody reactivity to the immunodominant epitopes of the caprine arthritis-encephalitis virus gp38 transmembrane protein associates with the development of arthritis. J Virol. 1994;68: 7139–47. Available: http://www.ncbi.nlm.nih.gov/pubmed/7933096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry LL, Wilkerson MJ, Hullinger GA, Cheevers WP. Depressed CD4+ T lymphocyte proliferative response and enhanced antibody response to viral antigen in chronic lentivirus-induced arthritis. J Infect Dis. 1995;171: 328–34. Available: http://www.ncbi.nlm.nih.gov/pubmed/7844368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript. Full genome sequences of newly assembled isolates are available on GenBank (MG554402-MG554414 and MH374283-MH374291.