Abstract
Objective
To classify and immunologically characterize persons with MS based on brain lesions and atrophy and their associated microRNA profiles.
Methods
Cerebral T2-hyperintense lesion volume (T2LV) and brain parenchymal fraction (BPF) were quantified and used to define MRI phenotypes as follows: type I: low T2LV, low atrophy; type II: high T2LV, low atrophy; type III: low T2LV, high atrophy; type IV: high T2LV, high atrophy, in a large cross-sectional cohort (n = 1,088) and a subset with 5-year lngitudinal follow-up (n = 153). Serum miRNAs were assessed on a third MS cohort with 2-year MRI phenotype stability (n = 98).
Results
One-third of the patients had lesion-atrophy dissociation (types II or III) in both the cross-sectional and longitudinal cohorts. At 5 years, all phenotypes had progressive atrophy (p < 0.001), disproportionally in type II (BPF −2.28%). Only type IV worsened in physical disability. Types I and II showed a 5-year MRI phenotype conversion rate of 33% and 46%, whereas III and IV had >90% stability. Type II switched primarily to IV (91%); type I switched primarily to II (47%) or III (37%). Baseline higher age (p = 0.006) and lower BPF (p < 0.001) predicted 5-year phenotype conversion. Each MRI phenotype demonstrated an miRNA signature whose underlying biology implicates blood-brain barrier pathology: hsa.miR.22.3p, hsa.miR.361.5p, and hsa.miR.345.5p were the most valid differentiators of MRI phenotypes.
Conclusions
MRI-defined MS phenotypes show high conversion rates characterized by the continuation of either predominant neurodegeneration or inflammation and support the partial independence of these 2 measures. MicroRNA signatures of these phenotypes suggest a role for blood-brain barrier integrity.
MS is an autoimmune disease of the CNS, characterized by recurrent episodes of inflammatory demyelination and accelerated global CNS tissue loss. MRI is the most useful clinical modality to diagnose and monitor MS-related pathology and is sensitive to both inflammatory and destructive pathologic processes as measured by T2-hyperintense lesion volume (T2LV) and whole-brain atrophy, respectively. There is increasing evidence that these 2 processes are at least partially independent.1 We have recently described an MRI-based, 4-phenotype classification that is based on the relationship between (1) brain volume loss and (2) brain T2LV.2 This MRI-based classification scheme showed that approximately one-fourth of the patients have a disassociation between atrophy and lesions (e.g., high-atrophy and low T2LV or low-atrophy/high T2LV). This disassociation is consistent with previous work, indicating that brain atrophy progresses somewhat independent of lesions.3–6 Moreover, recent work has also demonstrated serum immunologic biomarkers, which may uniquely be linked to destructive or lesional pathology, such as serum microRNAs (miRNAs),7 lipid antibodies,8 free hemoglobin,9 retinol binding protein,10 mitochondrial metabolites,11 and neurofilament light chains.12 CSF biomarkers have also been linked to MRI atrophy or T2 lesion volume including CXCL13,13 tau levels,14 oligoclonal bands,15 increased B-cell activation,16 glial fibrillic acidic protein,17 and the CSF-serum albumin quotient.18
With the goal of investigating the contributions of inflammation and neurodegeneration to MS pathophysiology, we established 2 major objectives for the present study: first, to characterize MRI-defined phenotypes in a large cross-sectional cohort and examine their (5-year) longitudinal stability and second, to identify differences in miRNA signatures among the MRI-phenotype groups. We additionally assessed the demographic, clinical, and MRI features associated with stability vs switching of the phenotypes over 5 years.
Methods
Cohort identification and patient characteristics
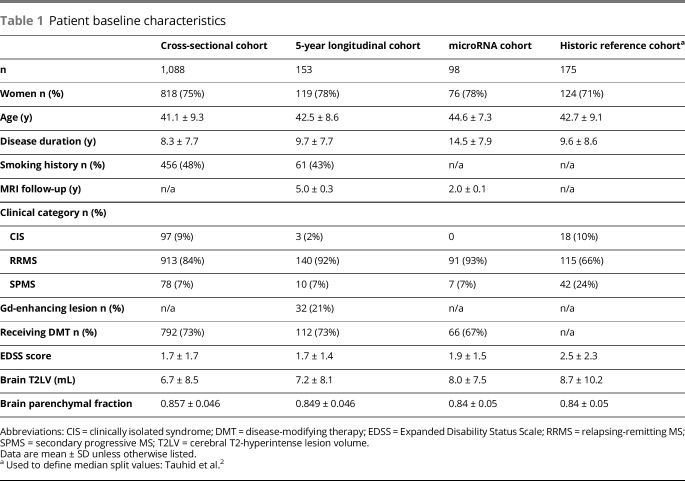
This was a single-center, retrospective longitudinal study of patients who met baseline inclusion criteria as follows: (1) diagnosis of MS on the relapsing-remitting (RR) spectrum, including clinically isolated syndrome, RR MS, or secondary progressive (SP) MS19; (2) age 18–60 years; (3) availability of brain MRI acquired at 1.5T by a consistent acquisition protocol; and (4) neurologic examination, including assessment of Expanded Disability Status Scale (EDSS) score20 within 6 months of neuroimaging. Patients with primary progressive MS were excluded. This initial group composed the cross-sectional cohort. Among these patients, additional cohorts were identified. A longitudinal cohort was also identified as those with 5-year follow-up clinical and MRI data (henceforth referred to as the 5-year longitudinal cohort). To identify relationships between MRI phenotypes and serum miRNAs, we defined a third cohort, which included only patients with MRI phenotype congruence/stability between baseline and 2-year follow-up who had baseline serum blood samples available. A cohort of 98 patients who met these criteria was identified, hereon referred to as the 2-year cohort. Patients' clinical and demographic characteristics are summarized in table 1, and the cohort selection process is depicted in figure e-1, links.lww.com/NXI/A93. After MRI quality control analysis, 3 patients in the 5-year cohort were excluded because of poor segmentation and/or motion artifact.
Table 1.
Patient baseline characteristics
Standard protocol approvals, registrations, and patient consents
This study was approved by our institutional local ethics committee on human experimentation. All patients provided written informed consent.
MRI acquisition and segmentation
All brain MRI scans were obtained on a fleet of 1.5T Signa GE scanners at the Brigham and Women's Hospital using similar acquisition parameters to produce axial dual echo images as follows: TR = 2,800–3,000 msec, TE1/TE2 = 30/80 msec, slice thickness = 3 mm (gapless), pixel size = 0.93 × 0.93 mm, producing 54 slices. Automated template-driven segmentation was performed to derive cerebral T2LV and brain parenchymal fraction (BPF),21 followed by expert manual correction. The number of gadolinium lesions was determined by an expert observer on axial T1-weighted spin-echo images, 5–7 minutes after an IV infusion of gadolinium contrast agent at a concentration of 0.1-mmoL/kg.
Classification of MRI-defined phenotypes
Patients were assigned MRI phenotypes based on a previously defined median split values of T2LV (4.21 mL) and BPF (0.8502) as described in our index cases, selected based on similarity to a broad cohort of patients with MS2:
Type I = mild atrophy (BPF > 0.8502) and low lesion burden (T2LV < 4.21 mL)
Type II = mild atrophy (BPF > 0.8502) and high lesion burden (T2LV > 4.21 mL)
Type III = moderate/severe atrophy (BPF < 0.8502) and low lesion burden (T2LV < 4.21 mL)
Type IV = moderate/severe atrophy (BPF < 0.8502) and high lesion burden (T2LV > 4.21 mL)
Thus, types I and IV had concordance between lesions and atrophy and types II and III a discordance.
Serum samples
Serum samples were collected in red-top Vacutainer tubes without additives, centrifuged at 2000 rpm for 10 minutes to separate the serum, and stored at −70°C until RNA extraction. Isolation of RNA was then performed (miRcury kit; Exiqon) and converted to complementary DNA using a synthesis kit (Exiqon). Prepared complementary DNA samples were stored at −20°C. We selected 186 candidate miRNAs based upon their relevance in our previous studies. Expression of these miRNAs was tested in prepared complementary DNA samples using a locked nucleic acid, SYBR green-based quantitative real time polymerase chain reaction method (LNA-SYBR-qRT-PCR). Normalization was performed using the mean expression of 4 miRNAs, with the best stability index determined by NormFinder software. Normalized cycle quantification (Cq) was calculated as mean Cq–assay Cq.
Statistical analysis and data interpretation
Mean values of group-level characteristics with continuous variables (EDSS score, T2LV, BPF, and disease duration) were compared using paired t-tests (baseline vs follow-up) and one-way analysis of variance (ANOVA) (baseline MRI phenotype comparison); ordinal and dichotomous variables (age, sex, smoking history, presence of gadolinium-enhancing lesion(s), and disease category) were compared using the Pearson chi-square test. A linear mixed effect model was used to compare MRI phenotype group differences over time; predictors of phenotype conversion were estimated using a linear/logistic regression or odds ratios.
MicroRNA expression was compared among the 4 groups using a proportional odds logistic regression model, with phenotype as the predictor and miRNA expression as the outcome. A proportional odds logistic regression model was used so that patients who had missing expression could be given a value lower than the smallest detected expression. If a significant difference between the groups was observed for the global test, pairwise group comparisons were completed. Differentially expressed miRNA pathways (at p < 0.05) were interpreted using miRPath v.3 with default settings and TarBase v7.0, accessed online between October 11, 2017 and October 13, 2017 at snf-515788.vm.okeanos.grnet.gr/.22 “Biologic directionality” of miRNAs was determined by comparing mean expression values between phenotypes: higher expression in a numerically higher phenotype was presumed to represent a “pathologic” association, and conversely, lower expression levels were presumed “protective.” Comparisons of types II and III were labeled “unknown” because of the lesion/atrophy dissociation. “Biologic MRI associations” were assigned based on the defined neuroimaging feature(s) to separate MRI phenotypes as follows: type I vs type II, “lesions”; type I vs type III, “atrophy”; type I vs type IV, “both”; type II vs type III, “unknown”; type II vs type IV, “atrophy”; and type III vs type IV, “lesions.”
All statistics were performed using the R (R-project.org) or STATA v14.0 (stata.com) software.
Data availability
Anonymized data will be shared on request from any qualified investigator.
Results
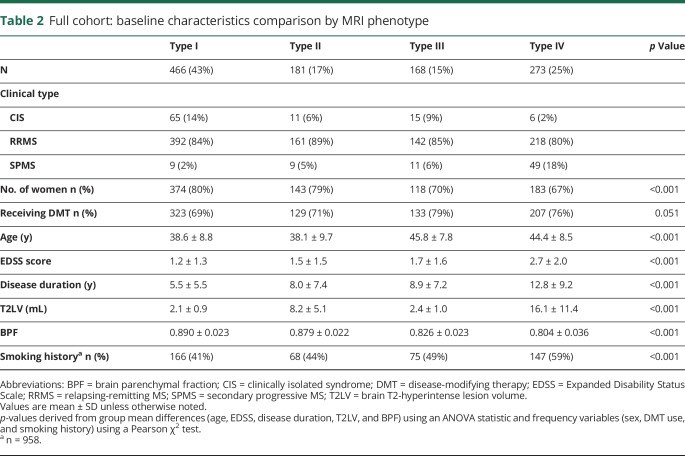
Baseline clinical, MRI, and demographic characteristics are summarized in table 1, with the inclusion of a comparison historical reference cohort initially used to define the median split values based on a broad representation of MS disease phenotypes similar to that of the New York State Multiple Sclerosis Consortium.2 Our median split criteria defined 4 MRI-based phenotypes (see Methods); data for the full cross-sectional cohort are summarized in table 2, with visual depiction of archetypal phenotypes in figure 1. In both the cross-sectional cohort (n = 1,088), as well as the 5-year longitudinal cohort (n = 153), ∼one-third of all patients (32% and 33%, respectively) were categorized as having dissociation between T2LV and atrophy (types II or III); the proportion of types II and III was very similar (33%) when using median split values obtained from the cross-sectional cohort instead of the historic cohort (data not shown). The overall unadjusted correlation between T2LV and BPF was −0.476 (p < 0.0001) in the longitudinal cohort and −0.538 in the cross-sectional cohort (p < 0.0001). Both of these associations remained significant after adjusting for age (p < 0.0001). We additionally analyzed the associations between BPF and T2LV using quartile splits of BPF; T2LV decreased as BPF increased: BPF quartile 1, T2LV 13.5 ± 12.4 mL; quartile 2, T2LV 6.1 ± 6.7 mL; quartile 3, T2LV 4.0 ± 4.1 mL; and quartile 4, T2LV 3.0 ± 2.5 mL; p < 0.001. In both cohorts, the following significant baseline group differences between phenotypes were observed by one-way ANOVA (p values all <0.001 for the cross-sectional cohort): sex, age, EDSS score, disease duration, T2LV, BPF, and smoking history (table 2). As shown in table e-1, links.lww.com/NXI/A93, notable baseline pairwise phenotype differences included (1) types III and IV both being older and with a higher proportion of men compared with types I and II (all p < 0.001); (2) disease duration, EDSS, and SPMS prevalence, all increased linearly from type I (mean disease duration 5.5 years, EDSS 1.2, SPMS 2%) to type IV (mean disease duration 12.8 years, EDSS 2.7, SPMS 18%, all p < 0.01) with the exception of pairwise comparison of types II and III, which were not significant for any of these comparisons (p > 0.05); (3) smoking history was significantly overrepresented in type IV compared with others (all p ≤ 0.05); (4) BPF decreased linearly from type I to type IV and differed significantly among all phenotypes (all p < 0.001); (5) T2LV differed significantly among all phenotypes (p < 0.001) except between types I and III.
Table 2.
Full cohort: baseline characteristics comparison by MRI phenotype
Figure 1. MRI phenotype 5-year conversion rates.

T2 FLAIR images showing changes in MS-MRI phenotypes over a 5-year follow-up period. Type 1 patients (33%) converted: 16% to type II, 12% to type III, and 5% to type IV; 46% of type II patients converted: 42% to type IV and 4% to type I. Types III and IV were predominantly stable (<10% interconversion). Higher age, higher T2LV, and lower BPF were predictive of phenotype conversion for types I and II. We observe conversion pathways, depicted by arrows: adaptive immunity/inflammatory-predominant changes characterize conversion from type I to type II, or type III to type IV, whereas degenerative-predominant changes are characterized by type I to type III or type II to type IV. Conversion of type I to type IV involves both pathways. BPF = brain parenchymal fraction; FLAIR = fluid-attenuated inversion-recovery; T2LV = T2-hyperintense lesion volume.
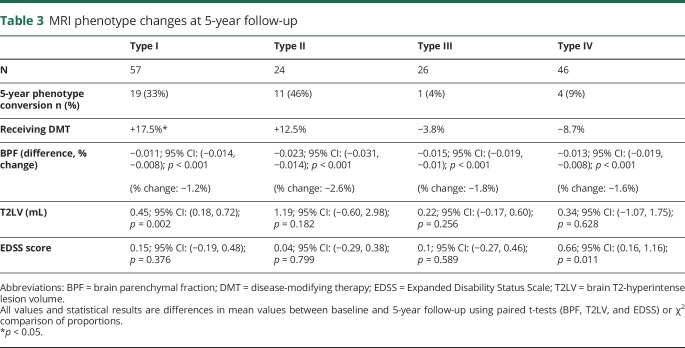
Longitudinal comparisons: Table 3 and figure 1 depict within-phenotype changes over 5 years, and table e-2, links.lww.com/NXI/A93 shows between-phenotype comparisons. All phenotypes demonstrated significant decreases in BPF (all p < 0.001); the 5-year rate of brain atrophy was highest in type II (vs type I: p < 0.001, vs type III: p = 0.056, and vs type IV: p = 0.013, figure e-2, links.lww.com/NXI/A93). T2LV significantly increased over 5 years only in type I (p < 0.01) and was not significantly different in between-group comparisons. EDSS score increased in types II and IV over 5 years, but the change was only significantly different when comparing group IV with group I (p = 0.048, figure e-3, links.lww.com/NXI/A93). In regard to 5-year phenotype conversion, type I was 67% stable, with 33% converting to type II (16%), type III (12%), or type IV (5%); type II was 54% stable, with 42% converting to type IV, none to type III, and 4% to type I. types III and IV were >90% stable and type III converted exclusively to type IV (4%). Predictors of 5-year conversion of MRI phenotype (types I and II combined, table e-3, links.lww.com/NXI/A93) included baseline lower BPF (p < 0.001) and older age (p = 0.008).
Table 3.
MRI phenotype changes at 5-year follow-up
In summary, clinical and demographic data generally differentiated all 4 MRI phenotypes from one another with the exception of types II and III, which were similar on most pairwise comparisons despite stark MRI differences. Higher phenotypes (e.g., III and IV) were characterized by more men, a history of smoking, higher age, and longer disease durations. Over the 5-year follow-up period, type I converted to either an inflammatory-predominant (type II) or degenerative-predominant (type III) phenotype. Types III and IV were terminal phenotypes that rarely interconvert.
miRNA study: 2-year longitudinal cohort with MRI-phenotype stability
The 2-year cohort was composed of 98 patients with MS (91 RR, 7 SP). Baseline characteristics are summarized in table 1. The MRI-defined phenotype breakdown was as follows: type I: n = 24; type II: n = 19, type III: n = 21, and type IV: n = 34. Forty-one percent (41%) of all patients were categorized as having dissociation between T2LV and atrophy (types II or III). Serum miRNA analysis (see Methods) of this cohort revealed 16 miRNAs (tables e-4 and e5, table 4, links.lww.com/NXI/A93) that significantly differed in their expression among the 4 MRI phenotypes at p < 0.05 after adjustment for age and sex; none survived corrections for multiple comparisons (table e-4). Among these 16 miRNAs, 15 overlapped with significant findings from our previous work examining correlations between MRI involvement and miRNAs in separate sets of patients with MS.7 Table e-4 shows pairwise group comparisons of the level of miRNA expression of these 16 miRNAs. Three of the miRNAs showed pairwise differences between phenotypes and were both internally consistent with phenotype definitions (see Methods), as well as externally consistent with our previous work7: hsa.miR.22.3p, hsa.miR.345.5p, and hsa.miR.361.5p. Both hsa.miR.22.3p and hsa.miR.345.5p are significantly and pathogenically upregulated in types II and IV compared with types I and III, suggesting an association with inflammation/lesions. Hsa.miR.361.5p was upregulated in type I compared with types II and III, suggesting a protective association against both lesions and atrophy. DIANA miRPath analysis revealed biological associations of these miRNAs (table 4, figure e-4, links.lww.com/NXI/A93); of the 16 significant miRNAs, 7 are involved in adherens junction and 4 in extracellular matrix functions; of the 3 notable miRNAs above, 2 seem to be involved in adherens junction function. A heatmap (figure 2) depicts miRNA expression levels differentiating the 4 MRI phenotypes.
Table 4.
Interpretation of differentially expressed miRNAs among the 4 MRI phenotypes
Figure 2. MicroRNA expression heatmap stratified by MRI phenotype.
MRI phenotypes are noted by color in the leftmost column (Type I = purple, Type II = green, Type III = red, Type IV = blue). The level of microRNA (miRNA) expression level is noted by color from blue (lowest/absent expression) to red (highest expression), and organized in numerical order from left to right. Each row represents an individual with MS.
Discussion
In this study, we present cross-sectional (n = 1,088) and longitudinal (n = 153) data using a novel MRI-based classification system of MS based on lesions and atrophy and demonstrate exploratory immunologic correlates of these MRI phenotypes using serum miRNA analysis (n = 98). The central findings of our work include (1) evidence for a dissociation between brain lesions and brain atrophy, as demonstrated by large numbers of patients classified as type II or type III; (2) longitudinal assessment showing dynamic changes in phenotypes and the rate and predictors of change of these MRI classifications; and (3) preliminary serum miRNAs that differentiate the MRI phenotypes and implicate potential targetable biological pathways involved in disease heterogeneity.
We first aimed at extending and validating previous findings from the development of an MRI classification of MS based on the relationship between brain lesions and atrophy.2 Using a large sample size, we show a high rate of patients with MS (32%), with a dissociation between whole-brain atrophy and the total volume of cerebral T2-hyperintense lesions (types II or III). A similar proportion was observed in our longitudinal cohort (33%, n = 153) and our miRNA cohort (41%, n = 98). Taken together, these observations provide evidence that these 2 pathologic processes are at least partially independent among a subset of persons with MS.
Over a 5-year follow-up period, the patients at the highest risk of disease progression as shown by conversion to more advanced phenotypes are types I (33% converting) and II (46% converting), with conversion risk at baseline linked to higher age and lower BPF (table e-3, links.lww.com/NXI/A93). These results are consistent with previous studies, showing that higher age is a risk factor for disease progression23,24 and that more brain atrophy at baseline predicts disability progression at 2- and 5-year follow-up periods.25,26 Similarly, a group who tested MRI classification systems like ours27 showed that 1 longitudinal model (the development of whole-brain atrophy and >4 new lesions) could predict short-term disability worsening. Another group classified 4 MRI phenotypes using gadolinium enhancement as a proxy for inflammation and BPF + T1-hypointense lesion volumes to determine neurodegeneration, finding that greater atrophy was associated with activated B-cell activity.16
Our findings provide insights into the relative contributions of inflammation and neurodegeneration in MS.1,6,28 Our data support partial independence of these processes, as reflected by individual patient phenotypes at baseline and the pathways leading to progression/conversions. Among the type I individuals at baseline who converted during the 5-year observation period (33%), roughly equal numbers proceed to type II, characterized by increased T2LV without significant global atrophy, or type III, characterized by high whole-brain tissue loss without much lesion progression. In contrast, type II converted nearly exclusively to type IV. This type I to type II to type IV “inflammation-then-degeneration” pathway (figure 1) supports arguments linking early inflammation to later neurodegeneration, mechanisms believed to be predominantly mediated by adaptive autoimmunity (inflammation), followed by innate (degeneration related) immune changes,29,30 and demonstrates that a portion of type II phenotypes are in a temporal transition phase with age and lower BPF being risk factors for subsequent conversion (table e-3, links.lww.com/NXI/A93). However, 54% of type II patients remained stable at follow-up, demonstrating that this temporal dispersion of dissociation is not complete. Of note, the 5-year change in T2LV was not significantly different in types II or IV, suggesting that these patients may have attained a stabilized T2 lesion burden plateau characteristic of later-stage MS.31
The 5-year brain atrophy rate was highest in patients who were type II at study entry, a finding we interpret as the result of the cascade of events linking the appearance of lesions to later (secondary) atrophy, which may take years to unfold.32,33 The patients may also have had “pseudo-expansion” of brain volume at the time of new or recent lesion appearance/expansion in the early stages of the disease, which would mask any atrophy present until the equilibration of fluid shifts.34 Type I patients are equally likely to proceed through an adaptive inflammation-dominant path (type II) as they are to an atrophy-dominant path (type III); this observation supports innate immunity or neurodegeneration as a predominant process in some patients, separate from inflammatory white matter lesions.1 This observation also raises the possibility that this type of individual may be biologically distinct from others; we speculate that type III may represent greater involvement of gray matter demyelination, cortical lesions, and tissue destruction related to leptomeningeal ectopic deposits or soluble inflammatory mediators not visible on conventional 1.5T MRI.35 Type III patients may also represent greater resistance to inflammatory processes, a greater responsiveness to their disease modifying therapy (DMT), or an enhanced reparative/remyelinating ability, reducing the load of T2-hyperintense lesions.36 Overall, our study lends insights into the inflammation-neurodegeneration debate by highlighting the dissociation between cerebral lesions and atrophy; we argue that although two-thirds of the patients have congruent measures of lesions and atrophy, the one-third that shows incongruence demonstrates that for at least a subset of patients, these processes are somewhat independent. Early stratification of MS into specific pathobiological pathways may offer novel translational insights into individually targeted therapeutics, especially for persons characterized by a more degenerative (type III) phenotype that may derive reduced benefit from traditional anti-inflammatory DMTs.
Of note, our data also showed a strong overrepresentation of smoking history, specifically in the type IV phenotype at baseline, a finding which corroborates previous studies demonstrating smoking as a risk factor for accelerated accumulation of T2LV, disability, atrophy, and conversion to SPMS.37–40
We characterized MRI phenotypes based on serum circulating miRNA profiles, using a 2-year cohort selected for age and sex balance, as well as stable MRI phenotype. Each MRI phenotype demonstrated a unique miRNA signature, and we found 16 miRNAs that differentiated the 4 phenotypes. Three of these miRNAs were both internally consistent (e.g., no contradictory expression-level data in pairwise phenotype comparisons) and externally valid as compared to the literature (table e-4, links.lww.com/NXI/A93): hsa.miR.22.3p, hsa.miR.345.5p, and hsa.miR.361.5p. DIANA miRPath analysis22 of these specific miRNAs revealed that hsa.miR.22.3p and hsa.miR.361.5p are both involved in blood-brain barrier function including adherens junctions (sealing the endothelial barrier), focal adhesion (of lymphocytes to the endothelium), and extracellular matrix integrity (endothelial-lymphocyte communication; table 4, figure e-4, links.lww.com/NXI/A93), a theme in microRNA regulation.41 Hsa.miR.22.3p is additionally associated with TGF-beta signaling, involved in the maturation of Th-17 cells believed to be important in the development of MS.42 Hsa.miR.345.5p demonstrated a pathologic association with T2LV in previous work,7 is upregulated in MS vs HC,43 and is dysregulated in both major depression and bipolar disorder.44 Of note, 4 of the miRNAs (hsa.miR.145.5p, hsa.miR.193a.5p, hsa.miR.195.5p, and hsa.miR.1972) were also consistently upregulated in type I, suggestive of a protective role in MS pathology. However, this protective aspect was not consistent with previous literature. Nonetheless we suggest these 16 miRNAs, especially hsa.miR.22.3p, hsa.miR.345.5p, and hsa.miR.361.5p, may be considered potentially relevant for future investigation.
Our study results are limited based on several factors. As a retrospective analysis (of prospective data), data are subject to systemic and selection biases, and we did not capture any dynamic treatment effects (substitution of 1 DMT with another, IV steroid use) or comorbid medical conditions that could contribute to miRNA or MRI results. However, we did compare our cross-sectional and longitudinal cohorts for major forms of selection bias; this did not reveal any significant baseline differences in age, sex, disease duration, BPF, T2LV, EDSS, and DMT usage (p > 0.05, data not shown). In addition, the MRI parameters used (T2LV, BPF) are unlikely to completely reflect the immunopathogenic process, especially at 1.5T. Spatial patterns of cerebral lesion topography have also been demonstrated to be important predictors of disability,45 and we did not model this process here; similarly, we did not quantify spinal cord disease, which could contribute to whole-brain atrophy through Wallerian degeneration. However, our results are more generalizable because of what would be observed in the routine clinical setting. The interpretation of biological associations of circulating (extracellular) miRNAs is an evolving process and requires further investigations; although some authors present evidence that these circulating miRNAs represent byproducts of cellular metabolism and death, there are also data supporting their role in cell-cell signaling as active paracrine or endocrine molecules.46,47 Last, the miRNA analysis was hypothesis-generating; none of our results remained significant after correction for multiple comparisons. Results may thus represent type I statistical errors and must be interpreted with caution. Results are of a correlative nature only and cannot be extrapolated to causation. They nonetheless provide insights into potentially important biological pathways, which may help explain MRI and clinical heterogeneity in MS.
Acknowledgment
The authors thank Svetlana Egorova, Sheena L. Dupuy, Mark Anderson, and Mariann Polgar-Turcsanyi for technical assistance.
Glossary
- BPF
brain parenchymal fraction
- Cq
cycle quantification
- EDSS
Expanded Disability Status Scale
- miRNA
microRNA
- SP
secondary progressive
- T2LV
T2-hyperintense lesion volume
Appendix 1. Author contributions

Study funding
This work was supported by a Clinical Research Training Fellowship in Multiple Sclerosis to Dr. Hemond from the American Academy of Neurology Institute and Sanofi Genzyme. This study was also supported by the Ann Romney Center for Neurologic Diseases.
Disclosure
C.C. Hemond received research support from American Academy of Neurology Institute and Sanofi Genzyme; B.C. Healy served on the editorial board of Statistical Methods in Medical Research and received research support from Merck Serono, Genzyme, Novartis, Verily Life Sciences, NIH, and NMSS; S. Tauhid served as the managing editor for the Journal of Neuroimaging; M.A. Mazzola reports no disclosures; F.J. Quintana served on the editorial boards of Biomedicine, Immunologia (Spain), and American Journal of Clinical and Experimental Immunology; served as an associate editor for Immunology (UK); served as an advisory board member for Seminars in Immunopathology; received research support from EMD Serono, Sanofi, Novartis, Teva, ONO Pharmaceutical, NIH, NMSS, the American Cancer Society, Israel Cancer Research Fund, NIH-NIAID, King Abdulaziz City for Sciences & Technology, Harvard Medical School, and BADERC; and holds stock or stock options in AnTolRx; R. Gandhi is employed by Sanofi Genzyme; received research support from Sanofi Genzyme, the NIH, and the Race to Erase MS; and holds stock or stock options in Sanofi; H.L. Weiner served on the advisory boards of The Guthy Jackson Charitable Foundation, Teva, Biogen, Novartis, Sanofi-Aventis, Tilos, CBridge Capital, Genentech, Genzyme, vTv Therapeutics, and MedDay; consulted for Biodextris, Biogen, Novartis, Serono, Teva, Sanofi Genzyme, Tilos, Tiziana Life Sciences, vTv Therapeutics, MedDay, Genentech, and CBridge Capital; and received research support from EMD Serono, Google Life Sciences, the NIH, and the NMSS; R. Bakshi received consulting fees from the advisory boards of Bayer, Biogen, Celgene, EMD Serono, Genentech, Guerbet, Sanofi Genzyme, and Shire; is Editor-in-Chief for the Journal of Neuroimaging; consulted for Bayer, Biogen, Celgene, EMD Serono, Genentech, Guerbet, Sanofi Genzyme, and Shire; and received research support from EMD Serono and SanofiGenzyme. Dr. Bakshi's spouse holds stock in Biogen. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NN.
References
- 1.Louapre C, Lubetzki C. Neurodegeneration in multiple sclerosis is a process separate from inflammation: yes. Mult Scler 2015;21:1626–1628. [DOI] [PubMed] [Google Scholar]
- 2.Tauhid S, Neema M, Healy BC, Weiner HL, Bakshi R. MRI phenotypes based on cerebral lesions and atrophy in patients with multiple sclerosis. J Neurol Sci 2014;346:250–254. [DOI] [PubMed] [Google Scholar]
- 3.Bermel RA, Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol 2006;5:158–170. [DOI] [PubMed] [Google Scholar]
- 4.Bakshi R, Neema M, Healy BC, et al. . Predicting clinical progression in multiple sclerosis with the magnetic resonance disease severity scale. Arch Neurol 2008;65:1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearney H, Altmann DR, Samson RS, et al. . Cervical cord lesion load is associated with disability independently from atrophy in MS. Neurology 2015;84:367–373. [DOI] [PubMed] [Google Scholar]
- 6.Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJ, Reynolds R, Martin R. Exploring the origins of grey matter damage in multiple sclerosis. Nat Rev Neurosci 2015;16:147–158. [DOI] [PubMed] [Google Scholar]
- 7.Regev K, Healy BC, Khalid F, et al. . Association between serum MicroRNAs and magnetic resonance imaging measures of multiple sclerosis severity. JAMA Neurol 2017;74:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakshi R, Yeste A, Patel B, et al. . Serum lipid antibodies are associated with cerebral tissue damage in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2016;3:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewin A, Hamilton S, Witkover A, et al. . Free serum haemoglobin is associated with brain atrophy in secondary progressive multiple sclerosis. Wellcome Open Res 2016;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokote H, Kamata T, Toru S, Sanjo N, Yokota T. Serum retinol levels are associated with brain volume loss in patients with multiple sclerosis. Mult Scler J Exp Transl Clin 2017;3:2055217317729688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazzarino GG, Amorini AM, Petzold A, et al. . Serum compounds of energy metabolism impairment are related to disability, disease course and neuroimaging in multiple sclerosis. Mol Neurobiol 2017;54:7520–7533. [DOI] [PubMed] [Google Scholar]
- 12.Kuhle J, Nourbakhsh B, Grant D, et al. . Serum neurofilament is associated with progression of brain atrophy and disability in early MS. Neurology 2017;88:826–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puthenparampil M, Federle L, Miante S, et al. . BAFF Index and CXCL13 levels in the cerebrospinal fluid associate respectively with intrathecal IgG synthesis and cortical atrophy in multiple sclerosis at clinical onset. J Neuroinflammation 2017;14:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietroboni AM, Schiano Di Cola F, Scarioni M, et al. . CSF β-amyloid as a putative biomarker of disease progression in multiple sclerosis. Mult Scler 2017;23:1085–1091. [DOI] [PubMed] [Google Scholar]
- 15.Farina G, Magliozzi R, Pitteri M, et al. . Increased cortical lesion load and intrathecal inflammation is associated with oligoclonal bands in multiple sclerosis patients: a combined CSF and MRI study. J Neuroinflammation 2017;14:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comabella M, Cantó E, Nurtdinov R, et al. . MRI phenotypes with high neurodegeneration are associated with peripheral blood B-cell changes. Hum Mol Genet 2016;25:308–316. [DOI] [PubMed] [Google Scholar]
- 17.Kassubek R, Gorges M, Schocke M, et al. . GFAP in early multiple sclerosis: a biomarker for inflammation. Neurosci Lett 2017;657:166–170. [DOI] [PubMed] [Google Scholar]
- 18.Uher T, Horakova D, Tyblova M, et al. . Increased albumin quotient (QAlb) in patients after first clinical event suggestive of multiple sclerosis is associated with development of brain atrophy and greater disability 48 months later. Mult Scler 2016;22:770–781. [DOI] [PubMed] [Google Scholar]
- 19.Polman CH, Reingold SC, Banwell B, et al. . Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 1983;33:1444–1452. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Warfield SK, Zou KH, et al. . Quantitative analysis of MRI signal abnormalities of brain white matter with high reproducibility and accuracy. J Magn Reson Imaging 2002;15:203–209. [DOI] [PubMed] [Google Scholar]
- 22.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. . DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillemin F, Baumann C, Epstein J, et al. . Older age at multiple sclerosis onset is an independent factor of poor prognosis: a population-based cohort study. Neuroepidemiology 2017;48:179–187. [DOI] [PubMed] [Google Scholar]
- 24.Scalfari A, Neuhaus A, Daumer M, Ebers GC, Muraro PA. Age and disability accumulation in multiple sclerosis. Neurology 2011;77:1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffery DR, Di Cantogno EV, Ritter S, Meier DP, Radue EW, Camu W. The relationship between the rate of brain volume loss during first 24 months and disability progression over 24 and 48 months in relapsing MS. J Neurol 2015;263:299–305. [DOI] [PubMed] [Google Scholar]
- 26.Lukas C, Minneboo A, de Groot V, et al. . Early central atrophy rate predicts 5 year clinical outcome in multiple sclerosis. J Neurol Neurosurg Psychiatry 2010;81:1351–1356. [DOI] [PubMed] [Google Scholar]
- 27.von Gumberz J, Mahmoudi M, Young K, et al. . Short-term MRI measurements as predictors of EDSS progression in relapsing-remitting multiple sclerosis: grey matter atrophy but not lesions are predictive in a real-life setting. PeerJ 2016;4:e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchinson M. Neurodegeneration in multiple sclerosis is a process separate from inflammation: no. Mult Scler 2015;21:1628–1631. [DOI] [PubMed] [Google Scholar]
- 29.Weiner HL. The challenge of multiple sclerosis: how do we cure a chronic heterogeneous disease? Ann Neurol 2009;65:239–248. [DOI] [PubMed] [Google Scholar]
- 30.Chitnis T, Weiner HL. CNS inflammation and neurodegeneration. J Clin Invest 2017;127:3577–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li DKB, Held U, Petkau J, et al. . MRI T2 lesion burden in multiple sclerosis: a plateauing relationship with clinical disability. Neurology 2006;66:1384–1389. [DOI] [PubMed] [Google Scholar]
- 32.Chard DT, Brex PA, Ciccarelli O, et al. . The longitudinal relation between brain lesion load and atrophy in multiple sclerosis: a 14 year follow up study. J Neurol Neurosurg Psychiatry 2003;74:1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudick RA, Fisher E, Lee JC, Simon J, Jacobs L. Use of the brain parenchymal fraction to measure whole brain atrophy in relapsing-remitting MS: Multiple Sclerosis Collaborative Research Group. Neurology 1999;53:1698–1704. [DOI] [PubMed] [Google Scholar]
- 34.Dalton CM, Chard DT, Davies GR, et al. . Early development of multiple sclerosis is associated with progressive grey matter atrophy in patients presenting with clinically isolated syndromes. Brain 2004;127:1101–1107. [DOI] [PubMed] [Google Scholar]
- 35.Zurawski J, Lassmann H, Bakshi R. Use of magnetic resonance imaging to visualize leptomeningeal inflammation in patients with multiple sclerosis. JAMA Neurol 2017;74:100–109. [DOI] [PubMed] [Google Scholar]
- 36.Stangel M, Kuhlmann T, Matthews PM, Kilpatrick TJ. Achievements and obstacles of remyelinating therapies in multiple sclerosis. Nat Rev Neurol 2017;13:742–754. [DOI] [PubMed] [Google Scholar]
- 37.Healy BC, Ali EN, Guttmann CR, et al. . Smoking and disease progression in multiple sclerosis. Arch Neurol 2009;66:858–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernán MA, Jick SS, Logroscino G, Olek MJ, Ascherio A, Jick H. Cigarette smoking and the progression of multiple sclerosis. Brain 2005;128:1461–1465. [DOI] [PubMed] [Google Scholar]
- 39.Degelman ML, Herman KM. Smoking and multiple sclerosis: a systematic review and meta-analysis using the Bradford Hill criteria for causation. Mult Scler Relat Disord 2017;17:207–216. [DOI] [PubMed] [Google Scholar]
- 40.Zivadinov R, Weinstock-Guttman B, Hashmi K, et al. . Smoking is associated with increased lesion volumes and brain atrophy in multiple sclerosis. Neurology 2009;73:504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Ramirez MA, Reijerkerk A, de Vries HE, Romero IA. Regulation of brain endothelial barrier function by microRNAs in health and neuroinflammation. FASEB J 2016;30:2662–2672. [DOI] [PubMed] [Google Scholar]
- 42.Okada H, Khoury SJ. Type17 T-cells in central nervous system autoimmunity and tumors. J Clin Immunol 2012;32:802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regev K, Paul A, Healy B, et al. . Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2016;3:e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maffioletti E, Cattaneo A, Rosso G, et al. . Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord 2016;200:250–258. [DOI] [PubMed] [Google Scholar]
- 45.Bodini B, Battaglini M, De Stefano N, et al. . T2 lesion location really matters: a 10 year follow-up study in primary progressive multiple sclerosis. J Neurol Neurosurg Psychiatry 2011;82:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol 2017;11:1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: a collision of two paradigms. Trends Biochem Sci 2016;41:883–892. [DOI] [PubMed] [Google Scholar]
- 48.Hommers L, Raab A, Bohl A, et al. . MicroRNA hsa-miR-4717-5p regulates RGS2 and may be a risk factor for anxiety-related traits. Am J Med Genet B Neuropsychiatr Genet 2015;168B:296–306. [DOI] [PubMed] [Google Scholar]
- 49.Bam M, Yang X, Zhou J, et al. . Evidence for epigenetic regulation of pro-inflammatory cytokines, interleukin-12 and interferon gamma, in peripheral blood mononuclear cells from PTSD patients. J Neuroimmune Pharmacol 2016;11:168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendes-silva AP, Pereira KS, Tolentino-Araujo GT, et al. . Shared biologic pathways between Alzheimer disease and major depression: a systematic review of MicroRNA expression studies. Am J Geriatr Psychiatry 2016;24:903–912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data will be shared on request from any qualified investigator.