Though not a household name like Zika virus, human cytomegalovirus (HCMV) causes permanent neurologic disability in one newborn child every hour in the United States, which is more than that for Down syndrome, fetal alcohol syndrome, and neural tube defects combined. There are currently no established effective measures to prevent viral transmission to the infant following HCMV infection of a pregnant mother. However, the glycoprotein B (gB)/MF59 vaccine, which aims to prevent pregnant women from acquiring HCMV, is the most successful HCMV vaccine tested clinically to date. Here, we used viral DNA isolated from patients enrolled in a gB vaccine trial who acquired HCMV and identified several impacts that this vaccine had on the size, distribution, and composition of the in vivo viral population. These results have increased our understanding of why the gB/MF59 vaccine was partially efficacious, and such investigations will inform future rational design of a vaccine to prevent congenital HCMV.
KEYWORDS: DNA sequencing, glycoprotein B, human cytomegalovirus, vaccine
ABSTRACT
Human cytomegalovirus (HCMV) is the most common congenital infection worldwide and a frequent cause of hearing loss and debilitating neurologic disease in newborn infants. Thus, a vaccine to prevent HCMV-associated congenital disease is a public health priority. One potential strategy is vaccination of women of child bearing age to prevent maternal HCMV acquisition during pregnancy. The glycoprotein B (gB) plus MF59 adjuvant subunit vaccine is the most efficacious tested clinically to date, demonstrating 50% protection against primary HCMV infection in a phase 2 clinical trial. Yet, the impact of gB/MF59-elicited immune responses on the population of viruses acquired by trial participants has not been assessed. In this analysis, we employed quantitative PCR as well as multiple sequencing methodologies to interrogate the magnitude and genetic composition of HCMV populations infecting gB/MF59 vaccinees and placebo recipients. We identified several differences between the viral dynamics in acutely infected vaccinees and placebo recipients. First, viral load was reduced in the saliva of gB vaccinees, though not in whole blood, vaginal fluid, or urine. Additionally, we observed possible anatomic compartmentalization of gB variants in the majority of vaccinees compared to only a single placebo recipient. Finally, we observed reduced acquisition of genetically related gB1, gB2, and gB4 genotype “supergroup” HCMV variants among vaccine recipients, suggesting that the gB1 genotype vaccine construct may have elicited partial protection against HCMV viruses with antigenically similar gB sequences. These findings suggest that gB immunization had a measurable impact on viral intrahost population dynamics and support future analysis of a larger cohort.
IMPORTANCE Though not a household name like Zika virus, human cytomegalovirus (HCMV) causes permanent neurologic disability in one newborn child every hour in the United States, which is more than that for Down syndrome, fetal alcohol syndrome, and neural tube defects combined. There are currently no established effective measures to prevent viral transmission to the infant following HCMV infection of a pregnant mother. However, the glycoprotein B (gB)/MF59 vaccine, which aims to prevent pregnant women from acquiring HCMV, is the most successful HCMV vaccine tested clinically to date. Here, we used viral DNA isolated from patients enrolled in a gB vaccine trial who acquired HCMV and identified several impacts that this vaccine had on the size, distribution, and composition of the in vivo viral population. These results have increased our understanding of why the gB/MF59 vaccine was partially efficacious, and such investigations will inform future rational design of a vaccine to prevent congenital HCMV.
INTRODUCTION
Human cytomegalovirus (HCMV) congenital infection affects 1 in 150 pregnancies (1) and is the most frequent nongenetic cause of sensorineural hearing loss and neurodevelopmental delay in infants worldwide (2). Additionally, HCMV is the most common infectious agent among allograft recipients, often causing end-organ disease such as hepatitis, pneumonitis, or gastroenteritis and predisposing the individual to graft rejection (3). It has been estimated that an efficacious HCMV vaccine would save the United States 4 billion dollars and 70,000 quality-adjusted life years annually; thus, HCMV vaccine development has remained a tier 1 priority of the National Academy of Medicine for the past 17 years (4).
The glycoprotein B (gB) plus MF59 adjuvant vaccine is the most efficacious HCMV vaccine platform trialed to date, demonstrating partial vaccine protection in multiple patient populations. In a cohort of HCMV-seronegative postpartum women, gB vaccination achieved a promising 50% vaccine efficacy (5). When subsequently repeated in a cohort of seronegative adolescent women, a similar level of vaccine protection was identified, though the protection was not statistically significant (6). Furthermore, in allograft recipients, the same gB vaccine reduced the duration of HCMV viremia and antiviral therapy (7). There are many hypothesized reasons why this vaccine did not achieve a higher level of protection, including rapidly waning antibody titers, poor inhibition of epithelial cell entry, or a postfusion gB conformation. We and others have observed that the gB/MF59 vaccine platform was particularly poor at eliciting heterologous neutralizing antibodies in diverse populations and hypothesize that nonneutralizing antibody responses may have played a role in vaccine protection (8–10). Previously, we reported population-level virus sequencing of HCMV hypervariable genes in this cohort, revealing no differences in viral genetic identity between vaccinees and placebo recipients (11). However, a critical question in the field remains whether vaccination exerts immune pressure resulting in any distinction between viral populations acquired by gB/MF59 vaccinees and placebo recipients.
With a genome consisting of 236 kbp (12) and encoding approximately 164 genes (13), HCMV has the most genetic material of any human virus. It is well established that HCMV is highly polymorphic between and within individuals, defined via a variety of sequencing methodologies, including restriction fragment length polymorphism analysis (14), targeted gene sequencing (11, 15–19), and whole-genome sequencing (20–22). Yet, the source of this diversity remains poorly understood. If multiple unique viral variants are identified in a single individual (so-called “mixed infection”), does this represent de novo mutations, simultaneous initial infection with multiple unique variants, or independent sequential infection events? Mixed infections have been frequently detected in both chronically HCMV-infected individuals (23) and immunocompromised hosts (16–18). Yet, we previously described that recently seroconverted women from the gB/MF59 vaccine trial predominantly had a single virus strain detected in all tissues and at time points up to 3.5 years (11) when evaluated by a traditional Sanger sequencing methodology, suggesting that mixed infections in healthy individuals may result from independent sequential infection events. Of note, the pathological relevance of mixed infection (detected by PCR) is purely theoretical, as the minor variant in a mixed population of viruses has never been successfully isolated in tissue culture.
One major limitation of traditional HCMV genotyping to quantify HCMV diversity is a lack of sensitivity to detect viral variants present at low frequency. Recent HCMV whole-genome next-generation sequencing (NGS) has suggested that there is remarkable intrahost diversity, stemming from the presence of low-frequency alleles representing minor viral variants (20–22). Thus, it has been established that HCMV likely exists within individual hosts as a heterogeneous population of related viral variants (22). Subsequent characterization of the intrahost composition and distribution of low-frequency viral variants has led to a recognition of unique viral populations between individual organs, representing anatomic compartmentalization of viral populations (21).
If HCMV diversity is due to the presence of distinct low-frequency viral variants, traditional sequencing methodologies may not be the most sensitive means to discern differences between intrahost viral populations. Thus, we applied a previously validated (24) sequencing methodology and analysis pipeline termed short NGS amplicon population profiling (SNAPP) to investigate the viral populations of recently seroconverted gB/MF59 vaccinees and placebo recipients. This technique, which employs sequencing of an approximately 550-bp region at tremendous read depth, has facilitated a more complete understanding of in vivo viral dynamics. We hypothesize that gB/MF59 vaccination restricted viral replication and dissemination throughout the body. The study of HCMV population dynamics, including viral load, pairwise genetic diversity, number of unique haplotypes (viral variants), and the characteristics of those variants, may yield a more comprehensive understanding of the mechanism of partial vaccine efficacy.
(This article was submitted to an online preprint archive [25]).
RESULTS
Viral load, number of haplotypes, and sequence diversity by vaccine group.
We obtained HCMV DNA extracted from whole blood, saliva, urine, and vaginal fluid of 32 trial participants following HCMV primary infection. Of these 32 participants, 11 were gB/MF59 vaccinees and 21 were placebo recipients. Samples were collected approximately monthly, though the sampling timeline was heterogeneous between trial participants, resulting in a total of 31 samples for vaccinees (mean of 2.82 samples per participant) and a total of 58 samples for placebo recipients (mean of 2.76 samples per participant). Considering data from all anatomic compartments, viral loads ranged from undetectable to 4.7 log10 DNA copies/ml. Around the time of seroconversion, the mean viral load in vaccine recipients was 0.85 times that in placebo recipients (2.86 log10 versus 3.45 log10 viral load, respectively), although this difference was not statistically significant in a regression analysis that accounted for multiple samples per patient (95% confidence interval [CI], 0.23 times to 3.07 times; P = 0.792).
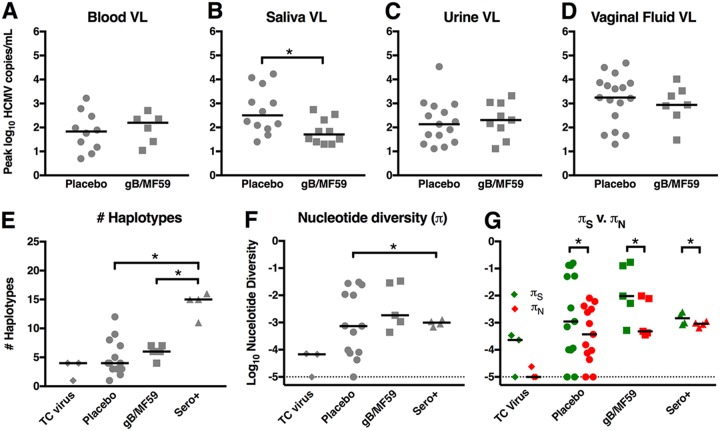
Peak viremia (Fig. 1A) and peak viral shedding in saliva, urine, and vaginal fluid (Fig. 1B to D) was identified for each patient and separated by vaccine group. The peak levels of viremia following primary HCMV acquisition were not significantly different between vaccinee and placebo recipients. Peak urine and vaginal fluid shedding were also not different between placebo and gB/MF59 vaccinees. However, saliva shedding was significantly higher in placebo recipients (P = 0.022, Friedman test plus post hoc exact Wilcoxon rank sum test), with median peak saliva shedding being 6.24 times higher than in gB vaccine recipients.
FIG 1.
Reduced peak saliva shedding, yet similar numbers of viral haplotypes and nucleotide diversity between HCMV-infected glycoprotein B vaccinees and placebo recipients. Peak plasma viral loads (A) as well as the peak magnitudes of virus shed in saliva (B), urine (C), and vaginal fluid (D) were compared between 11 gB vaccinees and 22 placebo recipients (samples not available from each compartment for each participant). Plasma (A), urine (C), and vaginal fluid (D) viral loads were not statistically different between HCMV-infected placebo recipients and gB/MF59 vaccinees, though there was reduced HCMV shedding in the saliva of vaccinees (B). Using SNAPP NGS data, the peak number of unique viral haplotypes (E) as well as peak nucleotide diversity (π) (F) were assessed for viral DNA amplified at the gB locus for 3 tissue culture virus isolates (TC virus), 13 placebo recipients, 5 gB/MF59 vaccinees, and 4 seropositive chronically HCMV-infected individuals (Sero+). (G) The magnitudes of nucleotide diversity resulting in synonymous (πS) versus nonsynonymous changes (πN) were compared. Horizontal bars indicate the median values for each group. *, P < 0.05 according to statistical tests: viral load, exact Wilcoxon rank sum test; haplotypes and π, Kruskal-Wallis test plus post hoc exact Wilcoxon rank sum test; πS versus πN, Wilcoxon signed-rank test.
Using a technique termed short NGS amplicon population profiling (SNAPP), approximately 550-bp variable gene regions encoding epitopes targeted by neutralizing antibodies were selected for gB and UL130 (membrane glycoprotein, component of pentameric complex). Gene regions were amplified by nested PCR and deep sequenced (see Fig. S1 in the supplemental material), and unique viral haplotypes were inferred by a modified SeekDeep analysis pipeline (26) as described in Materials and Methods. This pipeline is summarized graphically in Fig. S2. gB and UL130 haplotypes were inferred from a total of 14 placebo recipients and 6 gBF59 vaccinees following primary infection. Additionally, samples from 4 seropositive chronically HCMV-infected individuals were included for comparison (27). Sampling was heterogeneous, though frequently, samples representing multiple anatomic compartments and/or time points were sequenced per subject. DNA isolated from three tissue culture virus stocks were also amplified and sequenced as a genetically homogenous comparison. The peak numbers of viral haplotypes for each patient (including samples from all compartments) were similar between placebo and gB/MF59 vaccinees following primary HCMV infection at both the gB (Fig. 1E) and UL130 loci (see Fig. S3A). Interestingly, the number of gB viral haplotypes in chronically infected seropositive individuals was higher than in placebo recipients (P = 0.002, Kruskal-Wallis test plus post hoc exact Wilcoxon rank sum test) and in gB/MF59 vaccinees (P = 0.008, Kruskal-Wallis plus exact Wilcoxon rank sum test), with a median value noted to be 3.75 times and 2.5 times higher than in placebo and in gB/MF59 vaccinees, respectively. However, this trend was not evident at the UL130 locus.
The peak nucleotide diversity (π) for each patient was calculated for identified haplotypes at the gB (Fig. 1F) and UL130 loci (Fig. S3B). There was no statistical difference in gB π values between placebo recipients and vaccinees following primary HCMV infection, though seropositive individuals had 1.3-fold higher gB π in comparison to that of primary HCMV-infected placebo recipients (P = 0.011, Kruskal-Wallis test plus post hoc exact Wilcoxon rank sum test). π values attributable to synonymous mutations (πS) and nonsynonymous mutations (πN) were further compared within each group at the gB (Fig. 1G) and UL130 (Fig. S3B) loci. Again, there was no difference in πS or πN between infected placebo and vaccine recipients. However, gB πS significantly exceeded πN for primary HCMV-infected placebos (P = 0.004, Wilcoxon signed-rank test) and gB vaccinees (P = 0.001, Wilcoxon signed-rank test) as well as for seropositive chronically HCMV-infected individuals (P = 0.016, Wilcoxon signed-rank test), indicating that purifying selection was pervasive in the viral populations of each of these groups. Of note, the enhanced nucleotide diversity of seropositive individuals over that of acutely infected placebo recipients was not identified at the UL130 locus. Furthermore, at this locus, πS was also only significantly greater than πN in the gB/MF59 vaccinee subgroup (P = 0.006, Wilcoxon signed-rank test). Overall, these data suggest that the viral population in acutely infected vaccinees versus that in placebo recipients is similar though reduced compared to the viral population in seropositive chronically HCMV-infected individuals.
Viral load, number of HCMV haplotypes, and sequence diversity by anatomic compartment.
Viral load measured near the time of seroconversion was assessed for blood, saliva, urine, and vaginal fluid (see Table S1), as previously reported on for the placebo subset of patients (28). Samples were divided approximately equally between compartments: 22% from saliva, 27% from urine, 21% from vaginal fluid, and 29% from whole blood. By bivariate analysis, after accounting for multiple samples per patient, a regression analysis of log10 viral load on source indicated a statistically significant difference in viral copy number by anatomic compartment (P < 0.001), with vaginal virus shedding loads exceeding the viral loads in all other compartments. The mean viral load in vaginal swabs was 7.14 times that in oral swabs (95% CI, 1.05 to 50 times; P = 0.044), 14 times that in urine samples (95% CI, 33.3 to 100 times; P = 0.001), and 25 times that in whole blood samples (95% CI, 5.5 to 100 time; P < 0.001). Finally, we observed that the percentage of samples with detectable HCMV virus decreased over time in both gB vaccinees and placebo recipients. This decrease was observed in all body fluids, although levels in whole blood were already low around the time of seroconversion (see Table S2).
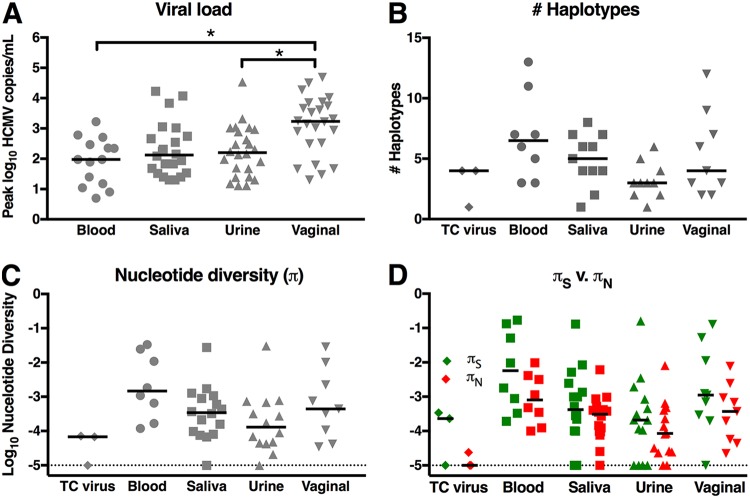
The peak HCMV loads for each patient in each anatomic compartment were compared (Fig. 2A). Vaginal HCMV shedding was of a higher magnitude than of HCMV viremia (P = 0.002, pairwise Wilcoxon signed-rank test), noted to be 17.95 times higher and 10.72 times higher than urine and saliva HCMV shedding loads, respectively (P = 0.001, pairwise Wilcoxon signed-rank test). There were no statistical differences in the peak numbers of viral haplotypes identified or peak nucleotide diversity between blood, saliva, urine, and vaginal fluid samples at either the gB (Fig. 2B and C) or UL130 (Fig. S3C and D) loci. Of note, the nucleotide diversity of HCMV in whole blood was 11.54-fold higher than that of HCMV shed in urine at the gB locus and 1.8-fold higher at the UL130 locus, which is consistent with previous observations (21) though not statistically significant. Finally, we observed that πs significantly exceeding πN in blood (P = 0.027, Wilcoxon signed-rank test), saliva (P = 0.008, Wilcoxon signed-rank test), urine (P = 0.011, Wilcoxon signed-rank test), and vaginal fluid (P = 0.010, Wilcoxon signed-rank test) at the gB locus (Fig. 2D), as well as in urine at the UL130 locus (P = 0.006, Wilcoxon signed-rank test) (Fig. S3D), again indicating purifying selection in these viral populations.
FIG 2.
High-magnitude viral shedding in vaginal fluid, yet similar numbers of unique viral variants and nucleotide diversity between anatomic compartments. Peak viral loads were compared between anatomic compartments for 11 gB/MF59 vaccinees, 22 placebo recipients, and 4 seropositive individuals (A), revealing high-magnitude HCMV shedding in vaginal fluid. Using SNAPP NGS data, the peak numbers of unique viral haplotypes (B) and peak nucleotide diversity (π) (C) were defined according to individuals for each anatomic compartment, resulting in data from 8 whole blood, 11 saliva, 10 urine, and 9 vaginal fluid samples from acutely infected gB vaccinees and placebo recipients as well as chronically HCMV-infected individuals. (G) The magnitudes of nucleotide diversity resulting in synonymous (πS) versus nonsynonymous changes (πN) were compared. Horizontal bars indicate the median values for each group. *, P < 0.05 according to statistical tests: viral load, Friedman test plus post hoc pairwise Wilcoxon signed-rank test; haplotypes and π, Kruskal-Wallis test plus post hoc exact Wilcoxon rank sum test; πS versus πN, Wilcoxon signed-rank test.
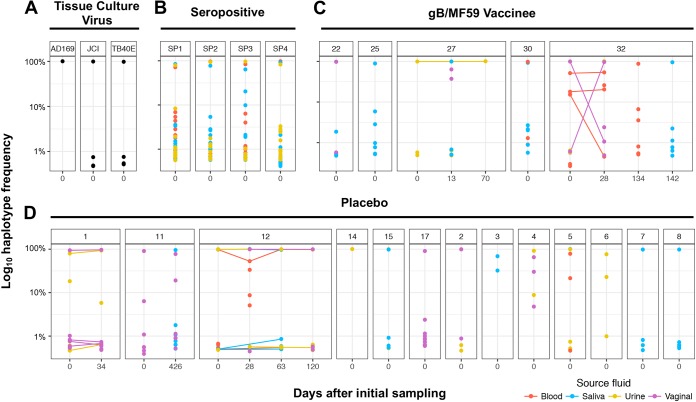
FIG 3.
Large numbers of low-frequency viral variants detected at gB locus in both primary HCMV-infected and chronically infected individuals. The relative frequency of each unique gB haplotype identified by SNAPP is displayed by individual patient and time point of sample collection. Tissue culture viruses (A) exhibited reduced population complexity by comparison. In primary HCMV-infected placebo recipients (C) and gB vaccinees (D), as well as chronically HCMV-infected women (B), there were typically one or more high-frequency haplotypes representing the dominant viral variants within the population, which were accompanied by haplotypes at very low frequency representing minor viral variants (<1% of viral haplotype prevalence). All haplotypes displayed exceeded the 0.44% threshold of PCR and sequencing error established for the SNAPP method (see Materials and Methods for detail).
Presence and persistence of low-frequency unique HCMV variants.
The relative frequency of unique viral haplotypes was identified for tissue culture virus stocks, chronically HCMV-infected individuals, and primary HCMV-infected gB/MF59 vaccinees and placebo recipients at the gB (Fig. 3) and UL130 (see Fig. S4) genetic loci. For all patients and at both loci, there was typically a single dominant viral variant, with a frequency approaching 100% (range, 54% to 99.6%). Along with this dominant variant, we found multiple low-frequency variants, often with a frequency of less than 1% of the population, that are genetically distinct. Some of these variants persist over time, suggesting that these identified variants are not simply a sequencing artifact. For example, longitudinal haplotypes identified in placebo recipients 1 and 12 indicate the persistence of both the dominant and low-frequency variants from one time point to the next.
Anatomic compartmentalization of HCMV populations in gB/MF59 vaccinees.
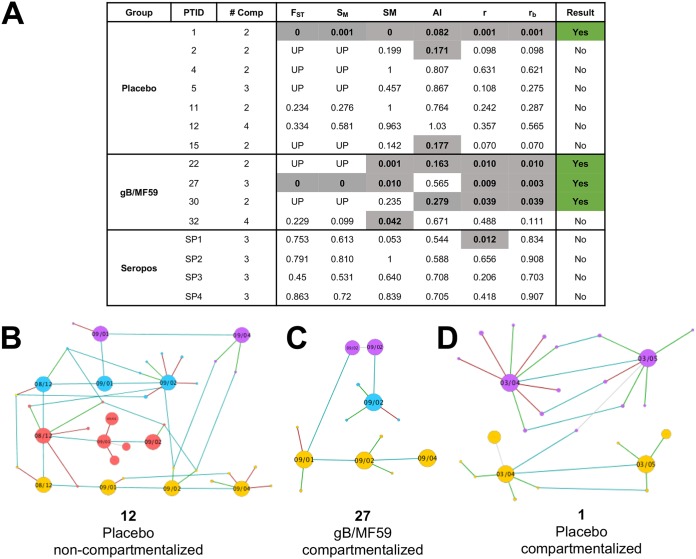
A panel of tests for genetic compartmentalization reliant upon 6 distinct distance- and tree-based methods was employed to assess the extent to which viral populations in different anatomic compartments of a single subject constitute distinct populations (29). We classified a viral population as compartmentalized when at least 3 of the 6 tests gave a statistically significant result. Given this definition, anatomic compartmentalization at the gB locus was observed for 1 of 7 placebo recipients, 3 of 4 gB vaccine recipients, and 0 of 4 seropositive chronically HCMV-infected individuals (Fig. 4A). These frequencies of genetic compartmentalization between placebo recipients and gB vaccinees approached statistical significance (P = 0.088, Fisher’s exact test), with increased compartmentalization in the vaccinee group. This same trend was not observed at the UL130 locus, as 2 of 9 placebo recipients, 1 of 4 gB vaccinees, and 2 of 3 seropositive individuals exhibited evidence of anatomic virus population compartmentalization (see Fig. S5). The pool of gB haplotypes for 3 representative individuals is shown chronologically and separated by anatomic compartment to demonstrate patients either lacking (Fig. 4B) or exhibiting (Fig. 4C and D) evidence of gB variant genetic compartmentalization.
FIG 4.
Evidence of viral genetic compartmentalization at gB locus in 3 of 4 gB vaccinees. (A) Table indicating the results of 6 distinct tests of genetic compartmentalization performed on the pool of unique gB haplotypes identified per patient, including Wright’s measure of population subdivision (FST), the nearest-neighbor statistic (Snn), the Slatkin-Maddison test (SM), the Simmonds association index (AI), and correlation coefficients based on distance between sequences (r) or number of phylogenetic tree branches (rb). For each test, >1,000 permutations were simulated. Significant test results suggesting genetic compartmentalization are shown in gray with bold text. Values for FST, Snn, SM, r, and rb represent uncorrected P values, with P < 0.05 considered significant. An AI of <0.3 was considered a significant result. The presence of three or more positive tests per patient was considered strong evidence for genetic compartmentalization, indicated in green. UP, underpowered (fewer than 5 haplotypes were present in each compartment, making FST and Snn error prone). (B to D) Networks of unique viral haplotypes by individual patient, with 1 patient lacking tissue compartmentalization (B) and 2 patients demonstrating strong evidence of viral genetic compartmentalization (D). Samples are organized chronologically from left to right, with blood shown in red, saliva in blue, urine in yellow, and vaginal fluid in purple. The size of each node reflects the relative prevalence of each haplotype. Light blue lines connect identical viral variants between time points and compartments, green lines connect variants with a synonymous mutation, and red lines those with a nonsynonymous mutation.
gB genotype analysis and force-of-infection modeling.
In addition to SNAPP, the full gB open reading frame (ORF) was amplified, fragmented, and sequenced by NGS to identify a gB consensus sequence for each unique sample (Fig. S1). Reassuringly, there was a high level of agreement of the gB genotype identified in primary HCMV-infected women between previously published Sanger sequencing data (11), full gB ORF NGS, and SNAPP (Table 1). As previously noted by Sanger sequencing, full gB ORF NGS indicated relatively few incidences of mixed infection between physiologic compartments or distinct time points. However, the SNAPP technique identified additional low-frequency viral variants corresponding to diverse gB genotypes, likely only discernible due to the enhanced sensitivity of this technique for low-frequency variants. Indeed, 11 of 18 (61%) trial participants had evidence of minor gB genotypes by SNAPP in comparison to 2 of 32 (6%) by Sanger sequencing. Yet, because the biologic significance of these low-frequency variants remains in question, we chose to proceed with genotype analysis based on more conventional Sanger sequencing and full gB ORF NGS.
TABLE 1.
Viral load and gB genotypes detected in clinical samples from placebo recipients and gB/MF59 vaccinees using different sequencing methodologies
| No. | Virus genotype |
Antibody titer |
Viral load by anatomic compartment (log10 range [n])a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sanger seq gB (subtype) | NGS full gB ORF | SNAPP gB | Endpoint gB (log10) | Neutralizing (log2) | Blood | Urine | Saliva | Vaginal fluid | |
| Placebo | |||||||||
| 1 | 4 (D2) | 4, 5 | 1, 4, 5 | 3.99 | 6 | —b | <2 (2) | — | 2.91–3.25 (2) |
| 2 | 3 (C1) | 3 | 1, 3 | 4.15 | 8 | — | 2.48 (1) | — | 4.50 (1) |
| 3 | 3 (C1) | 3 | 1, 3 | 4.27 | 8 | — | — | 3.05 (1) | |
| 4 | 1 (B2gaa) | 1 | 1, 5 | 4.40 | 8 | <2 (1) | <2–2.13 (2) | <2 (2) | <23.66 (2) |
| 5 | 2 (A1) | 2 | 1, 2, 5 | 3.24 | 3 | <2 (1) | <2 (1) | <2 (1) | 3.74 (1) |
| 6 | 3 (C1) | — | 1c | 4.06 | 7 | — | <2 (1) | — | — |
| 7 | 2 (A1) | — | 2 | 4.27 | 7 | — | 3.02 (1) | 2.25 (1) | — |
| 8 | 1 (B2gga) | 1 | 1 | 4.53 | 8 | <2 (3) | <2 (2) | <2–2.13 (2) | <2 (2) |
| 9 | 1 (B2gaa) | 1 | — | 3.68 | 6 | <2 (2) | <2 (2) | <2 (2) | <2–2.49 (2) |
| 10 | 1 (B2gaa) | 1 | — | 3.94 | 7 | <2 (2) | <2–2.63 (2) | <2–2.09 (2) | <2–3.87 (2) |
| 11 | 4, 1 (D2, B2gaa) | 4,1 | 4,1 | 4.29 | 4 | <2 (3) | <2 (2) | <2 (2) | <2 (2) |
| 12 | 1 (B2gga) | 1 | 1 | 4.56 | 7 | <2–2.78 (3) | 2.93–4.93 (4) | 3.15–4.07 (3) | 3.23–3.61 (2) |
| 13 | 1 (B1gga) | — | — | 3.00 | 1 | <2 (3) | <2 (2) | ||
| 14 | 1 (B2gaa) | 1 | 1 | 4.51 | 8 | <2 (2) | <2 (2) | <2 (2) | <2 (2) |
| 15 | 5 (E) | 5 | 1, 5 | 4.27 | 7 | — | <2 (3) | <2–4.23 (6) | <2 (1) |
| 16 | 5 (E) | — | — | 3.77 | 6 | — | <2 (1) | <2–3.83 (2) | 3.74–3.85 (2) |
| 17 | 5 (E) | 5 | 5 | 3.80 | 5 | <2 (2) | <2 (1) | 2.66 (1) | 4.69 (1) |
| 18 | 5 (E) | — | — | 2.30–3.22 (2) | 2.89 (1) | — | 3.15 (1) | ||
| 19 | 5 (E) | — | — | 3.87 | 3 | <2–2.47 (4) | 2.70–2.97 (2) | — | <2–4.27 (2) |
| 20 | 5 (E) | — | — | 4.21 | 8 | <2 (2) | 2.24 (1) | <2 (1) | 3.03 (1) |
| 21 | 5 (E) | — | — | 4.46 | 8 | <2 (1) | <2 (2) | <2–3.02 (2) | <2–3.22 (2) |
| gB/MF59 vaccinee | |||||||||
| 22 | 3 (C1) | 3 | 1, 3, 5 | 4.97 | 7 | — | 3.02 (1) | <2 (1) | 3.31 (1) |
| 23 | 5, 1 (E, B2gaa) | — | — | 4.99 | 6 | — | <2 (4) | <2 (5) | <2 (2) |
| 24 | 1 (B1gga) | — | — | 5.81 | 9 | <2 (1) | <2–2.47 (2) | <2 (2) | <2–2.90 (2) |
| 25 | 1 (B2gga) | 5c | 1 | 5.27 | 9 | <2 (2) | <2 (2) | <2–2.74 (2) | <2 (2) |
| 26 | 1 (B2gga) | — | — | 5.07 | 7 | <2 (2) | <2 (2) | <2–2.31 (2) | <2 (2) |
| 27 | 3 (C2) | 3 | 1, 3 | 5.59 | 10 | — | <2–2.16 (3) | <2–2.54 (2) | 2.52 (1) |
| 28 | 5 (E) | — | — | 5.15 | 8 | <2 (3) | 2.16–3.32 (2) | <2 (1) | <2–4.02 (2) |
| 29 | 5 (E) | — | — | 5.43 | 7 | — | 2.18–2.31 (2) | <2 (1) | — |
| 30 | 5 (E) | 5 | 1, 5 | 5.76 | 10 | <2–2.35 (2) | — | <2 (1) | — |
| 31 | 5 (E) | — | — | 4.75 | 8 | <2–2.71 (4) | <2 (2) | <2 (1) | <2–3.53 (2) |
| 32 | 5 (E) | 5 | 1, 5 | 6.52 | 10 | <2–2.34 (3) | <2–3.04 (2) | <2 (1) | <2–2.94 (3) |
Quantitative PCR limit of detection is 2 log10 copies/ml.
—, data omitted due to lack of available sample for given subject/anatomic compartment.
Sequence discrepant with Sanger sequencing. Result could not be confirmed due to sample limitation.
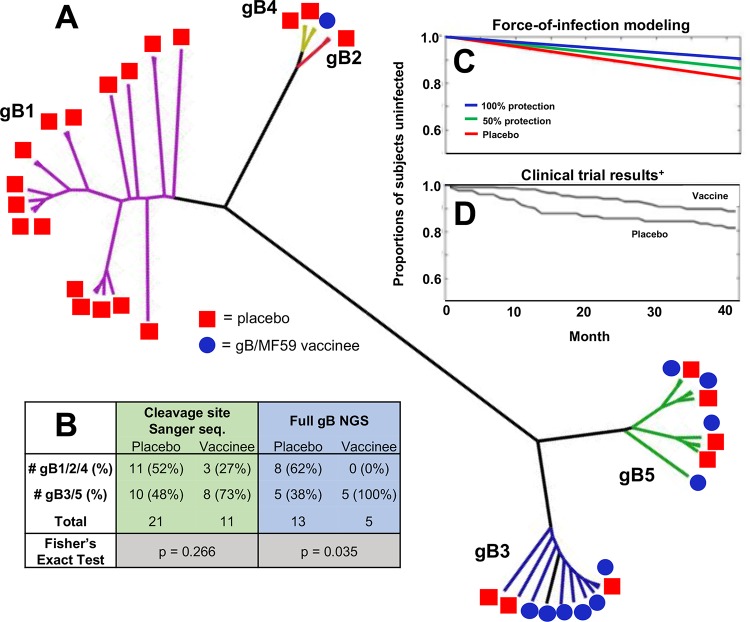
We previously reported no association between gB genotype and vaccination status using Sanger sequencing data (11). However, here we observed that gB/MF59 vaccinees relatively infrequently acquired gB1, gB2, and gB4 genotype viruses. Based on phylogenetic relatedness, we hypothesize that these 3 gB genotypes could be considered to comprise a gB1/2/4 genotype “supergroup” (Fig. 5A). Indeed, full gB ORF NGS data indicate that 8 of 13 placebo recipients acquired a gB1/2/4 virus compared with 0 of 5 vaccinees (Fig. 5B), which was a statistically significant result (P = 0.035, Fisher’s exact test). Sanger sequencing data show a similar trend with 11 of 21 placebo recipients acquiring a gB1/2/4 virus compared with 3 of 11 vaccinees, though this difference was not significant (P = 0.266, Fisher’s exact test).
FIG 5.
Possible protection against gB1/2/4 genotype supergroup viruses in gB/MF59 vaccinees. (A) Unrooted phylogenetic tree in a polar layout constructed using full gB open reading frame consensus sequences for each sequenced sample. Note, a consensus sequence was not defined for all samples from each subject, so this tree depicts multiple samples representing multiple time points/compartments for each trial participant. Samples with discrepant genotypes from Sanger sequencing not excluded. Clades representing gB genotypes are highlighted in different colors: gB1, purple; gB2, yellow; gB3, blue; gB4, red; gB5, green. (B) Numbers of distinct gB vaccinees and placebo recipients who acquired viral variants belonging to two supergroups of genetically similar gB genotypes (gB1/2/4 and gB 3/5) as defined by Sanger sequencing of the cleavage site (highlighted in green) and NGS of the full gB protein (highlighted in blue). (C) Force-of-infection modeling closely predicts observed gB/MF59 vaccine trial efficacy (D). Force-of-infection model iterations indicate that gB vaccinees are universally protected against acquisition of gB1/2/4 variants (blue line; consistent with full gB NGS data) or 50% protected (green line; consistent with Sanger sequencing data). Model makes assumption that (i) viruses belonging to the gB1/2/4 genotype supergroup represent 52% of the circulating virus pool and (ii) the HCMV force of infection is 5.7 per 100 person-years. (Panel D adapted from The New England Journal of Medicine [5].)
We sought to investigate whether vaccine-elicited protection against gB1/2/4 genotype viruses could have explained the partial vaccine efficacy observed in the gB/MF59 clinical trial (5) by modeling the HCMV force of infection (Fig. 5C). Assuming that the gB1/2/4 genotype supergroup comprises 52% of the circulating virus pool (11 of 21 acquired viruses among placebo recipients), we modeled the theoretical protection against gB1/2/4 variants assuming that vaccinees were either universally protected (blue line) or 50% protected (green line) in comparison to placebo recipients (red line). If we assume that vaccinees were ∼50% protected against gB1/2/4 variants (reflecting Sanger sequencing data, the most complete data set), the modeled vaccine efficacy is 26%. Thus, the force-of-infection model results are consistent with those observed in the clinical trial (5) but do not fully explain the observed 50% vaccine protection.
Viral load correlation to gB-binding antibodies, HCMV neutralization, and HCMV virus genotype.
A regression analysis was performed to assess a possible correlation between viral load and the magnitude of gB-binding or HCMV-neutralizing antibodies elicited by vaccination. For both binding and neutralization, separate analyses were performed using (i) all measured viral load values (see Fig. S7A and B) or (ii) only the highest viral load value in each participant (Fig. S7C and D). No statistically significant correlation between antibody responses and viral load was identified for either the highest measured viral load (gB antibodies: R2 = 0.0428, P = 0.26; neutralizing antibodies: R2 = 0.0035, P = 0.75) or when all viral load measurements were included (gB antibodies: R2 = 0.0035, P = 0.75; neutralizing antibodies: R2 = 0.0009, P = 0.71).
Additionally, we investigated the relationship between viral load and gB genotype in 30 women using Sanger sequencing-assigned gB genotypes (most complete data set). Two participants that did not have viral load measurments during seroconversion were excluded. Relatively high viral loads were observed among participants infected with gB5 genotype viruses in both vaccine and placebo recipients. To investigate further, we dichotomized genotype as gB5 versus all other gB genotypes and then performed a linear regression of log10 viral load on genotype. Around the time of seroconversion, a statistically significant association was found between log10 viral load and genotype, with the mean viral load in women with gB5 genotype viruses being 3.44 times that in women shedding non-gB5 genotype viruses (95% CI, 1.13 to 10.51; P = 0.031) (see Fig. S8). When considering all visits, this association was no longer statistically significant; however, this is perhaps a result of the decrease in viral load over time observed in all HCMV-infected participants.
DISCUSSION
Despite the partial efficacy demonstrated by gB/MF59 vaccination in multiple clinical trials (5–7), there has been little examination of the impact of this vaccine on the in vivo viral populations. In this investigation, we sought to employ highly sensitive, modern molecular approaches, including quantitative PCR and next-generation sequencing (NGS) to delve deeper into the question of whether there are discernible differences between viral populations acquired by gB/MF59 vaccinees and placebo recipients. Throughout the study, we identified several indications of vaccine-mediated effects on the viral population, including reduced viral shedding in the saliva of gB vaccinees, possible anatomic compartmentalization of gB variants in vaccinees, and a trend toward reduced acquisition of viruses with antigenic similarity to the vaccine construct among gB/MF59 vaccinees. Altogether, these findings suggest an impact of gB immunization on the intrahost viral population.
We identified that peak HCMV shedding in saliva was reduced by an order of magnitude in gB/MF59 vaccinees compared to that in those receiving placebo, though no difference was observed for peak systemic viral load or peak viral shedding in urine and vaginal fluid. This finding suggests that gB-elicited antibodies may have limited viral replication in salivary glands. gB/MF59 vaccination is known to elicit high titers of gB-specific IgG, IgA, and secretory IgA (S-IgA) in parotid saliva (30), which may have suppressed HCMV salivary replication and reduced saliva viral shedding. Of note, we also observed a trend toward reduced viral load in vaccinees when considering samples from all time points or those solely from the time point of seroconversion, though these comparisons were not statistically significant. Analogously, serum antibody gB binding and HCMV neutralization did not correlate with viral load at the peak or at the time of seroconversion. These data suggest altogether that gB/MF59 vaccination had a minimal quantifiable impact on in vivo HCMV replication. Indeed, our observation that gB5 genotype viruses were associated with increased viral load further suggests that host response may have a lesser impact on viral replication during acute infection than strain identity and viral pathogenicity. This phenomenon might be explained by the highly cell-associated nature of HCMV, reducing the likelihood of virion contact with circulating gB-specific antibodies. When shed in saliva, however, cell-free HCMV virions could be targeted by effector immunoglobulins.
A second possible impact of vaccination on the viral population was anatomic compartmentalization. We found that 3 of 4 gB vaccinees with viral DNA available from multiple compartments exhibited evidence of viral genetic compartmentalization at the gB locus, in contrast to only 1 of 7 placebo recipients. As previously described (11), we observed that the dominant viral haplotypes were identical between anatomic compartments in the majority of subjects. The evaluation of gB-specific compartmentalization was therefore only possible because of the ability of short NGS amplicon population profiling (SNAPP) to detect minor viral variants in clinical samples. Our data are consistent with HCMV whole-genome NGS indicating tissue-specific variants with intrahost variable single nucleotide polymorphisms (SNPs) present at a relatively low frequency (20, 21). The mechanism leading to compartmentalization in vaccinees is unclear, though it is possible this phenomenon stems from either neutral or positive selection in distinct anatomic compartments. Previously, the UL55 ORF was identified as having a high rate of plasma-associated SNPs, perhaps suggesting a role of gB variation in tissue tropism (22). One hypothesis for the observed genetic compartmentalization is that systemic gB-specific antibodies limited the dissemination of HCMV viruses between tissue compartments. Such a bottleneck might have reduced founder population size and increased the speed and likelihood of stochastic fixation of neutral SNPs and the formation of genetically distinct viral populations (31, 32). Alternatively, it is possible that local factors, including cell type and local antibody production/secretion at the site of HCMV replication, selected for “more fit” viral variants in each compartment. Yet, it remains puzzling why genetic compartmentalization is not detectable at the UL130 locus. Possible explanations include recombination between UL55 and UL130 loci and that in vivo diversity at the UL130 locus is low enough that we fail to identify compartmentalization given the genetic homogeneity.
Lastly, we observed an interesting trend that gB/MF59 vaccine recipients had reduced acquisition of gB1/2/4 genotype supergroup viruses. This finding was statistically significant when considering the genotypes identified by full gB ORF NGS, though merely a trend when considering a larger data set of the genotypes previously defined by Sanger sequencing (Fig. 5, Table 1) (11). Of note, the gB immunogen in this vaccine trial was based on the Towne strain (gB1 genotype prototypic sequence), suggesting the possibility of vaccine-mediated protection against viruses with genetic/antigenic similarity to the vaccine strain. As demonstrated by force-of-infection modeling, gB1/2/4 supergroup-specific protection partially explains the 50% vaccine efficacy observed in this phase 2 clinical trial. The concept of neutralization breadth has not been explored extensively for HCMV, though several studies have described strain-specific neutralization (33–35). It is important to place this finding in context. Low-frequency gB1/2/4 haplotypes were detectable in additional vaccinees by SNAPP (Table 1), suggesting there may not have been a true barrier to gB1/2/4 acquisition but perhaps merely that gB1/2/4 virus replication was restricted. Furthermore, since the sample size is not robust, it is possible that our observation may have been the result of sampling bias. Yet, despite the small sample size and statistically discrepant results, we believe it is compelling that these independent sequencing efforts completed nearly a decade apart and using diverse methods both indicate a similar trend toward reduced acquisition of viruses with an antigenically similar gB sequence to the vaccine strain among gB/MF59 vaccine recipients. While certainly not a definitive finding, our data raise the question of whether a genetically homogenous gB protein vaccine was simply insufficient to protect against antigenically diverse HCMV strains. Perhaps antigenic breadth should be a consideration for rational design of the next generation of HCMV vaccines.
One advantage of NGS over more traditional sequencing methodologies certainly is the ability to detect minor viral variants, which contribute to the diversity of the overall viral population (Fig. 3; see also Fig. S4 in the supplemental material). We discovered that numerous minor viral haplotypes, exceeding the threshold of PCR and sequencing error, were detectable in nearly all clinical samples tested though not in HCMV clinical and laboratory strains passaged in tissue culture. These results are consistent with HCMV whole-genome sequencing investigations that have suggested numerous genetic variants at <1% frequency in the viral population (20–22). Interestingly, seropositive women reliably had more gB haplotypes (Fig. 1E) than acutely infected vaccinated subjects, indicating a higher number of genetically distinct viral variants in seropositive chronically HCMV-infected individuals. This observation complements previous work demonstrating that recently seroconverted young women have very low incidence of mixed infection yet that multiple gB genotypes are almost universally detectable in chronically HCMV-infected individuals (23). Altogether, these data favor a model that mixed infections in healthy individuals result from independent sequential infection events. However, despite the enhanced sensitivity of NGS, this sequencing-based approach to profile viral populations cannot definitively distinguish superinfection.
Far and away, the largest limitation of this study was study design and sample availability. As a post hoc analysis of a vaccine trial in which none of the investigations were protocol specified, the findings must be interpreted cautiously. The scope of our investigation was restricted by (i) the original sampling timeline employed during the clinical trial, (ii) the availability of clinical samples, and (iii) the integrity of the DNA more than a decade following DNA extraction. Another limitation is that total cellular DNA was not quantified; therefore, viral loads could not be corrected using genomic DNA for normalization. Thus, reported saliva and vaginal fluid HCMV DNA viral load values do not represent absolute quantification but rather an approximation due to collection and processing methods. Viral loads between salivary and vaginal swabs were comparable, since they were both collected and processed in a similar manner, and may be lower than HCMV detected in whole blood and original urine fluids. Finally, as with any study based on PCR amplification and DNA sequencing, there is a potential for primer bias, contamination, and background error to obscure the results. We instituted several measures to increase data integrity. First, primers were designed and validated to prevent amplification bias (24). Additionally, PCR, sequencing, and analysis were completed in duplicates to reduce the likelihood of contamination affecting the results. An advantage of this investigation is that we were able to validate our two sequencing methodologies (SNAPP and full gB ORF NGS) by comparing observed gB genotypes with previously published data (11). The gB genotypes predicted by Sanger sequencing and NGS sequencing methodologies were identical for 88% of all samples. However, because of the relatively small cohort size and potential for sequencing error, our observed trends certainly merit further investigation.
Nonetheless, this investigation is the first to employ NGS of viral DNA from infected HCMV vaccinees and placebo recipients in an attempt to characterize the viral determinants of HCMV acquisition. Importantly, our data support notions in the HCMV field that the in vivo viral population has a level of genetic complexity that cannot be appreciated using conventional sequencing methodologies (20–22), though the biological relevance of this genetic diversity remains to be determined. Furthermore, our observation of reduced saliva shedding and a high rate of gB sequence compartmentalization in vaccinees suggests a definable impact of gB-elicited antibodies on viral population dynamics. Finally, the possible vaccine-elicited protection against gB1/2/4 supergroup viruses observed in this study is intriguing, implying that that antigen-specific immune responses may have played a role in vaccine protection. These findings, based on a small number of samples, are hypothesis generating and should be investigated in the future using a larger cohort to identify viral determinants and/or surrogate markers of vaccine protection. An enhanced understanding of the impact of anti-HCMV antibody-mediated immunity on viral populations could be harnessed for rational design of next-generation constructs to improve upon the partial efficacy of gB/MF59 vaccination.
MATERIALS AND METHODS
Study population.
The study population comprised 32 postpartum women who acquired HCMV infection while participating in a phase 2, randomized, double-blind, placebo-controlled clinical trial of an HCMV vaccine (5). Twenty-one women received placebo, while 11 received the gB/MF59 vaccine. Clinical trial participants were HCMV-seronegative, healthy postpartum women immunized with gB protein (based on Towne strain, gB1 genotype; Sanofi Pasteur) plus MF59 squalene adjuvant (Novartis) or placebo. Subjects were vaccinated on a 0-, 1-, and 6-month schedule and were screened for HCMV infection every three months for 3.5 years by using an antibody assay which detects seroconversion to HCMV proteins other than gB (36). Subjects with serologic evidence of infection were tested for HCMV in blood, urine, saliva, and vaginal swabs from 1 month to 3.5 years after seroconversion. Samples were collected from 30 women around the time of acute seroconversion (21 placebo and 9 vaccine recipients). Across all patients and time points, a total of 201 samples (oral, n = 50; urine, n = 58; vaginal, n = 45; and whole blood, n = 48) were collected during visits ranging from 1 to 36 months. The total numbers of samples per patient ranged from 1 to 12 (median, 7). While a maximum of 7 oral samples were available per patient, the maximum numbers of urine, vaginal, and whole blood samples were 4, 3, and 4, respectively. Aliquots of each specimen were stored at −80°C. A subset of samples was utilized for viral genetic analysis, dependent upon quantity and quality of the DNA remaining after extended freezer storage. Institutional review board (IRB) approval was obtained from University of Alabama at Birmingham and Johns Hopkins Hospital, and all subjects signed approved consent forms. The Duke University Health System determined that analysis of deidentified samples from these cohorts does not constitute human subjects research.
Tissue culture and virus growth.
Human retinal pigment epithelial (ARPE-19) cells (ATCC) were maintained for a maximum of 35 passages in Dulbecco’s modified Eagle medium-12 (DMEM-F12) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 1 mM sodium pyruvate, 50 U/ml penicillin, 50 μg/ml streptomycin and gentamicin, and 1% epithelial growth cell supplement (ScienCell). Human lung (MRC-5) fibroblasts (ATCC) were maintained for a maximum of 20 passages in DMEM containing 20% FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cell lines were tested for the presence of mycoplasma biannually. AD169 revertant virus (AD169r; a gift from Merck), as described previously (37), TB40/E-mCherry (a gift from N. Moorman) (38), and an HCMV clinical isolate from infant urine virus stocks were propagated with either MRC-5 or ARPE-19 cells in T75 culture flasks. Supernatant containing cell-free virus was collected when 90% of cells showed cytopathic effects and then was cleared of cell debris by low-speed centrifugation before passage through a 0.45-μm filter. Passage (P) histories for viral stocks are as follows: TB40/E, P3 MRC-5 and P7 ARPE; AD169r, P6 ARPE; and the clinical isolate, P4 MRC-5.
DNA extraction and quantitative PCR.
HCMV DNA was extracted from 400 μl of original samples (blood, urine, saliva, or vaginal swabs in culture medium) using the MagAttract Virus Mini M48 kit (Qiagen) on the BioRobot M48 instrument. The quantitative PCR assay is based on amplification of a 151-bp region from the US17 gene (28, 39). As previously reported, the limit of detection is 100 copies/ml (4 copies/reaction), with a measurable range of 2.0 to 8.0 log10 copies per ml. Total cellular DNA was not measured; thus, viral loads could not be corrected using genomic DNA for normalization. Thus, reported saliva and vaginal fluid DNA are an estimation only rather than an absolute concentration.
Antibody assays.
IgG to HCMV gB was measured using an enzyme-linked immunosorbent assay (ELISA) method. The vaccine antigen, a recombinant HCMV gB molecule (based on Towne strain, gB1 genotype; Sanofi Pasteur), was used to coat 96-well plates. Eight serial dilutions starting at 1:200 were tested. A horseradish peroxidase (HRP)-conjugated goat anti-human IgG (1:50,000 in HRP stabilizer; KPL) was used as the detection antibody, followed by development using 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate solution (TMB 2-component microwell peroxidase substrate kit; KPL). Endpoints were calculated based on the intersection between the straight line through the falling portion of the optical density-serum dilution curve and the cutoff optical density below which sera were negative for antibodies to gB. Neutralizing antibodies were measured using a plaque reduction assay with HCMV AD169 (ATCC CCL-171) and MRC-5 (ATCC VR-538) cells in 24-well tissue culture plates. Duplicate 2-fold dilutions of sera were tested starting at 1:8. Plaque counts were performed after a 7-day incubation, and the highest serum dilution with ≥50% reduction in plaque count was considered the endpoint titer.
Short NGS amplicon population profiling.
A flow chart detailing the sequencing strategy is shown in Fig. S1 in the supplemental material. Variable gene regions (∼550 bp in length), encoding epitopes targeted by neutralizing antibodies and flanked by conserved sequences, were identified for gB (UL55) and UL130. UL130 was selected as a control sequencing locus, as this protein is a target of potently neutralizing antibodies though was not included in the gB/MF59 vaccine. The gB amplicon included AD-4 and AD-5 epitopes (40), while the UL130 amplicon contained the majority of the coding sequence. gB and UL130 amplicons were amplified in duplicates by a nested PCR using the primers denoted in Table S3. Overhang regions were conjugated to PCR2 primers for subsequent Illumina index primer addition and sequencing: forward primer overhang, 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[locus]-3′; and reverse primer overhang, 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[locus]-3′. Template DNA extracted from primary fluids was added to 50 μl of a 1× PCR mixture containing 100 nM each primer, 2 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate (dNTP) mix (Qiagen), and 0.2 U/μl Platinum Taq polymerase. PCRs consisted of an initial 2-min denaturation at 98°C, followed by 35 PCR cycles (98°C for 10 s, 65°C for 30 s, and 72°C for 30 s) and a final 72°C extension for 10 min. Following each amplification step, products were purified using Agencourt AMPure XP beads (Beckman Coulter). Illumina Nextera XT index primers were added by 15 cycles of amplification. The indexed PCR product was run on a 1% agarose gel and then gel purified using a ZR-96 Zymoclean gel DNA recovery kit (Zymogen). The molar amount of each sample was normalized by real-time PCR using the KAPA library amplification kit (KAPA Biosystems). The library of individual amplicons was pooled, diluted to an end concentration of 14 pM, combined with 20% V3 PhiX (Illumina), and then sequenced by Illumina MiSeq using a 600-cycle V3 cartridge (Illumina).
Full glycoprotein B open reading frame PCR and sequencing.
A flow chart denoting the sequencing strategy is shown in Fig. S1. The full gB open reading frame (ORF) was amplified by nested PCR using the primers denoted in Table S3. Template DNA extracted from primary fluids was added to 50 μl of a 1× PCR mixture containing 100 nM each primer, 2 mM MgCl2, 200 μM (each) dNTP mix (Qiagen), and 0.2 U/μl Platinum Taq polymerase. PCRs consisted of an initial 2-min denaturation at 98°C, followed by 35 PCR cycles (98°C for 10 s, 65°C for 30 s, and 72°C for 3 min) and a final 72°C extension for 10 min. Following each amplification step, products were purified using Agencourt AMPure XP beads (Beckman Coulter). The PCR2 product was run on a 1% agarose gel and then gel extracted using the ZR-96 Zymoclean gel DNA recovery kit (Zymogen). Purified product was tagmented using the Nextera XT library prep kit (Illumina). Subsequently, Nextera XT index primers were added to the tagmented DNA by 15 cycles of amplification. The molar amount of each sample was normalized by real-time PCR using the KAPA library amplification kit (KAPA Biosystems). The library of individual amplicons was pooled, diluted to an end concentration of 14 pM, combined with 20% V3 PhiX (Illumina), and then sequenced by Illumina MiSeq using a 600-cycle V3 cartridge (Illumina).
Sanger sequencing.
An approximately 350-bp region of UL55 containing the furin cleavage site was sequenced as previously described (11).
SNAPP haplotype reconstruction and nucleotide diversity.
A data processing flow chart is shown in Fig. S2. First, raw paired-end reads were merged using the PEAR software under default parameters (41). The fused reads were then filtered using the extractor tool from the SeekDeep pipeline (http://baileylab.umassmed.edu/SeekDeep) (26), filtering sequences according to their length, overall quality scores, and presence of primer sequences. All filtered sequencing reads were included for subsequent haplotype reconstruction using the qluster tool from SeekDeep. This software accounts for possible sequencing errors by collapsing fragments with mismatches at low-quality positions. For each given sample, haplotypes had to be present in both of 2 sample replicates to be confirmed. On average, concordance between the replicates was quite high as assessed by linear regression correlation and slope of the relative frequencies of each haplotype (Fig. S6). Each gB haplotype was assigned to 1 of 5 described gB genotypes by assessing the shortest genetic distance (nucleotide substitutions) between the haplotype and reference gB genotype sequences (42). Nucleotide diversity (π) was computed as the average distance between each possible pair of sequences (43)
where L is the sequence length in nucleotides for π, N is the total number of reads in the sample, dij is the number of nucleotide differences between haplotypes i and j, and fi is the number of reads belonging to haplotype i. πN and πS were calculated as the average dS and dN between pairs of haplotypes weighted by the haplotype abundance:
where L is the sequence length in amino acids for πN and πS, N is the total number of reads in the sample, dSij is dS between haplotype i and j sequences, and fi is the number of reads belonging to haplotype i. Correlations were performed between various measures of viral population diversity (viral load, number of haplotypes, π, πS, and πN), and suggest that haplotypes, π, πS, and πN are somewhat related measures although are not directly equivalent (see Fig. S9).
Assessment of anatomic compartmentalization of virus populations.
A panel of tests using diverse analytical methods is hypothesized to be the most accurate means to infer tissue compartmentalization (29). Thus, we selected six tests employing both distance-based and tree-based algorithms. Wright’s measure of population subdivision (FST, distance based) compares the mean pairwise genetic distance between sequences from the same compartment to that of sequences from the same compartment (44). The nearest-neighbor statistic (Snn, distance based) measures how frequently the nearest neighbor to each sequence is in the same or different compartments (45). The Slatkin-Maddison test (SM, tree based) calculates the minimum number of migration events between compartments compared to the number of migration events expected in a randomly distributed population (46). The Simmonds association index (AI, tree based) examines the complexity of the phylogenetic tree structure (47). Finally, correlation coefficients (r and rb, tree based) correlate distances between sequences in a phylogenetic tree with the compartment of origin based either on the distance between sequences (r) or on the number of tree branches between sequences (rb). Distance-based tests used the TN93 distance matrix (48), and tree-based methods employed a neighbor-joining algorithm. Distance-based tests were not conducted for patients with fewer than 5 haplotypes per compartment, since this is known to produce unreliable results (29). All tests were conducted using HyPhy software (https://veg.github.io/hyphy-site) (version 2.22), with test statistics estimated from ≥100 permutations. For each test, a P value of <0.05 or an association index of <0.3 was considered statistically significant. The presence of three or more positive test results from these six test statistics was considered strong evidence for compartmentalization.
Phylogenetic trees and genotype assignment.
Nucleotide sequences of interest were aligned using the ClustalW algorithm (49) in Mega (version 6.06) (50). A neighbor-joining tree was constructed using the Los Alamos National Labs “neighbor treemaker” (https://www.hiv.lanl.gov/components/sequence/HIV/treemaker/treemaker.html) using the Felsenstein 84 distance method, and then the tree was plotted in FigTree (version 1.4.3). Full gB ORF sequences were assigned to gB genotypes based on phylogenetic proximity to reference gB sequences (42). To depict the breadth of diversity of all sequenced samples, a consensus sequence was derived for each sample with multiple time points/compartments per subject. Furthermore, samples were not excluded if the full gB ORF NGS-assigned genotype did not match the genotype assigned to SNAPP amplicons and/or previously published Sanger sequencing data (11). Samples were assigned to gB1/2/4 and gB3/5 supergroups based on phylogenetic proximity to reference gB sequences (see above). To reduce the chance of PCR contamination influencing the results, in the setting of mixed infection, an individual was only assigned to the gB1/2/4 genotype supergroup if all sequenced viruses belonged to this supergroup.
Force-of-infection modeling.
The cohort of women in the postpartum gB/MF59 vaccine trial were predominantly African-American (>70%) (5); thus, we utilized an HCMV force-of-infection estimate for non-Hispanic, African-American individuals of 5.7 per 100 persons (51). Additionally, we assumed that gB1/2/4 genotype supergroup viruses comprise 52% of the circulating virus pool, based on 11 of 21 placebo recipients acquiring gB1/2/4 viruses. Finally, we hypothesized that gB1-vaccinated individuals were universally protected against all circulating gB1/2/4 supergroup genotype viruses. Modeling was performed using Matlab, and the source code is included in the supplemental material.
Statistical analysis.
Descriptive displays and statistics were used to examine the patterns in viral load measurements by vaccination status (vaccine or placebo), source of measurement (type of body fluid), and gB genotype. Although there were multiple visits and measurements for study participants over time, the analyses of viral load distribution were initially restricted to samples collected at the first clinic visit following HCMV seroconversion. Analyses were performed to compare viral loads in vaccine versus placebo recipients, to determine whether viral copy numbers were different between body fluids, and whether specific genotypes were associated with higher viral load. A log10 transformation of viral load measurement was performed, and a value of 1 was added to each zero measurement (nondetectable) prior to transformation. Simple linear regression of log10 viral load was performed to assess the bivariate relationship between HCMV copy number and fluid type or genotype. The regression analysis accounted for the multiple fluid samples obtained from the same patient at the same visit by clustering and calculating robust standard errors to correlate observations within a single patient. To assess trends in viral load over time, a second analysis was performed using samples collected from all visits in all participants with measurements starting from the time of seroconversion. This descriptive analysis was performed to assess the kinetics of viral load over time in the different body fluids. Data were analyzed using STATA v10.
Additionally, in a separate analysis, the peak viral load was defined for each patient and zero-value measurements were excluded. Comparisons of peak viral load data between placebo and vaccine groups were performed using exact Wilcoxon rank sum tests. Comparisons of peak viral load between tissue compartments were performed using Friedman tests followed by pairwise Wilcoxon signed-rank tests. Comparisons of peak haplotype numbers and peak nucleotide diversity data between groups were performed using Kruskal-Wallis tests followed by exact Wilcoxon rank sum tests. Comparisons of peak haplotypes and peak nucleotide diversity data between tissue compartments were performed using pairwise Wilcoxon signed-rank tests. Comparisons of πS and πN within the same tissue compartment were performed using Wilcoxon signed-rank tests. The associations among different assays were calculated using Kendall’s tau. The Benjamini-Hochberg false-discovery rate (FDR) P value correction was used to correct for multiple comparisons (52). A P value of <0.05 (2 tailed) was considered significant for all analyses. All statistical tests were completed using SAS v9.4.
Data availability.
The full gB ORF and SNAPP sequence data used in the manuscript have been uploaded to GenBank under accession numbers MK157426 to MK158058. All other source data are available upon request.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Duke Viral Genetic Analysis core for assistance with sample preparation and sequencing. We also thank Nicholas Hathaway for the development of the SeekDeep analysis pipeline employed for haplotype analysis.
The gB/MF59 vaccine clinical trial was supported by grants to the University of Alabama at Birmingham from NIH/NIAID (P01AI043681 and U01AI063565) and by Sanofi Pasteur. This work was supported by an NIH/NICHD Director’s New Innovator grant to S.R.P (DP2HD075699), an NIH/NIAID R21 grant to S.R.P. (R21AI136556), an NIH/NIAID R08 grant to R.A.-B. (R08AI074907), and an NIH/NICHD F30 grant to C.S.N (F30HD089577).
The funders had no role in study design, data collection and interpretation, decision to publish, or the preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
C.S.N., R.A.-B., and S.R.P. designed the research; C.S.N., M.S., and M.F. performed the research; C.S.N., D.V.C, K.K., M.D.-W., and R.A.-B. analyzed the data; G.X., N.V., and M.D.-W. performed the statistical analysis of data; R.F.P, K.K., and R.A.-B. contributed samples and expertise; and C.S.N., R.A.-B., and S.R.P. wrote the paper.
S.R.P. provides consulting services to Pfizer Inc., Merck, and Moderna for their preclinical HCMV vaccine programs. R.F.P. is on the scientific advisory board for VBI vaccines. The other authors have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01695-18.
REFERENCES
- 1.Dollard SC, Grosse SD, Ross DS. 2007. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev Med Virol 17:355–363. doi: 10.1002/rmv.544. [DOI] [PubMed] [Google Scholar]
- 2.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramanan P, Razonable RR. 2013. Cytomegalovirus infections in solid organ transplantation: a review. Infect Chemother 45:260–271. doi: 10.3947/ic.2013.45.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratton KR, Durch JS, Lawrence RS (ed). 2000. Vaccines for the 21st century: a tool for decisionmaking. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 5.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, Corey L, Hill J, Davis E, Flanigan C, Cloud G. 2009. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 360:1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein DI, Munoz FM, Callahan ST, Rupp R, Wootton SH, Edwards KM, Turley CB, Stanberry LR, Patel SM, McNeal MM, Pichon S, Amegashie C, Bellamy AR. 2016. Safety and efficacy of a cytomegalovirus glycoprotein B (gB) vaccine in adolescent girls: a randomized clinical trial. Vaccine 34:313–319. doi: 10.1016/j.vaccine.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths PD, Stanton A, McCarrell E, Smith C, Osman M, Harber M, Davenport A, Jones G, Wheeler DC, O'Beirne J, Thorburn D, Patch D, Atkinson CE, Pichon S, Sweny P, Lanzman M, Woodford E, Rothwell E, Old N, Kinyanjui R, Haque T, Atabani S, Luck S, Prideaux S, Milne RS, Emery VC, Burroughs AK. 2011. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet 377:1256–1263. doi: 10.1016/S0140-6736(11)60136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson CS, Huffman T, Jenks JA, Cisneros de la Rosa E, Xie G, Vandergrift N, Pass RF, Pollara J, Permar SR. 2018. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc Natl Acad Sci U S A 115:6267–6272. doi: 10.1073/pnas.1800177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baraniak I, Kropff B, Ambrose L, McIntosh M, McLean GR, Pichon S, Atkinson C, Milne RSB, Mach M, Griffiths PD, Reeves MB. 2018. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A 115:6273–6278. doi: 10.1073/pnas.1800224115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CS, Herold BC, Permar SR. 2018. A new era in cytomegalovirus vaccinology: considerations for rational design of next-generation vaccines to prevent congenital cytomegalovirus infection. NPJ Vaccines 3:38. doi: 10.1038/s41541-018-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murthy S, Hayward GS, Wheelan S, Forman MS, Ahn JH, Pass RF, Arav-Boger R. 2011. Detection of a single identical cytomegalovirus (CMV) strain in recently seroconverted young women. PLoS One 6:e15949. doi: 10.1371/journal.pone.0015949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 13.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang ES, Huong SM, Tegtmeier GE, Alford C. 1980. Cytomegalovirus: genetic variation of viral genomes. Ann N Y Acad Sci 354:332–346. [DOI] [PubMed] [Google Scholar]
- 15.Bradley AJ, Kovacs IJ, Gatherer D, Dargan DJ, Alkharsah KR, Chan PK, Carman WF, Dedicoat M, Emery VC, Geddes CC, Gerna G, Ben-Ismaeil B, Kaye S, McGregor A, Moss PA, Pusztai R, Rawlinson WD, Scott GM, Wilkinson GW, Schulz TF, Davison AJ. 2008. Genotypic analysis of two hypervariable human cytomegalovirus genes. J Med Virol 80:1615–1623. doi: 10.1002/jmv.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis 39:155–161. doi: 10.1086/421496. [DOI] [PubMed] [Google Scholar]
- 17.Peek R, Verbraak F, Bruinenberg M, Van der Lelij A, Van den Horn G, Kijlstra A. 1998. Cytomegalovirus glycoprotein B genotyping in ocular fluids and blood of AIDS patients with cytomegalovirus retinitis. Invest Ophthalmol Vis Sci 39:1183–1187. [PubMed] [Google Scholar]
- 18.Sowmya P, Madhavan HN. 2009. Analysis of mixed infections by multiple genotypes of human cytomegalovirus in immunocompromised patients. J Med Virol 81:861–869. doi: 10.1002/jmv.21459. [DOI] [PubMed] [Google Scholar]
- 19.Hassan-Walker AF, Okwuadi S, Lee L, Griffiths PD, Emery VC. 2004. Sequence variability of the alpha-chemokine UL146 from clinical strains of human cytomegalovirus. J Med Virol 74:573–579. doi: 10.1002/jmv.20210. [DOI] [PubMed] [Google Scholar]
- 20.Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7:e1001344. doi: 10.1371/journal.ppat.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renzette N, Gibson L, Bhattacharjee B, Fisher D, Schleiss MR, Jensen JD, Kowalik TF. 2013. Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLoS Genet 9:e1003735. doi: 10.1371/journal.pgen.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renzette N, Pokalyuk C, Gibson L, Bhattacharjee B, Schleiss MR, Hamprecht K, Yamamoto AY, Mussi-Pinhata MM, Britt WJ, Jensen JD, Kowalik TF. 2015. Limits and patterns of cytomegalovirus genomic diversity in humans. Proc Natl Acad Sci U S A 112:E4120–E4128. doi: 10.1073/pnas.1501880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novak Z, Ross SA, Patro RK, Pati SK, Kumbla RA, Brice S, Boppana SB. 2008. Cytomegalovirus strain diversity in seropositive women. J Clin Microbiol 46:882–886. doi: 10.1128/JCM.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson CS, Cruz DV, Tran D, Bialas KM, Stamper L, Wu H, Gilbert M, Blair R, Alvarez X, Itell H, Chen M, Deshpande A, Chiuppesi F, Wussow F, Diamond DJ, Vandergrift N, Walter MR, Barry PA, Cohen-Wolkowiez M, Koelle K, Kaur A, Permar SR. 2017. Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight 2:94002. doi: 10.1172/jci.insight.94002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson CS, Vera Cruz D, Su M, Xie G, Vandergrift N, Pass RF, Forman M, Diener-West M, Koelle K, Arav-Boger R, Permar SR. 1995. Genetic signatures of human cytomegalovirus variants acquired by seronegative glycoprotein B vaccinees. bioRxiv doi: 10.1101/432922. [DOI] [PMC free article] [PubMed]
- 26.Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. 2018. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 46:e21. doi: 10.1093/nar/gkx1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehlinger EP, Webster EM, Kang HH, Cangialose A, Simmons AC, Barbas KH, Burchett SK, Gregory ML, Puopolo KM, Puopolo KP, Permar SR. 2011. Maternal cytomegalovirus-specific immune responses and symptomatic postnatal cytomegalovirus transmission in very low-birth-weight preterm infants. J Infect Dis 204:1672–1682. doi: 10.1093/infdis/jir632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forman MS, Vaidya D, Bolorunduro O, Diener-West M, Pass RF, Arav-Boger R. 2017. Cytomegalovirus kinetics following primary infection in healthy women. J Infect Dis 215:1523–1526. doi: 10.1093/infdis/jix188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarate S, Pond SL, Shapshak P, Frost SD. 2007. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol 81:6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JB, Adler SP, Hempfling S, Burke RL, Duliege AM, Starr SE, Plotkin SA. 1996. Mucosal antibodies to human cytomegalovirus glycoprotein B occur following both natural infection and immunization with human cytomegalovirus vaccines. J Infect Dis 174:387–392. [DOI] [PubMed] [Google Scholar]
- 31.Kimura M. 1962. On the probability of fixation of mutant genes in a population. Genetics 47:713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M, Ohta T. 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein M, Schoppel K, Amvrossiadis N, Mach M. 1999. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J Virol 73:878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban M, Britt W, Mach M. 1992. The dominant linear neutralizing antibody-binding site of glycoprotein gp86 of human cytomegalovirus is strain specific. J Virol 66:1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer H, Sundqvist VA, Pereira L, Mach M. 1992. Glycoprotein gp116 of human cytomegalovirus contains epitopes for strain-common and strain-specific antibodies. J Gen Virol 73:2375–2383. doi: 10.1099/0022-1317-73-9-2375. [DOI] [PubMed] [Google Scholar]
- 36.Dal Monte P, Pignatelli S, Mach M, Landini MP. 2001. The product of human cytomegalovirus UL73 is a new polymorphic structural glycoprotein (gpUL73). J Hum Virol 4:26–34. [PubMed] [Google Scholar]
- 37.Wang D, Li F, Freed DC, Finnefrock AC, Tang A, Grimes SN, Casimiro DR, Fu TM. 2011. Quantitative analysis of neutralizing antibody response to human cytomegalovirus in natural infection. Vaccine 29:9075–9080. doi: 10.1016/j.vaccine.2011.09.056. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor CM, Shenk T. 2011. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J Virol 85:3700–3707. doi: 10.1128/JVI.02442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka Y, Kanda Y, Kami M, Mori S, Hamaki T, Kusumi E, Miyakoshi S, Nannya Y, Chiba S, Arai Y, Mitani K, Hirai H, Mutou Y, Japan H, Oncology Clinical Study G. 2002. Monitoring cytomegalovirus infection by antigenemia assay and two distinct plasma real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant 30:315–319. doi: 10.1038/sj.bmt.1703661. [DOI] [PubMed] [Google Scholar]
- 40.Potzsch S, Spindler N, Wiegers AK, Fisch T, Rucker P, Sticht H, Grieb N, Baroti T, Weisel F, Stamminger T, Martin-Parras L, Mach M, Winkler TH. 2011. B cell repertoire analysis identifies new antigenic domains on glycoprotein B of human cytomegalovirus which are target of neutralizing antibodies. PLoS Pathog 7:e1002172. doi: 10.1371/journal.ppat.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou S. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188:388–390. [DOI] [PubMed] [Google Scholar]
- 43.Nelson CW, Hughes AL. 2015. Within-host nucleotide diversity of virus populations: insights from next-generation sequencing. Infect Genet Evol 30:1–7. doi: 10.1016/j.meegid.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hudson RR, Slatkin M, Maddison WP. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudson RR. 2000. A new statistic for detecting genetic differentiation. Genetics 155:2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slatkin M, Maddison WP. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol 75:11686–11699. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colugnati FA, Staras SA, Dollard SC, Cannon MJ. 2007. Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect Dis 7:71. doi: 10.1186/1471-2334-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full gB ORF and SNAPP sequence data used in the manuscript have been uploaded to GenBank under accession numbers MK157426 to MK158058. All other source data are available upon request.