Many plant viruses contain three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB), for intercellular movement. However, how the corresponding three proteins coordinate their functions remains obscure. In the present study, we provided multiple lines of evidence supporting the notion that PVX TGBp2 functions as the molecular adaptor bridging the interaction between the RdRp/dsRNA body and TGBp3 by forming “chain mail”-like structures in the RdRp/dsRNA body, which can also enhance viral replication. Taken together, our results provide new insights into the replication and movement of PVX and possibly also other TGB-containing plant viruses.
KEYWORDS: Potato virus X, TGB, TGBp2, movement, replication
ABSTRACT
Plant viruses usually encode one or more movement proteins (MP) to accomplish their intercellular movement. A group of positive-strand RNA plant viruses requires three viral proteins (TGBp1, TGBp2, and TGBp3) that are encoded by an evolutionarily conserved genetic module of three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB). However, how these three viral movement proteins function cooperatively in viral intercellular movement is still elusive. Using a novel in vivo double-stranded RNA (dsRNA) labeling system, we showed that the dsRNAs generated by potato virus X (PVX) RNA-dependent RNA polymerase (RdRp) are colocalized with viral RdRp, which are further tightly covered by “chain mail”-like TGBp2 aggregates and localizes alongside TGBp3 aggregates. We also discovered that TGBp2 interacts with the C-terminal domain of PVX RdRp, and this interaction is required for the localization of TGBp3 and itself to the RdRp/dsRNA bodies. Moreover, we reveal that the central and C-terminal hydrophilic domains of TGBp2 are required to interact with viral RdRp. Finally, we demonstrate that knockout of the entire TGBp2 or the domain involved in interacting with viral RdRp attenuates both PVX replication and movement. Collectively, these findings suggest that TGBp2 plays dual functional roles in PVX replication and intercellular movement.
IMPORTANCE Many plant viruses contain three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB), for intercellular movement. However, how the corresponding three proteins coordinate their functions remains obscure. In the present study, we provided multiple lines of evidence supporting the notion that PVX TGBp2 functions as the molecular adaptor bridging the interaction between the RdRp/dsRNA body and TGBp3 by forming “chain mail”-like structures in the RdRp/dsRNA body, which can also enhance viral replication. Taken together, our results provide new insights into the replication and movement of PVX and possibly also other TGB-containing plant viruses.
INTRODUCTION
Viruses must transport their genomes from the cell in which infection is initiated to adjacent cells to continue infection. Unlike their animal counterparts that enter neighboring cells via endocytosis (1), plant viruses move through the narrow channels in the plant cell membrane, termed plasmodesmata (PD) (2). The transport of viruses through PD is usually accomplished by one or more virus-encoded proteins named movement proteins (MPs). A group of positive-strand RNA plant viruses requires three viral MPs that are encoded by three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB). The TGB is an evolutionarily conserved genetic module that was found in nine genera within the families Alphaflexiviridae, Betaflexiviridae, and Virgaviridae and the unassigned genus Benyvirus (3, 4). TGB-encoded proteins, which are referred to as TGBp1, TGBp2, and TGBp3 according to the positions of their genes, are usually not absolutely required for viral genome replication, but they are crucial for the cell-to-cell and long-distance movement of the viruses (3, 4). However, how these three viral proteins coordinately accomplish virus intercellular movement requires further investigations.
Potato virus X (PVX) is type species in the genus Potexvirus within the family Alphaflexiviridae. The genome of PVX consists of a single-stranded, positive-sense RNA that encodes five open reading frames (ORFs). The largest ORF at the 5′ terminus of the genome (ORF1) encodes the viral RNA-dependent RNA polymerase (RdRp), which is the only viral protein that is absolutely required for viral replication (5, 6). ORF2 to -4 comprise the TGB module encoding the 25-kDa TGBp1, 12-kDa TGBp2, and 8-kDa TGBp3, respectively. ORF5 encodes the viral coat protein (CP) that is involved in viral movement and viral genome RNA encapsidation (7). TGBp1 can modify the PD size exclusion limit and move between cells (8–10). However, TGBp1 cannot target PD efficiently without the presence of TGBp2 and TGBp3 (4, 11–13). Additionally, TGBp1 also functions as an RNA helicase, a translational activator, and an RNA silencing suppressor (14–18). TGBp2 and TGBp3 are transmembrane proteins that localize to different subdomains of the endoplasmic reticulum (ER) and can induce ER-derived motile granules (19–21). TGBp2 contains two transmembrane motifs that adopt a “U”-shaped topology with both the N- and C termini in the cytosol (20, 22), whereas TGBp3 contains a single transmembrane domain with the N terminus in the ER lumen and C terminus exposed to the cytosol (19). PVX TGBp3 interacts with both TGBp1 and TGBp2 and can target both TGBp1 and TGBp2 from the cytosol to the peripheral area (11–13). However, the precise role of TGBp2 in PVX intercellular movement is poorly understood.
During infection, PVX can form large amorphous complexes in the perinuclear area, which are historically termed X-bodies. The X-bodies contain all five viral proteins, viral RNA, host factors, and host endomembrane and proteins (6, 23–25). Confocal, three-dimensional structured illumination (3D-SIM) superresolution microscopy and immune electron microscopy have shown that the X-body has a special layered structure, with TGBp1 at the most inner region as helically arranged aggregates and TGBp2/3 granules localized on the surface of TGBp1 aggregates; in contrast, encapsidated virions accumulate at the outermost face of the X-body (6, 20, 23, 26, 27). It has been proposed that viral RNA replication take places in the layer between TGBp1 aggregates and TGBp2/3 granules since abundant nonencapsidated viral RNAs have been observed in this area (6). Moreover, PVX also induces cap-like complexes at the entrances of PD that have a composition and structure similar to those of perinuclear X-bodies (11). These small X-body-like complexes at the entrances of PD are crucial for successful and efficient intercellular movement (11). These observations suggest that the amorphous bodies formed by PVX are complex virus-manufacturing factories per se that couple viral protein translation, RNA replication, particle encapsidation, and intercellular movement (6, 11). However, the precise replication sites within the amorphous bodies remain obscure.
Recently, we developed an in vivo double-stranded RNA (dsRNA) visualization system termed the dsRNA binding-dependent fluorescence complementation (dRBFC) assay (28). This system has already been utilized by our and other groups to determine the subcellular localizations of dsRNAs generated by plant viruses in several genera, including Potexvirus, Potyvirus, Hordeivirus, and Tombusvirus (28–30). The dsRNA is a replication intermediate of all RNA viruses (31). Thus, understanding the precise localization of the dsRNA and its relationship with other viral proteins could shed light on several basal virological questions, e.g., the precise structure of the viral replication site and how to efficiently couple viral replication and intercellular movement, and even uncover unsuspected roles of viral proteins. In the present study, we analyzed the dsRNA generated by PVX and its spatial localization with viral proteins, e.g., RdRp, TGBp2, and TGBp3, in living Nicotiana benthamiana cells. Our results revealed that TGBp2 colocalizes with dsRNA in the X-body via directly interacting with viral RdRp, and the recruitment of TGBp3 to the X-body is dependent on TGBp2. Finally, we found that TGBp2 can promote PVX replication. These results reveal an unexpected role of TGBp2 in PVX replication and intracellular trafficking of TGBp3 protein.
RESULTS
dsRNA colocalizes with viral RNA-dependent RNA polymerase in the X-body.
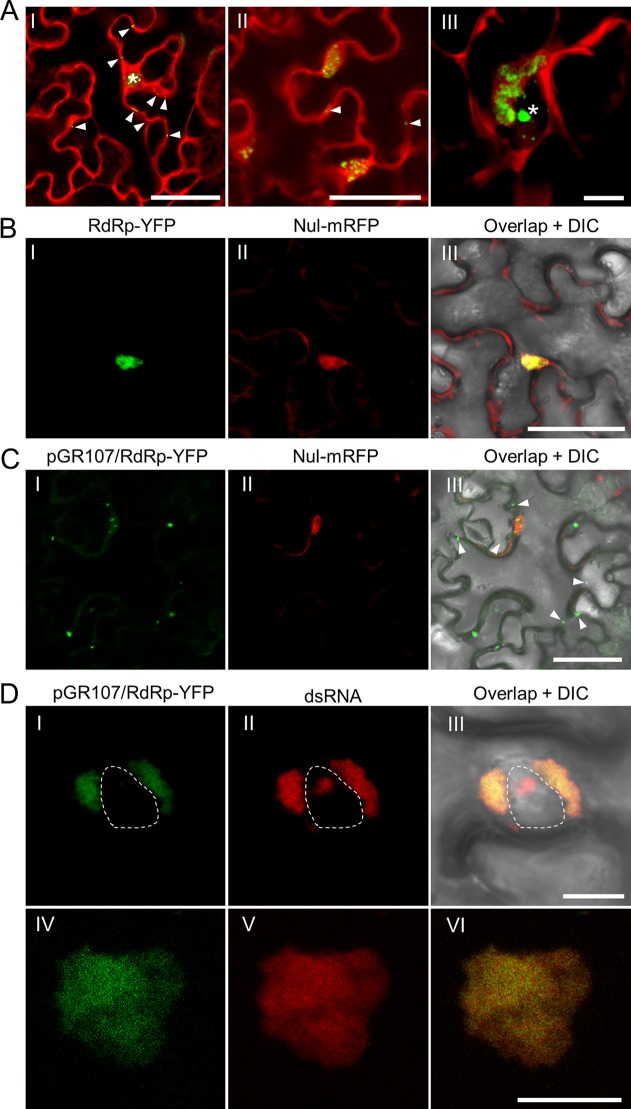
Abundant dsRNAs can be detected in PVX infected Nicotiana benthamiana epidermal cells either via the dRBFC assay or via transgenically expressed B2 protein, a dsRNA-binding protein encoded by Flock House virus (HFV) (28, 32). To further characterize these dsRNAs, a time course analysis was performed during PVX infection. Nicotiana benthamiana leaves were infiltrated with a modified PVX infectious clone, pGR.mCh, in which the CP was N-terminally fused to mCherry and a partial autoproteolytic 2A peptide (mCherry-2A-CP) (27), together with yellow fluorescent protein (YFP)-based dRBFC plasmids (28). The coinfiltrated leaf tissues were monitored by confocal microscopy from 36 to 72 h postinfiltration (hpi) at an interval of 12 h. Cells infected with pGR.mCh showed bright red fluorescence in the cytosol and in large perinuclear bodies (Fig. 1A). The red fluorescence in the cytosol was likely derived from free mCherry, whereas that in the large perinuclear bodies was mostly derived from the chimeric mCherry-2A-CP protein (27). The dsRNAs were found as small granules in the early infection stage (36 hpi) in the cytosol and in the nucleus. The cytoplasmic granules gradually gathered into large perinuclear irregularly shaped aggregates in the large perinuclear bodies during the late infection stage (Fig. 1A), suggesting that most cytoplasmic dsRNAs should be generated by PVX.
FIG 1.
In vivo visualization of dsRNA and RdRp in PVX infected N. benthamiana epidermal cells. (A) Visualization of the dsRNA during PVX infection at 36 (I), 48 (II), and 60 (III) hpi. The fluorescence from the dRBFC assay is shown in green, whereas the fluorescence from pGR.mCh is shown in red. The nucleus is indicated by a white asterisk and the peripheral dsRNA fluorescent foci are indicated by white arrowheads. Scale bar = 50 μm (I and II) or 10 μm (III). Note that the dRBFC signal in the nucleus represents endogenous d-bodies (28). (B and C) Subcellular localization of transiently expressed RdRp-YFP (green) in N. benthamiana epidermal cells in the absence of (B) or during (C) PVX infection at 48 hpi. The RdRp-YFP in panel C was expressed from pGR107/RdRp-YFP to ensure infection of PVX in the same cell. The cytoplasmic green fluorescent foci from RdRp-YFP are indicated by white arrowheads. The nuclei were labeled by a nuclear localization signal peptide (NLS)-tagged mRFP (Nul-mRFP). Scale bars = 50 μm. (D) Subcellular localization of perinuclear (I to III) or cytoplasmic (IV to VI) RdRp and dsRNA during PVX infection at 60 hpi; the RdRp-YFP (green) was expressed from pGR107/RdRp-YFP, and dsRNAs (red) were labeled by mRFP-based dRBFC plasmids. The nucleus is indicated by a white dashed line. All scale bars = 10 μm.
To further characterize the origination of these dsRNAs, we analyzed the colocalization between these dsRNAs and PVX RdRp during PVX infection. Due to repeated failures of the direct fusion of a fluorescent protein to the RdRp of a wild-type PVX infectious clone pGR107 (33), we alternatively directly inserted the expression cassette of pEarley101 (34) into the unique SfoI restriction site between the NOS terminator and the transfer DNA (T-DNA) left border in pGR107 to construct pGR107/Gateway-YFP. The DNA fragment encoding PVX RdRp was then inserted into the Gateway cassette to construct pGR107/RdRp-YFP. This plasmid allows the expression of a second copy of C-terminal YFP-tagged RdRp in the cell infected by PVX. This plasmid was then delivered into N. benthamiana leaves via agrobacterium infiltration alone or together with a set of monomer red fluorescent protein (mRFP)-based dRBFC assay plasmids, e.g., pdRBFC-B2-CmRFP and pdRBFC-NS1-NmRFP. As a control, RdRp-YFP was also transiently expressed from a binary vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter. In the absence of PVX infection, YFP fluorescence from RdRp-YFP was observed only in the nucleus (Fig. 1B). However, most RdRp was retained in the cytoplasm as perinuclear irregular aggregates or peripheric foci in PVX-infected cells (Fig. 1C and D), suggesting that cytoplasmic localization of PVX RdRp requires the presence of viral genome RNA or viral replication. Interestingly, the fluorescence from RdRp-YFP at the perinuclear area or in the cytosol completely overlapped the aggregation of dsRNA in the perinuclear area or in the cytosol (Fig. 1DI to DVI). These results thus confirmed that the large perinuclear and cytoplasmic dsRNAs were viral replication intermediates generated by viral RdRp, and they are referred to as RdRp/dsRNA bodies here.
Hierarchical arrangement of dsRNA, TGBp2, and TGBp3 in the PVX X-body.
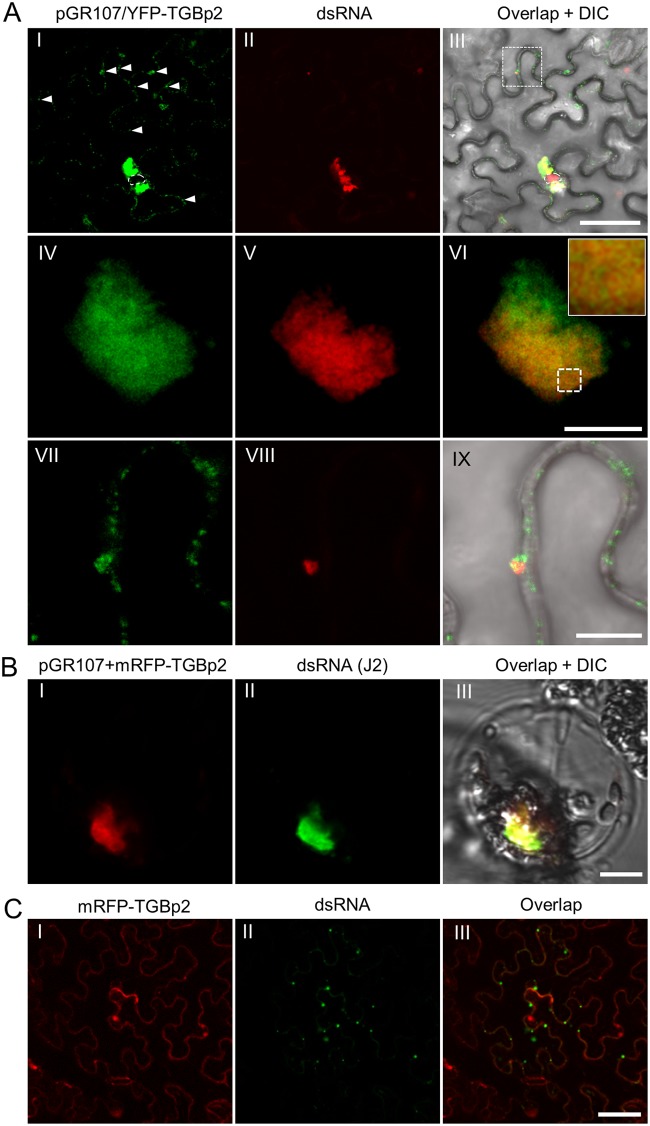
Previous studies have suggested that PVX replication might take place in the middle layer of the X-body, which contains abundant nonencapsidated vRNA, viral RdRp, and both TGBp2 and TGBp3 granules (6, 20, 26, 27, 35). Thus, we were interested in visualizing the relationship between the RdRp/dsRNA bodies and TGBp2 or TGBp3 in the X-body. To avoid disrupting the overlapping TGB ORFs in the PVX genome, an additional copy of fluorescent protein-tagged TGBp2 or TGBp3 was expressed using the strategy described above. TGBp2 was coexpressed as N-terminally YFP-tagged recombinant protein since a previous study has suggested that attaching an additional protein to the C terminus of TGBp2 might affect its function (6). pGR107/YFP-TGBp2 was coinfiltrated with mRFP-based dRBFC plasmids into N. benthamiana leaves. As a control, mRFP-TGBp2 was transiently expressed with dRBFC in the absence of virus infection. Consistent with previous reports (20, 21, 36), the majority of TGBp2 was accumulated as irregularly shaped perinuclear aggregations, with a small portion in the cytosol as small granules (Fig. 2AI). Interestingly, the TGBp2 aggregates were largely colocalized with RdRp/dsRNA bodies at low magnification (Fig. 2AI to AIII). Detailed inspection under a higher magnification showed that TGBp2 colocalized with RdRp/dsRNA bodies as a chain mail-like structure (Fig. 2AIV to AVI). Moreover, the small RdRp/dsRNA bodies at the cell periphery were also associated with TGBp2 (Fig. 2AVII to AIX). To further verify these results, we labeled the dsRNAs using the dsRNA-specific J2 antibody in N. benthamiana protoplasts transformed with pGR107 and mRFP-TGBp2. The results showed that mRFP-TGBp2 was also spread around the dsRNA aggregates (Fig. 2B). A previous study has shown that the TGBp2 of bamboo mosaic virus (BaMV), a potexvirus, shows nonspecific RNA binding activity (37). Therefore, it is possible that PVX TGBp2 also has nonspecific RNA binding activity that is involved in the localization of TGBp2 to the dsRNA generated by PVX RdRp. Thus, we transiently expressed N-terminally mRFP-tagged TGBp2 (mRFP-TGBp2) together with YFP-based dRBFC plasmids (28) in N. benthamiana epidermal cells. A nonfluorescent construct expressing Arabidopsis SGS3 (FLAG-4×Myc-AtSGS3) was also included to enhance the fluorescence of endogenous dsRNA bodies (28). The result showed that the transiently expressed TGBp2 granules were not colocalized with the endogenous dsRNA bodies (Fig. 2C), such as SGS3-RDR6 bodies (28), suggesting that the nonspecific RNA binding activity of PVX TGBp2, if any, was not sufficient to target itself to the endogenous dsRNA bodies.
FIG 2.
Subcellular localization of TGBp2 and dsRNA during PVX infection. (A) Confocal micrographs of YFP-TGBp2 (green) and dsRNA (red) during PVX infection in N. benthamiana epidermal cells at 48 hpi at lower (I to III) and higher (IV to IX) magnifications. YFP-TGBp2 was expressed from pGR107/YFP-TGBp2 and dsRNA was labeled using the mRFP-based dRBFC assay. The white arrowheads in frame I indicates the TGBp2-induced small granules. The nuclei are indicated by white dashed lines. The inset in frame VI is an enlargement of the dashed area to show the typical chain mail-like structure of TGBp2 on dsRNA. Scale bar = 50 μm (I to III) or = 10 μm (IV to IX). (B) Subcellular localization of dsRNA (green) and mRFP-TGBp2 (red) in a typical globular-like X-body in the N. benthamiana protoplast. The dsRNA was detected using J2 monoclonal antibodies. Scale bar = 10 μm. (C) Subcellular localization of dsRNA (green) and mRFP-TGBp2 (red) in the absence of PVX infection. Scale bar = 50 μm.
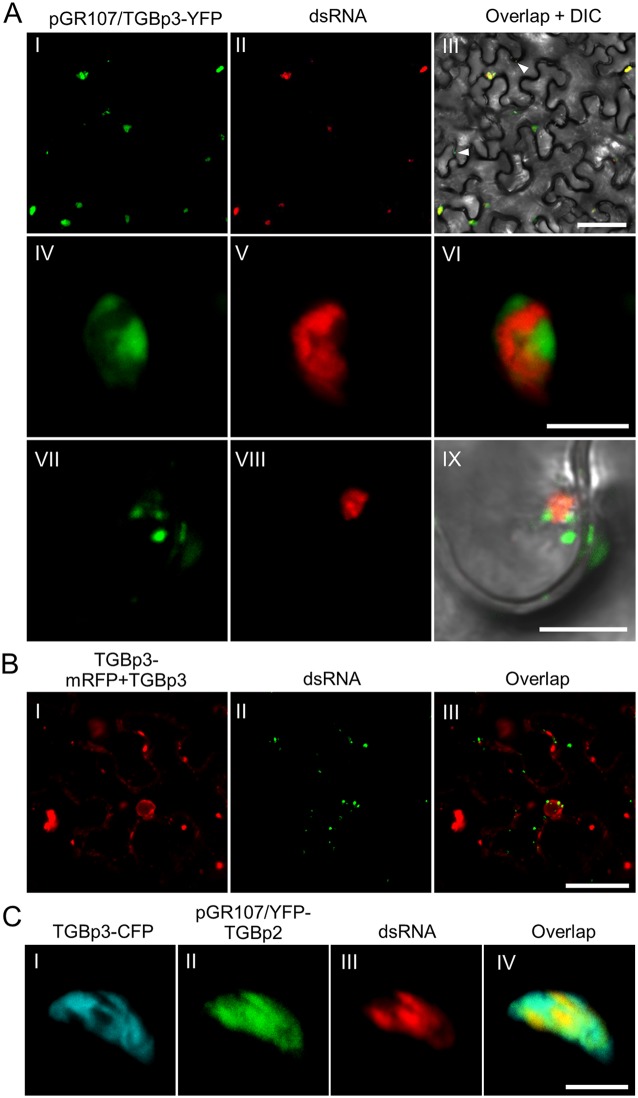
The relationship between TGBp3 and RdRp/dsRNA bodies during PVX infection was also analyzed using the same strategy. A PVX infectious clone expressing an additional C-terminally YFP-tagged TGBp3 (pGR107/TGBp3-YFP) was constructed and coinfiltrated with mRFP-based dRBFC plasmids into N. benthamiana leaves. In N. benthamiana epidermal cells infected with PVX, TGBp3-YFP clustered as aggregates with various size in the cytosol or at the cell periphery (Fig. 3AI). Interestingly, most TGBp3 aggregates were also associated with the RdRp/dsRNA bodies (Fig. 3AII and AIII). Detailed inspection showed that TGBp3-YFP, in contrast, condensed as isolated patches or clusters that closely neighbored the RdRp/dsRNA bodies (Fig. 3AIV to AVI). In the most recently infected cells, the small TGBp3-induced granules also localized alongside the small RdRp/dsRNA bodies (Fig. 3AVII to AIX). We also transiently expressed TGBp3-mRFP with mRFP-based dRBFC plasmids to analyze the colocalization of TGBp3 and dsRNA in the absence of PVX infection. Nontagged TGBp3 was also expressed to avoid the possible influence of mRFP on TGBp3 subcellular localization (11). The results showed that the transiently expressed TGBp3-mRFP did not display a distribution similar to that for endogenous dsRNA bodies in the absence of PVX infection (Fig. 3B).
FIG 3.
Subcellular localization of TGBp3 and dsRNA in PVX infected N. benthamiana cells. (A) Confocal micrographs of dsRNA (red) and TGBp3-YFP (green) in N. benthamiana leaf tissue during PVX infection at 48 hpi. TGBp3-YFP was expressed from pGR107/TGBp3-YFP, and the dsRNA was labeled using the mRFP-based dRBFC assay. The white arrowheads indicate TGBp3 foci at the cell periphery. Scale bar = 50 μm (I to III) or 10 μm (IV to IX). (B) TGBp3-mRFP is not associated with dsRNA in the absence of PVX infection. Scale bar = 10 μm. (C) Confocal micrographs of TGBp3-CFP (cyan), YFP-TGBp2 (green), and dsRNA (red) in N. benthamiana epidermal cells during PVX infection at 48 hpi. YFP-TGBp2 was expressed from pGR107/YFP-TGBp2. Scar bar = 10 μm.
We also coexpressed pGR107/YFP-TGBp2, and -TGBp3-CFP,and mRFP-based dRBFC plasmids to simultaneously label TGBp2, TGBp3, and the RdRp/dsRNA body in N. benthamiana epidermal cells during PVX infection. Consistent with previous reports (6, 12, 38), TGBp3 did not overlap with TGBp2, although they were colocalized in the same complex (Fig. 3C). Similarly, TGBp2 colocated with the RdRp/dsRNA body, whereas TGBp3 adjacent to the TGBp2-covered RdRp/dsRNA bodies localized as isolated patches or clusters (Fig. 3C).
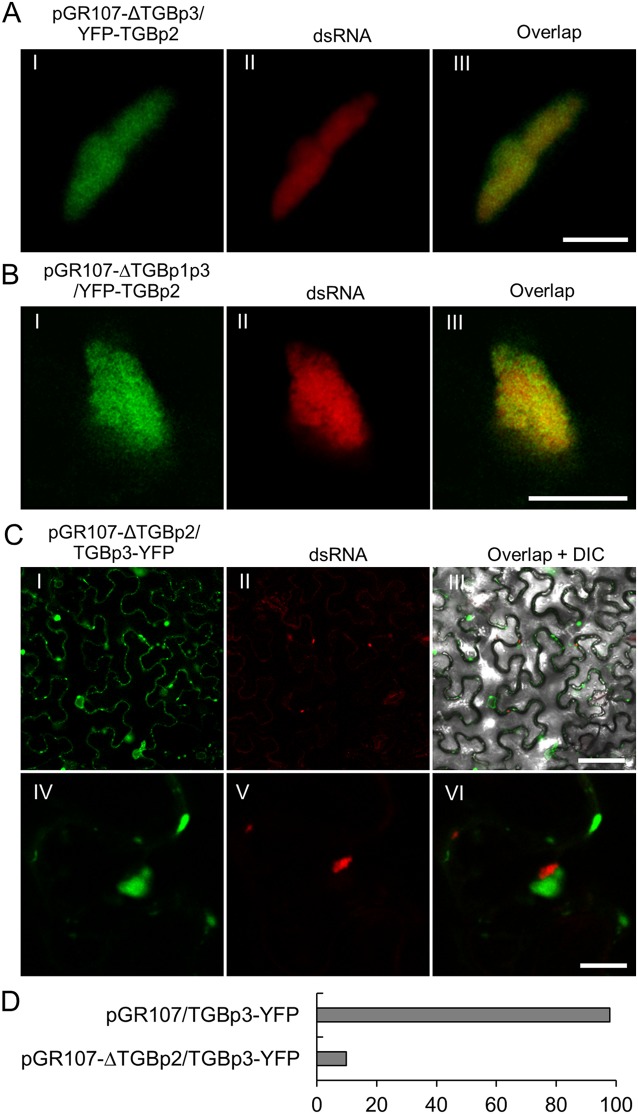
Recruitment of TGBp2 to the PVX X-body occurs independently of TGBp1 and TGBp3.
As TGBp2 can not localize to endogenous dsRNA bodies in the absence of viral replication, we suspected that TGBp2 might require another interacting viral protein(s), such as TGBp1 or TGBp3 (12, 13, 39, 40). Therefore, we deleted the TGBp3 in pGR107/YFP-TGBp2 by mutating residues 25 (Ser) and 26 (Leu) of TGBp3 into stop codons to construct a TGBp3 deletion PVX infectious clone, pGR107-ΔTGBp3/YFP-TGBp2. Infectivity analysis showed that this infectious clone failed to cause any symptoms, even at 30 days postinoculation (dpi), suggesting that TGBp3 was completely knocked out. We then coinfiltrated N. benthamiana leaves with pGR107-ΔTGBp3/YFP-TGBp2 and mRFP-based dRBFC assay vectors. Confocal microscopy showed that TGBp2 could still accumulate in the RdRp/dsRNA body (Fig. 4A), suggesting that the localization of TGBp2 to the RdRp/dsRNA bodies occurs independently of TGBp3. A previous study has also suggested that TGBp1 is the central orchestrator of the X-body, which is necessary and sufficient to remodel host actin and endomembranes to form the typical perinuclear X-body (6). Therefore, a TGBp1 and TGBp3 double-knockout PVX infectious clone, pGR107-ΔTGBp1p3/YFP-TGBp2, was constructed by mutating amino acids (aa) 6 (Ser) and 7 (Ser) of TGBp1 in pGR107-ΔTGBp3/YFP-TGBp2 into stop codons. This infectious clone was coinfiltrated with suitable dRBFC plasmids on N. benthamiana leaves. Interestingly, TGBp2 could still efficiently localize to the RdRp/dsRNA body in the cells infected by wild-type and TGBp3 knockout PVX (Fig. 4B). Taken together, these results suggest that the accumulation of TGBp2 in the dsRNA bodies occurs independently of both TGBp1 and TGBp3.
FIG 4.
Recruitment of TGBp2 to dsRNA occurs independently of TGBp1 and TGBp3. (A and B) Confocal micrographs of YFP-TGBp2 (green) and dsRNA (red) in N. benthamiana epidermal cells during infection by pGR107-ΔTGBp3/YFP-TGBp2 (A) or pGR107-ΔTGBp1p3/YFP-TGBp2 (B) at 48 hpi. The dsRNAs were labeled using the mRFP-based dRBFC assay. Scar bar = 10 μm. (C) Subcellular localization of TGBp3-YFP and dsRNA in N. benthamiana cells infected with a TGBp2-defective PVX infectious clone (pGR107-ΔTGBp2/TGBp3-YFP) at 48 hpi. Scar bar = 50 μm (I to III) or = 10 μm (IV to VI). A differential interference contrast (DIC) channel was included in the overlap micrograph to illustrate the outline of the cell. (D) Percentage of RdRp/dsRNA bodies associated with TGBp3 in N. benthamiana epidermal cells infected with pGR107/TGBp3-YFP or pGR107-ΔTGBp2/TGBp3-YFP.
To further analyze the role of TGBp2, a TGBp2 knockout PVX infectious clone coexpressing TGBp3-YFP (pGR107-ΔTGBp2/TGBp3-YFP) was constructed by mutating amino acids 10 (Ala) and 11 (Pro) of TGBp2 into stop codons. Interestingly, compared with the subcellular localization of TGBp3 during wild-type PVX infection (Fig. 3A), the subcellular of TGBp3 during pGR107-ΔTGBp2 infection was more diffusive, resembling the transient expression alone under the control of the CaMV 35S promoter (Fig. 3B and 4C). Moreover, we could hardly find TGBp3 granules associated with the RdRp/dsRNA bodies in the N. benthamiana epidermal cells infected with pGR107-ΔTGBp2 (Fig. 4CI to CIII). Very occasionally, TGBp3 granules could be found neighboring RdRp/dsRNA bodies; however, a clear distance was observed between the RdRp/dsRNA body and TGBp3 granule (Fig. 4CIV to CVI), suggesting that they are encountered incidentally due to cytoplasmic streaming. We directly compared the numbers of RdRp/dsRNA bodies associated with TGBp3 granules in N. benthamiana epidermal cells infected with the wild-type and TGBp2 knockout PVX infectious clones. The results showed that almost all (98% [n = 96]) RdRp/dsRNA bodies in pGR107/TGBp3-YFP-infected N. benthamiana epidermal cells were associated with TGBp3 granules, whereas less than 10% (n = 116) of the RdRp/dsRNA bodies in pGR107-ΔTGBp2/TGBp3-YFP-infected N. benthamiana epidermal cells were associated with TGBp3 granules (Fig. 4D). Taken together, these data suggest that the recruitment of TGBp3 into the X-body is dependent on TGBp2.
TGBp2 directly interacts with the replicase domain of viral RdRp.
We suspected that TGBp2 might directly interact with viral RdRp. To confirm this hypothesis, the bimolecular fluorescence complementation assay (BiFC) was applied (41). However, no yellow fluorescence could be detected in the N. benthamiana epidermal cells coinfiltrated with the N-terminal part of YFP (YN)-tagged TGBp2 (YN-TGBp2) and the C-terminal half of YFP (YC)-tagged full-length RdRp (YC-RdRp) or the reverse (YC-TGBp2 and YN-RdRp) under confocal microscopy. This finding could be due to the nuclear localization of transiently expressed RdRp and the cytoplasmic localization of TGBp2 (Fig. 1B and 2C). As an alternative, we divided RdRp into four functional domains: the N-terminal aa 1 to 340 as the methyltransferase domain (MET), aa 341 to 730 as the unstructured loop domain (UNL), aa 731 to 964 as the helicase domain (HEL), and the C-terminal aa 965 to 1421 as the replicase domain (REP) (Fig. 5A). First, we analyzed the subcellular localization of each domain based on the C-terminally YFP-tagged recombinant protein. The results showed that MET was mainly present as cytoplasmic granules, UNL localized mainly in the nucleus, HEL was mainly present as amorphous aggregates in the cytoplasm, and REP localized both in the cytoplasm and in the nucleus (Fig. 5B). These data suggested that the nuclear localization of RdRp is mainly determined by the UNL domain, whereas the MET and HEL domains have motifs for multimerization. These four domains were then fused to YC and coinfiltrated with YN-TGBp2 into N. benthamiana leaves. The results showed that the mild YFP fluorescence was successfully restored in the N. benthamiana epidermal cells expressing REP-YC and YN-TGBp2, whereas no yellow fluorescence was observed in the N. benthamiana epidermal cells expressing YN-TGBp2 and MET-YC, UNL-YC, or HEL-YC under the same conditions (Fig. 5C). Moreover, the YFP signal from REP-YC and YN-TGBp2 was detected exclusively in the cytoplasm, suggesting that TGBp2 might interact only with the cytoplasmic portion of REP (Fig. 5C). To further narrow down the interacting region, the REP domain was divided into two subdomains, the N-terminal REP (RepN) and C-terminal REP (RepC). Subcellular localization analyses showed that RepN was diffusively distributed both in the nucleus and cytoplasm, whereas the RepC domain was exclusively distributed in the cytoplasm and induced vesicle-like structures (Fig. 5D). These results suggested the presence of a transmembrane or membrane-associated motif in RepC and that the attachment of RepN would result in a loss of function of the transmembrane or membrane-associated motif in RepC. BiFC showed that RepN, but not RepC, could restore the YFP fluorescence, suggesting that TGBp2 interacted with a 166-aa peptide (aa 965 to 1131) in the REP domain (Fig. 5E). Consistently, the interaction between TGBp2 and RepN was observed exclusively in the cytoplasm (Fig. 5E).
FIG 5.
TGBp2 interacts with PVX RdRp. (A) Schematic representations of the PVX RdRp domains. MET, methyltransferase domain; UNL, unstructured loop domain; HEL, helicase domain; REP, replicase domain; RepN, N terminus of the REP domain; RepC, C terminus of the REP domain. The numbers represent the amino acid positions of the domain boundaries. (B) Subcellular localization of MET-YFP, UNL-YFP, HEL-YFP, and REP-YFP in N. benthamiana epidermal cells at 48 hpi. The insets show the typical cytoplasmic granules of MET-YFP and HEL-YFP, as well as the nuclear signal of UNL-YFP and REP-YFP. (C) BiFC for protein-protein interactions between TGBp2 and RdRp domains in N. benthamiana epidermal cells at 48 hpi. Micrographs were obtained using the same settings. Scale bars = 50 μm. (D) Subcellular localization of RepN-YFP and RepC-YFP in N. benthamiana epidermal cells at 48 hpi. The insets show the typical nuclear signal of RepN-YFP and cytoplasmic vesicles of RepC-YFP. The nuclei are indicated by white asterisks, and the typical vesicle-like structures are indicated by white arrowheads. Scale bars = 50 μm. (E) BiFC for protein-protein interactions between TGBp2 and RepN or RepC. Micrographs were obtained using the same parameters. Scale bars = 50 μm. (F) MYTH for protein-protein interaction between TGBp2 and RdRp domains. (G) Influence of TGBp2 on the subcellular localization of RdRp and REP domain in N. benthamiana epidermal cells at 48 dpi. RdRp-YFP and REP-YFP are shown in green, and mRFP-TGBp2 is shown in red. Scale bars = 50 μm.
To further confirm this possibility, the split-ubiquitin-based membrane yeast two-hybrid assay (MYTH) was also performed (42, 43). Yeast strain NMY51 was transformed with N-terminally LexA-VP16-Cub-tagged TGBp2 (LexA-VP16-Cub-TGBp2) plus C-terminally NubG-tagged MET (MET-NubG), UNL (UNL-NubG), HEL (HEL-NubG), RepN (RepN-NubG), or RepC (RepC-NubG). When TGBp2 was coexpressed with each of the five PVX RdRp fragments, we found that histidine auxotrophy was restored only when TGBp2 was cotransformed with RepN and not with HEL, UNL, MET, or RepC (Fig. 5F), further confirming that TGBp2 can interact with viral RdRp via RepN. To test whether the interaction between TGBp2 and RdRp changes the subcellular localization of the RdRp or REP domain, we transiently expressed mRFP-TGBp2 and RdRp-YFP or REP-YFP in N. benthamiana leaves. The confocal microscopy results showed that TGBp2 had no obvious influence on the subcellular localization of RdRp-YFP or REP-YFP (Fig. 5G). Taken together, these data suggest either that TGBp2 can interact only with the cytoplasmic portion of RdRp or that the TGBp2-RdRp interaction is not sufficient to retain RdRp in the cytoplasm in the absence of virus replication.
Both the central and C-terminal hydrophilic domains of TGBp2 are required for interactions with viral RdRp.
Potexviral TGBp2 contains two transmembrane motifs that separate it into three hydrophilic domains, the N-terminal domain (ND), middle domain (MD), and C-terminal domain (CD) (21, 22). To identify the motif responsible for the interaction with RdRp, a series of TGBp2 mutants were constructed (Fig. 6A). TGBp2ΔND and TGBp2ΔCD are ND and CD complete-deletion TGBp2 mutants, respectively, whereas TGBp2Δ6-11 and TGBp2Δ104-112 are ND and CD partial-deletion mutants, respectively. TGBp2Δ52-60 contains a 9-aa deletion in the MD domain. These TGBp2 mutants were transiently coexpressed with RepN-YC as N-terminally YN-tagged recombinant proteins in N. benthamiana epidermal cells. Confocal microscopy results showed that yellow fluorescence was visible in N. benthamiana epidermal cells expressing RepN and TGBp2Δ6-11, TGBp2ΔND, or TGBp2Δ104-112 but not in cells expressing RepN plus TGBp2ΔCD or TGBp2Δ52-60 (Fig. 6B), indicating that MD and CD are required for the interaction with RdRp. Noticeably, the fluorescence in N. benthamiana epidermal cells expressing TGBp2Δ104-112 and RepN was significantly lower than in those expressing wild-type TGBp2 and RepN, suggesting that the residues between aa 104 and 112 were partially required for the interaction with RepN. MYTH results showed that yeast transformants harboring RepN-NubG plus LexA-VP16-Cub-TGBp2ΔCD or LexA-VP16-Cub-TGBp2Δ52-60 failed to grow on synthetic defined (SD) medium lacking histidine, leucine, tryptophan, and adenine (SD-LWHA medium) (Fig. 6C). The yeast transformants harboring RepN-NubG and LexA-VP16-Cub-TGBp2Δ104-112 also failed to grow on SD-LWHA medium. Taken together, these results suggest that both the MD and CD are required for the interaction with PVX RdRp.
FIG 6.
The central and C-terminal hydrophilic domains of TGBp2 are required for interactions with viral RdRp. (A) Illustration of TGBp2 mutants. The two transmembrane motifs (TM1 and TM2) are shown in black. (B) BiFC for protein-protein interactions between RepN and TGBp2 mutants in N. benthamiana epidermal cells at 48 hpi. Micrographs were obtained using the same parameters. Scale bars = 50 μm. (C) MYTH for protein-protein interactions between RepN and TGBp2 mutants. (D) Subcellular localization of TGBp2 mutants. The insets show the typical morphology of ER-derived vesicles induced by TGBp2 or its mutants. Scare bar = 50 μm. (E) Subcellular localization of dsRNA and TGBp2 mutants during PVX infection. The nuclei are indicated by white asterisks. Scale bar = 50 μm.
To further confirm these results, we analyzed the subcellular localization of TGBp2ΔND, TGBp2Δ52-60, and TGBp2ΔCD in N. benthamiana epidermal cells as N-terminally YFP-tagged recombinant proteins. In the absence of PVX replication, YFP-TGBp2Δ52-60 was present as diffuse fluorescence or hollow vesicles in the cytoplasm, whereas YFP-TGBp2ΔND and YFP-TGBp2ΔCD exhibited morphologies very similar to that of YFP-TGBp2 (Fig. 2C and 6D). We also constructed PVX infectious clones in which the ND (aa 1 to 13), aa 52 to 60, or CD (aa 101 to 115) of TGBp2 was deleted and which coexpressing an N-terminally YFP-tagged TGBp2ΔND, TGBp2Δ52-60, or TGBp2ΔCD, respectively (pGR107-TGBp214-115/YFP-TGBp2ΔND, pGR107-TGBp2Δ52-60/YFP-TGBp2Δ52-60, and pGR107-TGBp21-100/YFP-TGBp2ΔCD) to analyze the subcellular localization of TGBp2 mutants in the context of PVX infection. During PVX infection, most YFP-TGBp2 proteins were clustered on the RdRp/dsRNA bodies in perinuclear areas as irregularly shaped aggregates in N. benthamiana epidermal cells (Fig. 6EI to EIII). YFP-TGBp2ΔND displayed subcellular localization similar to that of YFP-TGBp2 in N. benthamiana epidermal cells infected by pGR107-TGBp214-115/YFP-TGBp2ΔND (Fig. 6EIV to EVI). Interestingly, the yellow fluorescence of YFP-TGBp2Δ52-60 or YFP-TGBp2ΔCD was observed as diffuse fluorescence in the cytosol of the N. benthamiana epidermal cells infected by pGR107-TGBp2Δ52-60/YFP-TGBp2Δ52-60 or pGR107-TGBp21-100/-TGBp2ΔCD (Fig. 6EVII to EXII). Nevertheless, a small part of YFP-TGBp2ΔCD still gathered on the RdRp/dsRNA bodies in the perinuclear areas (Fig. 6EX to EXII), whereas YFP-TGBp2Δ52-60 was scarcely found colocalizing on RdRp/dsRNA bodies (Fig. 6EVII to EIX). Taken together, these results suggested that the MD of TGBp2 has a more important role than the CD in targeting TGBp2 itself to replication complexes, although both the MD and CD are required for this process.
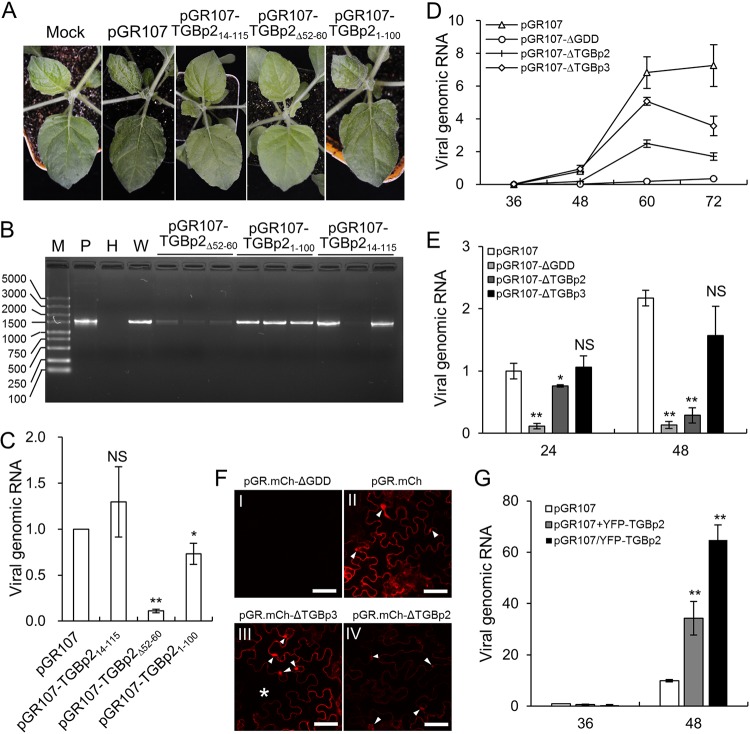
Deletion of the MD or CD affects both viral movement and replication.
As assisting viral movement is the principal role of TGBp2 (21), we suspected that the interaction with PVX RdRp was required for PVX movement. Therefore, PVX infectious clones in which the ND (aa 1 to 13), aa 52 to 60, or CD (aa 101 to 115) of TGBp2 was deleted (pGR107-TGBp214-115, pGR107-TGBp2Δ52-60, and pGR107-TGBp21-100) were constructed and their infectivity was compared with that of wild-type PVX on N. benthamiana seedlings. All N. benthamiana seedlings (n = 8) infiltrated with wild-type PVX (pGR107) developed typical viral symptoms on the upper leaves, e.g., leaf shrinking and vein clearing, as early as 5 dpi. Interestingly, all N. benthamiana seedlings (n = 16) that were infiltrated with pGR107-TGBp21-100 also developed symptoms of viral infection, although the symptoms always appeared 1 to 2 days later than those of pGR107, and most (11 out of 13) N. benthamiana seedlings infiltrated with pGR107-TGBp214-115 also developed viral symptoms at ∼12 dpi. In contrast, none of the N. benthamiana seedlings infiltrated with pGR107-TGBp2Δ52-60 developed viral symptoms, even at 20 dpi. Typical viral symptoms induced by wild-type PVX or these mutants on N. benthamiana seedlings at 8 dpi are shown in Fig. 7A. Reverse transcription-PCR (RT-PCR) was performed to confirm the presence of viral genomic RNA on systemic leaves at 10 dpi by amplifying an ∼1,500-nucleotide (nt) fragment comprising the TGBp2 coding region. Intriguingly, an amplicon of approximately 1,500 nt was detected in the symptomatic upper leaves of all N. benthamiana seedlings infiltrated with pGR107, pGR107-TGBp214-115, or pGR107-TGBp21-100 and in about 40% of N. benthamiana seedlings infiltrated with pGR107-TGBp2Δ52-60 (Fig. 7B). Moreover, the intensity of the pGR107-TGBp2Δ52-60 amplicon was consistently lower than that of the others (Fig. 7B). The amplified fragments were further confirmed by DNA sequencing. The results showed that all the amplified fragments were the target segment in the PVX genome. Intriguingly, the pGR107-TGBp2Δ52-60 and pGR107-TGBp2ΔCD progenies in all N. benthamiana seedlings retained the introduced mutations in TGBp2, whereas all pGR107-TGBp2ΔND progenies in the N. benthamiana systemic leaves with viral symptoms were restored to mimic the wild type. We further performed a time course analysis to determine the presence of virus in the systemic leaves by RT-PCR from 3 to 10 dpi. The results showed that viral RNAs of the wild type, pGR107-TGBp2Δ52-60, and pGR107-TGBp21-100 were detected at 4, 6, and 5 dpi at the earliest, respectively, whereas viral RNA of pGR107-TGBp214-115 was detected up to 10 dpi. These results suggest that pGR107-TGBp214-115 is completely defective in movement and pGR107-TGBp21-100 can still accomplish virus movement with a reduced efficiency, whereas the movement of pGR107-TGBp2Δ52-60 is remarkably attenuated.
FIG 7.
TGBp2 affects PVX replication. (A) Phenotype of wild-type PVX and its mutants containing various deletions in TGBp2 on N. benthamiana seedlings at 8 dpi. (B) RT-PCR detection of PVX genomic RNA on N. benthamiana systemic leaves at 10 dpi. M, DL5000 DNA marker (Vazyme Biotech Co., Ltd., Nanjing, China); P, positive plasmid; H, healthy N. benthamiana leaf; W, N. benthamiana leaf infected by wild-type PVX. (C) Quantitative RT-PCR analysis of the genomic RNA of pGR107, pGR107-TGBp2ΔND, pGR107-TGBp2Δ52-60, and pGR107-TGBp2ΔCD in N. benthamiana leaves at 48 hpi. The viral RNA level of wild-type PVX was normalized to 1. Bars represent SDs from three biological repeats. *, **, and NS, respectively, indicate P values of <0.01, <0.001, and >0.1 compared with the amount of wild-type PVX genomic RNA at the same time point by the Student t test. (D) Time course study of the replication of pGR107, pGR107-ΔGDD, and TGBp2 or TGBp3 knockout PVX infectious clones in N. benthamiana leaves. Bars represent the SEs from 5 biological repeats. (E) Accumulation of wild-type and TGBp2-mutated PVX in N. benthamiana protoplasts at 24 and 48 h posttransformation (hpt) The viral genomic RNA level of the wild type at 24 hpt was normalized to 1. Bars represent the SDs from three biological repeats. *, **, and NS, respectively, indicate P values of <0.01, <0.001, or >0.1 compared with amount of wild-type PVX genomic RNA at the same time point by the Student t test, respectively. (F) Comparison of the fluorescence of wild-type, replication-defective (ΔGDD), and TGBp2 or TGBp3 knockout PVX infectious clones in N. benthamiana leaves at 48 hpi. All micrographs were obtained using the same parameters. The typical irregularly shaped X-bodies are indicated by white arrowheads; the uninfected cell in image III is indicated by an asterisk. Scale bars= 50 μm. (G) Replication of wild-type PVX under the condition of overexpression of TGBp2. The viral genomic RNA level of wild-type at 36 hpt was normalized to 1. Bars represent SDs from three biological repeats.
As the intensities of the amplicons from N. benthamiana seedlings infiltrated with pGR107-TGBp2Δ52-60 were significantly lower than those amplified from plants infiltrated with pGR107, pGR107-TGBp214-115, or pGR107-TGBp21-100 (Fig. 7B), we suspected that the attenuated movement of pGR107-TGBp2Δ52-60 might be due to reduced viral replication. Therefore, quantitative RT-PCR (qRT-PCR) was performed to evaluate the viral RNA levels of pGR107, pGR107-TGBp2Δ52-60, and pGR107-TGBp21-100 in the systemic leaves. The results showed that the viral RNAs of pGR107-TGBp2Δ52-60 and pGR107-TGBp21-100 were reduced by ∼75 and 15% compared with that of pGR107. To confirm these results, N. benthamiana leaves were infiltrated with pGR107, pGR107-TGBp214-115, pGR107-TGBp2Δ52-60, or pGR107-TGBp21-100 at exactly the same concentration and the accumulation of viral genomic RNAs was evaluated 48 hpi. Consistently, qRT-PCR results showed that the viral RNAs of pGR107-TGBp2Δ52-60 and pGR107-TGBp21-100 were significantly lower than those of wild-type PVX at 48 hpi (Fig. 7C). In contrast, there were no significant differences between the viral RNAs of the wild type and pGR107-TGBp214-115 (Fig. 7C). Taken together, these results suggest that deletion of the ND of TGBp2 specially affects viral movement and deletion of the entire CD of TGBp2 influences viral movement possibly by distracting viral replication, whereas deletion of the 9 aa in the MD of TGBp2 affect both viral replication and movement.
TGBp2 is required for robust viral replication.
To further confirm the involvement of TGBp2 in PVX replication, we directly compared the replications of wild-type PVX and the TGBp2 knockout mutant (pGR107-ΔTGBp2) using a leaf disc assay. The TGBp3 knockout PVX infectious clone (pGR107-ΔTGBp3) was also included as a movement-defective PVX mutant. As a negative control, we also constructed a replication-defective PVX infectious clone by mutating three key residues (Gly-Asp-Asp) in RdRp into alanine (pGR107-ΔGDD) to indicate basal expression of the CaMV 35S promoter. These four PVX infectious clones were allowed to infiltrate N. benthamiana leaves at exactly the same concentration, and the accumulation of PVX genomic RNA was monitored from 36 to 72 h at 12-h intervals by qRT-PCR. At 36 hpi, a time point when Agrobacterium starts to inject the T-DNA into the cell, there was no obvious viral replication in any infectious clones compared with pGR107-ΔGDD (Fig. 7D). At 48 hpi, there was significantly more genomic RNA of the wild type than of pGR107-ΔGDD (Fig. 7D), suggesting that viral replication had occurred at this time point. The genomic RNA of pGR107-ΔTGBp3 was compatible with that of pGR107 at 48 hpi (Fig. 7D), suggesting that the replication of PVX was not affected by the deletion of TGBp3 and that there was no or very low, if any, intercellular movement at this time point. Interestingly, the genomic RNA levels of pGR107-ΔTGBp2 were only slightly higher than those of pGR107-ΔGDD at 48 hpi (Fig. 7D). During the late infection stage (60 and 72 hpi), the genomic RNAs of pGR107-ΔTGBp3 consistently increased at an intensity less than that of wild-type PVX but slightly decreased at 72 hpi. This result should be due to the smaller number of infectious cells caused by the prohibited intercellular movement ability of pGR107-ΔTGBp3, the gradual decrease in host resources for viral replication, and the enhanced host antiviral responses in those primary infectious cells. However, the genomic RNA levels of pGR107-ΔTGBp2 were consistently lower than those of pGR107 and pGR107-ΔTGBp3 at both 60 and 72 hpi (Fig. 7D). We also performed a replication assay in N. benthamiana protoplasts. The N. benthamiana protoplasts were transfected with pGR107, pGR107-ΔTGBp2, pGR107-ΔTGBp3, or pGR107-ΔGDD, and the accumulation of PVX genomic RNA was evaluated by qRT-PCR at 24 and 48 hpi. The results showed that the genomic RNA of pGR107-ΔTGBp2 was significantly lower than that of pGR107 or pGR107-ΔTGBp3 at both 24 and 48 hpi (Fig. 7E).
We also directly compared CP accumulation at a time point prior to vivid intercellular movement, i.e., 48 hpi. PVX CP is translated from a subgenomic RNA that is generated depending on viral replication, thus providing an opportunity to exclude the basal expression of the CaMV 35S promoter of PVX genome RNA. Therefore, the wild type, and TGBp2 or TGBp3 knockout and the replication-defective PVX infectious clones coexpressing mCherry-2A-CP were constructed (pGR.mCh, pGR.mCh-ΔTGBp2, pGR.mCh-ΔTGBp3, and pGR.mCh-ΔGDD, respectively). These PVX infectious clones were used to infiltrate N. benthamiana leaves, and the fluorescence intensity of mCherry on the infiltrated leaf area was monitored by confocal microscopy at 48 hpi. No red fluorescence could be detected from the leaves infiltrated with the replication-defective PVX infectious clone (pGR.mCh-ΔGDD) at 48 hpi (Fig. 7FI), further confirming that expression of the mCherry-2A-CP chimeric protein was dependent on viral replication. Red fluorescence was observed from leaves infected by the other three PVX infectious clones at 48 hpi (Fig. 7FII to FIV), consistent with previous results showing that neither TGBp2 nor TGBp3 is essential for PVX replication per se (5, 6, 44). However, in comparison to the bright red fluorescence from both pGR.mCh and pGR.mCh-ΔTGBp3 (Fig. 7FII and FIII), only weak red fluorescence was detected from pGR.mCh-ΔTGBp2 under the same conditions (Fig. 7FIV). We also tested whether overexpression of TGBp2 could enhance the replication of PVX. The N. benthamiana seedlings were infiltrated with pGR107, pGR107/YFP-TGBp2, or pGR107 plus YFP-TGBp2 at the same concentration to overexpress an additional copy of N-terminally YFP-tagged TGBp2 in cis and in trans. The accumulation of PVX genomic RNA was evaluated by qRT-PCR at 36 and 48 hpi. The results showed that there were no significant differences between the viral RNAs of the wild type, pGR107/YFP-TGBp2, and pGR107+YFP-TGBp2 at 36 hpi (Fig. 7G), suggesting that the viral loadings were equal between the three treatments. Interestingly, the viral genomic RNA levels in the N. benthamiana leaves overexpressing TGBp2 were significantly higher than in those infiltrated with pGR107 alone at 48 hpi (Fig. 7G). Taken together, these data confirm that TGBp2 is required for robust PVX replication.
DISCUSSION
In this study, the dsRNA in PVX-infected N. benthamiana epidermal cells was analyzed in detail using a novel in vivo dsRNA labeling system. We showed that the dsRNA was present as motile foci in the cytoplasm at the early stage of infection, which slowly clustered into large irregularly shaped aggregates in the perinuclear area (Fig. 1A). Moreover, the dsRNAs labeled using the dRBFC assay in the X-body colocalized with PVX RdRp (Fig. 1D). Since dsRNA is the intermediate for viral replication, these results thus confirmed the previous suggestion that robust replication occurs in X-bodies (6). The replication of many positive-stranded RNA viruses occurs in membranous inclusion bodies, vesicles, multivesicular bodies, or spherules that are remodeled from the plant cell endomembrane by viral protein(s) (45–48). RdRp is the only viral protein that is absolutely required for PVX replication (3, 4); however, a transmembrane domain or membrane-associated motif has not yet been revealed. In the present study, we found that the RepC domain localized exclusively in the cytoplasm and induced vesicle-like structures (Fig. 5D), indicating the presence of a transmembrane domain or membrane-associated motif within the C-terminal part of the REP domain and thus further confirming the previous suggestion that the replication of PVX also occurred in the membranous inclusion bodies formed by RdRp (26). Intriguingly, the full-length RdRp localized primarily to the nucleus when transiently expressed alone or together with TGBp2 (Fig. 1B and Fig. 5G), whereas it relocated to the cytoplasm during PVX infection (Fig. 1C). Moreover, different domains of RdRp displayed remarkably different subcellular localizations (Fig. 5B and D). These data suggest that PVX RdRp undergoes a sophisticated conformational conversion during PVX infection that results in its retention in the cytoplasm for viral replication. Interestingly, we found that REP-YFP localized in both the cytoplasm and nucleus, whereas RepC localized exclusively in the cytoplasm and induced the formation of vesicle-like structures (Fig. 5B and D), suggesting that the transmembrane or membrane-associated motif in the RepC domain might be wrapped by RepN. It is well known that the replication of positive-strand RNA viruses is usually coupled to translation of the replication protein (49–51). For instance, the RdRp of tobacco mosaic virus (TMV) cotranslationally binds to the 5′ untranslated region of genomic RNA to ensure rapid viral replication (52). The same phenomenon might also occur for PVX: as soon as it is translated, PVX RdRp immediately binds to the specific RNA motif in the viral genomic RNA outside the RdRp coding region to also result in exposure of the transmembrane domain or membrane-associated motif in the RepC domain for its retention in the cytoplasm to initiate viral replication. In the absence of PVX replication, the transmembrane domain or membrane-associated motif within the RepC domain is enclosed by RepN, resulting in its transfer to the nucleus via the strong nuclear localization signal in the UNL domain. However, further experiments are needed to fully validate this hypothesis.
TGBp2 is a small membrane protein that forms an ER-derived chain mail-like structure, as revealed by 3D-SIM superresolution microscopy (23). In this study, we showed that TGBp2 localized with RdRp/dsRNA body as the chain mail-like structure (Fig. 2A and B). Moreover, we found that TGBp2 directly interacted with the RepN domain of RdRp (Fig. 5). Notably, we failed to detect an interaction between TGBp2 and full-length RdRp by BiFC. As discussed above, a conformational conversion is needed to expose of the transmembrane or membrane-associated motif in the RepC domain. Thus, it is possible that RepN in the transiently expressed RdRp is inaccessible to TGBp2. Alternatively, the interaction of the two proteins might need the viral genomic RNA. BiFC and MYTH showed that the MD and CD are required for the interaction of TGBp2 with PVX RdRp (Fig. 6B and C). A TGBp2 mutant (TGBp2m2) that partially overlapped with the deletion in TGBp2Δ52-60 changed the morphology of TGBp2 (36). We also found deletion of aa 52 to 60 affects the cellular localization of TGBp2 (Fig. 6D). A PVX infectious clone harboring TGBp2m2 is defective in intercellular movement (36); in contrast, PVX containing TGBp2Δ52-60 can still accomplish viral movement with a very low efficiency (Fig. 7A and B). The reason for this difference is unclear at present; however, our results do suggest that the correct tertiary structure is essential for TGBp2 to interact with PVX RdRp. We are now trying to analyze the intercellular movement and long-distance movement of a PVX mutant harboring TGBp2Δ52-60. Our results also showed that the CD, but not the ND, was indispensable for the interaction with PVX RdRp. Intriguingly, deletion of the CD had very little, if any, influence on the cellular localization of TGBp2 (Fig. 6D). A topology study revealed both the ND and CD in the cytosol, whereas the MD was in the ER lumen (22). Thus, the MD impairs the interaction between TGBp2 and RdRp by influencing TGBp2 cellular localization, whereas the CD might directly interact with the RepN domain of RdRp. However, it is also possible that deletion of the CD affected the nonspecific RNA binding activity of PVX TGBp2, as a previous study based on BaMV TGBp2 showed that both the ND and CD may be involved in TGBp2 RNA binding (37). Interestingly, a PVX infectious clone harboring a mutant TGBp2 lacking the entire N-terminal domain was defective in movement but had a replication efficiency similar to that of wild-type PVX, whereas PVX infectious clones harboring a mutant TGBp2 lacking the 9 aa in the MD or the entire CD were attenuated in both movement and replication (Fig. 7A to C). These data suggest that the ND domain is primarily involved in viral movement, whereas the MD and CD, which are involved in interaction with PVX RdRp, are required for robust replication. Moreover, the replication of a TGBp2 deletion PVX infectious clone was greatly attenuated compared with that of wild-type PVX (Fig. 7D to F), and overexpression of TGBp2 enhanced PVX replication (Fig. 7G). Taken together, these data clearly confirm that TGBp2 is required for robust viral replication, in addition to functioning as a viral movement protein. Considering the specific localization of TGBp2 in the X-body, it is feasible that TGBp2 could provide an additional sheltered environment for PVX replication. However, it is also possible that the enhancement of PVX replication was due to the nonspecific RNA binding activity of TGBp2. We are currently attempting to explore the molecular mechanism responsible for this phenomenon.
During replication, PVX forms large amorphous complexes (X-bodies) that contain all five viral proteins, viral RNA, host factors, and host endomembrane and proteins (6, 23–25). We observed TGBp3 as isolated patches or clusters closely neighboring the RdRp/dsRNA bodies in the X-bodies (Fig. 3), which were clearly distinct from the chain mail-like structure of TGBp2 (Fig. 2). Moreover, our results showed that the localization of TGBp3 to the X-body was dependent on TGBp2 (Fig. 4C and D). Taken together, these results indicate that TGBp2 functions in bridging the interaction between the RdRp/dsRNA body and TGBp3. It has been proposed that plant viral replication and intercellular movement are tightly linked (53). For PVX, it is proposed that the two processes are coupled by the cap-like complexes at the entrances of PD (11). These cap-like complexes are small viral replication complex per se because they share the same components as the large perinuclear X-body and display vivid virus replication (11). However, how these complexes are formed has remained elusive. Based on our results and previous observations (11–13, 23), it is possible that TGBp1 is recruited to the PD mainly by TGBp3 and CP is inserted into PD by TGBp1 (11), whereas the RdRp/dsRNA bodies are transported to the PD via the bridging function of TGBp2. Interestingly, TGBp2 proteins of TGB-containing plant viruses from other families, e.g., potato mop-top virus (PMTV, a pomovirus), poa semilatent virus (PSLV, a hordeivirus), and barley stripe mosaic virus (BSMV, a hordeivirus), also associate with viral replication complexes (54, 55), possess nonspecific RNA binding activity (56), or interact with and rely on TGBp3 for its PD localization (39, 56, 57). This similar subcellular localization and protein-protein interaction network implies a conserved role for TGBp2 in TGB-containing viruses.
Previous studies have shown that the X-body has a layered structure with TGBp1 aggregates at its center, TGBp2/TGBp3 and nonencapsidated vRNA in the middle, and encapsidated virions at the cytoplasmic periphery (6, 23). Summarizing our results and these previous observations, a more sophisticated model of PVX X-body can be proposed: TGBp1 aggregates localize at the center of the X-body, followed by RdRp/dsRNA bodies; TGBp2 localizes on the surface of RdRp/dsRNA bodies, further enclosed by TGBp3; and nonencapsidated vRNA and encapsidated virions localize at the cytoplasmic periphery (Fig. 8). Additionally, a revised PVX intercellular movement model can be proposed based on our results and previous studies (Fig. 8). In this model, newly translated RdRp binds to viral genomic RNA in the cytoplasm to form the core replication units which subsequently recruit TGBp1, TGBp2, TGBp3, and CP to form typical cytoplasmic “X-bodies.” Due to the cytoplasmic streaming and/or other intracellular movement mechanisms, these X-bodies are gathered into a typical perinuclear X-body or transported by TGBp3 to the entrance of PD. It is also possible that the cap-like structure at the entrance of PD is assembled de novo via the recruitment of TGBp1 and TGBp2 by TGBp3, CP by TGBp1, and RdRp/dsRNA bodies by TGBp2.
FIG 8.
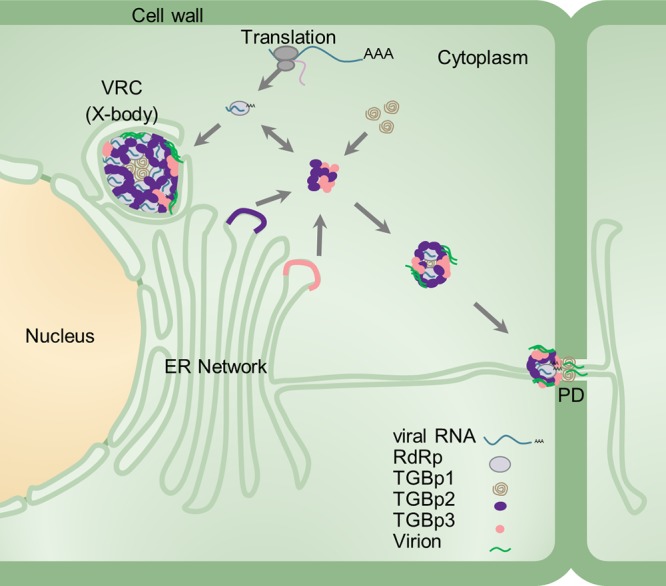
Schematic model of the PVX X-body. RdRp/dsRNA bodies, TGBp1, TGBp2, TGBp3, viral RNA, and virions are represented by gray spherules, brown helices, purple spherules, roseous spherules, cyan strings, and green streaks, respectively. The ribosome, nucleus, cytoplasm, cell wall, ER network, and PD are also indicated.
MATERIALS AND METHODS
Plant growth condition and virus inoculations.
The N. benthamiana plants were grown in pots in a growth chamber under a 16-h/8-h photoperiod and 60% humidity at 23°C. The agrobacterial infiltrations were performed as described previously (58). Overnight cultures of agrobacteria carrying the proper plasmid were washed twice with infiltration buffer (10 mM morpholineethanesulfonic acid [MES; pH 5.6], 10 mM MgCl, 100 μM acetosyringone) and used to infiltrate 3-week-old N. benthamiana leaves at an optical density at 600 nm (OD600) of 0.1 using a 1-ml needleless syringe. For PVX infectious clones, the Agrobacterium tumefaciens strain GV3101 containing pSoup helping plasmid was used (59).
Vector construction.
Transient-expression vectors were constructed using Gateway technology (Invitrogen, Shanghai, China). The full coding sequences of PVX RdRp, TGBp1, TGBp2, and TGBp3 and the coding sequence of the RdRp domains (MET [aa 1 to 341], UNL [aa 342 to 731], HEL [aa 732 to 965], REP [aa 966 to 1456], RepN [aa 966 to 1131], and RepC[aa 1132 to 1456]) were amplified with Phanta Super Fidelity DNA polymerase (Vazyme, Nanjing, China) using pGR107 (33) as the template and transferred into the entry vector pENTR/d-TOPO or vector pDONR221 (Invitrogen). TGBp2 mutants in the pENTR vector, including pENTR-TGBp2Δ6-11 (lacking aa 6 to 11), pENTR-TGBp2ΔND (lacking aa 1 to 15), pENTR-TGBp2ΔCD (lackig aa 101 to 115), pENTR-TGBp2Δ104-112 (lacking aa 104 to 112), and pENTR-TGBp2Δ52-60 (lacking aa 52 to 60) were constructed by Gibson assembly (New England BioLabs Inc., Shanghai, China). pEarleyGate-101, -102, 104 (34), and pGWB554 (60) were used to construct the C-terminally YFP-, cyan fluorescent protein (CFP)-, and mRFP-tagged and N-terminally YFP-tagged constructs, respectively. For BiFC, p35S-YN-Gateway, p35S-YC-Gateway, p35S-Gateway-YN, and p35S-Gateway-YC (61) were used to construct N- or C-terminal YN and YC-fused constructs. To construct a set of mRFP-based dRBFC assay plasmids, pDONR-B2, pDONR-NS1, and pDONR-VP35 were recombined into Gateway-compatible mRFP-based bimolecular fluorescence complementation vectors, pBaTl-ccdB-NmRFP and pBaTl-ccdB-CmRFP (62), by LR recombination to generate B2-NmRFP, B2-CmRFP, NS1-NmRFP, NS1-CmRFP, VP35-NmRFP, and VP35-CmRFP, respectively. Two Gateway-compatible MYTH plasmids (pBT3N-GW and pPR3C-GW) were constructed using the Gibson assembly method according to the manufacturer’s instruments. The fragment including attR1, the chloramphenicol resistance gene (CmR), the ccdB killer gene, and attR2 was amplified from pEarley104 and inserted into the multiple-cloning sites of pBT3-N and pPR3-C (Dualsystems Biotech AG, Schlieren, Switzerland). TGBp2, RDR, and their mutants in the pENTR vector were then inserted by LR recombination. To construct nontagged TGBp3, the full coding sequence of TGBp3 including the stop codon was amplified, transferred into the entry vector pDONR207, and then inserted into pEarley100 by BP and LR recombination, respectively.
A pGR107-based PVX infectious clone that included the two PUMHD-binding sites for PUMHD-based viral genomic RNA labeling and an N-terminal red fluorescent protein (mCherry) and 2A-fused CP (mCherry-2A-CP) (27), i.e., pGR.mCh, was constructed by replacing the sequence between the unique ClaI and XhoI restriction sites with the fragment from PVX.pum-Cherry-CP (27) released with the same restriction enzymes. The 2A peptide linker between mCherry and CP is derived from foot and mouth disease virus (FMDV), which leads to partial cotranslational cleavage of mCherry and CP in the overcoat PVX virus (63). To allow the expression of viral protein in the same cell as infected by PVX, the Gateway cassette of pEarley101 or pEarley104, including the CaMV 35S promoter, attR1, CmR, ccdB, and attR2, and the OCS terminator, was amplified and directly inserted into the unique SfoI restriction site between NOS terminator and T-DNA left border sequence in pGR107 using the InFusion method (Clontech, Shanghai, China) to construct pGR107/Gateway-YFP and pGR107/YFP-Gateway. The RdRp, TGBp3, TGBp2, TGBp2, or their mutants in the pENTR vector was then inserted into the Gateway cassette by the LR reaction.
To construct replication-defective (Gly-Asp-Asp to Ala-Ala-Ala) and TGBp1, TGBp2, or TGBp3 single- or double-knockout PVX mutants, a fragment from the unique AvrII site in RdRp to the unique HpaI site in CP (nt 4390 to 5899) was amplified with Phusion DNA polymerase and ligated into the pCR-Blunt vector (Invitrogen) to construct pCR-RdRp-CP. The three key residues (Gly-Asp-Asp) in RdRp for RNA polymerase activity were mutated to Ala, and amino acids 6 (Ser) and 7 (Ser) in TGBp1, amino acids 10 (Ala) and 11 (Pro) in TGBp2, or amino acids 25 (Ser) and 26 (Leu) in TGBp3 were mutated to stop codons using the QuickChange Mutagenesis II kit (Strategene). The insertions containing the appropriate mutations were released by AvrII and ClaI or NdeI and HpaI double digestion and inserted back into the appropriate PVX infectious clones treated with the same restriction enzymes. The PVX infectious clones harboring TGBp2ΔND, TGBp2Δ52-60, or TGBp2ΔCD were constructed by overlap PCR. The initiator codon (ATG) of TGBp2 was mutated to ACG, resulting in conversion of the 221st codon of TGBp1 (TAT) to TAC (both TAT and TAC encode Tyr), and the 14th codon for Ser (TCT) was mutated to ATG in the PVX infectious clone harboring TGBp2ΔND (pGR107-TGBp214-115); aa 52 to 60 of TGBp2 were completely deleted in PVX infectious clone harboring TGBp2Δ52-60 (pGR107-TGBp2Δ52-60), whereas the 101st codon (CAA) for Gln was mutated to a stop codon (TAA), which also resulted in conversion of the 7th codon of TGB3 (CTC) to CTT (both CTC and CTT encode Leu), in the PVX infectious clones harboring TGBp2ΔCD (pGR107-TGBp21-100). All plasmids were verified by DNA sequencing.
Confocal microscopy.
Confocal microscopy analysis was performed as described previously (28). The fluorescence signal in the infiltrated leaf area was monitored with a Leica TCS SP5 2 confocal laser scanning microscope (Leica, Germany). CFP was excited at 405 nm and collected between 465 and 485 nm, YFP was excited at 496 nm and collected between 520 and 535 nm, and mCherry or mRFP was excited at 543 nm and collected between 576 and 631 nm. The sequential mode was used when multiple fluorescent proteins were coexpressed, and each fluorescence signal was further confirmed separately to avoid signal cross-contamination.
N. benthamiana protoplast preparation and transfection.
The N. benthamiana protoplasts were prepared using well-expanded leaves from 4-week-old seedlings as described previously (58). Protoplasts were transfected with proper endotoxin-free plasmids via DNA-polyethylene glycol (PEG)-calcium transfection (28). The EndoFree Midi Plasmid Kit II (Tiangen Biotech Co., LTD, Beijing, China) was used to obtain the endotoxin-free plasmids according to the provided protocol.
Double-stranded RNA detection.
dsRNA was detected by both the binding-dependent fluorescence complementation assay (dRBFC assay) (28) and the J2 antibody-based immunofluorescence assay (64). For the dRBFC assay, agrobacteria carrying the two plasmids for the dRBFC assay, e.g., pdRBFC-B2-CmRFP and pdRBFC-NS1-NmRFP, and PVX infectious clone or transient-expression vectors were mixed in a ratio of 1:1:2 and used to coinfiltrate N. benthamiana leaves at a final OD600 of 0.1. The infiltrated leaf areas were monitored using confocal microscopy. The J2 antibody-based immunofluorescence assay was performed as described earlier (28).
RNA extraction and qRT-PCR.
The total RNA of N. benthamiana protoplasts was isolated using TRIzol reagent (Invitrogen) according to the provided protocol, whereas the total RNA of N. benthamiana leaf tissue was extracted using a RNAprep pure plant kit (Tiangen). Leaf tissues of 5 plants from the same treatment were pooled as a biological replicate to minimize variations within the same treatment. We used 500 μg (leaf tissue) or 200 μg (protoplasts) of total RNA as the template for first-strand cDNA synthesis using an oligo(dT)12-18 primer with the Superscript IV reverse transcriptase (Invitrogen). qRT-PCR was performed as described previously (65). In brief, qRT-PCR was performed in a 20-μl volume system containing 4 μl of 50-fold-diluted cDNA, 5 μM each primer, and 1× SYBR green master mix (Transgen) on a CFX96 Touch real-time PCR detection system (Bio-Rad). The genomic RNA of PVX was determined by amplification of a 257-bp fragment of the PVX CP gene, and the N. benthamiana ACTIN gene (NbActin; GenBank accession no. AY179605) was used as an internal control. All experiments were repeated at least three times.
MYTH.
MYTH was performed as described previously (58, 65). In brief, plasmids were introduced into yeast strain NMY51 (Waryong Biotech, Beijing, China) using the Super Yeast Transformation Kit II (Coolaber Biotech, Beijing, China) according to the provided protocol. Transformed yeast cells were plated onto SD medium lacking Trp and Leu and cultured for 2 days at 30°C. Several independent positive transformants were transferred to high-stringency selection plates lacking Trp, Leu, His, and Ade and incubated at 30°C for 3 to 4 days.
ACKNOWLEDGMENTS
We thank Karl J. Oparka (University of Edinburgh, UK) for providing the PVX infectious clone (PVX.pum-Cherry-CP) and Dirk Prüfer (Fraunhofer Institut für Molekularbiologie und Angewandte Ökologie, Aachen, Germany) for providing the mRFP-based BiFC Gateway plasmids. We also express special thanks to Jens Tilsner (University of St. Andrews, UK) for helpful advice regarding the confocal microscopy imaging.
This research was supported financially by the Natural Science Foundation of China (31671998) and the Natural Science Foundation of Heilongjiang Province (ZD2018002).
REFERENCES
- 1.Sieczkarski SB, Whittaker GR. 2002. Dissecting virus entry via endocytosis. J Gen Virol 83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 2.Schoelz JE, Harries PA, Nelson RS. 2011. Intracellular transport of plant viruses: finding the door out of the cell. Mol Plant 4:813–831. doi: 10.1093/mp/ssr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morozov SY, Solovyev AG. 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J Gen Virol 84:1351–1366. doi: 10.1099/vir.0.18922-0. [DOI] [PubMed] [Google Scholar]
- 4.Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. 2010. Varied movement strategies employed by triple gene block–encoding viruses. Mol Plant Microbe Interact 23:1231–1247. doi: 10.1094/MPMI-04-10-0086. [DOI] [PubMed] [Google Scholar]
- 5.Beck DL, Guilford PJ, Voot DM, Andersen MT, Forster RL. 1991. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology 183:695–702. doi: 10.1016/0042-6822(91)90998-Q. [DOI] [PubMed] [Google Scholar]
- 6.Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, Lacomme C, Santa Cruz S, Oparka KJ. 2012. The TGB1 movement protein of potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol 158:1359–1370. doi: 10.1104/pp.111.189605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz SS, Roberts AG, Prior DA, Chapman S, Oparka KJ. 1998. Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10:495–510. doi: 10.1105/tpc.10.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard AR, Heppler ML, Ju HJ, Krishnamurthy K, Payton ME, Verchot-Lubicz J. 2004. Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328:185–197. doi: 10.1016/j.virol.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 9.Angell SM, Davies C, Baulcombe DC. 1996. Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology 216:197–201. doi: 10.1006/viro.1996.0046. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Ding B, Baulcombe DC, Verchot J. 2000. Cell-to-cell movement of the 25K protein of potato virus X is regulated by three other viral proteins. Mol Plant Microbe Interact 13:599–605. doi: 10.1094/MPMI.2000.13.6.599. [DOI] [PubMed] [Google Scholar]
- 11.Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ. 2013. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201:981–995. doi: 10.1083/jcb.201304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SC, Wu CH, Wang CW. 2010. Traffic of a viral movement protein complex to the highly curved tubules of the cortical endoplasmic reticulum. Traffic. Traffic 11:912–930. doi: 10.1111/j.1600-0854.2010.01064.x. [DOI] [PubMed] [Google Scholar]
- 13.Schepetilnikov MV, Manske U, Solovyev AG, Zamyatnin AA Jr, Schiemann J, Morozov SY. 2005. The hydrophobic segment of potato virus X TGBp3 is a major determinant of the protein intracellular trafficking. J Gen Virol 86:2379–2391. doi: 10.1099/vir.0.80865-0. [DOI] [PubMed] [Google Scholar]
- 14.Atabekov JG, Rodionova NP, Karpova OV, Kozlovsky SV, Poljakov VY. 2000. The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271:259–263. doi: 10.1006/viro.2000.0319. [DOI] [PubMed] [Google Scholar]
- 15.Rodionova NP, Karpova OV, Kozlovsky SV, Zayakina OV, Arkhipenko MV, Atabekov JG. 2003. Linear remodeling of helical virus by movement protein binding. J Mol Biol 333:565–572. doi: 10.1016/j.jmb.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 16.Leshchiner AD, Solovyev AG, Morozov SY, Kalinina NO. 2006. A minimal region in the NTPase/helicase domain of the TGBp1 plant virus movement protein is responsible for ATPase activity and cooperative RNA binding. J Gen Virol 87:3087–3095. doi: 10.1099/vir.0.81971-0. [DOI] [PubMed] [Google Scholar]
- 17.Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, Yamaji Y, Namba S. 2014. In planta recognition of a double-stranded RNA synthesis protein complex by a potexviral RNA silencing suppressor. Plant Cell 26:2168–2183. doi: 10.1105/tpc.113.120535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voinnet O, Lederer C, Baulcombe DC. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–167. doi: 10.1016/S0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 19.Krishnamurthy K, Heppler M, Mitra R, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J. 2003. The potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309:135–151. doi: 10.1016/S0042-6822(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 20.Ju HJ, Samuels TD, Wang YS, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS, Verchot-Lubicz J. 2005. The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138:1877–1895. doi: 10.1104/pp.105.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra R, Krishnamurthy K, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J. 2003. The potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology 312:35–48. doi: 10.1016/S0042-6822(03)00180-6. [DOI] [PubMed] [Google Scholar]
- 22.Hsu HT, Chou YL, Tseng YH, Lin YH, Lin TM, Lin NS, Hsu YH, Chang BY. 2008. Topological properties of the triple gene block protein 2 of bamboo mosaic virus. Virology 379:1–9. doi: 10.1016/j.virol.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Linnik O, Liesche J, Tilsner J, Oparka KJ. 2013. Unraveling the structure of viral replication complexes at super-resolution. Front Plant Sci 4:6. doi: 10.3389/fpls.2013.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harries PA, Park JW, Sasaki N, Ballard KD, Maule AJ, Nelson RS. 2009. Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci U S A 106:17594–17599. doi: 10.1073/pnas.0909239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalla TA, Shepard JF. 1972. The structure and antigenic analysis of amorphous inclusion bodies induced by potato virus X. Virology 49:654–667. doi: 10.1016/0042-6822(72)90522-3. [DOI] [PubMed] [Google Scholar]
- 26.Bamunusinghe D, Hemenway CL, Nelson RS, Sanderfoot AA, Ye CM, Silva MAT, Payton M, Verchot-Lubicz J. 2009. Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393:272–285. doi: 10.1016/j.virol.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Tilsner J, Linnik O, Christensen NM, Bell K, Roberts IM, Lacomme C, Oparka KJ. 2009. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J 57:758–770. doi: 10.1111/j.1365-313X.2008.03720.x. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X, Deng P, Cui H, Wang A. 2015. Visualizing double-stranded RNA distribution and dynamics in living cells by dsRNA binding-dependent fluorescence complementation. Virology 485:439–451. doi: 10.1016/j.virol.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Xu K, Nagy PD. 2016. Enrichment of phosphatidylethanolamine in viral replication compartments via co-opting the endosomal Rab5 small GTPase by a positive-strand RNA virus. PLoS Biol 14:e2000128. doi: 10.1371/journal.pbio.2000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, Zhang Y, Yang M, Liu S, Li Z, Wang X, Han C, Yu J, Li D. 2017. The barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog 13:e1006319. doi: 10.1371/journal.ppat.1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son K-N, Liang Z, Lipton HL. 2015. Double-strand RNA is detected by immunofluorescence analysis in RNA and DNA virus infections including those by negative-strand RNA viruses. J Virol 89:9383–9392. doi: 10.1128/JVI.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monsion B, Incarbone M, Hleibieh K, Poignavent V, Ghannam A, Dunoyer P, Daeffler L, Tilsner J, Ritzenthaler C. 2018. Efficient detection of long dsRNA in vitro and in vivo using the dsRNA binding domain from FHV B2 protein. Front Plant Sci 9:70. doi: 10.3389/fpls.2018.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. 1999. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 35.Doronin SV, Hemenway C. 1996. Synthesis of potato virus X RNAs by membrane-containing extracts. J Virol 70:4795–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ju HJ, Brown JE, Ye C-M, Verchot-Lubicz J. 2007. Mutations in the central domain of potato virus X TGBp2 eliminate granular vesicles and virus cell-to-cell trafficking. J Virol 81:1899–1911. doi: 10.1128/JVI.02009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu HT, Tseng YH, Chou YL, Su SH, Hsu YH, Chang BY. 2009. Characterization of the RNA-binding properties of the triple-gene-block protein 2 of bamboo mosaic virus. Virol J 6:50. doi: 10.1186/1743-422X-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuels TD, Ju HJ, Ye CM, Motes CM, Blancaflor EB, Verchot-Lubicz J. 2007. Subcellular targeting and interactions among the potato virus X TGB proteins. Virology 367:375–389. doi: 10.1016/j.virol.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Solovyev AG, Stroganova TA, Zamyatnin AA Jr, Fedorkin ON, Schiemann J, Morozov SY. 2000. Subcellular sorting of small membrane-associated triple gene block proteins: TGBp3-assisted targeting of TGBp2. Virology 269:113–127. doi: 10.1006/viro.2000.0200. [DOI] [PubMed] [Google Scholar]
- 40.Gorshkova EN, Erokhina TN, Stroganova TA, Yelina NE, Zamyatnin AA Jr, Kalinina NO, Schiemann J, Solovyev AG, Morozov SY. 2003. Immunodetection and fluorescent microscopy of transgenically expressed hordeivirus TGBp3 movement protein reveals its association with endoplasmic reticulum elements in close proximity to plasmodesmata. J Gen Virol 84:985–994. doi: 10.1099/vir.0.18885-0. [DOI] [PubMed] [Google Scholar]
- 41.Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 42.Thaminy S, Auerbach D, Arnoldo A, Stagljar I. 2003. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res 13:1744–1753. doi: 10.1101/gr.1276503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stagljar I, Korostensky C, Johnsson N, te Heesen S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A 95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batten JS, Yoshinari S, Hemenway C. 2003. Potato virus X: a model system for virus replication, movement and gene expression. Mol Plant Pathol 4:125–131. doi: 10.1046/j.1364-3703.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- 45.Jin X, Cao X, Wang X, Jiang J, Wan J, Laliberté J-F, Zhang Y. 2018. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front Plant Sci 9:57. doi: 10.3389/fpls.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harak C, Lohmann V. 2015. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479–480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu K, Nagy PD. 2014. Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr Opin Virol 9:119–126. doi: 10.1016/j.coviro.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Laliberté J-F, Zheng H. 2014. Viral manipulation of plant host membranes. Annu Rev Virol 1:237–259. doi: 10.1146/annurev-virology-031413-085532. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Wimmer E, Paul AV. 2009. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta 1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novak JE, Kirkegaard K. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev 8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 51.Weiland JJ, Dreher TW. 1993. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc Natl Acad Sci U S A 90:6095–6099. doi: 10.1073/pnas.90.13.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamura-Nagaya K, Ishibashi K, Huang Y-P, Miyashita S, Ishikawa M. 2014. Replication protein of tobacco mosaic virus cotranslationally binds the 5′ untranslated region of genomic RNA to enable viral replication. Proc Natl Acad Sci U S A 111:E1620–E1628. doi: 10.1073/pnas.1321660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tilsner J, Oparka KJ. 2012. Missing links? The connection between replication and movement of plant RNA viruses. Curr Opin Virol 2:705–711. doi: 10.1016/j.coviro.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Cowan GH, Roberts AG, Chapman SN, Ziegler A, Savenkov EI, Torrance L. 2012. The potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Front Plant Sci 3:290. doi: 10.3389/fpls.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torrance L, Cowan GH, Gillespie T, Ziegler A, Lacomme C. 2006. Barley stripe mosaic virus-encoded proteins triple-gene block 2 and gammab localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J Gen Virol 87:2403–2411. doi: 10.1099/vir.0.81975-0. [DOI] [PubMed] [Google Scholar]
- 56.Cowan GH, Lioliopoulou F, Ziegler A, Torrance L. 2002. Subcellular localisation, protein interactions, and RNA binding of Potato mop-top virus triple gene block proteins. Virology 298:106–115. doi: 10.1006/viro.2002.1435. [DOI] [PubMed] [Google Scholar]
- 57.Lim HS, Bragg JN, Ganesan U, Ruzin S, Schichnes D, Lee MY, Vaira AM, Ryu KH, Hammond J, Jackson AO. 2009. Subcellular localization of the barley stripe mosaic virus triple gene block proteins. J Virol 83:9432–9448. doi: 10.1128/JVI.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng X, Xiong R, Li Y, Li F, Zhou X, Wang A. 2017. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 29:508–525. doi: 10.1105/tpc.16.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vain P, Afolabi AS, Worland B, Snape JW. 2003. Transgene behaviour in populations of rice plants transformed using a new dual binary vector system: pGreen/pSoup. Theor Appl Genet 107:210–217. doi: 10.1007/s00122-003-1255-7. [DOI] [PubMed] [Google Scholar]
- 60.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 61.Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EWT, Harada JJ, Rothstein SJ, Cui Y. 2010. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 62.Müller B, Noll GA, Ernst AM, Rüping B, Groscurth S, Twyman RM, Kawchuk LM, Prüfer D. 2010. Recombinant artificial forisomes provide ample quantities of smart biomaterials for use in technical devices. Appl Microbiol Biotechnol 88:689–698. doi: 10.1007/s00253-010-2771-4. [DOI] [PubMed] [Google Scholar]
- 63.Cruz SS, Chapman S, Roberts AG, Roberts IM, Prior DA, Oparka KJ. 1996. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci U S A 93:6286–6290. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng X, Wang A. 2017. The potyviral silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J Virol 91:e01478-16. doi: 10.1128/JVI.01478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]