Chronic infection with hepatitis B virus (HBV) is a major cause of liver disease and cancer in humans. Mammalian HBV-like viruses are also found in nonhuman primates, rodents, and bats. As for most viruses, HBV requires a successful interaction with a host receptor for replication. Cellular receptors are thus key determinants of host susceptibility as well as specificity. One hallmark of pathogenic virus-host relationships is the reciprocal evolution of host receptor and viral envelope proteins, as a result of their antagonistic interaction over time. The dynamics of these so-called “evolutionary arms races” can leave signatures of adaptive selection, which in turn reveal the evolutionary history of the virus-host interaction as well as viral pathogenicity and the genetic determinants of species specificity. Here, we show how HBV-like viruses have shaped the evolutionary history of their mammalian host receptor, as a result of their ancient pathogenicity, and decipher the genetic determinants of cross-species transmissions.
KEYWORDS: bats, genetic conflict, HBV, hepadnavirus, NTCP, positive selection, primates, receptor, rodents, virus-host interactions
ABSTRACT
Human hepatitis B virus (HBV) is a global health problem, affecting more than 250 million people worldwide. HBV-like viruses, named orthohepadnaviruses, also naturally infect nonhuman primates, rodents, and bats, but their pathogenicity and evolutionary history are unclear. Here, we determined the evolutionary history of the HBV receptors NTCP and GPC5 over millions of years of primate, rodent, and bat evolution. We use this as a proxy to understand the pathogenicity of orthohepadnaviruses in mammalian hosts and to determine the implications for species specificity. We found that NTCP, but not GPC5, has evolved under positive selection in primates (27 species), rodents (18 species), and bats (21 species) although at distinct residues. Notably, the positively selected codons map to the HBV-binding sites in primate NTCP, suggesting past genetic “arms races” with pathogenic orthohepadnaviruses. In rodents, the positively selected codons fall outside and within the presumed HBV-binding sites, which may contribute to the restricted circulation of rodent orthohepadnaviruses. In contrast, the presumed HBV-binding motifs in bat NTCP are conserved, and none of the positively selected codons map to this region. This suggests that orthohepadnaviruses may bind to different surfaces in bat NTCP. Alternatively, the patterns may reflect adaptive changes associated with metabolism rather than pathogens. Overall, our findings further point to NTCP as a naturally occurring genetic barrier for cross-species transmissions in primates, which may contribute to the narrow host range of HBV. In contrast, this constraint seems less important in bats, which may correspond to greater orthohepadnavirus circulation and diversity.
IMPORTANCE Chronic infection with hepatitis B virus (HBV) is a major cause of liver disease and cancer in humans. Mammalian HBV-like viruses are also found in nonhuman primates, rodents, and bats. As for most viruses, HBV requires a successful interaction with a host receptor for replication. Cellular receptors are thus key determinants of host susceptibility as well as specificity. One hallmark of pathogenic virus-host relationships is the reciprocal evolution of host receptor and viral envelope proteins, as a result of their antagonistic interaction over time. The dynamics of these so-called “evolutionary arms races” can leave signatures of adaptive selection, which in turn reveal the evolutionary history of the virus-host interaction as well as viral pathogenicity and the genetic determinants of species specificity. Here, we show how HBV-like viruses have shaped the evolutionary history of their mammalian host receptor, as a result of their ancient pathogenicity, and decipher the genetic determinants of cross-species transmissions.
INTRODUCTION
With approximately 257 million cases of chronic infections, human hepatitis B virus (HBV) infection is one of the most common viral infections and the leading cause of liver diseases worldwide. HBV is a member of the family Hepadnaviridae, which are ancient pathogens that naturally infect mammals (Orthohepadnavirus), birds (Avihepadnavirus), fishes (Metahepadnavirus), and amphibians (Herpetohepadnavirus) (1–3). Hepadnaviruses are host specific to each of these groups, suggesting an ancient virus-host association (2). However, the evolutionary history of orthohepadnaviruses as well as their pathogenicity in their mammalian hosts are poorly understood (4–6).
To date, orthohepadnaviruses have been identified in rodents, primates, and bats. However, their circulation seems predominant in the latter groups, which places primates and bats as two potential reservoirs of orthohepadnaviruses. Rodent orthohepadnaviruses have been reported in only three species of the Sciuridae family (i.e., the arctic ground squirrel, the ground squirrel, and the woodchuck). In primates, most orthohepadnaviruses have been isolated from hominoid species, including human, chimpanzee, gorilla, gibbon, and orangutan. Natural Orthohepadnavirus infections were also reported from two New World monkey species, the Woolly monkey (7) and the capuchin monkey (8). In contrast, bat orthohepadnaviruses are highly diverse and naturally infect several bat species from at least five divergent families (9–13), suggesting a long-term virus-host association. It has been further hypothesized that bats could be a source of primate HBVs (7). Phylogenetic analyses of the orthohepadnaviruses indicate few cross-species transmissions between distant primates but frequent interspecies circulations between hominoid species, as revealed by detections of human HBV genotypes in nonhuman species and by the genetic clustering of gorilla HBV within chimpanzee HBV strains and that of orangutan within gibbon HBV strains (14, 15). In bats, the occurrence of interspecies circulation between divergent bat species is contrasted by a clear virus-host association in the Rhinolophidae and Hipposideridae families (12).
Host proteins, hijacked by viruses for cellular entry, are key determinants for host susceptibility and species specificity (16–19). HBV has a liver tropism and requires a low-affinity attachment with the glypican 5 (GPC5) protein (20), followed by specific binding between the HBV preS1 domain and the cellular sodium taurocholate cotransporting polypeptide (NTCP) for entry into primate hepatocytes (21). In bats, experimental assays have shown that the tent-making bat orthohepadnavirus (TBHBV) was able to infect human primary hepatocytes using human NTCP (9). However, the entry pathway of bat orthohepadnaviruses, including their molecular interaction with NTCP, remains to be characterized.
NTCP is a multiple-transmembrane protein encoded by the solute carrier family 10 member 1 (SLC10A1) gene in humans and is functionally conserved for conjugated bile acid transport in mammals (22). Its three-dimensional (3D) structure has not been experimentally solved yet. However, mutagenesis studies have mapped the HBV-NTCP-binding determinants to amino acids 84 to 87 and 157 to 165 in NTCP (21, 23, 24). Replacing these motifs in mouse and crab-eating macaque NTCPs, two “HBV-resistant” species, with the human counterparts rendered these NTCPs functional receptors for HBV (21, 23, 24). NTCP thus appears to be a host limiting factor for HBV infection.
Such patterns of interaction between host proteins and orthohepadnaviruses can be studied in the context of virus-host evolution. Indeed, one hallmark of long-term relationships between pathogenic viruses and hosts is the reciprocal evolution of host and viral proteins, as a result of their antagonistic interaction over long periods of evolutionary time (25–28). In particular, pathogenic viruses and their host receptors can coevolve under a regime of evolutionary “arms races” where both partners reciprocally change over time for survival (25, 29). These arms races can leave evolutionary signatures at the exact sites of interaction, which are identifiable by estimating the rates of nonsynonymous substitutions (dN) over synonymous substitutions (dS) among orthologous genes (a dN/dS ratio of >1 indicates positive selection) (30). The study of these evolutionary interplays has been useful to assess the exact virus-host interfaces, the genetic factors underlying host range and viral cross-species transmissions (e.g., see references 29 and 31–35), as well as pathogenicity (36) in different systems.
Here, we examined whether orthohepadnaviruses have driven the evolution of their cellular receptors, GPC5 and NTCP, and we determined how the resulting genetic patterns may dictate virus host range. We show that NTCP, but not GPC5, has been under recurrent positive selection during primate, rodent, and bat evolution. Interestingly, the evolutionary fingerprints in primate NTCP overlap the known HBV-binding motifs, which may witness past genetic arms races with pathogenic orthohepadnaviruses. In rodents, the positively selected codons were both internal and external to the presumed HBV-binding sites. Noticeably, the latter were found to be highly variable across rodent phylogeny, which may contribute to the species specificity of rodent orthohepadnavirus. In bat NTCP, the genetic fingerprints are external to the presumed HBV-binding motifs, which may reflect either that pathogenic orthohepadnaviruses bind to other surfaces in bat NTCP or that orthohepadnaviruses have not been a selective pressure and that NTCP adaptation in bats results from another selective pressure (e.g., metabolism or diet). Finally, our findings support a model in which NTCP represents a genetic barrier for cross-species transmissions in primates. If bat orthohepadnaviruses use NTCP as a cellular receptor, this constraint may be less important in bats, which may have facilitated bat orthohepadnavirus circulation.
RESULTS AND DISCUSSION
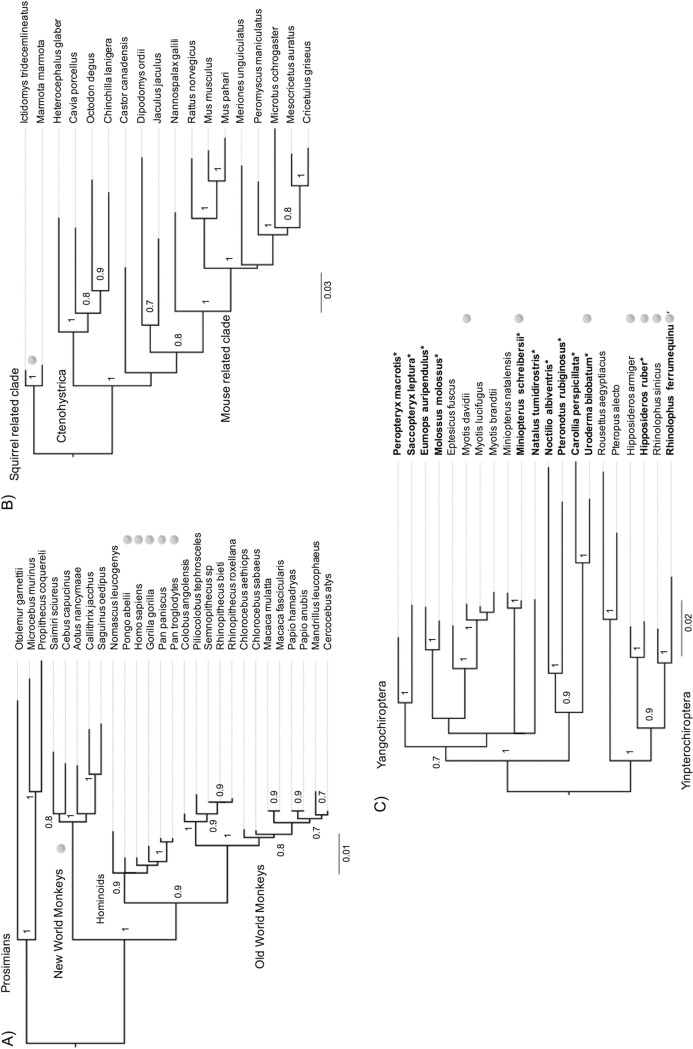
To characterize the evolutionary history of NTCP/SLC10A1 and GPC5 in primates, rodents, and bats, we retrieved their orthologous sequences from public databases and de novo sequenced the genes from additional species. NTCP sequences from 27 primate species, representing over 87 million years of divergence (37, 38), were retrieved, and the first exon of NTCP (containing one of the HBV-binding regions) was sequenced for two additional prosimian species and one New World monkey (Fig. 1A; see also Table S1 at https://figshare.com/articles/Table_S1_Information_on_the_species_and_the_sequences_used_for_evolutionary_analyses_in_primates_A_rodents_B_and_in_bats_C_/7315235). Similarly, available sequences of rodent NTCP were obtained for 18 species, spanning 65 million years of divergence (Fig. 1) (39). NTCP sequences were publicly available for only nine bat species, representing 5/19 bat families, which is too limited for robust evolutionary analyses (e.g., see reference 40), and our preliminary analyses based on the publicly available data were not statistically supported. We thus conducted an extensive sampling of bat species from key geographic locations (French Guyana, Metropolitan France, and Gabon) in order to cover a more substantial part of bat diversity. Species from 10 families of 19 were sampled, allowing us to significantly increase our sampling of NTCPs from bats. Through de novo sequencing of the NTCP/SLC10A1 genes from 12 divergent bat species, including 4 natural host species of orthohepadnaviruses, our samples now span 64 million years of bat divergence (41, 42) (Fig. 1C; see also Table S1 at https://figshare.com/articles/Table_S1_Information_on_the_species_and_the_sequences_used_for_evolutionary_analyses_in_primates_A_rodents_B_and_in_bats_C_/7315235).
FIG 1.
Phylogenetic analysis of primate, rodent, and bat NTCPs. The phylogenies were built using a PRANK alignment of orthologous nucleotide sequences of NTCPs from 27 primate (1,047 bp), 18 rodent (1,029 bp), and 21 bat (1,005 bp) species retrieved from public databases and newly sequenced in this study (in boldface type and highlighted by asterisks). The PhyML program was run using the substitution models GTR+I+G for primates and rodents and HKY+I+G for bats (see Materials and Methods for details). Node statistical supports are calculated with 1,000 bootstrap replicates (only values above 0.7 are shown). Virus pictograms indicate the species in which orthohepadnaviruses have been reported. In New World monkeys, orthohepadnaviruses have been reported from the woolly monkey and the capuchin monkey. However, NTCP sequences are not available for these species. The virus pictogram for New World monkeys indicates natural circulation of orthohepadnaviruses within this group. The bars indicate the number of nucleotide substitutions per site.
Pairwise amino acid identities of NTCP and GPC5 protein sequences (ranging from 81% to 96% [see Fig. S1A at https://figshare.com/articles/FIG_S1_NTCP_and_GPC5_protein_are_mostly_conserved_in_mammals/7315124]) revealed that they are mostly conserved in primates, rodents, and bats. This conserved property was also evident across mammalian species (78.1% pairwise amino acid identity [see Fig. S1A at https://figshare.com/articles/FIG_S1_NTCP_and_GPC5_protein_are_mostly_conserved_in_mammals/7315124]), which may reflect the overall pressure to maintain their structure and cellular functions. Notwithstanding, fitting the codon sequence alignments of NTCP and GPC5 to models that disallow positive selection (models M1 and M7, from the PAML Codeml package [43]) compared to those that allow for positive selection (M2 and M8, respectively), we found that primate, rodent, and bat NTCPs, but not GPC5s, have experienced significant and strong positive selection (Table 1). As GPC5 was shown to be a low-affinity attachment factor during the initial entry process of HBV (20), the contrasting evolutionary patterns observed between both proteins could reflect a difference in their molecular interaction affinity with HBV, with NTCP being the major and constraining receptor for HBV infection. As a result, pathogenic HBVs may have exerted a higher selective pressure on the NTCP protein than on GPC5.
TABLE 1.
Positive selection analyses of primate, rodent, and bat NTCP/SLC10A1 and GPC5 as well as of the orthohepadnavirus preS1 regiona
| Data set | Gene | M1 vs M2 P valueb | % positively selected sites in M2c | M2 ω (dN/dS ratio)d | M7 vs M8 P valueb | % positively selected sites in M8c | M8 ω (dN/dS ratio)d |
|---|---|---|---|---|---|---|---|
| Primates | |||||||
| All primate species | NTCP | 4E−03 | 2.61 | 3.87 | 2E−04 | 3.38 | 4.02 |
| Simians (hominoids, NWM, OWM) | NTCP | 6E−04 | 5.19 | 3.68 | 8E−05 | 2.91 | 5.74 |
| All primate species excluding hominoids | NTCP | 0.02 | 3.25 | 3.20 | 0.001 | 5.14 | 3.69 |
| Rodents | NTCP | 2E−08 | 2.10 | 4.16 | 8E−10 | 2.67 | 3.32 |
| Bats | NTCP | 2E−04 | 4.1 | 2.77 | 7E−08 | 7.31 | 2.20 |
| Primates | GPC5 | 1 | — | — | 0.20 | — | — |
| Rodents | GPC5 | 1 | 0.38 | ||||
| Bats | GPC5 | 1 | — | — | 1 | — | — |
| Hominoid HBV | HBV preS1 | 1 | — | — | 1 | — | — |
| Bat orthohepadnaviruses | HBV preS1 | 1 | — | — | 1 | — | — |
Results of the positive-selection analyses performed with PAML Codeml and comparing models that disallow positive selection (models M1 and M7; dN/dS ratio of ≤1) to models allowing for positive selection (M2 and M8). Positive selection in the primate NTCP/SLC10A1 gene was assessed for different data sets: the whole primate data set (n = 27), the simian primate data set (excluding the prosimians; n = 24), and the primate data set excluding hominoid species (n = 21). Evolutionary analyses were also carried out on rodents (18 species). For bats, the analyses were performed on 21 species, including the newly obtained sequences in this study. The GPC5 gene was analyzed for 20 primate, 19 rodent, and 7 bat species. The data shown were obtained with codon frequencies F61 and a starting omega dN/dS ratio of 0.4. Similar results were found with a codon frequency of F3*4 and a starting omega value of 1.5. NWM, New World monkeys; OWM, Old World monkeys.
P values generated from maximum likelihood ratio tests indicate whether the model that allows positive selection (models M2 and M8) better fits the data than the nearly neutral one (M1 and M7).
Percentage of codons evolving under positive selection (dN/dS ratio of >1). —, not applicable.
Average dN/dS ratio (ω) associated with the positively selected sites.
To assess which sites have been under positive selection in NTCP, we ran different site-specific models from HYPHY (44–47), and we used Bayesian empirical Bayes (BEB) posterior probabilities (PPs) at codon sites in PAML (models M2 and M8). Based on these analyses, we found different evolutionary patterns in primate, rodent, and bat NTCPs, involving distinct sites (Table 2).
TABLE 2.
Positively selected codons in primate, rodent, and bat NTCPsa
| Data set | Positively selected codons |
||||
|---|---|---|---|---|---|
| Codeml M2 (PP > 0.9) | Codeml M8 (PP > 0.9) | MEME (P < 0.1) | FUBAR (PP > 0.9) | REL (BF > 80) | |
| Primates | |||||
| All primate species | 157, 335 | 6, 9, 84, 157, 158, 303, 335, 341 | 142, 157, 196, 335 | 157, 158, 175, 335 | 157, 158, 303, 335 |
| Simians (hominoids, NWM, OWM) | 84, 157, 335 | 84, 157, 303, 335 | 84, 157, 335 | 84, 157, 161, 335 | 6, 84, 157, 158, 161, 303, 335 |
| Rodents | 33, 84, 157, 343 | 25, 33, 84, 157, 232, 343 | 20, 25, 107, 129, 161, 166, 192, 196, 200, 225, 232, 248, 303, 310, 343 | 25, 84, 107, 196, 232, 303, 332, 343 | 25, 107, 232, 330, 343 |
| Bats | 78, 203, 204 | 78, 112, 203, 204, 239, 247, 330 | 29, 33, 53, 61, 129, 140, 142, 165, 178, 294 | 29, 33, 107, 140, 192, 203, 204, 247, 294, 330 | 29, 33, 107, 192, 203, 204, 294, 330 |
Results from site-specific positive selection analyses. Codons with a high posterior probability (PP) of >0.9, a Bayes factor (BF) value of >80, or a significant P value (P < 0.1) were assigned to the class evolving under significant positive selection in models M2 and M8 from PAML Codeml and with MEME, FUBAR, and REL models from HyPhy (see Materials and Methods for details). Codons in bold are those that were identified by at least three models; those that are also underlined were found by all five methods. Codon numbering is based on the human NTCP sequence as a reference.
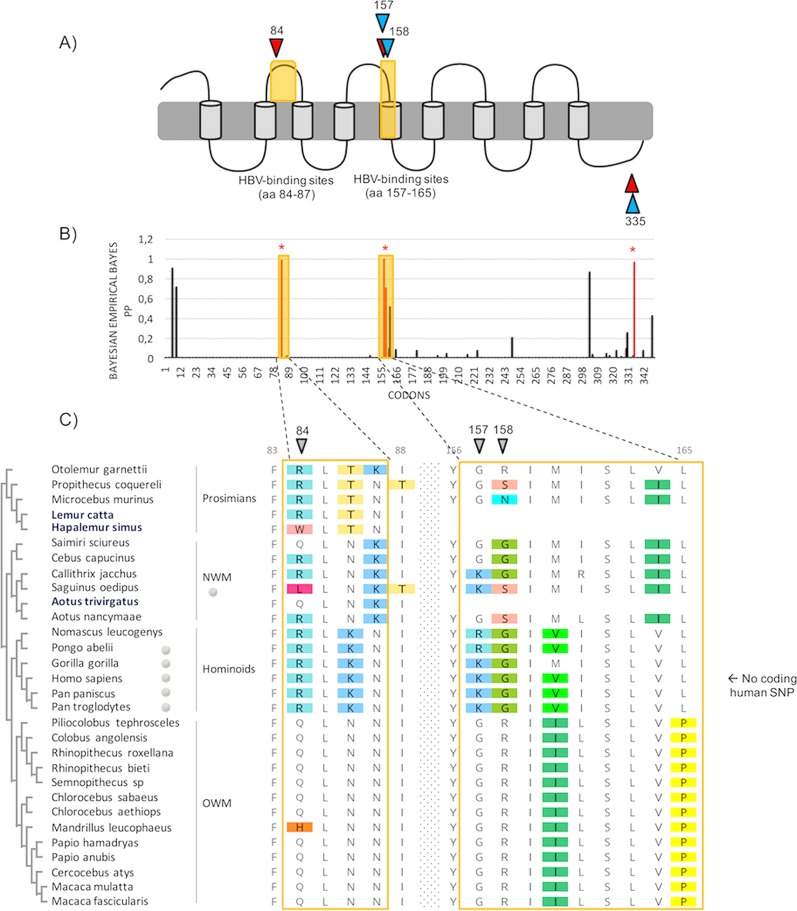
In primates, four codons (84, 157, 158, and 335) were found under positive selection (Table 2 and Fig. 2). Of these, the sites at positions 84, 157, and 158 correlate with amino acids that directly interact with the preS1 domain of the large envelope protein of HBV (Fig. 2A and C) (21, 23, 24). Functional studies through domain/point mutations have shown that species-specific differences at these HBV-binding sites of NTCP govern species susceptibility/resistance by impacting orthohepadnavirus receptor usage without affecting the bile acid transport function of NTCP (21, 23, 24, 48, 49). Combined with our findings, this supports a model of pathogen-driven selection, as observed in other systems in which signatures of diversification have been identified in housekeeping genes encoding proteins hijacked by viruses for cellular entry (e.g., see references 29 and 31–35). Given that NTCP is expressed only in hepatocytes, its role as a viral receptor is restricted to hepatotropic viruses, and hepadnaviruses seem to be the main driver candidates for such genetic patterns. Indeed, hepatitis C virus (HCV) has no direct binding interaction with NTCP (50). One cannot exclude a potential (additional) effect of hepatitis D virus (HDV), because this defective RNA pathogen depends on HBV envelope determinants to enter cells through NTCP, and chronic HBV/HDV infection causes more-severe liver diseases in humans than HBV monoinfection. However, at present, HDV has been described only in humans (51). Overall, this favors a model in which the adaptive fingerprints of primate NTCP are the result of Orthohepadnavirus selective pressure.
FIG 2.
Primate NTCP has evolved under positive selection, with positively selected sites mapping to the HBV-binding region. (A) Representation of NTCP with positively selected sites. A diagram representing the predicted domains of NTCP is shown (89), and the HBV-binding interfaces are highlighted in yellow. Specific codons found to be evolving under positive selection (PAML, Bayesian empirical Bayes [BEB] posterior probability [PP] of >0.9; MEME, P value of <0.1; FUBAR, PP of >0.9; REL, Bayes factor [BF] of >80), by at least three of the models used in this study (see Materials and Methods), are indicated in red and in blue for the primate (n = 27) and the simian primate (n = 24) data sets, respectively. aa, amino acids. (B) Histogram of the BEB PPs for a dN/dS ratio of >1 at each codon in an alignment of primate NTCP coding sequences. BEB values were inferred under the M8 site model implemented in PAML Codeml. The codons shown with stars are those identified as being under positive selection by at least three methods. (C) Amino acid alignment of the HBV-binding regions of NTCP from primates (27 publicly available species and 3 de novo-sequenced species). On the left, the cladogram of primate NTCP, with the newly sequenced species highlighted in blue, is presented. Amino acid alignment was performed with PRANK, and the color-coding is from Rasmol (Geneious, Biomatters). The positively selected sites, inferred by at least three methods, are indicated by the gray symbols. On the right, human SNP information is given for the HBV-binding regions in human NTCP. NWM and OWM, New and Old World monkeys, respectively. Codon numbering is based on the human NTCP sequence as a reference.
The four positively selected codons in primate NTCP have experienced recurrent selection for mutations that replace the encoded amino acids. These positions are therefore highly variable at the protein level (Fig. 2C), particularly in New World monkeys and prosimians. In contrast, the HBV-binding interfaces of NTCP are highly conserved within the hominoids (Fig. 2C). Evolutionary analyses of a primate NTCP alignment without the hominoid species still show a significant signal of adaptive evolution (Table 1). At the human population level, no polymorphism was reported at these HBV-binding sites (NCBI dbSNP [Single Nucleotide Polymorphism Database]). Notwithstanding, single nucleotide polymorphisms (SNPs), external to the HBV-binding determinants, have been reported in Asian and African populations (52–54). In particular, the S267F mutation has been shown to impair NTCP function and impact HBV infectivity (55, 56). Given the dual effect of this mutation, ascribing the presence of this polymorphism to an ongoing arms race with HBV is delicate although possible. Therefore, the absence of inter- and intraspecies variability in hominoid NTCP (Fig. 2C), despite the broad circulation of orthohepadnaviruses (4, 5), suggests that orthohepadnaviruses have no longer been a strong selective pressure, since or following their emergence in hominoids. Chronic HBV infection in humans and the related morbidity appear mostly after reproductive life (57), which supports this hypothesis. Although less likely, the passage of orthohepadnaviruses in hominoids may be too recent to track any signatures of evolutionary arms races in their NTCP sequences.
Altogether, these findings suggest that adaptive evolution of NTCP has occurred prior the hominoid diversification and that ancient and/or extinct pathogenic orthohepadnaviruses have circulated in primates. This model further implies that orthohepadnaviruses have been associated with primates much earlier, at least prior to the diversification of the simian primates, and for a far longer period during the Eocene (37, 38) than stated by previous hypotheses on HBV origin (4, 5). Our findings are also consistent with the recent discovery of another Orthohepadnavirus from a New World monkey, the capuchin monkey (CMHBV), and its basal position with woolly monkey HBV (WMHBV) in the phylogenetic tree (7, 8). On the other hand, orthohepadnaviruses have not yet been reported in the Old World monkeys and prosimians, except for isolated cases of cross-species transmissions of human HBVs in crab-eating macaques (58) and chacma baboons (59). Specific amino acid differences in the presumed HBV-binding interfaces of NTCPs may explain the supposed absence of HBV-like viruses in Old World monkeys (Fig. 2C) (49). Apart from hominoid species, primates are generally understudied for orthohepadnaviruses, which may bias the knowledge of their epidemiology and evolution. Our “indirect paleovirology” approach allows advancing knowledge on hepadnaviral evolutionary history in host species where viral surveys are complicated.
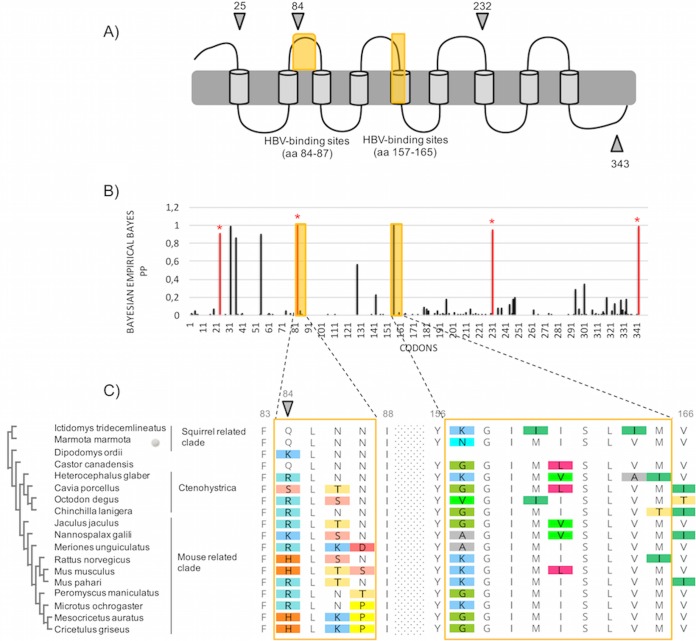
Likewise, information on rodent orthohepadnaviruses is very scarce, which makes the understanding of Orthohepadnavirus evolutionary history very difficult. By performing evolutionary analyses of rodent NTCP, we found that the evolutionary fingerprints are both internal and external to the presumed HBV-binding sites (Tables 1 and 2; see also Fig. 3) and that the presumed HBV-binding motifs in rodent NTCP have undergone rapid evolution (Table 2; see also Fig. 3C). Noticeably, codon 84 was found to be under significant positive selection, as in primates. It remains unclear whether all orthohepadnaviruses use NTCP as a cellular receptor. However, previous studies have shown that woodchuck NTCP supports human HBV infection, although at low levels (60), and that variability at positions 84 to 87 in mouse NTCP was the limiting factor for human HBV binding (23, 24). It is thus possible that rodent orthohepadnaviruses also use NTCP, although further work would be necessary to confirm this. In combination with our findings, it is tempting to speculate that the variability at the presumed HBV-binding sites in rodent NTCP reflect a past genetic arms race with ancient pathogenic orthohepadnaviruses. This is in accordance with the relatively high pathogenicity of woodchuck hepatitis virus (WHV) (61). This would also suggest that the Sciuridae are not the sole family infected by extinct and/or extant orthohepadnaviruses. Furthermore, the high variability, in particular at positions 84 and 157 (although codon 157 is supported by only two of the five evolutionary models used in this study [Fig. 3C and Table 2]), could be a key determinant of species specificity and explain, at least partly, the presumed restricted circulation of orthohepadnaviruses in rodents.
FIG 3.
Rodent NTCP has evolved under positive selection and displays similar patterns of evolution as those observed in primates. (A) Representation of NTCP (as in Fig. 2A) with positively selected sites during rodent evolution. Specific codons found to be evolving under significant positive selection by at least three of the models used in this study (see Materials and Methods) are indicated by the gray symbols. (B) Histogram of the BEB PPs for a dN/dS ratio of >1 at each codon in an alignment of rodent NTCP coding sequences. BEB values were inferred under the M8 site model implemented in PAML Codeml. The codons shown with stars are those identified as being under positive selection by at least three methods. (C) Amino acid alignment of the HBV-binding regions of rodent NTCP is presented along with the cladogram on the left, as in Fig. 2C. Codon numbering is based on the human NTCP sequence as a reference.
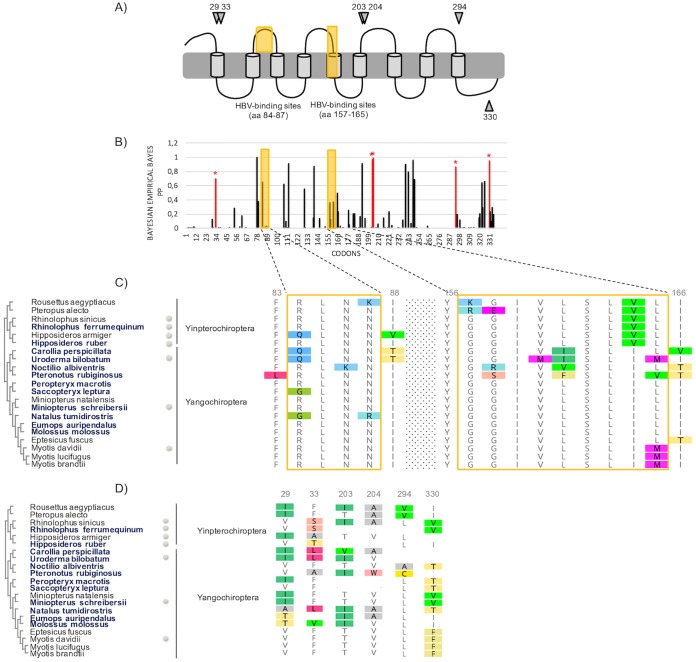
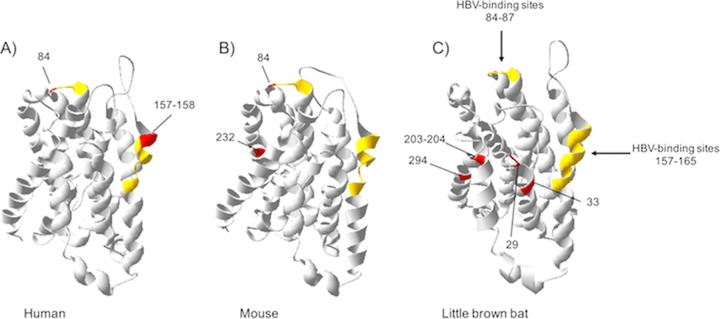
In bats, the signatures of diversifying selection in NTCP are scattered along the protein sequence (Fig. 4). Importantly, none of the six positively selected codons (29, 33, 203, 204, 294, and 330) map to the presumed HBV-binding motifs (Fig. 4). Instead, the latter are mostly conserved (Fig. 4C). The 3D homology model of bat NTCP also supports that the positively selected residues lie outside the presumed HBV-binding interface (Fig. 5). There are at least three possible explanations for such adaptation of bat NTCP.
FIG 4.
Strong signatures of positive selection in bat NTCP are observed, but none of the positively selected sites map to the presumed HBV-binding interface. (A) Representation of NTCP (as in Fig. 2A) with positively selected sites during bat evolution. Specific codons found to be evolving under significant positive selection by at least three of the models used in this study (see Materials and Methods) are indicated by the gray symbols. (B) Histogram of the BEB PPs for a dN/dS ratio of >1 at each codon in an alignment of bat NTCP coding sequences (from the PAML Codeml M8 model). The codons shown with stars are those identified as being under positive selection by at least three methods. (C) Amino acid alignment of the HBV-binding regions of NTCPs from 21 bat species is presented along with the cladogram on the left, as in Fig. 2C. The newly sequenced species are highlighted in boldface type. (D) Amino acid alignment of the positively selected sites (from panel A) in bat NTCP. Codon numbering is based on the human NTCP sequence as a reference.
FIG 5.
3D homology modeling of primate, rodent, and bat NTCPs highlighting the sites under positive selection and the HBV-binding determinants. The 3D modeling of NTCP was performed with the SWISS-MODEL program (87, 88) based on the crystal structure of ASBTYf (89). The amino acid NTCP sequences of human, mouse, and little brown bat were used as queries for modeling the structures of primate, rodent, and bat NTCPs, respectively. The models represent amino acids 27 to 308 for primates (A), rodents (B), and bats (C). The HBV-binding determinants are depicted in yellow, and the positively selected sites are highlighted red.
First, it is possible that bat orthohepadnaviruses bind to other NTCP surfaces in bats. It has been shown that tent-making bat orthohepadnavirus (TBHBV) was able to interact with human NTCP for entry into hepatocytes (9). Moreover, the preS1 domain of bat orthohepadnaviruses shows high sequence identity with the typical core signature of NTCP binding of primate HBV (NPLGFFP motif). This suggests that bat orthohepadnaviruses may use NTCP as a cellular receptor, but the molecular interactions have yet to be characterized. Otherwise, we cannot exclude that bat viruses use another receptor altogether. The study of pathogenic virus-host evolutionary interactions has the power to predict and identify the sites of interaction and has been successfully used to characterize binding interfaces of host-virus protein interactions (e.g., see references 62–66). In line with this, the positively selected codons identified in bat NTCP (Fig. 4D) could represent the binding surfaces of bat orthohepadnaviruses. If so, the signatures of rapid evolution in NTCP may be reminiscent of past genetic conflicts with orthohepadnaviruses during bat evolution. This hypothesis supposes not only a long-term coevolution but also that orthohepadnaviruses have been pathogenic to bats. Functional analyses are further required to decipher the implication of orthohepadnaviruses in bat NTCP evolution.
Second, the genetic fingerprints in bat NTCP may reflect a different selective pressure of orthohepadnaviruses in bat species. Indeed, if bat orthohepadnaviruses interact with the same NTCP surfaces, the absence of positive selection in this interface suggests that orthohepadnaviruses have not been strong drivers of bat NTCP evolution despite their presumed long-term association with chiropteran species (12, 67). Bats are assumed to host most zoonotic viruses asymptomatically (68). Specific immunological features may allow them to tolerate or enhance the antiviral response to efficiently control viral infection and/or pathogenesis (69–73), including orthohepadnavirus infections.
In the latter scenario, the observed signatures of positive selection in bat NTCP could have resulted from other pathogens, including pathogenic viruses or bacteria (e.g., see reference 74).
It is worth mentioning that the genetic fingerprints external to the presumed HBV-binding sites in primates and rodents could also be explained by the above-mentioned hypotheses. In particular, one cannot exclude that other sites (i.e., those under positive selection) are also involved in HBV binding.
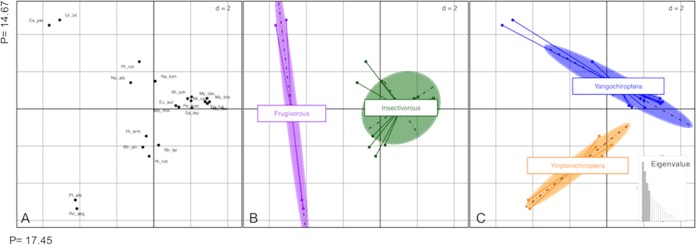
A third nonexclusive hypothesis is that the observed genetic fingerprints reflect adaptive changes related to bat metabolism. Indeed, as a major bile acid transporter, NTCP has a key role in mammalian enterohepatic circulation, digestion, and metabolic regulation (75). Given the huge diversity and adaptive radiations in bats into novel trophic niches (41), accompanied by extensive changes in various traits (such as diet), it is conceivable that the bat NTCP protein has been a target of molecular adaptation in relation to energy and/or dietary metabolism. To explore this hypothesis, we performed a principal-coordinate analysis (PCoA) on the genetic distances of bat NTCP sequences. The Euclidian distances were computed using the polymorphic sites from an alignment of bat NTCP protein sequences (Fig. 6). Projection of the PCoA data on the first two axes reveals a clear structuration of bat NTCP variability depending on diet (axis 1) and phylogeny (axis 2) (Fig. 6). This pattern suggests that bat NTCP may have, at least partly, evolved in response to the diversification of diet in bats, although more species are needed to further confirm the observed tendency. Such an evolutionary response to diet has been reported for the SLC2A4 gene, encoding the transmembrane glucose transporter 4 protein (GLUT4), in which the genetic footprints are due to the frugivorous diet of Old World fruit bats (76).
FIG 6.
Principal-coordinate analysis (PCoA) of bat NTCP highlights an effect of diet on NTCP evolution. PCoA was performed on the bat NTCP protein sequence alignment using the ADEGENET package in R (91). Only polymorphic sites were retained for the analysis. The pairwise distance between each sequence was calculated and then centered, and a planar representation of these distances was achieved by PCoA. The PCoA plots were generated using the first two principal coordinates (PCs), which explain a total of 33% of the variation. The graphics were obtained with the ADEGRAPHICS package available in R (92). The plot shows bat species grouping by similarities in polymorphic sites of the NTCP protein sequence. (A) Projection of species genetic distance, with each species label abbreviation. (B and C) A clear structure opposes frugivorous to insectivorous species on the first axis (B), while the second axis shows a tendency of phylogenetic groups opposing the Yinpterochiroptera to Yangochiroptera (C). Abbreviations: Ep_fus, Eptesicus fuscus; My_luc, Myotis lucifugus; My_dav, Myotis davidii; My_bra, Myotis brandtii; Mi_sch, Miniopterus schreibersii; Mi_nat, Miniopterus natalensis; Ur_bil, Uroderma bilobatum; Ca_per, Carollia perspicillata; Pt_rub, Pteronotus rubiginosus; Na_tum, Natalus tumidirostris; Mo_mol, Molossus molossus; Eu_aur, Eumops auripendulus; Pe_mac, Peropteryx macrotis; Sa_lep, Saccopteryx leptura; No_alb, Noctilio albiventris; Pt_ale, Pteropus alecto; Ro_aeg, Rousettus aegyptiacus; Rh_sin, Rhinolophus sinicus; Rh_fer, Rhinolophus ferrumequinum; Hi_arm, Hipposideros armiger; Hi_rub, Hipposideros ruber.
While a combination of coevolution and host switches has been described in mammalian orthohepadnaviruses (67, 77), the involved molecular factors/barriers are yet to be determined. Here, we suggest that NTCP may contribute to Orthohepadnavirus differential host specificity in mammals. In nonhominoid primates, the adaptive changes in NTCP may contribute to the current narrow host range of orthohepadnaviruses, while the conserved HBV-binding regions in hominoid NTCP (Fig. 2C) may allow viral interspecies circulation. This is further reflected on the virus side, where the determinants of NTCP interaction in the HBV preS1 domain are mostly conserved in hominoid viruses (see Fig. S2 at https://figshare.com/articles/FIG_S2_Comparative_amino_acid_variability_at_the_virus_and_host_interfaces_mammal_NTCP_and_orthohepadnavirus_preS1_domain/7321097). In addition, evolutionary analyses of a hominoid HBV preS1 alignment did not identify any significant adaptive signatures of positive selection (Table 1), supporting the absence of virus-host coevolutionary dynamics within the hominoids. In contrast, positive selection would be expected in the preS1 domain of orthohepadnaviruses from other primate clades. However, no viruses have been reported in prosimians, and only two orthohepadnaviruses have been reported in New World monkey species, which is not enough to seek signatures of positive selection. Likewise, orthohepadnaviruses have been reported in only three rodent species, thereby limiting the screen of positive selection on the virus side. In bats, while interspecies transmissions may have arisen on a background of an ancient Orthohepadnavirus-bat association, cross-species transmissions between highly divergent species seem more recurrent (12). The latter may be facilitated by biological and ecological factors specific to bats, including their large population sizes, high intra- and interspecies contact rates, or migration (78). If bat orthohepadnaviruses use the same NTCP motifs as those in primates, the conserved property of these sites in bat NTCP suggest that NTCP may be a weak genetic constraint in bats, which could increase the transmission of orthohepadnaviruses among diverse bat species and favor bat Orthohepadnavirus diversity.
Overall, this study brings insights on the pathogenicity and molecular determinants currently governing interspecies circulation of orthohepadnaviruses within mammals. While modern orthohepadnaviruses may have a mild pathogenicity in their hosts, our findings suggest that ancient pathogenic orthohepadnaviruses have been a selective pressure during mammalian evolution, particularly in primates and rodents. The contrasting genetic pattern in bats indicates a different Orthohepadnavirus-NTCP molecular interaction or may reflect adaptive changes associated with bat metabolism. Our study further points to NTCP as a genetic constraint for orthohepadnavirus cross-species transmissions in primates and rodents but less so in bats. This difference in NTCP genetic constraints may explain, at least partly, the difference in orthohepadnavirus diversity in mammals.
MATERIALS AND METHODS
Collection of NTCP and GPC5 orthologous sequences from public databases.
NTCP and GPC5 sequences from primates, rodents, and bats were obtained by tBLASTn searches of the nucleotide databases from GenBank (79) using human, mouse, and little brown bat NTCP or GPC5 protein sequences as queries, respectively. In total, 27 NTCP and 20 GPC5 sequences from hominoids, Old World monkeys, New World monkeys, and prosimians were obtained (see Table S1A at https://figshare.com/articles/Table_S1_Information_on_the_species_and_the_sequences_used_for_evolutionary_analyses_in_primates_A_rodents_B_and_in_bats_C_/7315235). NTCP and GPC5 sequences from 18 rodent species were obtained. For bats, NTCP and GPC5 sequences were publicly available for nine and seven species, respectively (see Tables S1B and C at https://figshare.com/articles/Table_S1_Information_on_the_species_and_the_sequences_used_for_evolutionary_analyses_in_primates_A_rodents_B_and_in_bats_C_/7315235).
Sampling of additional primate and bat species.
Prosimian peripheral blood mononuclear cells (PBMCs) were isolated using Histopaque 1077 from leftover blood samples (approximately 1 ml, from blood drawn for health purposes) from two Lemuridae prosimians (Lemur catta and Hapalemur simus) hosted at the Zoo de Lyon, France (Guillaume Douay). New World monkey cells from owl monkey (Aotus trivirgatus) (owl monkey kidney [OMK] cells) were obtained from CelluloNet Lyon and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum.
Bats were sampled in Gabon (Hipposideros cf. ruber), French Guiana (Peropteryx macrotis, Pteronotus rubiginosus, Carollia perspicillata, Natalus tumidirostris, Saccopteryx leptura, Eumops auripendulus, Molossus rufus, Noctilio albiventris, and Uroderma bilobatum), and France (Miniopterus schreibersii and Rhinolophus ferrumequinum). Importantly, M. schreibersii, R. ferrumequinum, U. bilobatum, and Hipposideros cf. ruber are known as natural hosts of hepadnaviruses (9–13). Authorization for bat capture in France and in French Guiana was provided by the Ministry of Ecology, Environment, and Sustainable Development over the period from 2015 to 2020 (approval no. C692660703 from the Departmental Direction of Population Protection [DDPP], Rhône, France). All methods (capture and animal handling) were approved by the Museum National d'Histoire Naturelle (MNHN), the Société Française pour l'Étude et la Protection des Mammifères (SFEPM), and the Direction de l'Environnement, de l'Aménagement et du Logement Guyane (DEAL-Guyane). African bat samples used here were collected in a previous study, which was approved by the Gabonese National Ethics Committee (authorization no. PROT/0020/2013I/SG/CNE).
Bats were captured using harp traps at the entrance of caves or mist nests hoisted on the forest floor and in the tree canopy. Captured bats were removed carefully from nests or harp traps as soon as possible to minimize injury or stress. Tissue samples for DNA analysis were collected from the wing membrane (patagium) using a 3-mm-diameter biopsy punch (Kai Industries, Gifu, Japan) and preserved in a 70% ethanol solution until DNA extraction. Bats were released after sampling.
Nucleic acid extraction and molecular identification.
Total genomic DNA (gDNA) was extracted from the primate cells and from the ethanol-preserved samples of bat punch specimens using a Macherey-Nagel NucleoSpin tissue kit according to the manufacturer’s protocol. To ensure proper identification of the sampled bat species, we amplified and sequenced the mitochondrial gene cytochrome b (Cytb), using the primers CytB-F and CytB-L/R (80). PCRs were carried out in a total volume of 25 μl containing 1× reaction buffer (DreamTaq DNA polymerase; Thermo Fisher Scientific), 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.2 μM each primer, 1 U of Taq polymerase (DreamTaq DNA polymerase; Thermo Fisher Scientific), and approximately 10 ng of extracted genomic DNA. Cycling conditions consisted of an initial denaturation step at 94°C for 3 min, followed by 35 cycles at 94°C for 30 s, 58°C for 45 s, and 72°C for 1 min and then a final extension step at 72°C for 3 min. PCR products with multiple bands were excised and purified from gel using the NucleoSpin gel and PCR clean-up kit from Macherey-Nagel. Single PCR products from each species were sequenced by a commercial company (GATC Biotech, Germany).
De novo sequencing of NTCP/SLC10A1 genes.
We amplified and sequenced the NTCP/SLC10A1 gene from the extracted gDNA. We used bat and primate NTCP alignments generated from publicly available sequences Specific primers targeting SLC10A1 intronic regions were designed to amplify and sequence each of the five exons (see Table S2 at https://figshare.com/articles/Table_S2_Primers_and_PCR_conditions_for_NTCP_amplification_in_bats_and_primates/7321100). We used the Qiagen Dream Taq kit under conditions for amplification presented in Tables S3 and S4 at https://figshare.com/articles/S3_Table_Primer_pairs_used_for_NTCP_amplification_of_each_species_/7321109 and https://figshare.com/articles/S4_Table_Mix_PCR_for_NTCP_amplification_/7321115. Gel purification was performed when necessary using the NucleoSpin gel and PCR clean-up kit from Macherey-Nagel. The exon sequences were assembled together to generate the whole coding sequence of NTCP.
Phylogenetic analyses of NTCP and GPC5.
NTCP and GPC5 sequences were aligned using webPRANK (81). Poorly aligned codons at the C terminus were removed for rodents and bats, leading to alignment lengths of 1,029 bp and 1,005 bp for rodents and bats, respectively. The overall pairwise amino acid identities were estimated for both proteins from eutherian mammal (NTCP, n = 55; GPC5, n = 44), bat (NTCP, n = 21; GPC5 n = 7), rodent (NTCP and GPC5, n = 18), and primate (NTCP, n = 27; GPC5, n = 20) species using MEGA v.7 (82). Using these same data sets, amino acid variability at each position was further assessed with the ConSurf Web server (83). The following approach was used for primate, rodent, and bat data sets. To account for potential confounding effects of recombination, we tested our data sets with the Genetic Algorithm for Recombination Detection (GARD) (84), which is available in the HYPHY package (46). Based on codon sequence alignments, we tested for the best substitution model using Smart Model Selection (SMS) in PhyML (85), which was used for subsequent phylogenetic analyses (GTR+Invariant sites [I]+Gamma [G] for primates and rodents and HKY+I+G for bats). A phylogenetic tree of NTCP and GPC5 orthologous sequences was constructed using the maximum likelihood method implemented in the ATGC-PhyML Web server (86). Node supports were tested using the bootstrap method through 1,000 replicates. For supplemental data sets, see https://figshare.com/articles/Dataset_S1_Nucleotide_Alignments_of_primate_rodent_and_bat_NTCP/6809618, https://figshare.com/articles/Dataset_S2_Nucleotide_Alignments_and_Phylogenetic_Trees_of_primate_rodent_and_bat_GPC5/6809657, and https://figshare.com/articles/Dataset_S3_Eutherian_NTCP_and_GPC5_codon_alignments/6809663.
Positive-selection analyses of NTCP and GPC5.
Detection of recurrent positive selection in primate, rodent, and bat NTCPs and GPC5s was carried out using five different methods. These include the Codeml program implemented in the PAML package (43), the Fast Unbiased Bayesian Approximation (FUBAR) that uses Bayesian inference to detect positive and negative selection at individual sites (44), and the Random Effects Likelihood (REL) and the Mixed Effects Model of Evolution (MEME) implemented through the HYPHY package (45, 47). Codeml allows both gene- and site-specific detection of positive selection by comparing constrained models that disallow positive selection (models M1 and M7; dN/dS ratio of ≤1) to unconstrained models allowing for positive selection (M2 and M8). We first ran the one-ratio model (M0) to check the parameters. The phylogenetic tree generated by this model was then used to run the other models under the following parameters: codon frequencies of F61 and F3*4 and starting omega (dN/dS ratio) values of 0.4 and 1.5. The percentage of sites exhibiting a significant signal of positive selection was estimated, as were the average dN/dS ratios of these sites. Likelihood ratio tests (LRTs) were performed to compare models M1 versus M2 and M7 versus M8, and posterior probabilities for sites were calculated according to the Bayesian empirical Bayes model (43). To ensure the robustness of results, we kept the sites that were significantly identified by at least three methods of the five used in this study (Table 2). Given the low number of prosimian samples and the longer branches of prosimians in the primate data set, we performed three different series of analyses for detection of positive selection: the first set included all the primate species (n = 27), the second data set comprised only simian species (hominoids, Old World monkeys, and New World monkeys; n = 24), and the last one included sequences of the first exons of NTCPs from the whole data set and the newly sequenced species (n = 30) (see Table S1 at https://figshare.com/articles/Table_S1_Information_on_the_species_and_the_sequences_used_for_evolutionary_analyses_in_primates_A_rodents_B_and_in_bats_C_/7315235).
Positive-selection analyses of HBV preS1.
The determinants of interaction with NTCP are located in the HBV preS1 domain of the large envelope protein. We thus performed positive-selection analyses on a codon sequence alignment of the preS1 domain, using models M1 versus M2 and M7 versus M8 of Codeml. Given the scarcity of Orthohepadnavirus sequences from primates, evolutionary analyses were restricted to the viruses naturally infecting hominoids and were therefore performed on 12 hominoid HBVs (human HBV genotypes A to H, chimpanzee HBV, gorilla HBV, gibbon HBV, and orangutan HBV). The same analysis was performed for bat orthohepadnaviruses (RBHBV China, HBVBV China, RBHVB Gabon, RBHBV Gabon, Lushi_RpHBV, Guizhou_MsHBV, Jiyuan_RfHBV, TBHBV, and LBHBV).
3D homology modeling of NTCP.
Human NTCP as well as the mouse and little brown bat NTCP protein sequences were used as queries for the SWISS-MODEL tool (https://swissmodel.expasy.org) (87, 88) to model the 3D homology of NTCPs. Models were inferred using the apical sodium-dependent bile acid transporter protein of Yersinia frederiksenii (ASBTYf) as a template (PDB accession no. 4N7W) (89). N-terminal residues 1 to 27 and C-terminal residues 309 to 349 could not be structurally predicted. As a result, the 3D model represents amino acids 28 to 308 for primate, rodent, and bat NTCP proteins. The protein structure was edited with Swiss PDB viewer (90).
Principal-coordinate analysis.
To explore what contributes to bat NTCP evolution, we carried out a principal-coordinate analysis (PCoA) on a bat NTCP protein sequence alignment. The latter was obtained by translating the codon alignment used for the positive-selection analyses, using MEGA v.7 (82). The polymorphic sites were extracted from the protein alignment, and pairwise Euclidian genetic distances between species were computed and centered, based on the polymorphic sites. PCoA was then performed on the genetic distance matrix using the ADEGENET package in R (91). The PCoA plots were generated using the first two principal coordinates (PCs), which explain a total of 33% of the variation. The graphics were obtained with the ADEGRAPHICS package available in R (92).
Accession number(s).
The exon sequences have been deposited in GenBank under accession no. MK131104 to MK131149.
ACKNOWLEDGMENTS
We thank Patrice André, Andrea Cimarelli, Rossana Colon-Thillet, Marie Delattre, Nels Elde, Manolo Gouy, and Mégane Wcislo for their comments on the manuscript. We also thank Anne-Béatrice Dufour for her help on PCoA. We are particularly grateful to the Poitou-Charentes association as well as the volunteers and field workers who have helped us during the field sessions: V. Alt, M. Bely, G. Chagneau, M. Dorfiac, S. Dufour, C. Gizardin, G. Leblanc, M. Leuchtmann, E. Loufti, A. Le Guen, and L. Trebucq. We thank Guillaume Douay and the Zoo de Lyon for their collaboration and their gift of leftover blood samples from prosimians. We thank A. Cimarelli, head of the Host-Pathogen Interaction during Lentiviral Infection team at the CIRI Lyon. We thank all the contributors of publicly available genome sequences.
This work is funded by the ANR LABEX ECOFECT (ANR-11-LABX-0048 of the Université de Lyon, within the program Investissements d’Avenir [ANR-11-IDEX-0007] operated by the French National Research Agency). L.E. is supported by the CNRS and by grants from amfAR (Mathilde Krim Phase II Fellowship no. 109140-58-RKHF), the Fondation pour la Recherche Médicale (FRM Projet Innovant no. ING20160435028), the FINOVI (“recently settled scientist” grant), the ANRS (no. ECTZ19143), and a JORISS incubating grant. D.P. is supported by the CNRS, the European Regional Development Fund (ERDF), and the ANR EBOFAC.
L.E. and D.P. conceptualized and supervised the study. S.J., J.-B.P., B.N., F.-L.C., L.E., and D.P. determined the methodology. S.J., J.-B.P., A.D.B., B.N., C.R., L.E., and D.P. performed investigations. S.J., L.E., and D.P. performed the formal analysis of the data. L.E. and D.P. were project administrators. F.-L.C., L.E., and D.P. acquired funding. J.-B.P., B.N., L.E., and D.P. acquired resources. S.J., L.E., and D.P. wrote the original draft and revised the paper; all authors reviewed and edited the paper.
REFERENCES
- 1.Dill JA, Camus AC, Leary JH, Di Giallonardo F, Holmes EC, Ng TF. 2016. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J Virol 90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Borold J, Salzburger W, Kaderali L, Briggs JAG, Bartenschlager R. 2017. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22:387.e6–399.e6. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh A, Weber CC, Kehlmaier C, Braun EL, Green RE, Fritz U, Ray DA, Ellegren H. 2014. Early Mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet 10:e1004559. doi: 10.1371/journal.pgen.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Littlejohn M, Locarnini S, Yuen L. 2016. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb Perspect Med 6:a021360. doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locarnini S, Littlejohn M, Aziz MN, Yuen L. 2013. Possible origins and evolution of the hepatitis B virus (HBV). Semin Cancer Biol 23:561–575. doi: 10.1016/j.semcancer.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Souza BF, Drexler JF, Lima RS, Rosario MDO, Netto EM. 2014. Theories about evolutionary origins of human hepatitis B virus in primates and humans. Braz J Infect Dis 18:535–543. doi: 10.1016/j.bjid.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanford RE, Chavez D, Brasky KM, Burns RB, Rico-Hesse R. 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci U S A 95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Carvalho Dominguez Souza BF, Konig A, Rasche A, de Oliveira Carneiro I, Stephan N, Corman VM, Roppert PL, Goldmann N, Kepper R, Muller SF, Volker C, de Souza AJS, Gomes-Gouvea MS, Moreira-Soto A, Stocker A, Nassal M, Franke CR, Rebello Pinho JR, Soares M, Geyer J, Lemey P, Drosten C, Netto EM, Glebe D, Drexler JF. 2018. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J Hepatol 68:1114–1122. doi: 10.1016/j.jhep.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Drexler JF, Geipel A, Konig A, Corman VM, van Riel D, Leijten LM, Bremer CM, Rasche A, Cottontail VM, Maganga GD, Schlegel M, Muller MA, Adam A, Klose SM, Carneiro AJ, Stocker A, Franke CR, Gloza-Rausch F, Geyer J, Annan A, Adu-Sarkodie Y, Oppong S, Binger T, Vallo P, Tschapka M, Ulrich RG, Gerlich WH, Leroy E, Kuiken T, Glebe D, Drosten C. 2013. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc Natl Acad Sci U S A 110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Yang XL, Li W, Zhu Y, Ge XY, Zhang LB, Zhang YZ, Bock CT, Shi ZL. 2017. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol J 14:40. doi: 10.1186/s12985-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He B, Fan Q, Yang F, Hu T, Qiu W, Feng Y, Li Z, Li Y, Zhang F, Guo H, Zou X, Tu C. 2013. Hepatitis virus in long-fingered bats, Myanmar. Emerg Infect Dis 19:638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie F-Y, Lin X-D, Hao Z-Y, Chen X-N, Wang Z-X, Wang M-R, Wu J, Wang H-W, Zhao G, Ma RZ, Holmes EC, Zhang Y-Z. 2018. Extensive diversity and evolution of hepadnaviruses in bats in China. Virology 514:88–97. doi: 10.1016/j.virol.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Zhang F, Xia L, Hu T, Chen G, Qiu W, Fan Q, Feng Y, Guo H, Tu C. 2015. Identification of a novel orthohepadnavirus in Pomona roundleaf bats in China. Arch Virol 160:335–337. doi: 10.1007/s00705-014-2222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Margolis HS, Purcell RH, Ebert J, Robertson BH. 2000. Identification of hepatitis B virus indigenous to chimpanzees. Proc Natl Acad Sci U S A 97:1661–1664. doi: 10.1073/pnas.97.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grethe S, Heckel J-O, Rietschel W, Hufert FT. 2000. Molecular epidemiology of hepatitis B virus variants in nonhuman primates. J Virol 74:5377–5381. doi: 10.1128/JVI.74.11.5377-5381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webby R, Hoffmann E, Webster R. 2004. Molecular constraints to interspecies transmission of viral pathogens. Nat Med 10:S77. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baranowski E, Ruiz-Jarabo CM, Domingo E. 2001. Evolution of cell recognition by viruses. Science 292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 18.Baranowski E, Ruiz-Jarabo CM, Pariente N, Verdaguer N, Domingo E. 2003. Evolution of cell recognition by viruses: a source of biological novelty with medical implications. Adv Virus Res 62:19–111. doi: 10.1016/S0065-3527(03)62002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffin JM. 2013. Virions at the gates: receptors and the host-virus arms race. PLoS Biol 11:e1001574. doi: 10.1371/journal.pbio.1001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verrier ER, Colpitts CC, Bach C, Heydmann L, Weiss A, Renaud M, Durand SC, Habersetzer F, Durantel D, Abou-Jaoudé G, López Ledesma MM, Felmlee DJ, Soumillon M, Croonenborghs T, Pochet N, Nassal M, Schuster C, Brino L, Sureau C, Zeisel MB, Baumert TF. 2016. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 63:35–48. doi: 10.1002/hep.28013. [DOI] [PubMed] [Google Scholar]
- 21.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anwer MS, Stieger B. 2014. Sodium-dependent bile salt transporters of the SLC10A transporter family: more than solute transporters. Pflugers Arch 466:77–89. doi: 10.1007/s00424-013-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 24.Yan H, Peng B, He W, Zhong G, Qi Y, Ren B, Gao Z, Jing Z, Song M, Xu G, Sui J, Li W. 2013. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol 87:7977–7991. doi: 10.1128/JVI.03540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerson NR, Sawyer SL. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol 19:286–294. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enard D, Cai L, Gwennap C, Petrov DA. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5:e12469. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 28.Sironi M, Cagliani R, Forni D, Clerici M. 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet 16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. 2013. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demogines A, Farzan M, Sawyer SL. 2012. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J Virol 86:6350–6353. doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelber JT, Demogines A, Harbison CE, Allison AB, Goodman LB, Ortega AN, Sawyer SL, Parrish CR. 2012. Evolutionary reconstructions of the transferrin receptor of caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog 8:e1002666. doi: 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng M, Ndungo E, Kaczmarek ME, Herbert AS, Binger T, Kuehne AI, Jangra RK, Hawkins JA, Gifford RJ, Biswas R, Demogines A, James RM, Yu M, Brummelkamp TR, Drosten C, Wang L-F, Kuhn JH, Müller MA, Dye JM, Sawyer SL, Chandran K. 2015. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. Elife 4:e11785. doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyerson NR, Sharma A, Wilkerson GK, Overbaugh J, Sawyer SL. 2015. Identification of owl monkey CD4 receptors broadly compatible with early-stage HIV-1 isolates. J Virol 89:8611–8622. doi: 10.1128/JVI.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demogines A, Truong KA, Sawyer SL. 2012. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol Biol Evol 29:445–449. doi: 10.1093/molbev/msr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Compton AA, Hirsch VM, Emerman M. 2012. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MPC, Silva A, O’Brien SJ, Pecon-Slattery J. 2011. A molecular phylogeny of living primates. PLoS Genet 7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecon-Slattery J. 2014. Recent advances in primate phylogenomics. Annu Rev Anim Biosci 2:41–63. doi: 10.1146/annurev-animal-022513-114217. [DOI] [PubMed] [Google Scholar]
- 39.Fabre P-H, Hautier L, Dimitrov D, Douzery EJ. 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol Biol 12:88. doi: 10.1186/1471-2148-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBee RM, Rozmiarek SA, Meyerson NR, Rowley PA, Sawyer SL. 2015. The effect of species representation on the detection of positive selection in primate gene data sets. Mol Biol Evol 32:1091–1096. doi: 10.1093/molbev/msu399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teeling EC, Springer MS, Madsen O, Bates P, Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 42.Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, May-Collado LJ. 2011. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr 3:RRN1212. doi: 10.1371/currents.RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 44.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 47.Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 48.Lempp FA, Wiedtke E, Qu B, Roques P, Chemin I, Vondran FWR, Le Grand R, Grimm D, Urban S. 2017. Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 66:703–716. doi: 10.1002/hep.29112. [DOI] [PubMed] [Google Scholar]
- 49.Müller SF, König A, Döring B, Glebe D, Geyer J. 2018. Characterisation of the hepatitis B virus cross-species transmission pattern via Na+/taurocholate co-transporting polypeptides from 11 New World and Old World primate species. PLoS One 13:e0199200. doi: 10.1371/journal.pone.0199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verrier ER, Colpitts CC, Bach C, Heydmann L, Zona L, Xiao F, Thumann C, Crouchet E, Gaudin R, Sureau C, Cosset FL, McKeating JA, Pessaux P, Hoshida Y, Schuster C, Zeisel MB, Baumert TF. 2016. Solute carrier NTCP regulates innate antiviral immune responses targeting hepatitis C virus infection of hepatocytes. Cell Rep 17:1357–1368. doi: 10.1016/j.celrep.2016.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abbas Z, Jafri W, Raza S. 2010. Hepatitis D: scenario in the Asia-Pacific region. World J Gastroenterol 16:554–562. doi: 10.3748/wjg.v16.i5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho RH, Leake BF, Roberts RL, Lee W, Kim RB. 2004. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. J Biol Chem 279:7213–7222. doi: 10.1074/jbc.M305782200. [DOI] [PubMed] [Google Scholar]
- 53.Li N, Zhang P, Yang C, Zhu Q, Li Z, Li F, Han Q, Wang Y, Lv Y, Wei P, Liu Z. 2014. Association of genetic variation of sodium taurocholate cotransporting polypeptide with chronic hepatitis B virus infection. Genet Test Mol Biomarkers 18:425–429. doi: 10.1089/gtmb.2013.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su Z, Li Y, Liao Y, Cai B, Chen J, Zhang J, Li L, Ying B, Tao C, Zhao M, Ba Z, Zhang Z, Wang L. 2016. Polymorphisms in sodium taurocholate cotransporting polypeptide are not associated with hepatitis B virus clearance in Chinese Tibetans and Uygurs. Infect Genet Evol 41:128–134. doi: 10.1016/j.meegid.2016.03.039. [DOI] [PubMed] [Google Scholar]
- 55.Lee HW, Park HJ, Jin B, Dezhbord M, Kim DY, Han K-H, Ryu W-S, Kim S, Ahn SH. 2017. Effect of S267F variant of NTCP on the patients with chronic hepatitis B. Sci Rep 7:17634. doi: 10.1038/s41598-017-17959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Xu G, Gao Z, Zhou Z, Guo G, Li D, Jing Z, Sui J, Li W. 2018. The p.Ser267Phe variant of sodium taurocholate cotransporting polypeptide (NTCP) supports HBV infection with a low efficiency. Virology 522:168–176. doi: 10.1016/j.virol.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Blumberg BS, London WT. 2000. Hepatitis B virus and the prevention of primary cancer of the liver, p 406–412. In Blumberg BS. (ed), Hepatitis B and the prevention of primary cancer of the liver, vol 4 World Scientific, London, United Kingdom. [Google Scholar]
- 58.Dupinay T, Gheit T, Roques P, Cova L, Chevallier-Queyron P, Tasahsu S, Le Grand R, Simon F, Cordier G, Wakrim L, Benjelloun S, Trépo C, Chemin I. 2013. Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island. Hepatology 58:1610–1620. doi: 10.1002/hep.26428. [DOI] [PubMed] [Google Scholar]
- 59.Dickens C, Kew MC, Purcell RH, Kramvis A. 2013. Occult hepatitis B virus infection in chacma baboons, South Africa. Emerg Infect Dis 19:598–605. doi: 10.3201/eid1904.121107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu L, Hu H, Liu Y, Jing Z, Li W. 2017. Woodchuck sodium taurocholate cotransporting polypeptide supports low-level hepatitis B and D virus entry. Virology 505:1–11. doi: 10.1016/j.virol.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Popper H, Roth L, Purcell RH, Tennant BC, Gerin JL. 1987. Hepatocarcinogenicity of the woodchuck hepatitis virus. Proc Natl Acad Sci U S A 84:866–870. doi: 10.1073/pnas.84.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. 2012. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Compton AA, Emerman M. 2013. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog 9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rasche A, Souza B, Drexler JF. 2016. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr Opin Virol 16:86–94. doi: 10.1016/j.coviro.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Brook CE, Dobson AP. 2015. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol 23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pavlovich SS, Lovett SP, Koroleva G, Guito JC, Arnold CE, Nagle ER, Kulcsar K, Lee A, Thibaud-Nissen F, Hume AJ, Muhlberger E, Uebelhoer LS, Towner JS, Rabadan R, Sanchez-Lockhart M, Kepler TB, Palacios G. 2018. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell 18:1098.e18–1110.e18. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, Tachedjian M, Zhu Y, Zhou P, Jiang X, Ng J, Yang L, Wu L, Xiao J, Feng Y, Chen Y, Sun X, Zhang Y, Marsh GA, Crameri G, Broder CC, Frey KG, Wang LF, Wang J. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, Wang LF, Shi ZL, Zhou P. 2018. Dampened STING-dependent interferon activation in bats. Cell Host Microbe 23:297–301. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De La Cruz-Rivera PC, Kanchwala M, Liang H, Kumar A, Wang LF, Xing C, Schoggins JW. 2018. The IFN response in bats displays distinctive IFN-stimulated gene expression kinetics with atypical RNASEL induction. J Immunol 200:209–217. doi: 10.4049/jimmunol.1701214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JH, Mok L, Michalski WP, Mendenhall IH, Tachedjian G, Wang LF, Baker ML. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc Natl Acad Sci U S A 113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barber MF, Elde NC. 2014. Escape from bacterial iron piracy through rapid evolution of transferrin. Science 346:1362–1366. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab 17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen B, Han X, Zhang J, Rossiter SJ, Zhang S. 2012. Adaptive evolution in the glucose transporter 4 gene Slc2a4 in Old World fruit bats (family: Pteropodidae). PLoS One 7:e33197. doi: 10.1371/journal.pone.0033197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geoghegan JL, Duchêne S, Holmes EC. 2017. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog 13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altringham JD. 2011. Bats: from evolution to conservation. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 79.NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Irwin DM, Kocher TD, Wilson AC. 1991. Evolution of the cytochrome b gene of mammals. J Mol Evol 32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 81.Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A 102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 85.Lefort V, Longueville J-E, Gascuel O. 2017. SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 87.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 88.Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Bruni R, Quick M, Zhou M. 2014. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505:569–573. doi: 10.1038/nature12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johansson MU, Zoete V, Michielin O, Guex N. 2012. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics 13:173. doi: 10.1186/1471-2105-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 92.Julien-Laferriere A, Siberchicot A, Dray S. 2015. The Adegraphics package.