Endogenous retroviruses are genomic traces of past infections present in all vertebrates. Most of these elements degenerate over time and become nonfunctional, but the mouse genome still contains several families with full infection abilities. The GLN retrovirus is one of them, and its members encode particles that are able to infect only mouse cells. Here, we identified the cellular protein used as a receptor by GLN for cell entry. It is SLC19A1, the reduced folate carrier. We show that GLN infection is limited to mouse cells due to both a mutation in the mouse gene preventing the glycosylation of SLC19A1 and also other residues conserved within the rat but not in the hamster and human proteins. Like all other gammaretroviruses whose receptors have been identified, GLN uses a member of the solute carrier superfamily for cell entry, highlighting the role of these proteins for retroviral infection in mammals.
KEYWORDS: Env, ERV, GLN, endogenous retrovirus, gammaretrovirus, receptor
ABSTRACT
Approximately 10% of the mouse genome is composed of endogenous retroviruses belonging to different families. In contrast to the situation in the human genome, several of these families correspond to recent, still-infectious elements capable of encoding complete viral particles. The mouse GLN endogenous retrovirus is one of these active families. We previously identified one fully functional provirus from the sequenced genome of the C57BL/6 mouse strain. The GLN envelope protein gives the infectious viral particles an ecotropic host range, and we had demonstrated that the receptor was neither CAT1 nor SMIT1, the two previously identified receptors for mouse ecotropic retroviral envelope proteins. In this study, we have identified SLC19A1, the reduced folate carrier, as the cellular protein used as a receptor by the GLN retrovirus. The ecotropic tropism exhibited by this envelope is due to the presence or absence of an N-linked glycosylation site in the first extracellular loop as well as the specific amino acid sequence of the extracellular domains of the receptor. Like all the other retroviral envelope proteins from the gammaretrovirus genus whose receptors have been identified, the GLN envelope protein uses a member of the solute carrier superfamily as a receptor.
IMPORTANCE Endogenous retroviruses are genomic traces of past infections present in all vertebrates. Most of these elements degenerate over time and become nonfunctional, but the mouse genome still contains several families with full infection abilities. The GLN retrovirus is one of them, and its members encode particles that are able to infect only mouse cells. Here, we identified the cellular protein used as a receptor by GLN for cell entry. It is SLC19A1, the reduced folate carrier. We show that GLN infection is limited to mouse cells due to both a mutation in the mouse gene preventing the glycosylation of SLC19A1 and also other residues conserved within the rat but not in the hamster and human proteins. Like all other gammaretroviruses whose receptors have been identified, GLN uses a member of the solute carrier superfamily for cell entry, highlighting the role of these proteins for retroviral infection in mammals.
INTRODUCTION
In enveloped viruses, an essential step in the viral life cycle is binding and fusion of the virus with the host cell. In gammaretroviruses, this fusion of virus and host cell membranes is mediated by the viral envelope protein (Env). When Env comes into contact with a specific receptor or receptors expressed on the host cell envelope, it changes conformation, leading to fusion of the cell and viral membranes. This releases the contents of the virus into the host cell, where the viral genetic material can integrate into the host genome and produce new viral particles. It is the interaction between the viral Env and the cellular receptor that primarily determines the host range that a virus can infect. Some viral Envs, such as vesicular stomatitis virus G protein (VSV-G), have a very wide tropism. The receptors recently identified to be LDLR and other members of this family (1) are widely expressed, and viruses pseudotyped with this glycoprotein can infect a broad range of cell types across a diverse range of species. In contrast, other Envs can have a host range limited to a specific cell type and/or a particular species (e.g., HIV-1, which infects primarily macrophages and CD4+ T cells through the CD4 receptor and CCR5 and CXCR4 coreceptors).
The first gammaretroviral receptor identified was CAT-1, also known as SLC7A1, the receptor for ecotropic murine leukemia virus (E-MLV) (2). CAT-1 is a 14-pass transmembrane protein that transports cationic amino acids. E-MLV is a mouse ecotropic virus; i.e., it can infect only cells of mouse origin. Regions within the third extracellular loop of mouse CAT-1, which contains 2 glycosylation sites, were found to be essential for its use as a receptor (3). Glycosylation of mouse CAT-1, however, is not essential for its function as a receptor for E-MLV (4, 5), although deglycosylation of rat and hamster cells allowed infection by this virus. The ecotropism of the virus is therefore due to both the glycosylation state of the receptor as well as other specific residues. Other receptors used by gammaretroviruses have since been identified and are all members of the SLC (solute carrier) superfamily (reviewed in reference 6).
Endogenous retroviruses originate from an ancestral infection of the germ line by an exogenous retrovirus. Occasionally, viral genes can be adopted for host cellular functions, but for the most part, these endogenous viruses lose their ability to code for viral proteins over time and are no longer able to code for infectious viruses. In contrast to humans, there are multiple active endogenous retroviruses in the mouse genome. Previously, we searched for functional copies among members of the endogenous gammaretrovirus GLN family (7). We identified one provirus in the C57BL/6 mouse strain that is still able to produce fully infectious viral particles (8). GLN is ecotropic, infecting cells from the mouse species only, but it utilizes a different receptor than those used by the other 2 previously described mouse ecotropic viral Envs, E-MLV and M813 MLV, which utilize CAT-1 (SLC7A1) and SMIT-1 (SLC5A3), respectively.
Here, using a cDNA library screen, we have identified the receptor used by the mouse GLN retrovirus as SLC19A1, a reduced folate transporter, which has not been described previously to act as a receptor for any other virus.
RESULTS
In this study, we searched for the receptor for the mouse GLN endogenous retrovirus, using a gain-of-function cDNA library screen adapted from the one described previously by Bianchi et al. (9) (Fig. 1A). Nonpermissive human 293T cells were transfected with pools of approximately 100 cDNAs from a normalized mouse brain cDNA library and allowed to express for 48 h before being exposed to green fluorescent protein (GFP)-expressing virions pseudotyped with GLN Env. Seventy-two hours after virus exposure, the 293T cells were analyzed for GFP expression by flow cytometry (Fig. 1B). We identified one positive cDNA pool (out of approximately 200, corresponding to roughly 20,000 genes). This positive pool was used to transform bacteria. Pools and subpools were then iteratively generated and screened using the same protocol until we isolated a unique positive colony. We identified a single gene in the original pool, which when expressed in 293T cells made them permissive to infection by GLN Env (Fig. 1C). This cDNA corresponded to the SLC19A1 gene that encodes the reduced folate carrier (10). This is the first time that this protein has been identified as a virus receptor, although the closely related family member SLC19A2 (a thiamine transporter) has been described as a receptor for 2 other gammaretroviruses, koala retrovirus type B (KoRV-B) (11, 12) and feline leukemia virus type A (FeLV-A) (13).
FIG 1.
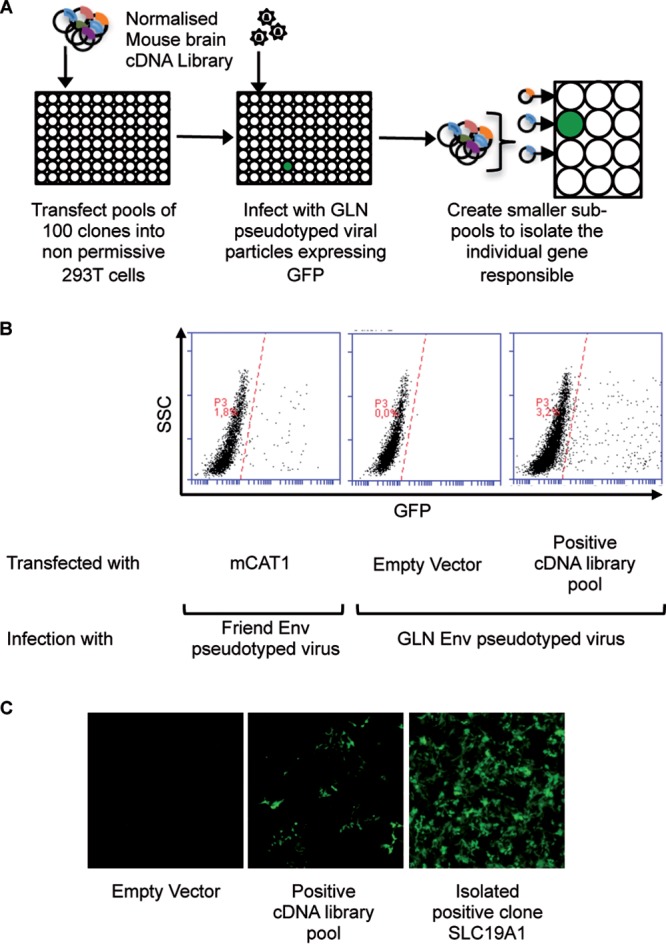
cDNA library screen to identify the GLN cellular receptor. (A) Schematic representation of the screening method. Nonpermissive 293T cells were transfected with pools of 100 cDNAs from a normalized mouse brain library and then exposed to GFP-expressing GLN-pseudotyped virions 48 h later. Seventy-two hours after viral exposure, the 293T cells were analyzed for GFP expression by fluorescence microscopy and flow cytometry. Positive pools were retransformed into competent bacteria and used to generate subpools until the unique positive clone was identified. (B) Flow cytometric analysis of the cDNA screen showing the cDNA pool containing the GLN receptor. Cells transfected with mouse CAT-1 (mCAT1) and infected with Friend Env-pseudotyped viruses were used as positive controls for the assay. SSC, side scatter. (C) Fluorescence microscopy images of GFP-positive 293T cells after infection with GLN-pseudotyped viruses when transfected with the positive cDNA pool and the isolated cDNA (SLC19A1) responsible for enabling infection.
We then sought to confirm the specificity of SLC19A1 as the receptor for GLN. To date, all identified gammaretrovirus receptors are multipass transmembrane proteins called SLC transporters (reviewed in reference 6). In some cases (such as gibbon ape leukemia virus [GALV], FeLV-C, and members of the human endogenous retrovirus W [HERV-W] interference group), gammaretroviruses have been shown to be able to utilize more than one protein to enter cells, but the different receptors of a given virus always transport the same solute. We tested the ability of GLN Env to infect cells expressing other SLC transporters. We investigated SLC19A2, one of the two thiamine transporters which belong to the same SLC family and are the proteins most closely related to SLC19A1, as well as SLC46A1, which is less closely related at the amino acid level but is able to transport folate like SLC19A1 (Fig. 2A). These SLC genes were inserted into a lentiviral vector that was used to transduce 293T cells. Stable cell populations expressing the genes of interest were generated by selection with hygromycin and then exposed to GFP-expressing virions pseudotyped with GLN Env or control Envs (4070A amphotropic MLV, shortened to Ampho and VSV-G). We show that GLN Env-pseudotyped viral particles can infect 293T cells expressing the SLC19A1 gene but not the closely related gene SLC19A2 or the other folate transporter, SLC46A1 (Fig. 2B). SLC19A1 alone was found to render human 293T cells permissive to infection by GLN Env and importantly had no effect on viruses pseudotyped with other Envs (Fig. 2C), demonstrating the specificity of SLC19A1 on GLN Env-pseudotyped viruses. GLN is therefore likely to use just the one protein for cellular entry, with neither the most closely related transporter, SLC19A2, nor SLC46A1, the other SLC transporter for folate, being able to function as a receptor, confirming its specificity as the receptor for the GLN retrovirus.
FIG 2.
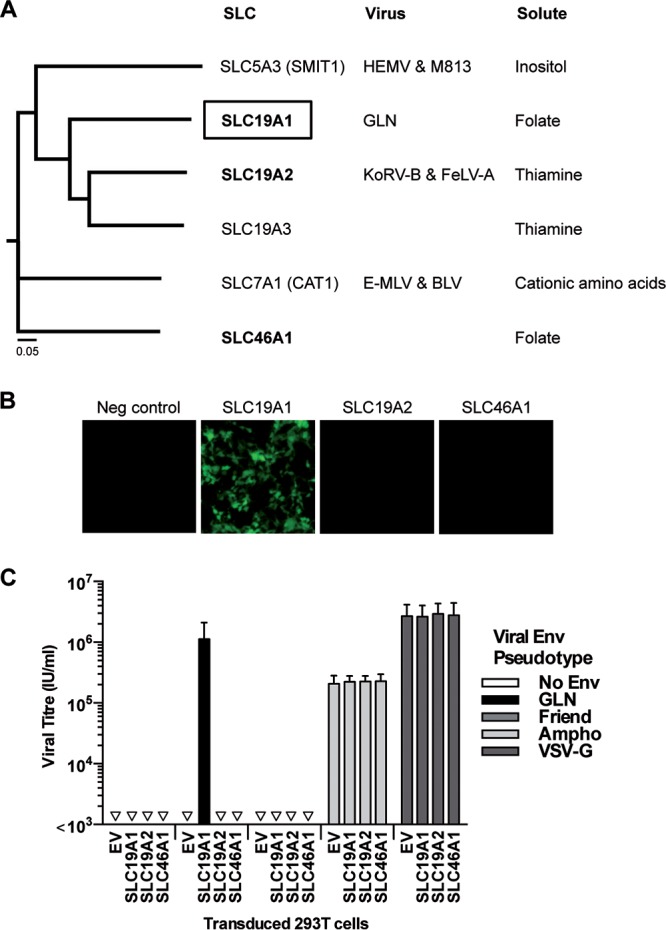
Specificity of SLC19A1 as the receptor for GLN. (A) Neighbor-joining phylogenetic tree without distance corrections showing the GLN receptor SLC19A1 and its similarities to other SLC proteins at the amino acid level. A closely related SLC19 family member, SLC19A2, and the other folate transporter, SLC46A1 (in boldface type), were tested for their ability to render 293T cells permissive to infection by GLN. HEMV, hortulanus endogenous murine leukemia virus; BLV, bovine leukemia virus. (B) 293T cells were transduced to express SLC19A1, SLC19A2, SLC46A1, or an empty vector (EV) control. After selection with hygromycin, the cell populations were exposed to GLN-pseudotyped viruses, and fluorescence was analyzed 72 h later by microscopy. Images are from one infection assay representative of results from 3 independent experiments. (C) Viral titers of different pseudotyped viruses on the same transduced cells were measured using flow cytometry (n = 3; the standard deviations [SD] are represented by the error bars).
GLN is an endogenous retrovirus, and as such, an ancestral virus had managed to infect the germ line of the mouse. If the ancestral virus used SLC19A1 to enter the mouse genome, it should be expressed in the germ line (e.g. ovary or testis, or early embryos). We measured the levels of SLC19A1 transcripts in different mouse tissues by reverse transcription-quantitative PCR (RT-qPCR). The expression pattern of SLC19A1 in mice is broad, and we detected its expression in all mouse tissues tested (Fig. 3). Importantly, this included the male and female reproductive organs as well as the developing embryo, consistent with a receptor for an endogenized retrovirus.
FIG 3.
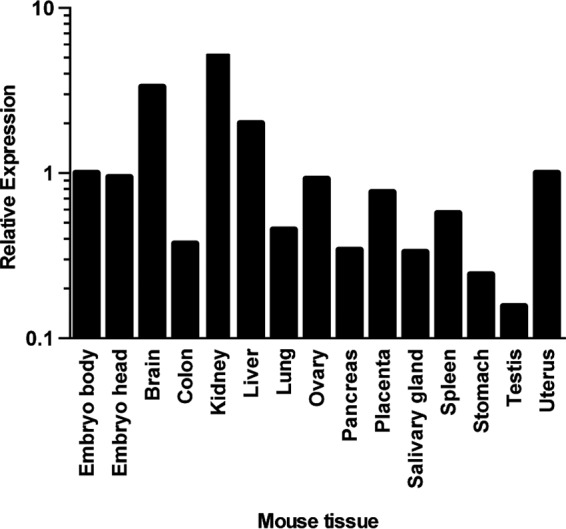
Relative expression levels of SLC19A1 in different mouse tissues. RT-qPCR was used to measure the expression of SLC19A1 transcripts from pooled RNA obtained from multiple organs. SLC19A1 transcript levels were normalized to the level of the housekeeping gene RPLPO and are shown relative to levels for the embryo body. The lowest expression level of SLC19A1 is in the testis (threshold cycle [CT], 27) compared to the housekeeping gene RPLP0 (CT, 20). This measurement was performed once. Bars represent average values from 2 technical replicates.
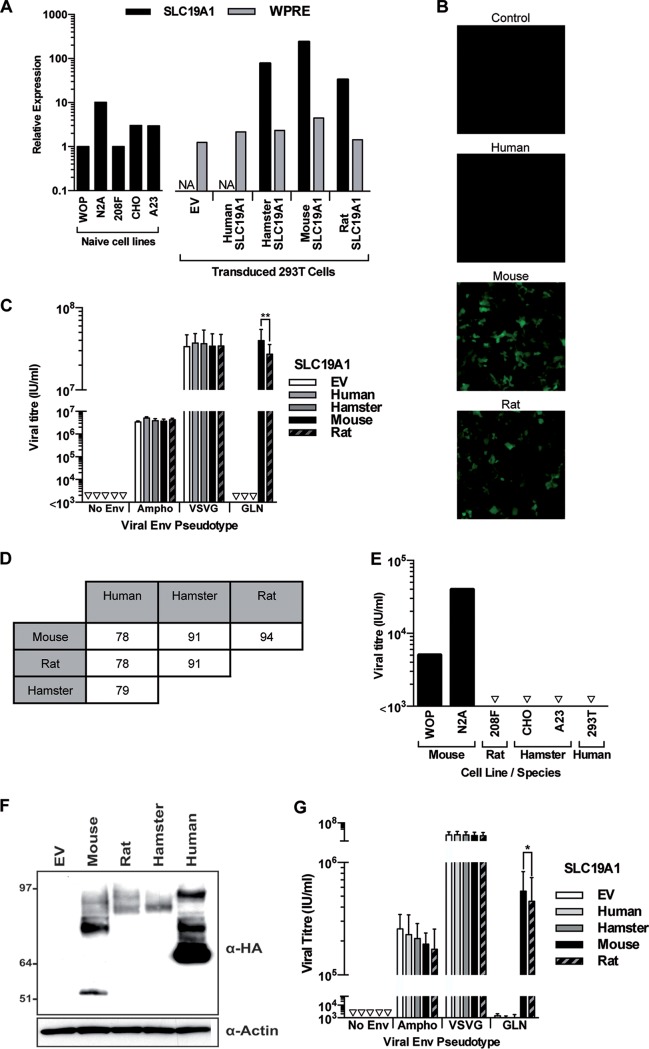
When we originally described the GLN retrovirus, we tested its tropism using different cell lines and found that it was an ecotropic virus, with a tropism restricted solely to mouse cells (8). We decided to determine if this observed tropism could be explained by differences in the sequences of the SLC19A1 genes of the different species. We therefore tested if the mouse SLC19A1 gene alone is able to make 293T cells permissive to GLN infection. SLC19A1 cDNAs from human, rat, and hamster were cloned into the lentiviral vector and used to generate stable cell populations as described above. We checked by RT-qPCR using primers in the common woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) part of the lentiviral transcript that all constructs were expressed at similar levels (Fig. 4A, gray bars). As described earlier, mouse SLC19A1 is enough to render 293T cells sensitive to infection by viral pseudotypes with GLN Env on their surface. In contrast, hamster and human versions of the cDNA did not render 293T cells permissive to infection by GLN-pseudotyped virions, as expected from the tropism observed in vivo. In the same assay, 293T cells expressing rat SLC19A1 could be infected by GLN Env-pseudotyped virions although clearly less efficiently (1.5- to 2-fold-lower viral titers; P < 0.01 by a t test) than cells expressing the mouse gene (Fig. 4B and C). This is not completely unexpected, since the rat version is the closest to the mouse protein, and they share 94% amino acid similarity (Fig. 4D), but is in contradiction with the strict ecotropism that we had initially observed (8) and reproduced here in the same experiment (rat 208F cells versus mouse WOP or N2A cells) (Fig. 4E). In order to verify that each protein is properly expressed, we generated hemagglutinin (HA)-tagged versions of SLC19A1. These constructs were all detected by Western blotting, although the human protein was expressed at a significantly higher level (Fig. 4F). The profile of the proteins on the blot is complex. This could be due to the stable overexpression of SLC19A1 leading to misfolded proteins, and it is mostly consistent with data from previous publications (14, 15). These HA-tagged proteins behaved in the same way as the untagged counterparts when tested for their ability to make 293T cells permissive to infection by GLN-pseudotyped virions (Fig. 4G), indicating that the differences that we observed were not due to variations in the expression levels but were due to the intrinsic properties of the proteins.
FIG 4.
Specificity of SLC19A1 as the cellular receptor for GLN. 293T cells were transduced to express SLC19A1 from different species before being exposed to GFP-expressing pseudotyped viruses. (A) RT-qPCR was used to measure the relative expression of SLC19A1 from different rodent cell lines and the exogenous expression of the SLC19A1 transgenes in stably transduced 293T cells. NA, not applicable, as the SLC19A1 qPCR oligonucleotides are able to detect only rodent versions of the transcript (100% homology with mouse, rat, and hamster sequences) and were not used to analyze empty vector- and human SLC19A1-transduced cells. Therefore, relative expression of the transgene was also assessed by detection of the 3′ untranslated region (UTR) (WPRE) present in all lentiviral constructs. Levels of both transcripts were normalized to the level of the housekeeping gene RPLPO. Expression levels of SLC19A1 and WPRE are shown relative to those in WOP and empty vector-transduced 293T cells, respectively. This measurement was performed once using one set of cells that were used for the experiment in panel B. Bars represent average values from 2 technical replicates. (B and C) Infection by GLN was assessed by fluorescence (B), and the titers of viruses pseudotyped with Ampho, VSV-G, GLN Envs, or a no-Env control were calculated by flow cytometry (C) (**, P < 0.01 by a paired t test; n = 3 except for GLN infections [n = 6]; the SD are represented by the error bars). (D) Amino acid similarity of the SLC19A1 sequences between different species. (E) Demonstration of the species tropism of GLN by infection of cell lines of different rodent species as well as of the human 293T cell line after transduction with mouse SLC19A1. Note that the differences in the titers observed between the mouse WOP and N2A cell lines is likely due to the difference in SLC19A1 expression levels (panel A), since we ensured by sequencing that both cell lines express identical versions of the SLC19A1 gene. Shown are data from one representative experiment out of three. (F) 293T cells were transduced with 3′ HA-tagged versions of the SLC19A1 genes. Western blotting was used to detect expression of the protein using an anti-HA antibody. The image shown is representative of results from 3 independent experiments. (G) 293T cells expressing the HA-tagged versions of the SLC19A1 constructs were exposed to GFP-expressing pseudotyped viruses. (*, P < 0.05 by a paired t test; n = 4).
The apparent discrepancy between the tropisms seen in the different cell lines and the transfected 293T cells cannot be accounted for by a lack of expression of SLC19A1 in rat cells, since RT-qPCR showed similar levels of expression between the mouse WOP cell line and the rat 208F cells that we used in this experiment (Fig. 4A, black bars). One possible explanation is that lentiviral transduction results in an abnormally high expression level of the cDNA, rendering the rat copy weakly active as a receptor, whereas it does not allow infection when expressed at a normal level. Indeed, stably transduced 293T cells express 20-fold-higher levels of SLC19A1, as measured by RT-qPCR, than the rat 208F cell line (Fig. 4A, black bars). The difference could be even greater at the protein level due to the optimized leader sequence present in the lentiviral vector.
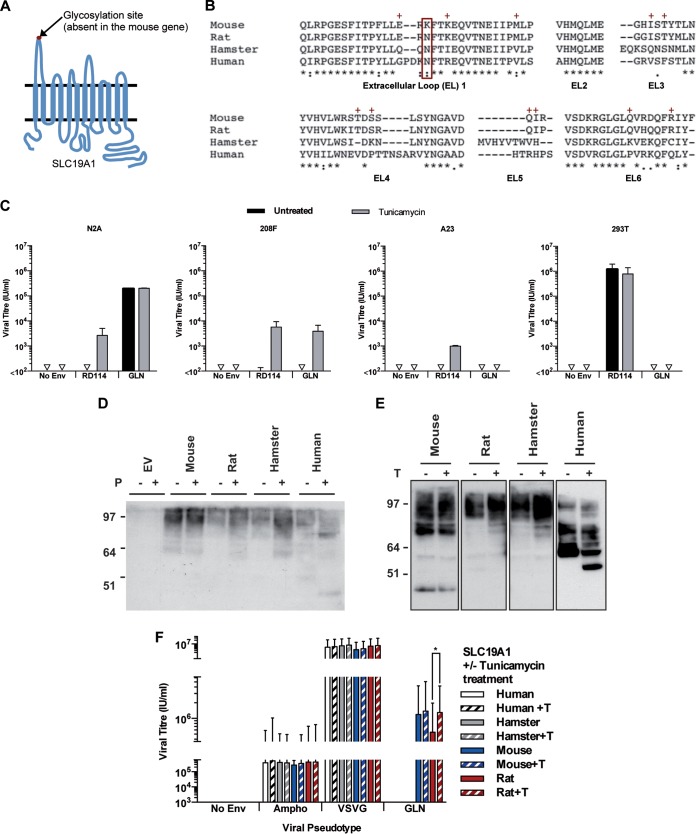
SLC19A1 is a 12-pass transmembrane protein with 6 extracellular loops and has a canonical MFS (major facilitator superfamily) fold (Fig. 5A) (16–18). As described above, GLN Env recognizes specifically the mouse version of the protein and not the hamster or human one. The rat protein, which is most closely related to the mouse protein, can also be used as a receptor but only when overexpressed, suggesting an intermediate phenotype. Figure 5B shows a multiple-sequence alignment of the amino acid residues of the extracellular loops of SLC19A1 from these species, which are likely to be the domains recognized or not by GLN because of their extracellular localization. Interestingly, the sequences of all species, except mouse, contain a single potential N-linked glycosylation site located at position N56 (N58 when referenced to the human gene), within the first extracellular loop (mutation N to K in the mouse protein) (Fig. 5A and B) (reviewed in reference 19). This site has been demonstrated to be glycosylated in the human gene (15, 20) and could be the determinant allowing infection of mouse cells but not those from other species. We tested if this is the case by treating different cell lines with tunicamycin, an N-linked glycosylation inhibitor (shown to be able to deglycosylate the human SLC19A1 protein in [15]), prior to infection with retroviral particles pseudotyped with GLN Env or RD114 Env (known to be sensitive to glycosylation in rodent cells [21]) (Fig. 5C). As expected, the RD114-pseudotyped particles could infect human 293T cells, and rodent cells only after treatment with tunicamycin, proving that the deglycosylation was effective. Among untreated cells, only mouse N2A cells could be infected by the GLN Env-carrying particles, in agreement with the above-described results. Interestingly, tunicamycin treatment rendered rat 208F cells sensitive to infection by GLN Env, indicating that the glycosylation of the N56 residue of SLC19A1 is enough to prevent infection by GLN Env. In contrast, the human and hamster cell lines remained refractory to infection by GLN Env even after tunicamycin treatment.
FIG 5.
Effects of glycosylation on GLN tropism. (A) Schematic of SLC9A1 showing the 12 transmembrane domains and the 6 extracellular loops. A putative N-linked glycosylation site is present in the first extracellular loop of all species except for mouse. (B) Clustal analysis of the amino acid sequences of SLC19A1 showing the amino acid similarities of the six extracellular loops between mouse, rat, hamster, and human proteins. The glycosylation site at N56 (boxed in red) is absent in the mouse protein. The red + symbols indicate residues shared by mouse and rat proteins but not human and hamster proteins. (C) Titers of viruses pseudotyped with RD114 or GLN envelopes were measured on different cell lines with or without 16 h of tunicamycin (0.2 μg/ml) pretreatment to remove N-linked glycosylation sites (n = 3; the SD are represented by the error bars). (D) 293T cells were transduced to express HA-tagged SLC19A1 from different species. Cell lysates were treated with PNGase F (shortened to P) to remove N-linked glycans before probing for SLC19A1 by Western blotting using an anti-HA antibody. (E) Cells were treated with tunicamycin (shortened to T) before collection of cell lysates, and SLC19A1 proteins were detected by Western blotting using an anti-HA antibody. (F) The titers of viruses pseudotyped with different Envs were measured on 293T cells expressing HA-tagged SLC19A1 from different species with and without tunicamycin pretreatment.
To further analyze the role of the glycosylation of SLC19A1 in our model of GLN Env-mediated infection, we then utilized HA-tagged proteins. Lysates obtained from 293T cells transduced with the HA-tagged versions of the proteins were treated with peptide-N-glycosidase F (PNGase F) to enzymatically remove N-linked glycosylation. Western blotting (Fig. 5D) shows that the mouse protein is unaffected by PNGase F treatment, while the proteins from the other 3 species all migrate at a lower molecular weight than the untreated proteins, consistent with the loss of sugar residues leading to a decrease in the apparent molecular weight. Additionally, cell lysates extracted from cells treated with tunicamycin showed similar results (Fig. 5E). Tunicamycin treatment of rat SLC19A1 restored viral titers to the level of the mouse protein and had no effect on the proteins from the other 2 species (Fig. 5F), consistent with the above-described data.
Since the mouse and rat proteins are equally efficient as receptors when deglycosylated, its is not the K amino acid in the mouse protein that is preferred over N for Env binding. More likely, the sugar moieties added on the N amino acid in the rat protein may block access to other residues via steric hindrance, as previously observed for other retroviral Envs and their receptors (22). Altogether, these data show that residues (likely to be located on the first extracellular loop, although we cannot rule out the possibility that other loops may be involved) shared between the mouse and rat proteins (Fig. 5B), but not the hamster and human ones, are involved in SLC19A1 recognition and binding by GLN Env and that N-linked glycosylation of the rat protein at position 56 prevents or limits it from being used as a receptor in rat cells.
DISCUSSION
In this study, we have identified the receptor of the mouse endogenous GLN retrovirus as another member of the SLC family, SLC19A1. Neither SLC19A2, a closely related SLC, nor the other folate transporter, SLC46A1, is able to render cells permissive to GLN, suggesting that SLC19A1 is the sole receptor used by this virus. Interestingly, we found that cells overexpressing the rat protein could be infected by GLN, whereas rat cells expressing endogenous levels of the protein could not. Using tunicamycin, we also showed that preventing glycosylation of endogenous rat SLC19A1 renders it functional for infection mediated by GLN Env. This indicates that, unlike the hamster or human version of SLC19A1, the rat one is nearly usable by GLN Env under natural conditions. It is possible that the overexpression obtained by transducing the cells augments the membrane concentration of rat SLC19A1 compared to that in normal cells and therefore increases the total avidity for GLN Env even if the affinity is lower than that of the mouse protein. Another possibility is that the overexpression caused by transduction overloads the cell glycosylation machinery, allowing for unglycosylated/incompletely glycosylated SLC19A1 to be expressed at the cell surface. The use of HA-tagged versions of the SLC19A1 proteins shows that, when deglycosylated, the mouse and rat proteins are equal in their abilities to act as receptors for GLN. Altogether, the data suggest that a site other than the K/N56 position is a strong determinant for the ability of GLN Env to utilize SLC19A1 as a receptor. This site is expected to be conserved between mouse and rat but not in the hamster and human versions of the protein. Glycosylation at position 56 is also involved in the GLN Env-SLC19A1 interaction, playing an inhibitory role.
To date, gammaretroviral Envs (also called transmembrane [TM] class 1 retroviral Envs [6]) have been found to uniquely use members of the SLC family for infection of their hosts, and this study adds another family member to this list. The SLC family, despite their name and numbering system, is a collection of diverse proteins, which transport a wide variety of solutes. SLCs can be active, ion-coupled, or exchanger transporters, or even simply passive channels. They are classified into 4 main groups, designated groups α, β, γ, and δ (plus so-called unclassified SLCs) (23), and receptors for gammaretroviruses have been found in groups α, β, and γ as well as among ungrouped SLCs. SLCs from different groups have limited sequence homology and can have very different structures. For example, members of the MFS (or the α group, to which SLC19A1 belongs) share an organization, with their 12 transmembrane domains arranged in 2 sets of 6-helix bundles separated by a large flexible hinge which allows the 2 halves of the protein to fold onto itself, forming the central cavity through which solutes are transported. The binding site for gammaretroviral Envs in this case is expected to be found within the extracellular loops between the transmembrane domains, as was demonstrated previously for the human T cell leukemia virus (HTLV) receptor SLC2A1, another MFS member (24). In comparison, SLC1A5, the receptor for numerous gammaretroviral Envs (e.g., RD114, baboon endogenous virus [BaEV], and HERV-W), is one of the unclassified SLCs. Its structure was recently solved and shown to be a homotrimer, with each monomer containing 8 transmembrane domains as well as 2 intramembrane hairpin loops (25). Surprisingly enough, the Env binding site is not found within the extracellular loops but is located inside a transmembrane domain (variable in length and sequence among SLC1 family members) from which a loop protrudes like an “antenna” into the extracellular space. The diversity of structures among SLCs that are used as receptors by retroviruses suggests that there may not be a specific domain shared by SLCs that is recognized by gammaretroviral Envs, and the model where small modifications within Env proteins could enable them to recognize different receptors is probably false. What, then, is the reason why SLCs are apparently the preferred choice of receptor for gammaretroviruses? First, SLCs are the second most common membrane protein after G-protein-coupled receptors, with more than 400 family members, most of which are located on the plasma membrane. Second, they are mostly ubiquitously expressed, increasing the probability that they will be present on potential target cells of the virus. Finally, they are widespread among all kingdoms, from bacteria to vertebrates, and are mostly conserved within mammals. The use of SLCs as receptors therefore increases the likelihood of horizontal transfers that may help gammaretroviruses find new hosts and thrive as either exogenous or even endogenous retroviruses, as exemplified by the recent acquisition of KoRV in wild koalas, likely from overseas rodents.
MATERIALS AND METHODS
Plasmids.
MLV-derived retroviral particles were produced using plasmids CMVi (MLV core proteins) and CNCG (GFP-containing MLV-derived viral RNA) and a phCMV-based expression vector for a viral Env protein (VSV G protein, 4070A amphotropic Env, ecotropic Friend Env, and RD114 Env [26, 27]). The expression plasmid for GLN Env was described previously (called GLN-2 Env in reference 8). HIV-1-derived lentiviral particles were produced using plasmid 8.91 (HIV-1 core proteins [28]), the phCMV VSV G expression plasmid, a pDual lentiviral vector coexpressing GFP or a SLC (or nothing in the case of the empty vector control), and the hygromycin resistance gene (29).
Open reading frames (ORFs) for SLC19A1 were obtained from the mouse brain cDNA library (reference number 10104-N; Express Genomics), cloned directly from CHO cell cDNA (hamster gene), or bought (from Dharmacon for the rat gene and R&D Systems for the human one) and cloned into a pDual lentiviral vector via PCR. ORFs for mouse SLC19A2 and SLC46A1 genes were similarly bought (Dharmacon) and subcloned into both expression vectors. 3′ HA-tagged versions of the SLC19A1 genes were also generated by PCR. All plasmids were checked by sequencing.
Cells.
Human 293T cells (ECACC 12022001), mouse N2A (ECACC 89121404) and WOP (Cellausaurus CVCL-3433) cells, rat 208F cells (ECACC 85103116), and hamster CHO cells (ECACC 85050302) were grown at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), 100 mg/ml streptomycin, and 100 U/ml penicillin.
cDNA library screening.
A normalized mouse brain cDNA library was prepared mostly as described previously (9). Briefly, the library was plated to obtain approximately 100 colonies per plate. The colonies on each plate were pooled and used for DNA extraction to generate 100 colony pools that were used for the screening. Two hundred such pools were used to transfect 293T cells seeded in 96-well plates using Fugene 6. The transfected cells were infected with GLN Env-pseudotyped retroviral particles 48 h after transfection and checked for fluorescence another 72 h later. Only one pool was found to give a positive signal and was used to transform competent bacteria and subsequently subpooled until the individual cDNA responsible for the positive signal was isolated.
Viral infections.
Lentiviral and retroviral particles were produced by transiently transfecting 293T cells using JetPrime (Polyplus Transfection) with a 3/2/1 ratio of the different plasmids (lentiviral vector, viral core proteins, and viral Env protein). Supernatants were collected at 3 days posttransfection and filtered (0.45-μm filters) before use. All infections using retroviral particles were performed in the presence of Polybrene (final concentration, 8 μg/ml). GFP expression was assessed at 3 days posttransduction, when expression is at a maximum level.
RT-qPCR analysis.
Total RNA was extracted using the RNeasy extraction kit and QIAshredder (Qiagen), treated with DNase I (Ambion), and reverse transcribed using random hexamers (Applied biosystems). Quantitative PCR (qPCR) was performed using the QuantiFast SYBR green PCR kit (Qiagen) on an ABI Prism 7000 system. The efficacies of the PCRs were checked for each primer pair, and transcript levels were normalized to the level of RPLPO using the ΔΔCT method.
Generation of a phylogenetic tree.
Amino acid sequences of the mouse SLC proteins were obtained from the NCBI gene database, and multiple-sequence alignment analysis was performed with Clustal Omega (30) to generate a neighbor-joining tree without distance corrections.
Western blotting.
Cells were lysed in a solution containing phosphate-buffered saline (PBS) and 1% NP-40 complemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Cell lysates were treated or not with PNGase F (Biolabs), subjected to SDS-PAGE under reducing conditions using the NuPAGE system (precast Novex 4 to 12% Bis-Tris gels; Life Technology), and transferred onto nitrocellulose membranes with a semidry system. The HA-tagged versions of SLC19A1 were detected using an anti-HA antibody from Roche (clone 3F10, diluted 1/5,000), and an antibody specific for β-actin (catalog number 3700; Cell Signaling Technology) was used when required to ensure equal loading of the different samples after stripping the membranes using ReBlot Plus strong (Merck Millipore). Horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare or Dako) and ECL Plus reagent (GE Healthcare) were used for chemiluminescence detection.
ACKNOWLEDGMENT
J.T. was supported by a postdoctoral fellowship from the Ligue National contre le Cancer.
REFERENCES
- 1.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. 2013. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A 110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 3.Miller DG, Miller AD. 1992. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol 66:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Klamo E, Kuhmann SE, Kozak SL, Kavanaugh MP, Kabat D. 1996. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J Virol 70:6884–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiden MV, Farrell K, Wilson CA. 1994. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol 68:626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwood AD, Ishida Y, O’Brien SP, Roca AL, Eiden MV. 2018. Transmission, evolution, and endogenization: lessons learned from recent retroviral invasions. Microbiol Mol Biol Rev 82:e00044-17. doi: 10.1128/MMBR.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itin A, Keshet E. 1986. A novel retroviruslike family in mouse DNA. J Virol 59:301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribet D, Harper F, Esnault C, Pierron G, Heidmann T. 2008. The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J Virol 82:4413–4419. doi: 10.1128/JVI.02141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi E, Doe B, Goulding D, Wright GJ. 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon KH, Lanpher BC, Chiu J, Kelley K, Cowan KH. 1994. A novel cDNA restores reduced folate carrier activity and methotrexate sensitivity to transport deficient cells. J Biol Chem 269:17–20. [PubMed] [Google Scholar]
- 11.Xu W, Stadler CK, Gorman K, Jensen N, Kim D, Zheng H, Tang S, Switzer WM, Pye GW, Eiden MV. 2013. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A 110:11547–11552. doi: 10.1073/pnas.1304704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shojima T, Yoshikawa R, Hoshino S, Shimode S, Nakagawa S, Ohata T, Nakaoka R, Miyazawa T. 2013. Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J Virol 87:9943–9948. doi: 10.1128/JVI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothem L, Berman B, Stark M, Jansen G, Assaraf YG. 2005. The reduced folate carrier gene is a novel selectable marker for recombinant protein overexpression. Mol Pharmacol 68:616–624. doi: 10.1124/mol.105.013540. [DOI] [PubMed] [Google Scholar]
- 15.Wong SC, Zhang L, Proefke SA, Matherly LH. 1998. Effects of the loss of capacity for N-glycosylation on the transport activity and cellular localization of the human reduced folate carrier. Biochim Biophys Acta 1375:6–12. doi: 10.1016/S0005-2736(98)00118-7. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson PL, Flintoff WF. 1999. Topological and functional analysis of the human reduced folate carrier by hemagglutinin epitope insertion. J Biol Chem 274:16269–16278. doi: 10.1074/jbc.274.23.16269. [DOI] [PubMed] [Google Scholar]
- 17.Liu XY, Matherly LH. 2002. Analysis of membrane topology of the human reduced folate carrier protein by hemagglutinin epitope insertion and scanning glycosylation insertion mutagenesis. Biochim Biophys Acta 1564:333–342. doi: 10.1016/S0005-2736(02)00467-4. [DOI] [PubMed] [Google Scholar]
- 18.Cao W, Matherly LH. 2004. Analysis of the membrane topology for transmembrane domains 7-12 of the human reduced folate carrier by scanning cysteine accessibility methods. Biochem J 378:201–206. doi: 10.1042/BJ20031288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganapathy V, Smith SB, Prasad PD. 2004. SLC19: the folate/thiamine transporter family. Pflugers Arch 447:641–646. doi: 10.1007/s00424-003-1068-1. [DOI] [PubMed] [Google Scholar]
- 20.Matherly LH, Czajkowski CA, Angeles SM. 1991. Identification of a highly glycosylated methotrexate membrane carrier in K562 human erythroleukemia cells up-regulated for tetrahydrofolate cofactor and methotrexate transport. Cancer Res 51:3420–3426. [PubMed] [Google Scholar]
- 21.Marin M, Lavillette D, Kelly SM, Kabat D. 2003. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J Virol 77:2936–2945. doi: 10.1128/JVI.77.5.2936-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tailor CS, Lavillette D, Marin M, Kabat D. 2003. Cell surface receptors for gammaretroviruses. Curr Top Microbiol Immunol 281:29–106. [DOI] [PubMed] [Google Scholar]
- 23.Hoglund PJ, Nordstrom KJ, Schioth HB, Fredriksson R. 2011. The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol Biol Evol 28:1531–1541. doi: 10.1093/molbev/msq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manel N, Battini JL, Sitbon M. 2005. Human T cell leukemia virus envelope binding and virus entry are mediated by distinct domains of the glucose transporter GLUT1. J Biol Chem 280:29025–29029. doi: 10.1074/jbc.M504549200. [DOI] [PubMed] [Google Scholar]
- 25.Garaeva AA, Oostergetel GT, Gati C, Guskov A, Paulino C, Slotboom DJ. 2018. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat Struct Mol Biol 25:515–521. doi: 10.1038/s41594-018-0076-y. [DOI] [PubMed] [Google Scholar]
- 26.Dewannieux M, Collins MK. 2008. Spontaneous heteromerization of gammaretrovirus envelope proteins: a possible novel mechanism of retrovirus restriction. J Virol 82:9789–9794. doi: 10.1128/JVI.02696-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemaitre C, Tsang J, Bireau C, Heidmann T, Dewannieux M. 2017. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog 13:e1006451. doi: 10.1371/journal.ppat.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 29.Perro M, Tsang J, Xue SA, Escors D, Cesco-Gaspere M, Pospori C, Gao L, Hart D, Collins M, Stauss H, Morris EC. 2010. Generation of multi-functional antigen-specific human T-cells by lentiviral TCR gene transfer. Gene Ther 17:721–732. doi: 10.1038/gt.2010.4. [DOI] [PubMed] [Google Scholar]
- 30.Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]