Negative-strand RNA viruses (NSVs) include the most pathogenic viruses known. New methods to monitor their evolutionary trends are urgently needed for the development of antivirals and vaccines. The protein translation machinery of the host cell is currently recognized as a main genomic regulator of RNA virus evolution, which works especially well for positive-strand RNA viruses. However, this approach fails for NSVs because it does not consider the unique mechanism of their viral RNA synthesis. For NSVs, the viral RNA-dependent RNA polymerase (vRdRp) must gain access to the genome sequestered in the nucleocapsid. Our work suggests a paradigm shift that the interactions between the RNA genome and the nucleocapsid protein regulate the activity of vRdRp, which selects codon usage.
KEYWORDS: nucleocapsid, sequestered genome, viral polymerase
ABSTRACT
Negative-strand RNA viruses (NSVs) include some of the most pathogenic human viruses known. NSVs completely rely on the host cell for protein translation, but their codon usage bias is often different from that of the host. This discrepancy may have originated from the unique mechanism of NSV RNA synthesis in that the genomic RNA sequestered in the nucleocapsid serves as the template. The stability of the genomic RNA in the nucleocapsid appears to regulate its accessibility to the viral RNA polymerase, thus placing constraints on codon usage to balance viral RNA synthesis. By in situ analyses of vesicular stomatitis virus RNA synthesis, specific activities of viral RNA synthesis were correlated with the genomic RNA sequence. It was found that by simply altering the sequence and not the amino acid that it encoded, a significant reduction, up to an ∼750-fold reduction, in viral RNA transcripts occurred. Through subsequent sequence analysis and thermal shift assays, it was found that the purine/pyrimidine content modulates the overall stability of the polymerase complex, resulting in alteration of the activity of viral RNA synthesis. The codon usage is therefore constrained by the obligation of the NSV genome for viral RNA synthesis.
IMPORTANCE Negative-strand RNA viruses (NSVs) include the most pathogenic viruses known. New methods to monitor their evolutionary trends are urgently needed for the development of antivirals and vaccines. The protein translation machinery of the host cell is currently recognized as a main genomic regulator of RNA virus evolution, which works especially well for positive-strand RNA viruses. However, this approach fails for NSVs because it does not consider the unique mechanism of their viral RNA synthesis. For NSVs, the viral RNA-dependent RNA polymerase (vRdRp) must gain access to the genome sequestered in the nucleocapsid. Our work suggests a paradigm shift that the interactions between the RNA genome and the nucleocapsid protein regulate the activity of vRdRp, which selects codon usage.
INTRODUCTION
The negative-strand RNA virus (NSV) family includes some of the most well-known human pathogens, such as Ebola virus, influenza virus, and respiratory syncytial virus (1–4). The virally encoded RNA-dependent RNA polymerase (vRdRp) of NSV must recognize the nucleocapsid and unpack the sequestered genomic RNA from the nucleocapsid to use it as the template for viral RNA synthesis (5, 6). On the other hand, translation of viral proteins completely relies on the translation machinery of the host cell. This host reliance should direct the evolution of the viral codon usage bias (CUB) toward that of the host. This is certainly an effective way to optimize virus growth if translation is the major barrier for the virus, as observed in positive-strand RNA viruses (7).
However, there is no such coevolutionary virus-host relationship in the genome of NSVs. The genome of NSVs maintains a constant CUB, in spite of a high mutation rate by vRdRp (8). The calculated mutation rate for Paramyxoviridae is high enough to completely randomize the viral sequence 6 times per year, while the observed mutation rate is only about 6 nucleotide changes per year (8–11). Furthermore, it has also been noted that the viral CUB can be significantly different from the host CUB (8). While this is often attributed to the suppression of CpG codons for evading the host immune system (12), the correlation is not ubiquitously distributed throughout all NSVs. Other requirements for virus growth may place constraints on the evolution of NSVs. One factor could be the nucleotide content that is related to interactions of the genome with other proteins. Vesicular stomatitis virus (VSV) is a prototypic NSV that carries five viral genes: nucleocapsid (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large protein (L). A study showed that by altering the codon pair bias score in a portion of the L protein (polymerase) gene, which changes CUB, the virulence of VSV was attenuated in mice without changing the efficiency of viral protein translation (13).
Since NSV vRdRp needs to open the nucleocapsid to access the sequestered genomic RNA for transcription/replication, we propose that the stability of the genomic RNA in the nucleocapsid plays a regulatory role in the ability of the polymerase to successfully carry out viral RNA synthesis. As shown in Fig. 1A, the nucleocapsid is formed through intricate cross-molecular interactions between adjacent subunits, and the accessibility of the genomic RNA could vary with the local sequence (14, 15). During both transcription and replication, one “stable” sequence could cause the nucleocapsid template to tighten and reduce the processivity of vRdRp, whereas a different “unstable” sequence could cause the nucleocapsid template to loosen and increase the processivity of vRdRp or the rate of vRdRp dissociation from the nucleocapsid (Fig. 1B). To verify this mechanism, in situ activities of VSV vRdRp were correlated with altered codon usage using minigenome assays. The results show that the balance between purines and pyrimidines in the genome sequence plays an essential role in regulating the polymerase activity. The requirements for transcriptional/replicational control constrain codon usage of NSVs, which explains why NSVs maintain their independent genomic stability despite reliance on the host machinery for viral protein translation.
FIG 1.
(A) Ribbon representation of three nucleocapsid protein subunits constructed from the structure reported under PDB accession number 3PTX (45). Shown in red is the N-terminal arm, which interacts with adjacent subunits. Shown in blue and magenta are the N lobe and the C lobe, respectively. Shown in yellow is the C-terminal loop, which also interacts with adjacent subunits. Furthermore, the backbone of the RNA is shown as a tan line sandwiched between the N and C lobes. (B) Cartoon representation of a tight or loose interaction of the genomic RNA in the nucleocapsid. This would regulate the accessibility of the sequestered RNA to vRdRp and cause possible disassociation or stalling of vRdRp. (C) Stick representation of nine nucleotides (adenosine) encapsidated in the nucleocapsid. Shown in red are the first 4 nucleotides that have bases stacked with each other. Shown in blue are nucleotides 5, 7, and 8 that also have bases stacked. Nucleotide 6 is shown in yellow and does not directly interact with other nucleotides.
RESULTS
Effects of changing the codon usage bias on transcription/replication.
In order to investigate the effect that CUB has on transcription/replication, CUB of the VSV genome was compared to those of human and sand fly, two VSV hosts (Table 1). Furthermore, a similar analysis was extended to the genomes of Ebola virus and influenza virus. Compared to CUB in human, codons highlighted in green in Table 1 are more frequently used in VSV, and codons in red are less frequently used. It is obvious that CUB of VSV does not match that of human or sand fly. A similar CUB pattern was also observed in Ebola virus and influenza virus, indicating that this phenomenon could be ubiquitous throughout the NSV family.
TABLE 1.
Codon usage frequencies of the hostsa
Codon usage frequencies of the hosts were downloaded from the GenScript codon usage frequency table tool and calculated for the VSV genome using the Sequence Manipulation Suite from bioinformatics.org. Changes in the usage fraction were defined as significant if the difference was at least ±Δ0.14. Green indicates an increased fraction in NSV compared to human, whereas red indicates a decreased fraction. Changes of nucleotides in VSV versus human codons are marked. For Lutzomyia longipalpis, fly codons are marked yellow if their fraction differs from that of VSV or blue if it is the same as that of VSV (at least ±Δ0.04) but differs from human. A.A., amino acid; EBOV, Ebola virus: IFV, influenza virus.
A minigenome system was designed to quantitate in situ activities of vRdRp on VSV nucleocapsid (Fig. 2A). The N gene was selected to represent the VSV genome. There are long stretches of green and short interspersed patches of red throughout the N gene (Fig. 2B). This minigenome system is useful for comparable measurements of the vRdRp activities when the nucleotide content of the genome is altered. For comparisons with the wild-type (wt) minigenome (16), one synthetic genome was recoded such that codons with low usage frequency (in red) were changed to those with high usage frequency (named the high-usage genome), and another synthetic genome was recoded such that codons with high usage frequency (in green) were changed to those with low usage frequency (named the low-usage genome). During the NSV life cycle, three species of RNA are produced: mRNA (during transcription), cRNA (during replication), and vRNA (during replication). In the minigenome assays, each RNA species was quantitated by quantitative reverse transcription-PCR (RT-qPCR). It is important to note that the units on the y axis in Fig. 2C are log2 units for ease of presentation, and the fold reduction was calculated as described below. In Fig. 2C, top, the full-length RNA levels from genomes with different CUBs were compared. For mRNA, an ∼744-fold reduction was observed for the high-usage genome relative to the wt genome, and an ∼750-fold reduction was observed for the low-usage genome. For cRNA, reductions of ∼39-fold and ∼55-fold were observed, respectively, and for vRNA, reductions of ∼5.5-fold and ∼1.8-fold were observed, respectively, for the high- and low-usage genomes. These observations clearly indicate that the activity of vRdRp is severely compromised during transcription and replication by just switching CUBs.
FIG 2.
(A) Vector for the minigenome based on the N gene of VSV. Under the control of the T7 promoter, the vector expresses the vRNA copy of the N gene, flanked by the leader and trailer sequences. (B) Highlighted sequence of the N gene showing the location of the high-frequency codons, in green, or the low-frequency codons, in red. Furthermore, highlighted in cyan is the placement of 3 stop codons to prevent translation of the mRNA transcripts. Also shown, in magenta, is the PCR tag utilized in qPCR to identify mRNA transcripts transcribed only from the minigenome. (C, top) Relative fold decreases in RNA levels from all the minigenomes quantitated by in situ vRdRp activity assays. N stands for the wt genome, H stands for the high-usage genome, and L stands for the low-usage genome. Notice the log2 axis for ease of viewing the fold differences on similar scales. (Bottom) Fold increases in levels of shortened RNAs using primers that correspond to the beginning of each viral RNA moiety, as ratios to the full-length RNA moieties. Each experiment was carried out in triplicate, and the error bars represent the standard deviations between the experiments. (D) Construction of the chimeric minigenomes. Each minigenome was constructed based on the golden ratio, with the darker color representing the wt genome and gray representing the high- or low-usage genome. Each chimeric genome is shown in panel B as NH1 or NL1, etc. All sequences are presented in the supplemental material.
To further dissect which specific step in viral RNA synthesis is more affected by a change in CUB, eight chimeric minigenomes between the wt genome and either the high- or low-usage genome were constructed (Fig. 2D) based on the golden ratio. The golden ratio can be defined as the ubiquitously found ratio of 1.618 and is defined as the ratio between two Fibonacci numbers (17). This ratio has been found throughout nature, including the arrangement of leaves on the stem of a plant and even the spirals of a sunflower (17). This ratio has also been found in the human genome within the frequencies of different nucleotides (18). It is for this reason, when designing the chimeric minigenomes, that the golden ratio was chosen as the natural ratio to split the genome to investigate the sequence properties. Among these chimeras, N1, N2, and N3 show statistically significant reductions in mRNA levels, while N4 shows a non-statistically significant increase. In the chimeric genome, N1, N2, and N3 all have an altered CUB either in the transcription or replication initiation region (3′-end regions) or near the termination region (5′-end region) of the genomic sequence, whereas N4 has the wt CUB in both. Defects in elongation by vRdRp could also result in reductions of mRNA levels, as described below, but it may not be the main factor because no significant mRNA reduction was observed with the N4 chimeric genome. This would indicate that for transcription of mRNA, the CUB is more stringent, especially for initiation and termination by vRdRp. This is consistent with the requirements for transcription by VSV vRdRp because viral transcription must be initiated and terminated at the correct position in order to balance the levels of all five mRNAs in the polycistronic genome. For cRNA and vRNA, on the other hand, it seems that elongation is the most affected activity of vRdRp since the initiation and termination of replication are more dependent on the 3′ and 5′ noncoding sequences (named the leader and trailer sequences, respectively), neither of which was changed in the chimeric genome. The largest reduction in genomic RNA levels was observed when the genomes with a large portion of altered CUB (N2 and N4) were used as the template. After initiation of replication, the processivity of vRdRp is dependent on CUB when it copies the entire genome.
It is possible that the reduction in levels of full-length RNAs was due to premature termination. Another set of primers was designed to quantify RNAs from any possible early termination during transcription and replication. The RNAs quantitated by these primers correspond to total RNAs of the shortened RNAs, in addition to the full-length RNAs. As shown in Fig. 2C, levels of the total mRNA transcripts were increased by ∼19-fold and ∼35-fold, respectively, for the high- and low-usage genomes. Early termination of viral RNA transcription was clearly increased by the altered sequence content. It is also interesting that the level of total mRNA was twice that of the full-length mRNA when the wt genome was used, indicating the common presence of early termination in transcription. An increase in shortened mRNA transcripts was observed across all of the chimeric minigenomes. This effect is exacerbated across the high-usage chimeras, especially for N2, N3, and N4. On the other hand, an increase in levels of shortened cRNA and vRNA was observed for the low-usage chimeras, but there was less of an increase in shortened mRNA transcripts than in those of the high-usage chimeras.
Role of the purine/pyrimidine ratio in nucleocapsid stability.
In the nucleocapsid, the bases of the sequestered RNA stack in a consistent manner in each subunit (Fig. 1C) (14). The first four bases stack with each other, the fifth base stacks with bases 7 and 8, and the sixth base flips out of the stacked bases. The strength of π-stacking interactions is in the order purine-purine>purine-pyrimidine>pyrimidine-pyrimidine (19). We therefore hypothesized that sequences more rich in stacked purines would yield a more stable nucleocapsid, which is consistent with the structures of nucleocapsid-like particles (NLPs) that encapsidate poly(rA) (20, 21).
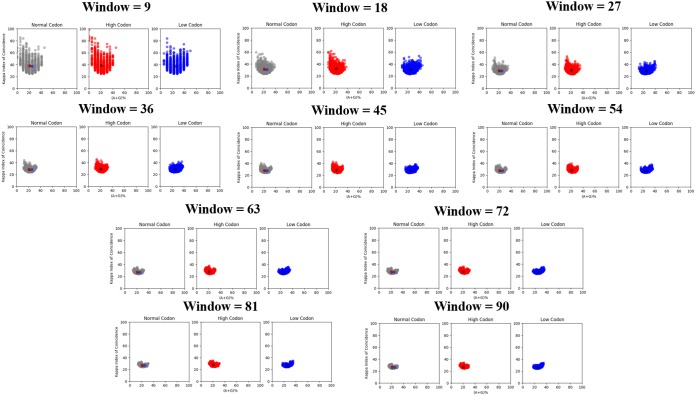
The sequence analysis technique kappa index of coincidence (KIC) was often used to identify different classes of promoters in eukaryotes (22). KIC measures the probability of finding a repeating pattern in a nucleotide sequence. A high KIC indicates a structured sequence pattern with a high level of nucleotide repeats. When plotted against percent A+G content (A+G%), this provides valuable insight into regions that have a highly clustered purine/pyrimidine content. In this case, the purine content (A+G%) versus KIC was plotted for the wt and high- and low-usage genomes. Since KIC is sensitive to simple or short tandem repeats, and to window sizes, the calculation was carried out using various window sizes (Fig. 3) (22). Analysis of the template strand for mRNA synthesis revealed that there is a slightly lower degree of purine content for the high-usage genome than for the wt genome when the window size was 9 or 18 nucleotides. The more significant indication was that the lower A+G% in the high-usage genome was associated with a higher KIC, shown by a large tail in the top left portion of the graph. The high KIC suggests that pyrimidines were more clustered in the high-usage genome, leading to more pyrimidine stacking (23). As the window size increases, the left portion of the graph was more densely packed, further indicating the presence of larger stretches of pyrimidines. At the same time, the center of mass in each plot with smaller window sizes is consistently associated with higher KIC scores for the high-usage genome, consistent with higher pyrimidine clustering at the local sequence level. When larger window sizes were used, the center of mass shifted slightly more to the right, almost overlapping that of the wt genome (on the x axis), indicating that the clustered pyrimidine contents of the wt and high-usage genomes match more closely at a global level. This confirms that local purine/pyrimidine clustering may play an important role in regulating vRdRp activities in transcription. For the low-usage genome, on the other hand, there is a significant tail to the right of the graph and a right shift of the center of mass throughout all window sizes. This is indicative of higher purine content and clustering, while the overall clustering of purines and pyrimidines matches that of the wt genome.
FIG 3.
Calculated A+G% versus KIC. Each panel is a different window size ranging from 9 to 90 nucleotides as multiples of 9 due to the number of nucleotides in each N subunit. The wt genome is plotted in gray, the high-usage genome is plotted in red, and the low-usage genome is plotted in blue. The centers of mass are plotted in their respective color for each panel, and all three centers of mass are plotted on the wt plot. The KIC is a measurement of the probability of a repeating sequence within a set window. It is plotted against A+G% here for the purposes of investigating the purine/pyrimidine distribution throughout the wt, high-codon-usage, and low-codon-usage sequences. Altering these base-stacking patterns changes the distribution of the plot and corresponds to the decrease in vRdRp activity.
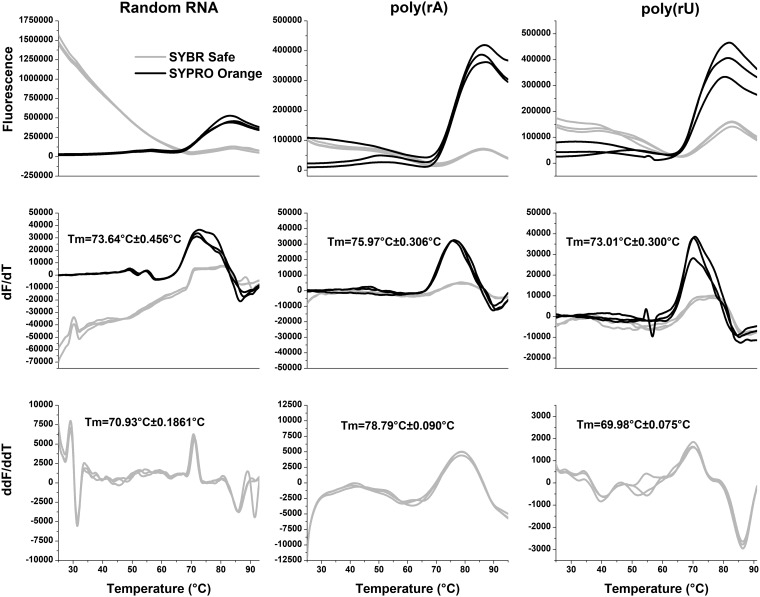
To verify that the stability of the nucleocapsid is related to the purine/pyrimidine content, thermal shift assays (TSAs) were carried out with recombinantly purified NLPs (20, 24, 25). A selected piece of RNA could be reconstituted in the NLP by removal of the original RNA with RNase A and subsequent incubation with the selected RNA (20). As previously shown (20), TSA can be effectively used to measure the stability of the NLP. Utilizing two different fluorescent dyes, SYBR Safe and SYPRO orange, to target the RNA and protein, respectively, melting curves of NLPs reconstituted with poly(rA) and poly(rU) sequences were determined (Fig. 4). SYBR Safe fluoresces when it intercalates DNA/RNA. The RNA in the NLP is highly structured and has significant base-stacking interactions, which allows for a clear observance of SYBR Safe fluorescence when incubated with the NLP. As the temperature increases and the RNA becomes unstructured, the fluorescence decreases. The temperature at which the rate of fluorescence is changing the fastest, which we call Tfree and is shown in the second derivative graph, indicates the point at which the RNA has been released from the NLP. SYPRO orange has been used to detect protein melting and has become an invaluable tool to screen compounds against protein targets. For both the melting temperature (Tm) and Tfree, which are measured by SYPRO orange and SYBR Safe, respectively, an increase in the melting temperature in comparison with controls indicates a stabilization of the protein or encapsidated RNA, correspondingly. The results showed that the incorporation of poly(rA) in the NLP increased the stability by 2.33°C compared to the recombinantly purified NLP with a random RNA sequence (20) and by 2.96°C compared to the NLP with poly(rU). Furthermore, incorporation of poly(rA) in the NLP delayed the thermo-release of RNA by 7.86°C compared to the recombinantly purified NLP with a random RNA sequence and by 8.81°C compared to the NLP with poly(rU) (24). Both of these experiments depict a large stabilization of the NLP by poly(rA). This is consistent with data from a previous study in which a polyamide (UMSL1011) that stabilizes the RNA in NLPs delayed RNA thermo-release by 2.04°C (24). These experimental results confirmed that nucleocapsid stability is dependent on the purine content of sequestered RNA, which may change the accessibility of the genomic RNA in the nucleocapsid to vRdRp to modulate its transcription/replication activities.
FIG 4.
Thermal shift assays. A thermal shift assay was carried out on the randomly incorporated RNA in recombinantly expressed NLP, poly(rA), and poly(rU) in reconstituted NLPs. SYBR Safe, which monitors RNA thermo-release from the NLP, is shown in gray, while SYPRO orange, which monitors protein denaturation, is shown in black. Each trace is representative of data from a separate experiment. The top panels indicate the raw melting data. The middle panels are the first derivative graphs with the corresponding average Tm (from the SYPRO orange trace) and standard deviation calculated from the peak of each trace. The bottom panels show the second derivative of the SYBR Safe trace and the average Tfree and standard deviation calculated from the peak of each trace, which is representative of the thermal release of RNA.
DISCUSSION
The high mutation rate of vRdRp has been related to virus evolvability and replication (26). In positive-strand RNA viruses, the faster genome replication and optimal codon usage select viruses with good fitness (27–29). Since RNA viruses rely on the host cell for protein translation, the CUB of positive-strand RNA viruses is similar to that of the host cell since its genomic RNA is also used as the mRNA for protein translation (30, 31). In NSVs, however, their CUB does not match that of the host cell despite their reliance on host protein translation. This phenomenon has previously been attributed to limiting CpG to evade the host’s immune system and to the concept of codon constellation (8, 12, 31–33). More importantly, analyses of codon usages revealed that CUB of NSVs did not result from transitional selection (34–36). To identify other factors of CUB selection in NSVs, viral proteins that have close interactions with the coding regions of the viral genome are considered. One of the unique mechanisms that control viral genome replication of NSVs is that the template used by vRdRp in viral RNA synthesis is the nucleocapsid. The entire viral RNA genome is completely sequestered by the N protein (14, 37), and its access to vRdRp may regulate viral RNA synthesis in NSVs (20, 38).We hypothesize that access to the genomic RNA sequestered in the nucleocapsid constrains codon usage by NSVs, which is a determinantal factor in CUB selection during NSV evolution.
These constraints were examined in this study by evaluating in situ activities of VSV vRdRp using a minigenome assay. Our data showed that vRdRp became highly dysregulated with large decreases in transcription and replication when the codon usage in the VSV minigenome was deoptimized (Fig. 2C). Changes in viral RNA synthesis were not related to protein translation since all viral proteins were produced by other vectors. This clearly confirmed that the nucleotide sequence of the viral genome in the nucleocapsid plays an essential role in regulating viral RNA synthesis by vRdRp. In an effort to identify which steps are more affected by changes of codon usage, eight chimeric minigenomes were also created. The results showed that the regions corresponding to initiation and termination of gene transcription had more-dramatic detrimental effects on vRdRp activities when CUB was deoptimized (Fig. 2C and D). In the context of the virus genome, this is consistent with the finding that vRdRp is required to accurately initiate and terminate mRNA transcription in order to balance the transcript products from the polycistronic genome (39). For replication, elongation of cRNA and vRNA was most limited by CUB changes because the ends of the genome, which regulate the initiation and termination of genome replication by vRdRp, were not changed.
The consequence of CUB changes was analyzed for nucleotide contents and clustering. Since the crystal structures of VSV NLPs suggest that purines have more-stable base stacking (14, 20), an increase or decrease in A+G% and their clustering may alter the access of the sequestered genomic RNA to vRdRp. A+G% was calculated and compared with KIC for high- and low-usage genomes (Fig. 3). Since KIC is a measure of randomness, a deviation from the wt sequence could potentially deregulate viral RNA synthesis. The results showed that pyrimidine clustering was increased in the high-usage genome and that purine clustering was increased in the low-usage genome. An abnormal stability of the nucleocapsid could not maintain the precise activities of vRdRp. To further corroborate the correlation of A+G% and their clustering with levels of viral RNA synthesis, thermal shift assays were used to measure the stability of the nucleocapsid when different RNA sequences were encapsidated. The results showed that NLPs were stabilized by the presence of purine sequences and destabilized by the presence of pyrimidine sequences in comparison with the recombinantly prepared NLP that encapsidates random RNAs. These data directly support that the nucleotide content and base clustering of the NSV genome, as a consequence of CUB selection, regulate the vRdRp accessibility of the genomic RNA sequestered in the nucleocapsid. The regional accessibility of the genomic RNA may be increased by reducing the A+G content or clustering, but early termination could also be increased. Evolution of NSVs results in their own CUB that balances mRNA transcripts and efficient genome replication.
Insights on the evolutionary principles of CUB in NSVs have great implications in managing outbreaks of highly pathogenic viruses, such as Ebola virus, influenza virus, and rabies virus. Analyses of codon usage in these viruses (12, 36, 40, 41) found that these NSVs constantly mutate in different hosts, but their CUB was not coincident with that of the host. Our conclusion that optimal viral RNA synthesis regulated by the nucleocapsid is an additional determinant factor in CUB selection of NSVs will lead to an improved algorithm for mapping the evolutionary trajectory of these pathogens. This will greatly help in the design of detection methods, vaccines, and antiviral treatments for emerging NSVs.
MATERIALS AND METHODS
Minigenome plasmid.
The pN, pP, and pL plasmids were provided by Asit K. Pattnaik at the University of Nebraska—Lincoln. The template for the minigenome was constructed as previously described (42). To insert the N gene into the minigenome template, the Luc gene was removed from the minigenome using BamHI. The N gene was amplified from pN using primers shown in Table 2, with BamHI cleavage sites on each side. This fragment was then cloned into the minigenome vector. In order to create a PCR tag for the mRNA transcribed from the minigenome, mutations corresponding to primers shown in Table 2 were introduced using the Q5 site-directed mutagenesis kit (New England Biolabs) according to the manufacturer’s protocol. Furthermore, 3 stop codons were added after the start codon to prevent translation utilizing primers shown in Table 2, using the QuikChange II mutagenesis kit (Agilent Technologies) according to the manufacturer’s protocol. High-codon-usage, low-codon-usage, and chimeric minigenomes were synthesized by GenScript (Piscataway, NJ) and cloned into the minigenome vector using the BamHI restriction enzyme sites.
TABLE 2.
Primers for minigenome construction
Minigenome assay.
BSR-T7 cells (a gift from Biao He, University of Georgia) were maintained in Dulbecco modified Eagle medium (DMEM) containing 5% fetal bovine serum (FBS) with 100 U of penicillin and 20 U of streptomycin (43). Plasmids were then transfected into the cells in 60-mm plates after cells reached approximately 95% confluence. Briefly, cells were washed twice with Opti-MEM serum-free medium (Invitrogen) and then transfected with 2 μg of pN-minigenome, 1.5 μg of pN, 1 μg of pP, and 0.5 μg of pL using Lipofectamine 2000 (Invitrogen) (42). The cells were then incubated for 5 h at 37°C, and fresh DMEM with 5% FBS was then added. At 48 h posttransfection (hpt), RNA was extracted from the cells using TRIzol reagent (Ambion by Life Technologies) according to the manufacturer’s protocol. Immediately after RNA isolation, 1 μg of RNA was used for RT-PCR using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The primers for RT-PCR were specific for mRNA, cRNA, and vRNA, as outlined in Table 3. Quantitative PCR (qPCR) was performed on a QuantStudio 3 real-time PCR system using the primers in Table 4 and PowerUp SYBR green master mix according to the manufacturer’s protocol (Thermo Fisher Scientific) (see Fig. S1 in the supplemental material for an outline of primer recognition). The data were analyzed by the 2−ΔΔCT method as described previously (16). Furthermore, mock, template-free, no-RT-PCR, and template-only controls did not yield a CT (threshold cycle) value (data not included) after 40 cycles, indicating specific amplification of the selected genes (16). In addition, melting analysis revealed a single moiety for mRNA, cRNA, and vRNA for every minigenome vector.
TABLE 3.
Primers used for RT-PCR to make cDNA transcripts from selected RNAs
TABLE 4.
Primers used during qPCR for the different viral RNAs produced during the VSV life cycle
Sequence analysis.
The kappa index of coincidence was calculated as described previously, with the modification of plotting the x axis as A+G% (22). All calculations were performed using Python (Python Software Foundation, version 3.5 [https://www.python.org/]), and the script is available at http://github.com/rgumpper/Biochemical_Tools. Data plots were drawn utilizing the open-source plotting library for Python, Matplotlib (44). Sliding windows of 9, 18, 27, 36, 45, 54, 63, 72, 81, and 90 nucleotides were used for KIC calculations to examine the difference between local and global genome structures. A window size divisible by 9 was used as the nucleocapsid encapsidates 9 nucleotides per N subunit.
Expression/purification of NLPs and thermal shift assay.
The NLPs were expressed and purified as previously described (25). Briefly, the plasmid containing pET N/P was transformed into BL21(DE3) cells for protein expression. Starter cultures were allowed to grow at 37°C in LB broth overnight in the presence of ampicillin. The starter culture was then used to inoculate 1-liter cultures, again in the presence of ampicillin, and allowed to grow while shaking at 37°C until the A600 reached an optical density (OD) of 0.6. Protein expression was then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the culture was allowed to grow for an additional 16 h at 24°C. The cells were then harvested; resuspended in a solution containing 20 mM Tris (pH 8.0), 500 mM NaCl, 5 mM imidazole, and 5 mM β-mercaptoethanol; and sonicated. After an initial Ni-nitrilotriacetic acid (NTA) purification step (according to Novagen’s protocol), size exclusion was carried out with a HiLoad 16/100 Superdex 200 prep-grade column equilibrated with 50 mM Tris (pH 7.5) and 300 mM NaCl. This produced NLPs which contain randomly encapsidated RNAs. The NLP was then reconstituted with either poly(rA) or poly(rU) purchased from Midland (Midland, TX) as described previously (20). Briefly RNase A (1 mg/ml) was incubated with purified NLPs overnight at 42°C to remove the encapsidated RNA from the recombinant protein-RNA complex. Excess RNA and RNase A were removed via size exclusion. Before reconstitution with poly(rA) and poly(rU), an RNase inhibitor (1 U/μl) (Qiagen) was added to the empty NLP and incubated at room temperature for 30 min. The respective RNA was then added in an excess of a 1:5 molar ratio (NLP to RNA), and an oscillation of temperature was then carried out between 42°C and room temperature in 15-min intervals a total of four times. Excess RNA was removed by further size exclusion purification, and the RNA composition was checked by the 260/280-nm ratio on a UV-visible (UV-vis) spectrophotometer. NLPs containing RNA from recombinant purification, poly(rA), or poly(rU) were then subjected to TSA as previously described (24).
Data availability.
All data are available in the main text or the supplemental material, and the Python 3.5 code used for the KIC sequence analysis is available on a publicly available repository (http://www.github.com/rgumpper).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Apurv Patel for assistance in cloning the various chimeric minigenomes.
This work was funded in part by an NIH grant (R01 AI106307).
M.L., R.H.G., and W.L. were responsible for designing and performing experiments and analyzing the data. M.L. and R.H.G. wrote and prepared the manuscript.
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01775-18.
REFERENCES
- 1.Bont L, Checchia PA, Fauroux B, Figueras-Aloy J, Manzoni P, Paes B, Simões EAF, Carbonell-Estrany X. 2016. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in Western countries. Infect Dis Ther 5:271–298. doi: 10.1007/s40121-016-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appolinario CM, Jackson AC. 2015. Antiviral therapy for human rabies. Antivir Ther 20:1–10. doi: 10.3851/IMP2851. [DOI] [PubMed] [Google Scholar]
- 3.Balmith M, Faya M, Soliman ME. 2017. Ebola virus: a gap in drug design and discovery—experimental and computational perspective. Chem Biol Drug Des 89:297–308. doi: 10.1111/cbdd.12870. [DOI] [PubMed] [Google Scholar]
- 4.Wu W, Liu S. 2017. The drug targets and antiviral molecules for treatment of Ebola virus infection. Curr Top Med Chem 17:361–370. [DOI] [PubMed] [Google Scholar]
- 5.Emerson SU, Wagner RR. 1972. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol 10:297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emerson SU, Yu Y. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 15:1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rima BK. 2015. Nucleotide sequence conservation in paramyxoviruses; the concept of codon constellation. J Gen Virol 96:939–955. doi: 10.1099/vir.0.070789-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Dai W, Wang Y, Lu C, Fan H. 2013. Analysis of synonymous codon usage patterns in torque teno sus virus 1 (TTSuV1). Arch Virol 158:145–154. doi: 10.1007/s00705-012-1480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rima BK, Earle JA, Baczko K, ter Meulen V, Liebert UG, Carstens C, Carabana J, Caballero M, Celma ML, Fernandez-Munoz R. 1997. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J Gen Virol 78:97–106. doi: 10.1099/0022-1317-78-1-97. [DOI] [PubMed] [Google Scholar]
- 12.Deka H, Chakraborty S. 2016. Insights into the usage of nucleobase triplets and codon context pattern in five influenza A virus subtypes. J Microbiol Biotechnol 26:1972–1982. doi: 10.4014/jmb.1605.05016. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Yang C, Tekes G, Mueller S, Paul A, Whelan SP, Wimmer E. 2015. Recoding of the vesicular stomatitis virus L gene by computer-aided design provides a live, attenuated vaccine candidate. mBio 6:e00237-15. doi: 10.1128/mBio.00237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Green TJ, Tsao J, Qiu S, Luo M. 2008. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J Virol 82:674–682. doi: 10.1128/JVI.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Sumpter DJT. 2018. Is the golden ratio a universal constant for self-replication? PLoS One 13:e0200601. doi: 10.1371/journal.pone.0200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagishi ME, Shimabukuro AI. 2008. Nucleotide frequencies in human genome and Fibonacci numbers. Bull Math Biol 70:643–653. doi: 10.1007/s11538-007-9261-6. [DOI] [PubMed] [Google Scholar]
- 19.Friedman RA, Honig B. 1995. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J 69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green TJ, Rowse M, Tsao J, Kang J, Ge P, Zhou ZH, Luo M. 2011. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol 85:2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milles S, Jensen MR, Communie G, Maurin D, Schoehn G, Ruigrok RW, Blackledge M. 2016. Self-assembly of measles virus nucleocapsid-like particles: kinetics and RNA sequence dependence. Angew Chem Int Ed Engl 55:9356–9360. doi: 10.1002/anie.201602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagniuc P, Ionescu-Tirgoviste C. 2012. Eukaryotic genomes may exhibit up to 10 generic classes of gene promoters. BMC Genomics 13:512. doi: 10.1186/1471-2164-13-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagniuc P, Cristea PD, Tuduce R, Ionescu-Tirgoviste C, Gavrila L. 2012. DNA patterns and evolutionary signatures obtained through kappa index of coincidence. Rev Roum Sci Tech Electrotech Energ 57:100–109. [Google Scholar]
- 24.Gumpper RH, Li W, Castaneda CH, Scuderi MJ, Bashkin JK, Luo M. 2018. A polyamide inhibits replication of vesicular stomatitis virus by targeting RNA in the nucleocapsid. J Virol 92:e00146-18. doi: 10.1128/JVI.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green TJ, Macpherson S, Qiu S, Lebowitz J, Wertz GW, Luo M. 2000. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J Virol 74:9515–9524. doi: 10.1128/JVI.74.20.9515-9524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duffy S. 2018. Why are RNA virus mutation rates so damn high? PLoS Biol 16:e3000003. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzsimmons WJ, Woods RJ, McCrone JT, Woodman A, Arnold JJ, Yennawar M, Evans R, Cameron CE, Lauring AS. 2018. A speed-fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol 16:e2006459. doi: 10.1371/journal.pbio.2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butt AM, Nasrullah I, Tong Y. 2014. Genome-wide analysis of codon usage and influencing factors in chikungunya viruses. PLoS One 9:e90905. doi: 10.1371/journal.pone.0090905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauring AS, Acevedo A, Cooper SB, Andino R. 2012. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma XX, Feng YP, Liu JL, Zhao YQ, Chen L, Guo PH, Guo JZ, Ma ZR. 2014. The characteristics of synonymous codon usage in the initial and terminal translation regions of encephalomyocarditis virus. Acta Virol 58:86–91. doi: 10.4149/av_2014_01_86. [DOI] [PubMed] [Google Scholar]
- 31.Ma MR, Hui L, Wang ML, Tang Y, Chang YW, Jia QH, Wang XH, Yan W, Ha XQ. 2014. Overall codon usage pattern of enterovirus 71. Genet Mol Res 13:336–343. doi: 10.4238/2014.January.21.1. [DOI] [PubMed] [Google Scholar]
- 32.Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. 2014. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng X, Virk N, Chen W, Ji S, Ji S, Sun Y, Wu X. 2013. CpG usage in RNA viruses: data and hypotheses. PLoS One 8:e74109. doi: 10.1371/journal.pone.0074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar CS, Kumar S. 2017. Synonymous codon usage of genes in polymerase complex of Newcastle disease virus. J Basic Microbiol 57:481–503. doi: 10.1002/jobm.201600740. [DOI] [PubMed] [Google Scholar]
- 35.Kumar N, Bera BC, Greenbaum BD, Bhatia S, Sood R, Selvaraj P, Anand T, Tripathi BN, Virmani N. 2016. Revelation of influencing factors in overall codon usage bias of equine influenza viruses. PLoS One 11:e0154376. doi: 10.1371/journal.pone.0154376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristina J, Moreno P, Moratorio G, Musto H. 2015. Genome-wide analysis of codon usage bias in ebolavirus. Virus Res 196:87–93. doi: 10.1016/j.virusres.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 38.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball LA, Pringle CR, Flanagan B, Perepelitsa VP, Wertz GW. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J Virol 73:4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gun L, Haixian P, Yumiao R, Han T, Jingqi L, Liguang Z. 2018. Codon usage characteristics of PB2 gene in influenza A H7N9 virus from different host species. Infect Genet Evol 65:430–435. doi: 10.1016/j.meegid.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Cai Y, Zhai X, Liu J, Zhao W, Ji S, Su S, Zhou J. 2018. Comprehensive analysis of codon usage on rabies virus and other lyssaviruses. Int J Mol Sci 19:E2397. doi: 10.3390/ijms19082397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nayak D, Panda D, Das SC, Luo M, Pattnaik AK. 2009. Single-amino-acid alterations in a highly conserved central region of vesicular stomatitis virus N protein differentially affect the viral nucleocapsid template functions. J Virol 83:5525–5534. doi: 10.1128/JVI.02289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das SC, Nayak D, Zhou Y, Pattnaik AK. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J Virol 80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 45.Schrodinger LLC. 2015. The PyMOL Molecular Graphics System, version 1.8. Schrodinger LLC, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplemental material, and the Python 3.5 code used for the KIC sequence analysis is available on a publicly available repository (http://www.github.com/rgumpper).