Only noncp pestivirus strains are capable of establishing life-long persistent infections to generate the virus reservoir in the field. The molecular basis for this biotype is only partially understood and only investigated in depth for BVDV-1 strains. Temporal control of viral RNA replication correlates with the noncp biotype and is mediated by limiting amounts of cellular DNAJC14 that activate the viral NS2 protease to catalyze the release of the essential replicase component NS3. Here, we demonstrate that several species of noncp pestiviruses depend on DNAJC14 for their RNA replication. Moreover, all cp pestiviruses, in sharp contrast to their noncp counterparts, replicate independently of DNAJC14. The generation of a cp BVDV in the persistently infected animal is causative for onset of mucosal disease. Therefore, the observed strict biotype-specific difference in DNAJC14 dependency should be further examined for its role in cell type/tissue tropism and the pathogenesis of this lethal disease.
KEYWORDS: DNAJC14, NS2-3 cleavage, chaperone, host factor, pestivirus, polyprotein processing, positive-strand RNA virus, viral protease
ABSTRACT
Pestiviruses like bovine viral diarrhea virus (BVDV) are a threat to livestock. For pestiviruses, cytopathogenic (cp) and noncytopathogenic (noncp) strains are distinguished in cell culture. The noncp biotype of BVDV is capable of establishing persistent infections, which is a major problem in disease control. The noncp biotype rests on temporal control of viral RNA replication, mediated by regulated cleavage of nonstructural protein 2-3 (NS2-3). This cleavage is catalyzed by the autoprotease in NS2, the activity of which depends on its cellular cofactor, DNAJC14. Since this chaperone is available in small amounts and binds tightly to NS2, NS2-3 translated later in infection is no longer cleaved. As NS3 is an essential constituent of the viral replicase, this shift in polyprotein processing correlates with downregulation of RNA replication. In contrast, cp BVDV strains arising mostly by RNA recombination show highly variable genome structures and display unrestricted NS3 release. The functional importance of DNAJC14 for noncp pestiviruses has been established so far only for BVDV-1. It was therefore enigmatic whether replication of other noncp pestiviruses is also DNAJC14 dependent. By generating bovine and porcine DNAJC14 knockout cells, we could show that (i) replication of 6 distinct noncp pestivirus species (A to D, F, and G) depends on DNAJC14, (ii) the pestiviral replicase NS3-5B can assemble into functional complexes in the absence of DNAJC14, and (iii) all cp pestiviruses replicate their RNA and generate infectious progeny independent of host DNAJC14. Together, these findings confirm DNAJC14 as a pivotal cellular cofactor for the replication and maintenance of the noncp biotype of pestiviruses.
IMPORTANCE Only noncp pestivirus strains are capable of establishing life-long persistent infections to generate the virus reservoir in the field. The molecular basis for this biotype is only partially understood and only investigated in depth for BVDV-1 strains. Temporal control of viral RNA replication correlates with the noncp biotype and is mediated by limiting amounts of cellular DNAJC14 that activate the viral NS2 protease to catalyze the release of the essential replicase component NS3. Here, we demonstrate that several species of noncp pestiviruses depend on DNAJC14 for their RNA replication. Moreover, all cp pestiviruses, in sharp contrast to their noncp counterparts, replicate independently of DNAJC14. The generation of a cp BVDV in the persistently infected animal is causative for onset of mucosal disease. Therefore, the observed strict biotype-specific difference in DNAJC14 dependency should be further examined for its role in cell type/tissue tropism and the pathogenesis of this lethal disease.
INTRODUCTION
Bovine viral diarrhea virus (BVDV) is a member of the genus Pestivirus in the family Flaviviridae (1). BVDV and other pestiviruses, such as classical swine fever virus (CSFV), represent important pathogens causing significant economic damage in livestock industries worldwide (2). The single-stranded RNA genome is approximately 12.3 kb long, has positive polarity, and comprises a single long open reading frame (ORF) which is flanked by 5′ and 3′ untranslated regions (UTRs) (3, 4). Translation of the pestiviral RNA genome results in the production of a polyprotein encompassing in the N-terminal third Npro along with all structural proteins and in the remaining C-terminal part the nonstructural (NS) proteins. The first protein of the ORF, Npro, is an autoprotease (5), which releases itself from the remainder of the polyprotein and thereby generates the N terminus of the core protein (C). The core protein, in concert with the envelope glycoproteins Erns, E1, and E2, together with the viral RNA represent the major components of the virion (4, 6–8). Recent morphological and biochemical data indicated that BVDV particles show a low envelope glycoprotein content of E1 and E2, with both envelope proteins being apparently less abundant than Erns (6). Cellular proteases mediate all additional cleavages required to generate mature C, Erns, E1, and E2, as well as to release the hydrophobic protein p7 (9). Mature p7 is required for the generation of infectious viral progeny and has been suggested to function as a viroporin (10, 11). NS2 is an autoprotease that is responsible for NS2-3 cleavage in cis to generate NS2 and the NS3 N terminus (12–14), an activity for which NS2 of noncp pestiviruses requires the activating cellular chaperone DNAJC14 (also designated Jiv) (15, 16). Furthermore, NS2 has, typically as uncleaved NS2-3, an essential, but not well-characterized, function in virion morphogenesis for which the NS2 cysteine protease activity is not required (16–18). However, it was recently demonstrated that BVDV strains could be adapted to an alternative NS2-3-independent packaging pathway involving free NS2 and NS3 (19–21). NS3 is a multifunctional protein with an N-terminal chymotrypsin-like serine protease domain (22, 23) and a C-terminal helicase and NTPase domain, which is essential for viral RNA replication (24–26). Cleavage between NS3 and NS4A, as well as all downstream cleavages in the pestiviral polyprotein, is catalyzed by this NS3 serine protease domain, which is assisted in this function by its cofactor, NS4A (27–29). In complex with NS3, NS4A also holds critical functions in RNA replication and virion morphogenesis (16, 21, 30). NS4B is a hydrophobic membrane protein and an essential component of the pestiviral replicase (25, 31, 32). NS5A is a phosphorylated protein of unknown but essential function within the viral replication complex that can be complemented in trans and has been reported to modulate NS5B RNA-dependent RNA polymerase (RdRp) activity (32–34). NS5B represents the viral RdRp (35–39).
A special feature of pestiviruses is that they exist as two different biotypes, noncytopathogenic (noncp) and cytopathogenic (cp) viruses, in cell culture (reviewed in reference 2). In general, cp BVDV strains evolve from noncp ancestor viruses by RNA recombination in the course of a persistent infection that is developing into lethal mucosal disease (40–42). While noncp viruses replicate in their host cells without detectable damage or reduced cell viability, the replication of the cp variants induces apoptosis leading to death of the infected cells, which can be observed as a cytopathic effect (CPE) (43–45). The observation that most isolates acquired from the field are noncp pestiviruses highlights the fact that only the noncp biotype can establish persistence upon fetal infection (46).
This ability of noncp pestiviruses to establish long-term persistent infections rests on their ability to strictly control RNA replication via the complex mechanism of regulating the NS2 autoprotease activity and, thus, NS2-3 cleavage by the limited activating host factor DNAJC14 (see below) (14, 15). In contrast, the efficient generation of NS3 is a hallmark of cp virus strains and their cytopathogenicity (47–49). Different cp pestivirus strains show highly diverse genome organizations and use various strategies for the generation of NS3 (2). The unifying feature of all of them is an upregulation of NS3 release and a putative independence of NS3 expression from the endogenous DNAJC14 level.
In numerous cp BVDV and cp BDV strains, a cell-derived RNA insert, previously termed cINS or Jiv (for J-domain protein interacting with viral protein), has been identified (49–52). In the BVDV strain NADL, a Jiv insertion of 90 amino acids (aa) (Jiv90) is located within the NS2 protein (49). In the context of an infectious cDNA clone of BVDV NADL, it was demonstrated that the Jiv90 fragment of DNAJC14 is essential for efficient NS2-3 cleavage and is the reason for the cp phenotype of the virus (49).
Further experiments extended those observations and showed that DNAJC14 is essential for the temporal regulation of NS2-3 cleavage in noncp BVDV-infected cells by stably binding to NS2. Its intracellular level could be correlated with NS2-3 cleavage efficiency as well as with viral cytopathogenicity (15, 50). By investigating BVDV CP7 and NCP7 infections in cells with experimentally altered DNAJC14 levels, the following model emerged: DNAJC14 stably binds to NS2 in the context of newly translated viral polyproteins, thereby activating the autoprotease in NS2 to cleave NS2-3 (53). According to the current model, DNAJC14 remains bound to NS2 after NS2-3 cleavage and cannot be recycled. Since endogenous DNAJC14 is only present in small amounts in bovine or swine culture cells, ongoing translation of the viral polyprotein leads to a situation where newly translated NS2-3 proteins will not have access to functionally available DNAJC14 at later time points of infection. Thus, NS2-3 cleavage will not occur efficiently, resulting in the production of uncleaved NS2-3 (15). Because NS3 is an essential constituent of the viral RNA replicase which cannot be functionally replaced by NS2-3, the shift in polyprotein processing from NS3 to NS2-3 production correlates with a massive downregulation of viral RNA replication (15). This downregulation is a prerequisite for the noncp biotype of pestiviruses in cell culture and an essential property for the establishment of persistent infections in vivo. In cp pestiviruses the Jiv-dependent temporal regulation of NS2-3 cleavage is believed to be overwritten by NS3-generating alterations in genome organization, rendering cp viruses independent of the cellular DNAJC14 pool. Stimulation of the NS2 protease by Jiv90 was also shown for CSFV (16). Moreover, by introducing sequences encoding the DNAJC14 Jiv-90 domain between the Npro and capsid gene of noncp CSFV Alfort (termed CSFV Alfort-Jiv), the DNAJC14-dependent deregulation of NS2-3 cleavage could also be observed for CSFV (54). With CSFV Alfort-Jiv virus it could be shown that increased NS2-3 cleavage and the concomitant expression of increasing NS3 amounts resulted in enhanced viral RNA replication as well as viral cytopathogenicity (54).
In contrast to pestiviruses where DNAJC14 acts as a proviral host factor to support viral genome replication, DNAJC14 has been reported to restrict flaviviral replication by affecting flaviviral polyprotein processing and replication complex (RC) assembly (55–57). These different outcomes in recruiting chaperone proteins nicely illustrate the interplay of viruses with their host proteins to complete their multiplication cycle. Chaperones are involved in a remarkable variety of cellular processes, including the folding of newly synthesized proteins, refolding of misfolded or aggregated proteins, the translocation of secretory proteins across membranes, protein complex assembly and disassembly, and interference with host antiviral responses (58–61). Accordingly, a picture emerges in which viruses divert chaperones from their normal cellular function to play similar roles during viral infection.
DNAJC14-dependent regulation is well established for BVDV-1 and experimentally indicated for Jiv-90 expressing pestiviruses, including CSFV (16, 54, 62). However, several open questions existed: (i) it remains to be determined which noncp pestivirus species require this cochaperone as an essential host factor for their replication, (ii) it is unclear if all types of cp pestivirus genomes replicate in the absence of DNAJC14 expression, and (iii) it is unknown if DNAJC14 has additional functions in the pestiviral life cycle, such as virion morphogenesis or during replicase assembly, similar to what has been reported for flaviviruses (55, 56).
The first question is especially important, since numerous novel pestiviruses have been isolated from different animal species and the existence of at least seven additional pestivirus species in addition to the established pestivirus species BVDV-1, BVDV-2, CSFV, and BDV has been proposed (63–70). These species are represented by closely related Hobi-like viruses from cattle and buffalo (pestivirus H), Aydin-like pestiviruses from sheep and goat (pestivirus I), and the cp giraffe pestivirus together with a related noncp pestivirus (pestivirus G), as well as by the more distantly related pronghorn antelope virus (pestivirus E), Bungowannah virus (pestivirus F), and other recently identified, even more distantly related pestiviruses detected in rat (pestivirus J) and pigs (atypical porcine pestivirus; pestivirus K). Consequently, it has been suggested that we term the established pestivirus species BVDV-1, BVDV-2, CSFV, and BDV pestivirus A to D, respectively (71). While most of these distantly related pestiviruses (such as pronghorn antelope virus or giraffe pestivirus) replicate in cell lines of bovine, ovine, and porcine origin and, thus, show a typical pestiviral cell tropism in vitro, other even more distantly related pestiviruses, such as Bungowannah virus, exhibit a rather unique cell tropism in vitro and could replicate in cell lines of vervet, monkey, mouse, human, and even bat origin (72). With the MDBK and SK6 DNAJC14-knockout (KO) cells newly established in this study, it was possible to demonstrate that the replication of noncp pestiviruses from six investigated diverse pestivirus species (A to D, F, and G) depends on cellular DNAJC14. Furthermore, we demonstrated that virion morphogenesis, as well as the assembly process of the minimal replicase NS3-NS5B, is not affected by the functional knockout of DNAJC14.
RESULTS
Generation of functional MDBK and SK6 DNAJC14-KO cells by CRISPR/Cas9 technology.
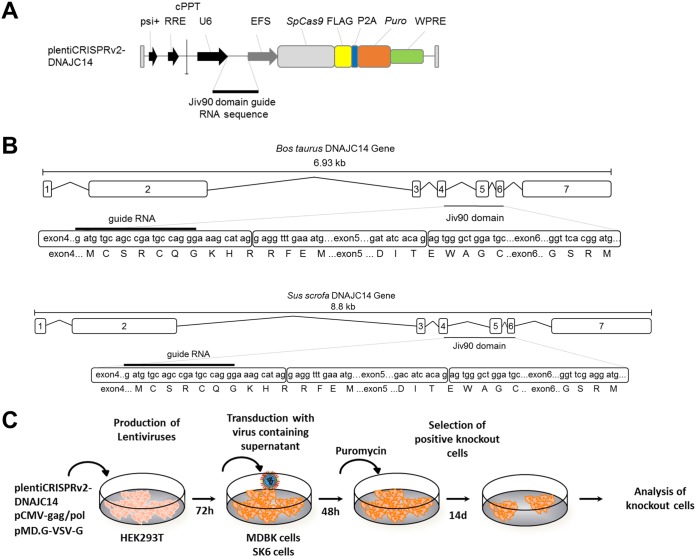
We generated bovine MDBK and porcine SK6 DNAJC14-KO cell clones using CRISPR/Cas9 technology to further investigate the role of DNAJC14 in pestiviral replication. Accordingly, we created different plentiCRISPRv2-DNAJC14 expression plasmids encoding a human codon-optimized Cas9 protein from Streptococcus pyogenes and DNAJC14-specific guide RNAs that can be expressed in a U6 promoter-dependent fashion (Fig. 1A, upper) (73). A 90-amino-acid portion of the DNAJC14 protein (Jiv90) was shown to be essential for stimulating BVDV and CSFV NS2-3 cleavage and for the subsequent establishment of pestiviral replication complexes (15, 50). Consequently, a DNAJC14-specific guide RNA sequence was designed which specifically targeted the N-terminal region of the Jiv90 domain of bovine and porcine DNAJC14 (Fig. 1B). This DNAJC14-specific guide RNA sequence was used to obtain the plentiCRISPRv2-DNAJC14 plasmid (Fig. 1A), which was employed for the production of lentiviral particles. Lentiviral transduction of naive MDBK or SK6 cells was followed by the selection of DNAJC14-guide RNA expressing MDBK or SK6 cells by puromycin treatment (Fig. 1C). Since endogenous DNAJC14 expression is very low in naive MDBK and SK6 cells and the detection of endogenous bovine and porcine DNAJC14 is further compromised by the cross-reactivity of the available DNAJC14-specific antibodies with other cellular proteins of similar size (data not shown), surviving puromycin-resistant MDBK or SK6 cells could not be screened initially by Western blotting or fluorescence-activated cell sorting (FACS) using DNAJC14-specific antibodies. Instead, putative MDBK DNAJC14-KO cells were functionally screened for their inability to support the replication of noncp BVDV-1 strains (NCP7 and NCP8) without affecting the replication of a cp BVDV strain (BVDV-1 NADL), thereby exhibiting a characteristic DNAJC14-specific phenotype (Fig. 1C). In the case of putative SK6 DNAJC14-KO cells, the functional screening involved infection of these cells with CSFV Alfort-Tübingen. Subsequently, single-cell MDBK or SK6 DNAJC14-KO clones were derived by dilution cloning.
FIG 1.
CRISPR/Cas9-mediated genome disruption of bovine and porcine DNAJC14. (A) Scheme of the plentiCRISPRv2-DNAJC14 vector. (B) Illustration of the Bos taurus DNAJC14 gene with intron and exon composition (upper) and of the Sus scrofa DNAJC14 gene. The sgRNA sequence (5′-ATGATGTGCAGCCGATGCCA-3′) and the protospacer adjacent motif (PAM) targeting the Jiv90 domain of the bovine DNAJC14 gene in exon 4 is indicated below the gene composition as a black bar. (C) Scheme of DNAJC14-KO cell generation.
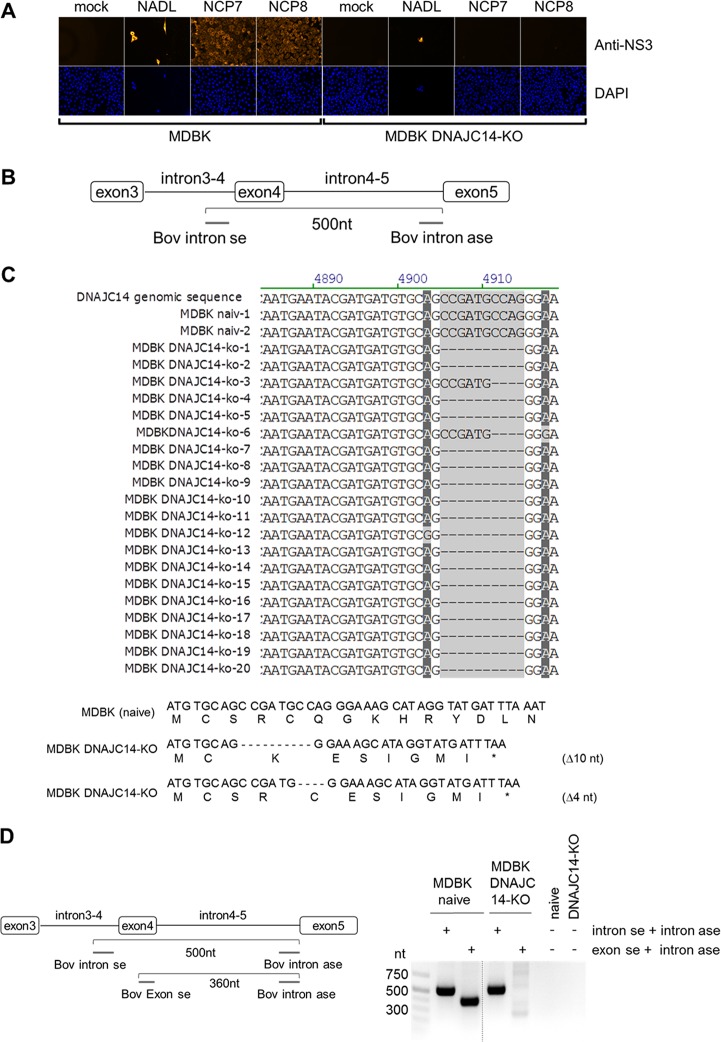
Functional MDBK DNAJC14-KO cells support the replication of cp BVDV strain NADL but not of noncp BVDV strains NCP7 and NCP8.
The functional MDBK DNAJC14-KO cell screening was performed with the noncp BVDV-1a strains NCP7 and NCP8 and the cp BVDV-1 strain NADL, encoding the critical Jiv90 fragment, as a cellular insertion in the NS2 gene; thus, it should replicate independently of the endogenous DNAJC14 (49). Based on this screening scheme, several puromycin-resistant putative MDBK DNAJC14-KO cell clones were challenged with the noncp BVDV strain NCP7 or NCP8 at a multiplicity of infection (MOI) of 3, and virus replication was monitored at 48 h postinfection (hpi) by NS3-specific immunofluorescence (IF) assay to determine if the infected cell clones exhibit a functional DNAJC14 KO phenotype (Fig. 2A). In parallel, infection of the potential MDBK DNAJC14-KO cell clones was done with cp BVDV-1 strain NADL (MOI of 3) to confirm that these cell clones still support cp BVDV replication, thereby excluding functional off-target effects by the CRISPR-Cas9 treatment (Fig. 2A). Infection of parental MDBK cells served as a reference for cp and noncp BVDV replication (Fig. 2A). The positive BVDV-NS3-specific IF and the strong onset of CPE confirmed that the cp BVDV strain NADL replicated in naive MDBK cells as well as in all MDBK DNAJC14-KO cell clones tested (Fig. 2A). In contrast, several of the MDBK DNAJC14-KO cell clones did not detectably support the replication of noncp BVDV-1 strains NCP7 and NCP8 (Fig. 2A shows a representative infection experiment with DNAJC14-KO cell clone 2). The initial KO efficiency of putative MDBK DNAJC14-KO cells was estimated at approximately 50% to 60% (based on NS3-specific IF assay of putative MDBK DNAJC14-KO cells infected with NCP7 at 48 hpi) prior to dilution cloning of puromycin-resistant cells. After three rounds of dilution cloning of puromycin-resistant cells, all cell clones tested showed a functional DNAJC14-KO. The MDBK DNAJC14-KO cell clone (clone 2) with a functional DNAJC14-KO phenotype was further characterized to determine the DNAJC14-specific gene alterations introduced by CRISPR/Cas9 treatment. PCR analysis of genomic DNA isolated from naive MDBK cells and MDBK DNAJC14-KO cell clone 2 was conducted by intron-specific primers flanking DNAJC14 exon 4 sequences (Fig. 2B). Sequence analysis of the PCR-amplified fragments of genomic DNA derived from naive MDBK cells confirmed that wild-type DNAJC14 sequence could be detected (MDBK naiv-1 and naiv-2) (Fig. 2C). Among the 20 PCR amplicons derived from DNAJC14-KO cell clone 2, 18 amplicons exhibited a deletion of 10 nucleotides (nt) (DNAJC14-KO amplicons 1, 2, 4, 5, and 7 to 20) and 2 DNAJC14-KO amplicons (DNAJC14-KO clones 3 and 6 showed a deletion of 4 nucleotides (Fig. 2C, top). No wild-type DNAJC14 sequence could be identified in the DNAJC14-specific PCR amplicons derived from DNAJC14-KO clone 2, establishing the efficient alteration of DNAJC14 genomic sequence by CRISPR/CAS9 (Fig. 2C). Both detected DNAJC14 genomic sequence alterations in the MDBK DNAJC14-KO cell clone were predicted to result in a frameshift at the deletion site and in the generation of DNAJC14 mRNAs encoding an altered amino acid sequence and premature termination codon within the Jiv90 domain which would produce, when translated, C-terminally truncated DNAJC14 proteins lacking 82 or 79 amino acids of the Jiv90 domain (Fig. 2C, bottom). To further validate MDBK DNAJC14-KO clone 2, we performed a seminested PCR analysis. To this end, genomic DNA from MDBK DNAJC14-KO clone 2 cells was purified and used for seminested PCR with DNAJC14 intron- and exon-specific primers (Fig. 2D). The 3′ end of the DNAJC14 exon-specific primer employed in the seminested PCR is complementary to the sequence in which the genomic alterations have been detected; consequently, no nested PCR product was expected for the MDBK DNAJC14-KO cells (Fig. 2D, left). The PCR results confirmed that no wild-type DNAJC14 sequence could be amplified from genomic DNA of MBDK DNAJC14-KO cell clone 2 (Fig. 2D, right). Thus, we succeeded in obtaining MDBK DNAJC14-KO cells by using the CRISPR/Cas9 system.
FIG 2.
Establishment and functional characterization of MDBK DNAJC14-KO cells. (A, upper) Naïve MDBK cells and MDBK DNAJC14-KO cell clones were infected with the indicated viruses (NCP7, NCP8, and NADL) at an MOI of 3 and analyzed by NS3-specific IF assay at 72 hpi. (Lower) Cell nuclei were visualized by DAPI staining. (B) Scheme of the genomic PCR amplification depicting the relative positions of the intron-specific primer flanking exon 4 of the bovine DNAJC14 genomic DNA. (C, upper) Analysis of PCR amplicons obtained from PCR amplification of genomic DNA isolated from naive MDBK cells (2 amplicons, naive 1 and 2) and MDBK DNAJC14-KO cell clone 2 (20 amplicons, MDBK DNAJC14-KO amplicons 1 to 20). Sequences surrounding the DNAJC14-specific guide RNA-mediated genome alteration are shown. Bovine DNAJC14 genomic DNA sequence is shown at the top as a reference. (Lower) Bovine DNAJC14 DNA sequence and amino acid sequence are depicted for naive MDBK cells (top) and MDBK DNAJC14-KO cell clone 2 with the two identified guide RNA-mediated DNAJC14 genome alterations (Δ10 and Δ4). The depicted amino acid sequence starts with the beginning of the Jiv90 domain. Positions of the stop codons resulting from deletions introduced by the DNAJC14 guide RNA are indicated by a star. (D) Seminested PCR analysis to confirm MDBK DNAJC14-KO phenotype. (Left) Scheme of the seminested PCR assay depicting the position of intron- and exon-specific primers on the DNAJC14 genomic DNA. The gel-purified intron-specific amplicons (approximately 500 nt) were used as the template for seminested PCR using DNAJC14 exon- and intron-specific primers (product size, 360 nt) to further verify KO cell clones. (Right) Agarose gel analysis of seminested PCR products. The size standard is given on the left.
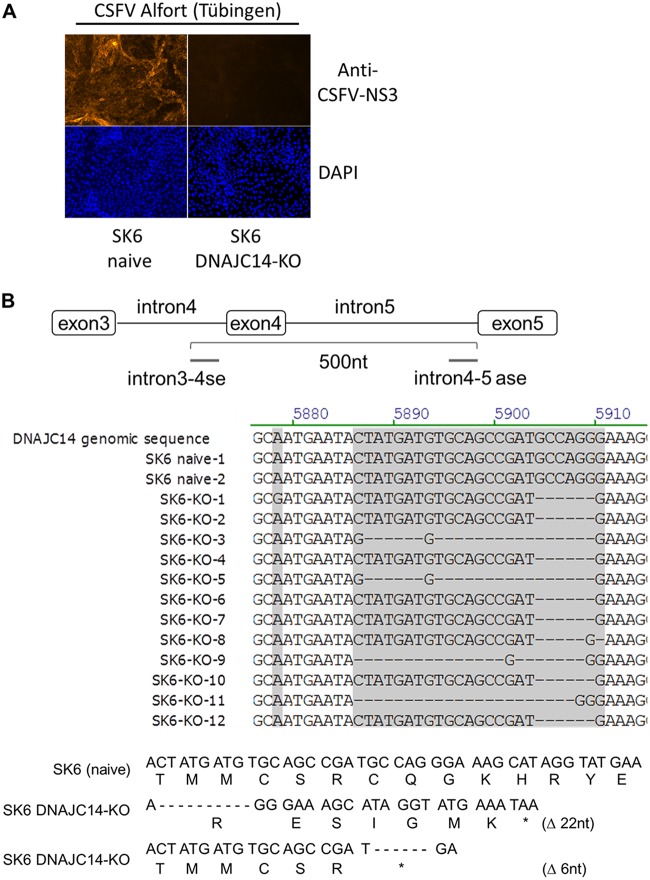
Functional SK6 DNAJC14-KO cells are defective in supporting the replication of noncp CSFV strain Alfort-Tübingen.
Since the DNAJC14-specific guide RNA target sequence located at the start of the Jiv90 domain is conserved between bovine and porcine DNAJC14, we could employ the same plentiCRISPRv2-DNAJC14 plasmid to generate SK6 cells with a functional DNAJC14 knockout (Fig. 1B). SK6 DNAJC14-KO cells were also expected not to support noncp CSFV replication. Hence, after lentiviral transduction and the selection of DNAJC14 guide RNA expressing SK6 cells by puromycin treatment, potential SK6 DNAJC14-KO cells were experimentally screened for a functional DNAJC14 knockout by infecting them with noncp CSFV strain Alfort at an MOI of 3 (Fig. 3A). Parental SK6 cells served as a reference. SK6 DNAJC14-KO cell clones were identified based on the absence of detectable CSFV replication 72 hpi by an anti-CSFV-E2 IF assay (Fig. 3A), and single-cell SK6 DNAJC14-KO clones were derived by dilution cloning. Treatment of SK6 cells with the DNAJC14-specific guide RNA resulted in an estimated initial KO efficiency of approximately 40% to 50% (based on NS3-specific IF assay of putative SK6 DNAJC14-KO cells infected with CSFV) prior to dilution cloning of puromycin-resistant cells. Three rounds of dilution cloning of puromycin-resistant SK6 DNAJC14-KO cells were performed to achieve a functional KO in all SK6 DNAJC14-KO cell clones tested. The PCR analysis of genomic DNA isolated from the putative SK6 DNAJC14-KO cell clone 9 was conducted by intron-specific primers flanking porcine DNAJC14 exon 4 sequences (Fig. 3B, top). The examination of 12 PCR amplicons from this cell clone by DNA sequencing revealed that 4 amplicons had a deletion of 22 nucleotides (SK6 DNAJC14-KO amplicons 3, 5, 9, and 11), and 8 PCR products showed a deletion of 6 nucleotides (SK6 DNAJC14-KO amplicons 1, 2, 4, 6 to 8, 10, and 12) (Fig. 3B, top). Sequence comparison with DNAJC14-specific PCR amplicons derived from genomic DNA isolated from parental SK6 cells further confirmed that no wild-type porcine DNAJC14 sequence could be identified in DNAJC14-KO clone 9, verifying the efficient genomic alteration of the porcine DNAJC14 gene by CRISPR/CAS9. The identified DNAJC14 genomic sequence alterations were predicted to result in the generation of nonfunctional DNAJC14 mRNAs with premature termination codons within the Jiv90 domain (Fig. 3B, bottom). Accordingly, we also obtained functional SK6 DNAJC14-KO cells.
FIG 3.
Molecular characterization of SK6 DNAJC14-KO cells. (A) Naïve SK6 cells and SK6 DNAJC14-KO cell clones were infected with CSFV (Alfort-Tübingen) at an MOI of 3 and analyzed by NS3-specific IF assay at 72 hpi (top). (Lower) Cell nuclei were visualized by DAPI staining. (B, top) Scheme of genomic PCR amplification with the intron-specific primer flanking exon 4 of the porcine DNAJC14 genomic DNA. Analysis of PCR amplicons obtained from PCR amplification of genomic DNA isolated from naive SK6 cells (2 amplicons, SK6 naive 1 and 2) and SK6 DNAJC14-KO cell clone 9 (12 amplicons, SK6 DNAJC14-KO amplicons 1 to 12) is shown. Sequences surrounding the guide RNA-mediated genome alteration of the DNAJC14 gene are shown. (Middle) Porcine DNAJC14 genomic DNA sequence is shown as a reference. Porcine DNAJC14 Jiv90 DNA and amino acid sequence is depicted for naive SK6 cells and SK6 DNAJC14-KO cell clone 9 with the two identified guide RNA-mediated DNAJC14 genome alterations (Δ22 and Δ6). (Lower) Depicted amino acid sequence starts with the beginning of the Jiv90 domain. Positions of the stop codons resulting from deletions introduced by the DNAJC14 guide RNA are indicated by a star.
noncp pestiviruses depend on DNAJC14 expression for their replication.
The functional importance of the temporal NS2 activation by DNAJC14 for noncp pestiviruses is established for BVDV-1 and experimentally indicated for Jiv90-expressing CSFV but is unproven for other species. Interestingly, in vitro studies with different pestiviruses showed that the cell culture tropism of Bungowannah virus differs remarkably from those of other distantly related pestiviruses. It was suggested that the replication machinery together with the viral envelope is responsible for this broad cell tropism in vitro (72). Along these lines, the successful establishment of bovine and porcine DNAJC14-KO cells enabled us to evaluate whether replication of other pestivirus species depends on functional DNAJC14. Consequently, naive MDBK and MDBK DNAJC14-KO 2 cells were infected with a selection of noncp pestivirus isolates comprising species BVDV-1b NCP7, BVDV-1a SD-1, BVDV-1b ncp-4584, BVDV-2 61/120, and BVDV-2 C58644 and pestivirus isolates PG-2, BDV-1 X818, and BDV-2 reindeer (Fig. 4B). The selection of noncp pestivirus isolates used here represents the established species BVDV-1, BVDV-2, and border disease virus (BDV), as well as pestivirus strain PG-2, which belongs to the tentative species of giraffe-like viruses (pestivirus G) (Fig. 4A). As a control, these cells were also infected with cp strain BVDV giraffe, which encodes a DNAJC14 mRNA fragment in its NS2 gene (Fig. 4B). Moreover, we used our newly generated SK6 DNAJC14-KO cells to test the DNAJC14 dependency of the noncp pestivirus isolates CSFV 2.3 (Alfort-Tübingen), CSFV 2.3 (Diepholz), and Bungowannah virus, which represents one of the recently described novel tentative pestivirus species whose origin and host reservoir are still not determined (Fig. 4C). Virus infections were performed in parallel in the respective naive MDBK and SK6 cells to ensure productive genome replication of all tested virus isolates. Cells were infected with the different pestivirus isolates at an MOI of 1. Replication was monitored by IF assays at 72 hpi (Fig. 4). As expected, the infection of MDBK DNAJC14-KO 2 cells with cp strain BVDV giraffe did result in a productive infection, with onset of CPE comparable to that of naive MDBK cells (Fig. 4B). In contrast, infection of MDBK or SK6 DNAJC14-KO cells with different noncp pestivirus isolates did not result in detectable virus replication at 72 hpi for any of these tested noncp virus isolates (Fig. 4). Since all noncp isolates used in this study did productively replicate in the parental MDBK or SK6 cells (Fig. 4), these results demonstrated that all noncp pestiviruses we analyzed in this series of experiments depend on DNAJC14 for their replication. Accordingly, we could expand the functional importance of the DNAJC14-dependent temporal NS2 activation not only for other noncp pestivirus isolates belonging to BVDV-2 or BDV species but also for the Bungowannah virus (Fig. 4B), representing a highly distinct pestivirus compared to the established pestivirus species BVDV-1, BVDV-2, CSFV, and BDV (64, 74).
FIG 4.
Infection of DNAJC14-KO cells demonstrates that replication of noncp pestiviruses depends on DNAJC14. (A) Phylogenetic tree (modified from reference 109) representing the four established pestivirus species BVDV-1, BVDV-2, BDV, and CSFV, together with the additional tentative pestivirus species recently proposed by the Flaviviridae Study Group of the International Committee on Taxonomy of Viruses (71). Letters in brackets represent the pestivirus species Pestivirus A to K according to the recent proposal for a revised pestivirus nomenclature (71). Pestivirus species used for the experiments in panel B are BVDV-1, BVDV-2, BDV, CSFV, giraffe, and Bungowannah (highlighted in boldface letters). Asterisks indicate pestiviruses for which no isolates exist or are not available. (B) Naïve MDBK (upper) and MDBK DNAJC14-KO clone 2 cells (lower) were infected with noncp pestivirus isolates BVDV-1b NCP7, BVDV-1a SD-1, BVDV-1b ncp-4584, BVDV-2 61/120, BVDV-2 C58644, and PG-2, the latter of which belongs to the pestivirus species giraffe (pestivirus G), BDV-1 X818, BDV-2 reindeer, and, as a control, cp strain giraffe (pestivirus G). (C) Naïve SK6 (upper) and SK6 DNAJC14-KO clone 9 (lower) cells were infected with noncp pestivirus CSFV strains Alfort-Tübingen and Diepholz (Dieph.) and noncp pestivirus strain Bungowannah. All infections were performed in parallel in the respective DNAJC14-KO cell lines with the indicated pestivirus isolates at an MOI of 1. Viral genome replication was monitored by the anti-NS3-based IF assay at 72 hpi (depicted as yellow fluorescence). Cell nuclei were counterstained with DAPI. All infections were performed in three independent experiments, and a representative data set is shown.
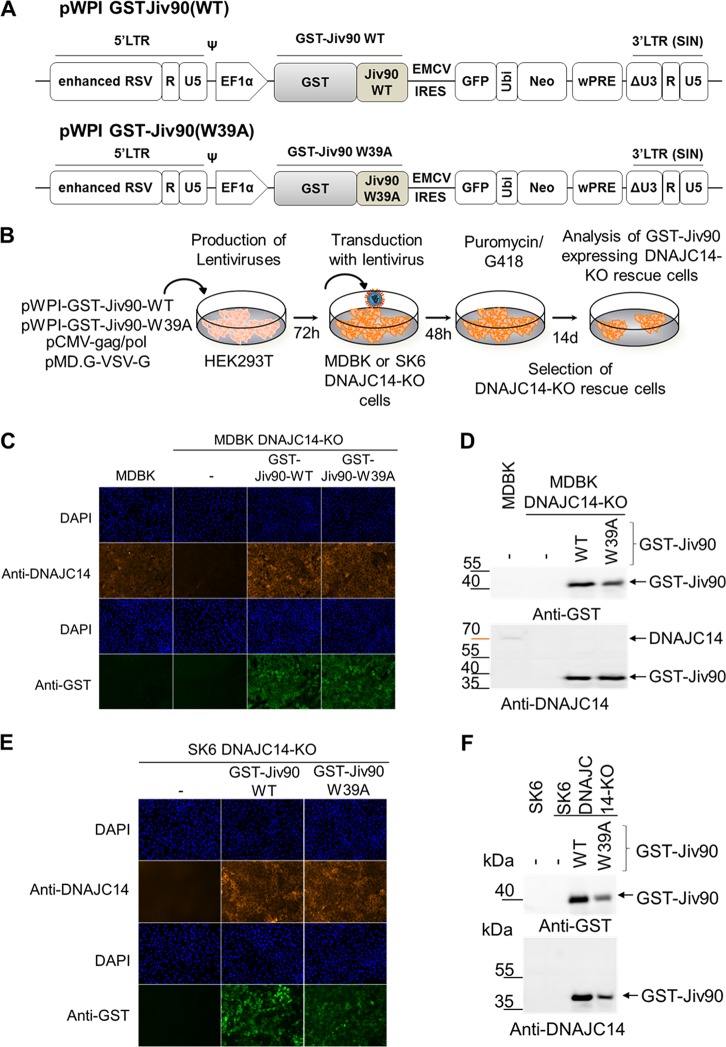
Establishment of MDBK and SK6 DNAJC14-KO rescue cell lines expressing GST-Jiv90(WT) or GST-Jiv90(W39A).
To exclude off-target effects and to further confirm the specific function of the isolated Jiv90 domain of DNAJC14 in the pestiviral life cycle, we conducted gene rescue experiments in our MDBK and SK6 DNAJC14-KO cell lines. Accordingly, we established rescue cell lines expressing either glutathione S-transferase (GST)-Jiv90(WT) or its inactive derivative, GST-Jiv90(W39A), resistant to the DNAJC14 guide RNA due to escape mutations in the guide RNA binding site (Jiv90Esc) based on the validated functional MDBK and SK6 DNAJC14-KO cell lines by lentiviral transduction of pWPI-GST-Jiv90Esc(WT) or pWPI-GST-Jiv90Esc(W39A) expression plasmid, followed by simultaneous puromycin/G418 selection (Fig. 5A and B). The rescue cell lines expressed comparable levels of either GST-Jiv90(WT) or GST-Jiv90(W39A), as confirmed by anti-GST- and anti-Jiv90-specific IF assay (Fig. 5C and E) or Western blot analysis (Fig. 5D and F).
FIG 5.
Generation and validation of MDBK and SK6 DNAJC14-KO rescue cell lines expressing GST-Jiv90(WT) or GST-Jiv90(W39A). (A) Scheme of the lentiviral expression constructs pWPI-GST-Jiv90(WT) and pWPI-GST-Jiv90(W39A). LTR, long terminal repeat. (B) Schematic of the experimental approach used for the generation of the MDBK and SK6 DNAJC14-KO rescue cells. (C) Analysis of GST-Jiv90 expression in MDBK DNAJC14-KO rescue cells by IF. DNAJC14-KO cells and DNAJC14-KO rescue cells expressing GST-Jiv90(WT) or GST-Jiv90(W39A) were stained with anti-DNAJC14 (recognizing epitope SARYCAECNR in the Jiv90 domain) or anti-GST-specific antibodies. Cell nuclei were counterstained with DAPI. (D) Western blot analysis of GST-Jiv90 expression in MDBK DNAJC14-KO rescue cells. Protein lysates obtained from naive MDBK and MDBK DNAJC14-KO cells and MDBK DNAJC14-KO rescue cells expressing either GST-Jiv90(WT) or GST-Jiv90(W39A) were separated by SDS-PAGE and analyzed with anti-GST (top) or anti-DNAJC14 (bottom). The positions of DNAJC14 GST-Jiv90(WT) and GST-Jiv90(W39A) are indicated on the right, and the protein marker positions are given on the left. (E) Analysis of GST-Jiv90 expression in SK6 DNAJC14-KO rescue cells by anti-GST- and anti-DNAJC14-specific IF. (F) Western blot analysis of GST-Jiv90 expression in SK6 DNAJC14-KO rescue cells expressing either GST-Jiv90(WT) or GST-Jiv90(W39A). Protein lysates obtained from naive SK6, SK6 DNAJC14-KO cells, and SK6 DNAJC14-KO rescue cells expressing either GST-Jiv90(WT) or GST-Jiv90(W39A) were separated by SDS-PAGE and analyzed with anti-GST (top) or anti-DNAJC14 (bottom). The positions of DNAJC14 GST-Jiv90(WT) and GST-Jiv90(W39A) are indicated on the right, and the protein marker positions are given on the left.
DNAJC14 gene rescue experiments with MDBK and SK6 DNAJC14-KO cells confirm that only the Jiv90 domain is required for the pestiviral life cycle.
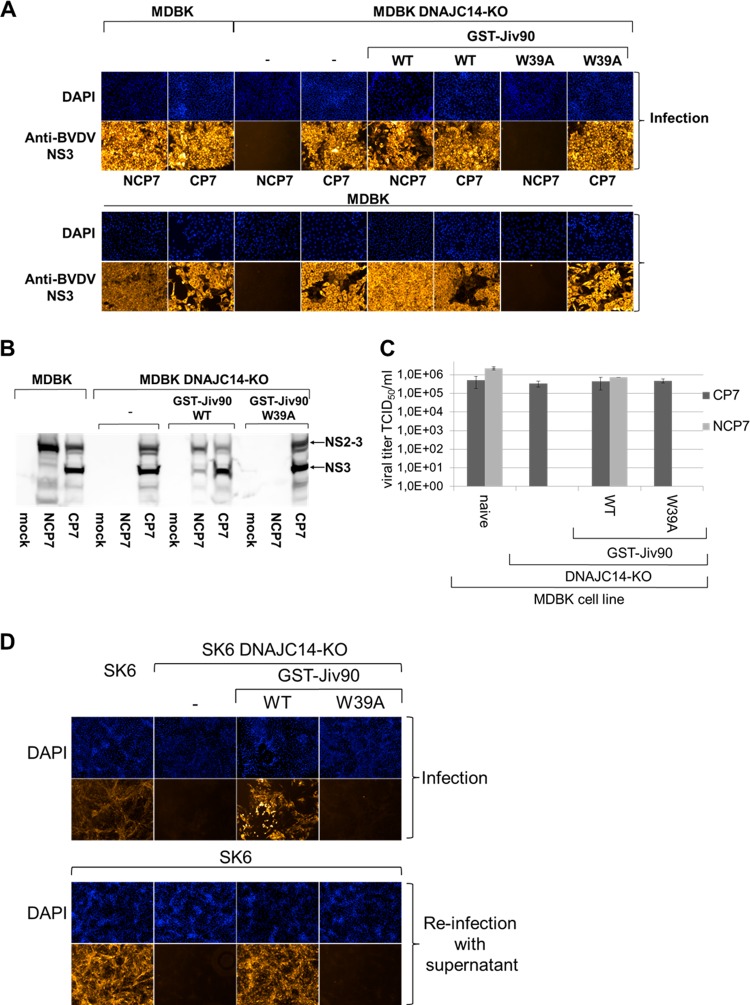
The DNAJC14 protein is a pivotal host factor for noncp pestiviruses, as demonstrated by the inability of different noncp pestiviruses to replicate in DNAJC14-KO cells (Fig. 4). In order to confirm the specific function of the Jiv90 domain in the pestiviral life cycle, we used MDBK DNAJC14-KO cells expressing either GST-Jiv90(WT) or GST-Jiv90(W39A) and infected them with BVDV-1b strains NCP7 and CP7 at an MOI of 3. NCP7 and CP7 infections of naive MDBK and MDBK DNAJC14-KO cells, respectively, served as references. As expected, CP7 replication was comparable between the parental MDBK cells, MDBK DNAJC14-KO cells (also see below), and the two MDBK DNAJC14-KO rescue cell lines (Fig. 6A, upper). In contrast, NCP7 replication was only supported in the parental MDBK cells and the MDBK DNAJC14-KO GST-Jiv90(WT) rescue cell line but not in MDBK DNAJC14-KO cells or in the MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cell line (Fig. 6A, upper). This observation was further strengthened by reinfection experiments of naive MDBK cells with supernatants obtained 72 hpi from the initial NCP7 and CP7 infection of all four cell lines. These reinfection experiments confirmed that MDBK DNAJC14-KO cells and the MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cell line do not support noncp NCP7 infection (Fig. 6A, lower). Furthermore, NCP7 infections in MDBK DNAJC14-KO GST-Jiv90(WT) rescue cells resulted in a detectable CPE comparable to that of CP7 infection, while NCP7 infection of parental MDBK cells did not (Fig. 6A, upper). This change in the biotype was not detectable when the supernatant of NCP7 infection of DNAJC14-KO GST-Jiv90(WT) rescue cells was used to reinfect parental MDBK cells, in contrast to a reinfection of naive MDBK cells with the supernatant from CP7 infection of DNAJC14-KO GST-Jiv90(WT) rescue cells (Fig. 6A, lower). In agreement with these functional observations, our Western blot analysis of the NS2-3 cleavage in all four MDBK cell lines confirmed the expected regulation of NS2-3 cleavage by DNAJC14 (Fig. 6B). The overexpression of the functional Jiv90(WT) domain in MDBK DNAJC14-KO Jiv90(WT) cells led to deregulated NS2-3 cleavage, resulting in increased NS3 release in MDBK DNAJC14-KO Jiv90(WT) cells infected with NCP7 compared to naive MDBK cells (Fig. 6B), leading to a characteristic steady-state level of NS2-3 cleavage in the infected MDBK DNAJC14-KO Jiv90(WT) rescue cells. Furthermore, higher steady-state levels of functional Jiv90 produced in Jiv90-overexpressing MDBK DNAJC14-KO Jiv90(WT) rescue cells did not result in complete NS2-3 cleavage in either NCP7- or CP7-infected MDBK DNAJC14-KO Jiv90(WT) rescue cells (Fig. 6B), which is also in agreement with earlier observations (15). To further strengthen these observations, we determined the virus titers of NCP7 and CP7 infections in those four different MDBK cell lines (Fig. 6C). Compared to naive MDBK cells, overexpression of the functional Jiv90 domain of DNAJC14 in MDBK DNAJC14-KO GST-Jiv90(WT) rescue cells did not have a negative impact on the viral titer, confirming the establishment of a characteristic steady-state level of NS2-3 and NS3 that supports effective virus production (Fig. 6C).
FIG 6.
Expression of GST-Jiv90(WT) functionally rescues the DNAJC14 gene knockout in MDBK and SK6 cells and supports noncp pestivirus replication in MDBK and SK6 DNAJC14-KO rescue cells. (A, upper) Immunofluorescence analysis after infection of different target cells. Naïve MDBK, MDBK DNAJC14-KO, MDBK DNAJC14-KO GST-Jiv90(WT), and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells were infected with CP7 and NCP7 viruses at an MOI of 0.1. At 72 hpi, immunofluorescence analysis was performed using a monoclonal anti‐BVDV NS3 antibody. Nuclei are DAPI stained. (A, lower) Immunofluorescence analysis of the reinfection experiment. Cell culture supernatants from CP7 or NCP7 infection of naive MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells were used to infect naive MDBK cells with 500 μl of filtered supernatant. At 72 hpi, immunofluorescence analysis was performed using a monoclonal anti‐BVDV NS3 antibody. Cell nuclei were counterstained with DAPI. (B) Western blot analysis of NS2-3 cleavage in MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells infected with noncp BVDV NCP7 and cp BVDV CP7. The lysate separated in each lane represents about 5 × 105 cells. Lysates were prepared at 72 hpi. Positions of NS3 and NS2-3 are marked with arrows. Depicted is a representative Western blot of three independent experiments. (C) Viral titer analyses of MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells infected with noncp BVDV NCP7 and cp BVDV CP7. Cell culture supernatants collected at 72 hpi (MOI of 0.1) were analyzed for infectious virus by limiting dilution assay (TCID50/ml). Mean values and standard deviations of viral titers from three experiments are depicted. (D, upper) Naïve SK6 and SK6 DNAJC14-KO cell as well as SK6 DNAJC14-KO GST-Jiv90(WT) and SK6 DNAJC14-KO GST-Jiv90(W39A) rescue cells were infected with CSFV Alfort-Tübingen at an MOI of 0.1. At 72 hpi, immunofluorescence analysis was performed using a monoclonal anti‐CSFV NS3 antibody. Nuclei are DAPI stained. (D, lower) Immunofluorescence analyses of reinfection experiment with naive SK6 cells. Cell culture supernatants from CSFV infection of naive SK6 and SK6 DNAJC14-KO cells as well as SK6 DNAJC14-KO GST-Jiv90(WT) and SK6 DNAJC14-KO GST-Jiv90(W39A) rescue cells were used to infect naive SK6 cells with 500 μl of filtered supernatant. At 72 hpi, immunofluorescence analysis was performed using a monoclonal anti‐E2 antibody. Cell nuclei were counterstained with DAPI. Depicted is a representative data set from three independent experiments.
These functional observations could be confirmed by infection experiments of the respective SK6 DNAJC14-KO rescue cell lines with noncp CSFV Alfort-Tübingen (Fig. 6D). Again, only the parental SK6 and the SK6 DNAJC14-KO GST-Jiv90(WT) rescue cell line supported replication of noncp CSFV, while both SK6 DNAJC14-KO cells and the SK6 DNAJC14-KO GST-Jiv90(W39A) rescue cell line did not (Fig. 6D). Taken together, the capacity of the MDBK cells and SK6 DNAJC14-KO GST-Jiv90(WT) rescue cell line in supporting noncp BVDV and CSFV replication confirmed the specific function of the Jiv90 domain and that this domain is sufficient to regulate RNA replication and viral biotype protease activation.
DNAJC14 gene knockout and the overexpression of the Jiv90 domain do not affect the assembly of a functional minimal pestiviral replicase.
While DNAJC14 acts as a proviral host factor for pestiviral genome replication, this cellular chaperone also was recently shown to act as a flavivirus restriction factor, most likely by modulating the assembly of flaviviral replication complexes (RCs) (55, 56).
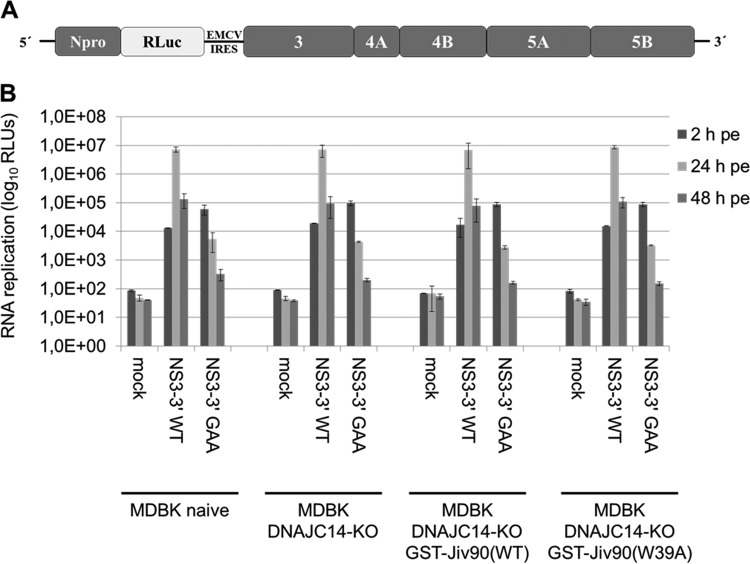
Whether DNAJC14 also has the ability to modulate the functional assembly of pestiviral RCs could not be unambiguously determined without a functional MDBK DNAJC14-KO cell line. To analyze this quantitatively, we used a bicistronic NS3-5B replicon (Bici-388 RLuc IRES-NS3-5B) encoding BVDV Npro and Renilla luciferase in the first ORF and the minimal pestiviral replicase (NS3-5B) in its second open reading frame (Fig. 7A) (25, 33). Accordingly, these replicons are independent from the need to undergo DNAJC14-stimulated NS2-3 cleavage to assemble functional replicase complexes. The Bici-388 RLuc IRES-NS3-5B/WT RNA was electroporated into the parental MDBK and MDBK DNAJC14-KO cells and the respective MDBK DNAJC14-KO rescue cell lines expressing GTS-Jiv90(WT) or GST-Jiv90(W39A). The rescue cell lines were included to investigate if the overexpression of GST-Jiv90(WT) and GST-Jiv(W39A) in cells with a functional DNAJC14 knockout affects RNA replication of the BVDV NS3-5B replicon. Bici-388 RLuc IRES-NS3-5B/GAA RNA, a mutant replicon containing point mutations (GDD to GAA) abolishing RdRp, served as a control (25). RLuc activities were determined at 2, 24, and 48 h postelectroporation (hpe). At 2 hpe, similar values were obtained for both RNAs in all cell lines, demonstrating similar electroporation efficiencies. At 24 hpe, comparable maximum values of RLuc activity were detected for the Bici-388 RLuc IRES-NS3-5B/WT replicon in all four cell lines analyzed. The observed subsequent decrease of RLuc activity at 48 hpe is due to the onset of the replication-induced CPE (Fig. 7B). As expected, the mutant replicon with a defective RdRp (GAA) did not replicate (Fig. 7B).
FIG 7.
Assessment of the capacity of the BVDV NS3-3′ replicon to support assembly of a minimal replicase and viral RNA replication in DNAJC14-KO and DNAJC14-KO GST-Jiv90 rescue cells by luciferase assay. (A) Schematic representation of the BVDV Bici-388 RLuc-NS3-3′ replicon. (B) In vitro-transcribed BVDV Bici-388 RLuc-NS3-3′/WT and BVDV Bici-388 RLuc-NS3-3′/GAA replicon RNA (2 μg) was electroporated into naive MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells, and cells were collected at 2, 24, and 48 hpe to determine luciferase activity. Mean values from three independent experiments are shown. Error bars indicate standard deviations. Mock, no RNA electroporated; WT, wild type; GAA, polymerase-inactive mutant; RLUs, relative light units; RLuc, Renilla luciferase.
The comparable replication efficiencies of the BVDV NS3-5B replicon in parental MDBK and MDBK DNAJC14-KO cells and MDBK DNAJC14-KO rescue cells demonstrate that the DNAJC14 gene knockout does not affect the assembly of a functional minimal pestiviral replicase and that the overexpression of GST-Jiv90(WT) or GST-Jiv(W39A) in the MDBK DNAJC14-KO cells does not modulate the RNA replication efficiency (Fig. 7B). Furthermore, these results also confirmed that no additional off-target genome alterations were introduced by the CRISPR/CAS9 system, which could impair replication of noncp pestivirus strains and thereby confound our functional DNAJC14 analysis.
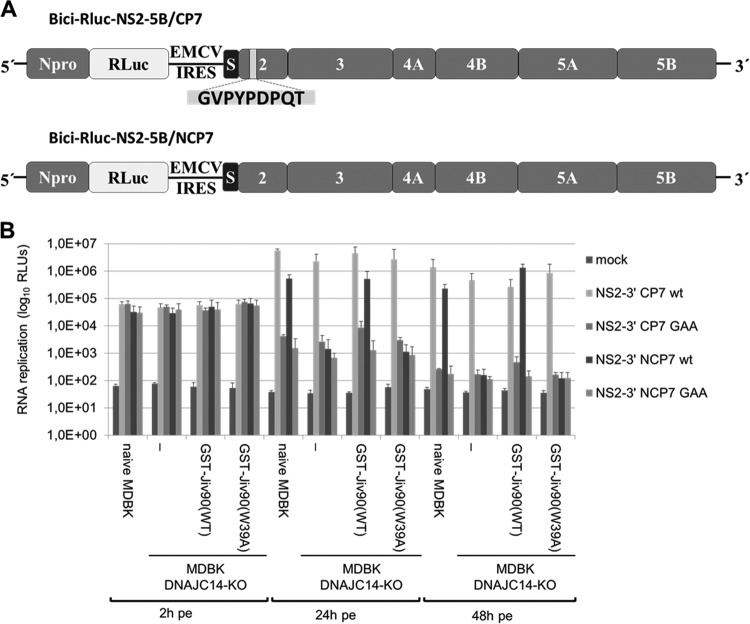
Analysis of viral RNA synthesis of noncp and cp NS2-5B RNA replicons in MDBK DNAJC14-KO cells.
In contrast to the DNAJC14 dependency of noncp pestiviruses, pestiviral cp strains use various strategies to generate NS3 with the aim of upregulating NS3 release (2). This led to the hypothesis that cp strains are independent of the cellular DNAJC14 pool for their genome replication. To confirm that these noncp and cp characteristics of NS3 release can be recapitulated in our MDBK DNAJC14-KO and rescue cell lines, we used noncp (Bici-388 RLuc IRES-NS2-5B/NCP7) and cp (Bici-388 RLuc IRES-NS2-5B/CP7) NS2-5B replicons based on the NCP7 and CP7 BVDV sequence (75). These bicistronic NS2-5B replicons encode BVDV Npro and Renilla luciferase (RLuc) in the first ORF and the BVDV NS2-5B/NCP7 or the BVDV NS2-5B/CP7 polyprotein, respectively, in its second open reading frame (Fig. 8A). These replicons differ only with respect to their NS2 sequence: compared to NCP7 NS2, the CP7 NS2 gene encompasses a 27-nucleotide (9-aa) insertion that was shown to be critical for efficient cleavage at the NS2/3 site by NS2 (75). Accordingly, the BVDV NS2-5B/CP7 BVDV replicon should undergo efficient NS2-3 cleavage independent of endogenous DNAJC14, while the BVDV NS2-5B/NCP7 BVDV replicon is expected to be dependent on the temporal regulation of NS2-3 cleavage by DNAJC14 to assemble functional replicase complexes. BVDV NS2-5B/NCP7 GAA and BVDV NS2-5B/CP7 GAA mutant replicons containing point mutations (GDD to GAA) abolishing RdRp served as controls. These replicon RNAs were electroporated into naive MDBK, MDBK DNAJC14-KO, and MDBK DNAJC14-KO rescue cells and analyzed for their ability to replicate in these cell lines. At 2 hpe, similar RLuc values were obtained for all four NS2-5B replicon RNAs in the different cell lines, demonstrating similar electroporation efficiencies. As expected, the NS2-5B/CP7 WT replicon replicated in all four cell lines with similar efficiencies after 24 h (Fig. 8B). At 48 hpe, the replication efficiencies for this replicon were slightly reduced compared to the 24-hpe values in all four cell lines due to the onset of a CPE (Fig. 8B). In contrast, the NS2-5B/NCP7 WT replicon only replicated in naive MDBK cells and MDBK DNAJC14-KO GST-Jiv90(WT) rescue cells but not in MDBK DNAJC14-KO cells and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells, as expected (Fig. 8B). The observation that the NS2-5B/NCP7 WT replicon exhibited a lower replication efficiency than the NS2-5B/CP7 WT replicon in naive MDBK cells at 24 h and 48 hpe argues that the DNAJC14-mediated temporal regulation mechanism of NS2-3 cleavage during the replication of the NS2-5B/NCP7 WT replicon contributes to this lower replication efficiency and is in line with the lower RNA replication efficiency known for NCP7 infections than CP7 infections (14). In MDBK DNAJC14-KO rescue cells expressing GST-Jiv90(WT), the NS2-5B/NCP7 WT replicon showed robust replication similar to (24 hpe) or even better than (48 hpe) that of the NS2-5B/NCP7 WT replicon in naive MDBK cells (Fig. 8B). The BVDV NS2-5B/NCP7 GAA and BVDV NS2-5B/CP7 GAA mutant replicons with a defective RdRp did not replicate, as expected. Taken together, these results show that the replication of noncp pestiviruses can be enhanced by the isolated Jiv90 domain also in the absence of DNAJC14. Moreover, these data demonstrate that the CP7-specific 9-aa insertion renders the NS2 protease independent of DNAJC14 and, thus, allows the replication of this bicistronic RNA.
FIG 8.
Analysis of viral RNA synthesis of noncp and cp BVDV NS2-5B RNA replicons in MDBK DNAJC14-KO cells. (A) Scheme of the bicistronic reporter replicons Bici RLuc NS2-5B/CP7 (top) and Bici RLuc NS2-5B/NCP7 (bottom). The 9-aa insertion in NS2 of Bici RLuc NS2-5B/CP7 is indicated as a light gray box within the NS2 gene. S (black box) depicts the C-terminal portion of p7 (38 aa) that acts as a signal sequence for translocation of NS2-3 (10). (B) Cells were electroporated with 1 μg of the corresponding in vitro-transcribed RNAs and incubated for the indicated times at 2, 24, and 48 hpe, and luciferase activity was determined. Measurements were carried out in triplicate, and experiments were repeated at least 3 times. Data from one representative experiment are shown. Error bars represent standard deviations. Mock, no RNA electroporated; GAA, NS5B mutation, replication deficient; WT, wild type; RLUs, relative light units; RLuc, Renilla luciferase.
All types of cp pestivirus genomes replicate in the absence of DNAJC14 expression.
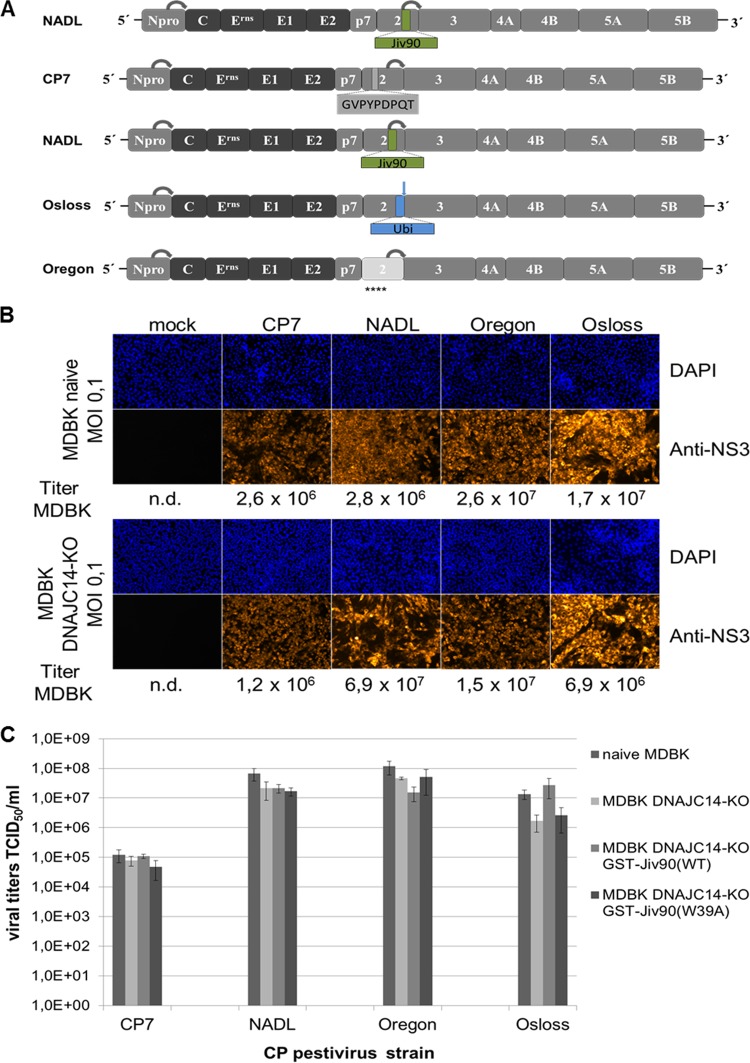
The genomes of cp pestiviruses show astonishingly diverse genome organizations (reviewed in references 2 and 76). Despite the differences in their genome alterations, the salient feature of all of them is an upregulation of NS3 expression and possibly an independence of NS3 expression from the endogenous DNAJC14 level, as demonstrated above for a replicon carrying NS2 of cp BVDV-1 strain CP7. To test the latter hypothesis, MDBK DNAJC14-KO cells were infected with different cp BVDV strains representing different prototypes of cp genomes (Fig. 9A). In the cp BVDV-1 strain NADL, the NS2 protease is activated by a 90-aa fragment of the cellular DNAJC14 (Jiv90) located in the C-terminal part of NS2 (77). Another type of cp BVDV strains, such as strain Osloss, has insertions of cell-derived sequences coding for ubiquitin or ubiquitin-like proteins just upstream of the NS3 gene (78). Since in those polyproteins cellular proteases generate the C terminus of the cell-derived ubiquitin/ubiquitin-like protein and thereby the N terminus of NS3, complete cleavage can be expected. Accordingly, DNAJC14 should not be required for the replication of this virus type. As described above, strain CP7 has a 9-amino-acid insertion located in the central part of NS2, which is associated with activation of the NS2 protease, responsible for NS2-3 cleavage (75). For BVDV-1 strain Oregon, it was described that an accumulation of amino acid exchanges in NS2 is responsible for upregulated NS2-3 processing by the NS2 protease. Accordingly, this strain exhibits a cp phenotype with the least obvious alterations in the viral genome (79, 80). In experiments executed in parallel, these four pestiviral cp virus isolates were used to infect naive MDBK cells and MDBK DNAJC14-KO cells at an MOI of 0.1, followed by NS3-specific IF at 72 hpi (Fig. 9B). The respective cell culture supernatants obtained at 72 hpi were subsequently used to determine virus titers on naive MDBK cells. The results showed that the DNAJC14 gene knockout does not affect viral genome replication and virion morphogenesis for all four tested cp virus isolates (Fig. 9B). These results demonstrate that all known cp pestivirus types replicate in the absence of endogenous functional DNAJC14, confirming their independence from this host factor. In principle, overexpression of the functional Jiv90 domain of DNAJC14 might render NS2-3 cleavage too efficient and, thus, could have a negative impact on the viral titer of certain pestivirus strains. To address this question, we determined the virus titers of the four prototype cp pestiviruses in naive MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cell lines (Fig. 9C). While the viral titer for NADL, Oregon, and Osloss strains were slightly reduced in MDBK DNJAC14-KO cells compared to those in naive MDBK cells, overexpression of the functional Jiv90 domain of DNAJC14 in MDBK DNAJC14-KO GST-Jiv90(WT) rescue cells did not have a negative impact on the viral titer, suggesting the establishment of a characteristic steady-state level of NS2-3 and NS3 supporting robust virus production (Fig. 9C). Together, these findings prove DNAJC14 is a pivotal cellular cofactor for the replication of various noncp pestiviruses and establish the independence of different types of cp pestiviruses from the cellular host factor DNAJC14.
FIG 9.
Analysis of genome replication of different types of cp pestivirus genomes in the absence of DNAJC14 expression. (A) Schematic representation of genome organizations of replicating cp pestiviruses. The ORF depicting the encoded proteins as well as the flanking 5′- and 3′-UTRs are indicated. The N-terminal autoprotease Npro and the nonstructural proteins p7-NS5B are highlighted as gray boxes, and the structural proteins are indicated as black boxes. BVDV CP7 encompasses an insertion of 9 amino acids (highlighted in gray) in NS2. BVDV NADL NS2 contains the minimal fragment of the cellular Jiv-mRNA (Jiv90; highlighted in green) located in the NS2. Both insertions within NS2 are critical for high-efficiency cleavage at the NS2/3 site by the NS2 cysteine protease (indicated by a gray arrow). The genome of BVDV Osloss contains an insertion of a ubiquitin (Ubi) coding sequence (indicated in blue) at the NS2/NS3 gene border. Efficient cleavage at the NS2/3 site is mediated by cellular ubiquitin hydrolases (indicated by a blue arrow). NS2 of BVDV Oregon (highlighted in light gray) is characterized by the presence of several amino acids (indicated by stars below the NS2 box) that are spread over the protease domain and deviate from the pestiviral NS2 consensus sequence. Npro cleavage is indicated by a gray arrow. (B) Analysis of viral replication after infection of naive (upper) and DNAJC14-KO (lower) MDBK cells with the indicated cp BVDV strains CP7, NADL, Oregon, and Osloss at an MOI of 0.1. (Lower) At 72 hpi, immunofluorescence analysis was performed using a monoclonal anti‐BVDV NS3 antibody. (Upper) Nuclei were DAPI stained. At 72 hpi, viral supernatants were harvested and analyzed for the presence of infectious viruses in naive MDBK cells. Viral titers were determined as TCID50/ml and are given below the IF assay for each infection. Data are representative of three independent experiments. n.d., not determined. (C) Viral titer analyses. MDBK and MDBK DNAJC14-KO cells as well as MDBK DNAJC14-KO GST-Jiv90(WT) and MDBK DNAJC14-KO GST-Jiv90(W39A) rescue cells were infected with cp pestivirus strains CP7, NADL, Osloss, and Oregon. Cell culture supernatants collected at 72 hpi (MOI of 0.1) were analyzed in MDBK cells for infectious virus by limiting dilution assay (TCID50/ml). Mean values and standard deviations of viral titers from three experiments are depicted.
DISCUSSION
Positive-strand RNA viruses from the family Flaviviridae, like pestiviruses, employ viral nonstructural proteins not only in RNA replication but also in virion morphogenesis, highlighting the multifunctional nature of many of these proteins (4). Their structural and functional complexities are most likely the reason for their often observed dependency on chaperones for their folding and function (81, 82).
In the pestiviral life cycle, the temporal regulation of NS2-3 cleavage by the NS2-interacting and activating cellular cofactor DNAJC14 is of critical importance to switch between the cleaved form, required for RNA replication, and uncleaved NS2-3, essential for virion morphogenesis. This DNAJC14-dependent regulation of NS2-3 cleavage is a salient feature of the noncp biotype and highly relevant for the remarkably successful long-term persistence of ruminant pestiviruses in their hosts (2). This temporal regulation by DNAJC14 is well established for BVDV-1 and has been indicated for CSFV by studies demonstrating that DNAJC14 can also stimulate the NS2-3 cleavage of CSFV (16, 54, 62). However, it remained an open question whether noncp viruses from other pestivirus species also require DNAJC14 as an essential host factor for their replication. With the newly established MDBK and SK6 DNAJC14 knockout cells, it was now possible to investigate whether the replication of all noncp pestivirus species for which cell culture systems are available depends on cellular DNAJC14 (Fig. 4). Accordingly, the results of the present study demonstrate that, in addition to BVDV-1 and CSFV, noncp pestivirus isolates belonging to the species BVDV-2 or BDV, as well as members of other tentative species, depend on DNAJC14 (Fig. 4). Interestingly, the DNAJC14 dependency could even be demonstrated for the Bungowannah virus (Fig. 4B), which is only very distantly related to the four established pestivirus species, BVDV-1, BVDV-2, BDV, and CSFV (74). Other distantly related tentative pestivirus species could not be included in our analysis, as virus isolates either do not exist (rat pestivirus and bat pestivirus) or were not available to us (Linda virus). Together, these observations confirm that the RNA replication of noncp strains from six distinct pestivirus species (e.g., A to D, F, and G) depends on cellular DNAJC14. Therefore, the observed strict DNAJC14 dependency of these virus isolates suggests that they replicate in cells of different origin, provided that they express a functional form of DNAJC14. This assumption is supported by previous culture-based studies, which have demonstrated that the distantly related Bungowannah virus is growing in a range of different cells, extending from humans to bats (64, 72). In fact, in cells from Rhinolophus sinicus (Chinese rufous horseshoe bat), a homologue of DNAJC14 with a functional Jiv90 domain can be found (83 and data not shown). Interestingly, the unique in vitro tropism depended in this case on the replication machinery, e.g., the ability to use DNAJC14-mediated NS2 activation, together with the viral envelope (72). In the future, it will be of particular interest to determine the DNAJC14 dependency of the recently discovered atypical porcine pestivirus (APPV), since its NS2 sequence differs dramatically from that of the pestiviruses investigated in this study (63, 65).
For cp pestiviruses it has been proposed that they do not depend on DNAJC14 with respect to RNA replication (14, 15). This assumption was based on genome alterations rendering these viruses, with regard to their NS3 release, independent of the NS2 protease. This is the case for all cp pestiviruses with insertions of cell-derived sequences located just upstream of the NS3 gene, coding for ubiquitin, ubiquitin-like proteins (e.g., SUMO or NEDD8), or proteins with ubiquitin-like folds (e.g., LC3 or GATE-16) (2). In these polyproteins, cellular proteases generate the C terminus of the inserted cellular proteins and thereby the N terminus of NS3 (2). However, for other cp variants with more subtle genome alterations, such as the BVDV strain CP7 or Oregon, the DNAJC14 dependency could be proven unequivocally only by a DNAJC14 knockout. This experimental proof could now be provided with the DNAJC14-KO cells and demonstrated that all different cp pestiviruses with various kinds of genome alterations can replicate in the absence of DNAJC14 (Fig. 9). This finding also has implications for the development of the so-called mucosal disease in cattle persistently infected by noncp BVDV. Causative for the outbreak of this lethal disease is the sporadic emergence and rapid spread of a cp BVDV variant in such an animal. Possibly, the independence of these cp virus mutants from DNAJC14 makes cell types/tissues, which do not support noncp BVDV replication, accessible for them. This might explain in part the explosive spread of the cp virus mutant which is connected to the onset of mucosal disease.
Interestingly, the activation of the NS2 autoprotease in noncp pestiviruses differs from the activation process of the closely related HCV NS2 autoprotease, which employs the NS3 protease domain as an essential cofactor for NS2 protease activation instead of the cellular DNAJC14 (84). However, both viral NS2 autoproteases require activation by an essential cofactor to promote NS2-3 cleavage, indicating that both proteases need cofactor-dependent assistance to adapt an active protease conformation. Interestingly, both activating cofactors contain putative zinc fingers. The reason for this dependency is not well established. In the case of pestiviruses, one attractive hypothesis is that this activation step ensures the timely order of molecular events leading first to the DNAJC14-supported NS2-mediated release of NS3, allowing efficient replicase assembly to occur (15, 21, 53). At later time points, without accessible DNAJC14, NS2-3 no longer can adopt the active protease conformation and, as NS2-3 is part of a larger protein complex including structural and nonstructural proteins, supports virion morphogenesis (15, 53). The molecular mechanisms leading to these active conformations required for NS2-3 cleavage are still poorly understood (see below). Structural information regarding their individual precleavage conformation will be essential for a better understanding of this critical step in the life cycle of these viruses.
Our experiments using the GST-Jiv90(WT) rescue cell lines confirm that the Jiv90 domain of DNAJC14 (i.e., in the absence of the full-length protein) is also sufficient to activate the NS2 proteases of different pestiviruses (Fig. 6), most likely by its interaction with the two Jiv90-binding domains in NS2 (53). Our experiments reemphasize that, mechanistically, the cochaperone function of the DNAJC14 J-domain (essential for the association between J-domain proteins with chaperones of the Hsp70 family [85]) is not required for NS2 activation, as proposed earlier (53). The observation that the W39A variant of Jiv90 is unable to activate NS2 despite comparable binding to NS2 (15) suggests that either differences in the folding of the Jiv90 domain render Jiv90(W39A) unable to activate the NS2 autoprotease or wild-type Jiv90 is able to recruit other essential but unknown host cell component(s) to the NS2-3/Jiv90(WT) complex that are not recruited to a NS2-3/Jiv90(W39A) complex. The tools established in this study will be of help to address this question.
The strong DNAJC14 dependency of noncp pestiviruses is mechanistically interesting and provides another example to the growing list of viral proteins which are functionally activated and/or folded by the cellular Hsp90/Hsp70 chaperone systems (86). The importance of the cellular Hsp70 chaperone system in the flaviviral life cycle was recently dissected for the related Dengue virus (DENV) (87). In this study, a direct role for Hsp70 and DNAJB11 in viral RNA synthesis and for Hsp70 and DNAJB6 in virion production was shown (87). While for DENV several DNAJ cochaperones act as proviral factors, DNAJC14 has been shown to act as a broadly active flavivirus restriction factor by interfering with steps required for viral RC formation when it is massively overexpressed (55–57, 87). A similar role during the formation of the minimal pestiviral RC was not observed, since the replication of BVDV NS3-5B replicon RNA in DNAJC14-KO cells was not affected, demonstrating that DNAJC14 is not regulating steps leading to the assembly of the minimal pestiviral replicase (Fig. 7). Accordingly, the minimal set of RC components does not require DNAJC14 for the acquisition of the folded, functional state they possess in the pestiviral replicase. Since other DNAJ cochaperones, such as DNAJB11, recently have been demonstrated to directly regulate flaviviral RNA replication together with the cytosolic Hsp70 (87), it will be interesting to determine if other DNAJ cofactors are required for the assembly of the minimal pestiviral RC.
Interestingly, for yellow fever virus (YFV) it was recently shown that the antiviral activity exerted by DNAJC14 when overexpressed is based on its ability to reduce YFV NS3/4A/2K cleavage, thereby inhibiting YFV RC formation and replication (57). Together with its critical function in modulating pestiviral NS2-3 cleavage, this observation suggests that DNAJC14 modulates processing at viral polyprotein cleavage sites that have to be tightly regulated in order to generate appropriate levels of uncleaved versus cleaved proteins that can serve different functions (14, 57, 62, 88). These differences point to a scenario where the relative levels of chaperones and cochaperones could determine the overall effect on various steps in the viral life cycle. Along these lines, increased levels of DNAJC14 have been demonstrated to cause a deregulation of pestiviral RNA replication that in turn leads to a switch in the biotype from noncp to cp (2, 14). In contrast, elevated levels of DNAJC14 inhibit flaviviral replication (55). Accordingly, essential processes of the viral life cycle may become favored for different viruses as a result of an increased level of a particular cochaperone, while other processes might be suppressed or even activated by the lack of critical chaperone components.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK; ATCC CCL 22) and 293T cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were grown in Dulbecco’s minimum essential medium (DMEM) with 10% horse serum (Fisher Scientific GmbH, Schwerte, Germany), 100 U/ml penicillin, and 100 μg/ml streptomycin. SK6 cells were kindly provided by J. D. Tratschin (Institute of Virology and Immunoprophylaxis, Mittelhäusern, Switzerland) (89) and were cultivated in MEM with 10% fetal calf serum (Thermo Fisher Scientific GmbH, Schwerte, Germany), 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were kept at 37°C and 5% CO2. BVDV-1a strain NADL and BVDV-1b strains NCP7 and CP7 were obtained from E. Dubovi (Cornell University College of Veterinary Medicine, Ithaca, NY) (90). BVDV-1b strains NCP8, CP8 (51), and ncp-4584 (91), BVDV-1a SD-1 (92), CSFV 2.3 Alfort-Tübingen (93), CSFV 2.3 Diepholz (94), BDV-1 X818 (95), BDV-2 reindeer (68), and cp pestivirus strain giraffe (68) and noncp strain PG-2 (96), which both belong to the pestivirus species G, have been described previously. BVDV-2 strains 61/120 and C58644 were obtained from/during routine diagnosis. Bungowannah virus (64) was kindly provided by Peter Kirkland (Elizabeth Macarthur Agricultural Institute, Department of Primary Industries, Menangle, New South Wales, Australia).
Plasmid constructs.
BVDV replicon constructs. The plasmids pBici-388 RLuc-NS3-3′ and pBici-388 RLuc-NS2-3′ have been described (33). The bicistronic Bici-RLuc NS2-3′/NCP7 replicon was constructed as follows. A sequence encoding the C-terminal 38 aa of p7, the entire NS2, and the N terminus of NS3 was amplified using the primers BVDV BsmBI-p7-CT se (5′-CGTCTCCCATGCTAAGAGAGGAAAACACCAAAAAATGGGTC-3′) and BVDV NS3 AgeI ase (5′-ACCGGTTTCCAATCCCCTCCTTACCTTTAGTAGTGCTG-3′), and the PCR product was cloned into pGEM-T vector (Promega, Madison, WI), generating pGEM-T BsmBI-p7-NS2(NCP7)-NS3-AgeI. In addition, the sequence comprising the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES) flanked by FseI and BsmBI restriction sites was amplified using primers RLuc-FseI se (5′-GTACATCAAGAGCTTCGTGGAGCGCGTGCTGAAGAACGAGCAGTAATGGCCGGCC-3′) and IRES-BsmBI ase (5′-GCAGAGCCATGGTATTATCATCGTGTTTTTCAAAGGAAAACCACGTCCCC-3′), with Bici-RLuc NS3-3′ replicon DNA as the template, and cloned into the pGEM-T vector to obtain pGEM-T FseI-IRES-BsmBI. pBici-RLuc NS2-3′/NCP7 was generated by ligating FseI-IRES-BsmBI and BsmBI-p7-NS2(NCP7)-NS3-AgeI fragments into the pBici-RLuc NS3-3′ vector cleaved with FseI and AgeI restriction enzymes.
DNAJC14 guide RNA expression vector. A 20-bp guide sequence (5′-ATGATGTGCAGCCGATGCCAGGG-3′) targeting a DNA sequence within exon 4 of the DNAJC14 gene was selected from predicted high-specificity protospacer adjacent motif (PAM) target sites in the bovine and porcine DNAJC14 gene sequences using the CCTop-CRISPR/Cas9 target online predictor (97). Two complementary oligonucleotides (5′-CACCATGATGTGCAGCCGATGCCAGGG-3′ and 5′-AAACTGGCATCGGCTGCACATCAT-3′) containing the DNAJC14 target sequence and BsmBI ligation sequence were used. One hundred μmol of each oligonucleotide was phosphorylated by T4 polynucleotide kinase (New England Biolabs), annealed, and ligated into the BsmBI-digested plentiCRISPR-v2 vector to generate plentiCRISPRv2-DNAJC14 (73, 98–100).
Lentiviral pWPI-GST-Jiv90Esc plasmid for generation of DNAJC14-KO rescue cell lines expressing GST-Jiv90. To generate pWPI GST-Jiv90(WT)Esc and pWPI GST-Jiv90(W39A)Esc, the GST-Jiv90(WT) or GST-Jiv90(W39A) coding sequence was amplified by PCR using primer SbfI-GST se (5′-CCTGCAGGATGAGCCCTATACTAGGTTATTGGAAAATTAAGGGCC-3′) and primer MluI-Jiv90 ase (5′-ACGCGTCACATCCGTGAACCAAATGAGATGTGATAAGG-3′) (SbfI and MluI sites are underlined) with pcite-GST-Jiv90(WT) or pcite-GST-Jiv90(W39A) as the template DNA (15) and cloned into pGEM-T to generate pGEM-SbfI-GST-Jiv90(WT)-MluI or pGEM-SbfI-GST-Jiv90(W39A)-MluI, respectively. QuikChange mutagenesis was performed with pGEM-SbfI-GST-Jiv90-MluI and pGEM-SbfI-GST-Jiv90(W39A)-MluI to introduce guide RNA escape mutations in the guide RNA binding site (Jiv90Esc) (ATGATGTGTAGTAGGTGTCAAGGCAAACACCGAAGATTT; changed positions are underlined) to obtain pGEM-T-SbfI-Jiv90Esc(WT)-MluI and pGEM-T-SbfI-Jiv90Esc(W39A)-MluI, respectively. The GST-Jiv90Esc sequences were cloned into the lentiviral plasmid vector pWPI msc GUN (a kind gift from T. Pietschmann, Twincore, Hannover, Germany) via SbfI and MluI restriction sites, resulting in the lentiviral plasmids pWPI GST-Jiv90Esc(WT) and pWPI GST-Jiv90Esc(W39A), containing a green fluorescent protein (GFP)-ubiquitin-neomycin phosphotransferase fusion protein as a selectable marker (101).
Production of lentiviral particles and transduction of 293T cells.
Lentiviral particles were produced by transient cotransfection of 2 × 106 293T cells with the guide RNA-carrying plentiCRISPRv2-DNAJC14 vector, a packaging vector (pCMVΔR8.91) (102), and an envelope vector expressing the vesicular stomatitis virus glycoprotein (VSV-G) (pMD.G) (103) using polyethylenimine (PEI) as a transfection reagent according to the manufacturer’s protocol (Polysciences, Inc., Warrington, PA). In detail, 4.5 μg of plentiCRISPRv2-DNAJC14 and 1.5 μg of pCMVΔR8.91 3 μg of pMD.G were mixed with 45 μl PEI solution and cotransfected into 293T cells. At 72 h posttransfection, viral supernatants were collected and passed through 0.2-μm-pore-size filters to ensure removal of any viral aggregates. Lentiviral particles were used for the transduction of MDBK or SK6 cells following selection with puromycin for 2 weeks.
Generation of stable MDBK and SK6 DNAJC14-KO GST-Jiv90 rescue cell lines.
For the generation of stable MDBK and SK6 DNAJC14-KO rescue cells expressing GST-Jiv90, lentiviral gene transfer was used as described for the generation of MDBK and SK6 DNAJC14-KO cells. Briefly, MDBK and SK6 DNAJC14-KO cells in which endogenous DNAJC14 expression was knocked out using plentiCRISPRv2-DNAJC14 expressing a guide RNA specific to the 5′ end of the Jiv90 domain of bovine and porcine DNAJC14 were used for the generation of the respective GST-Jiv90Esc(WT) and Jiv90Esc(W39A) rescue cell lines. Jiv90 expression was restored in these cells by lentiviral transduction with either pWPI GST-Jiv90Esc(WT) or pWPI GST-Jiv90Esc(W39A). Lentiviral particles were generated by PEI-mediated cotransfection of pCMV ΔR8.91, pMD.G, and either pWPI GST-Jiv90Esc(WT) or pWPI GST-Jiv90Esc(W39A) at a ratio of 3∶1∶3 into 293T cells. Lentiviral particles were collected 48 h posttransfection and used to transduce MDBK and SK6 DNAJC14-KO cells using a Polybrene transduction protocol. Selection was carried out in the presence of 1 mg/ml G418 (Thermo Fisher Scientific GmbH, Schwerte, Germany).
Clonal isolation of cell lines.
Isolation of clonal MDBK DNAJC14-KO and SK6 DNAJC14-KO cell lines with specific DNAJC14 gene modifications and the MDBK DNAJC14-KO GST-Jiv90 and SK6 DNAJC14-KO GST-Jiv90 rescue cell lines expressing GST-Jiv90(WT) or GST-Jiv90(W39A) was achieved by serial dilutions, followed by an expansion period to establish clonal cell lines.
Functional testing of MDBK and SK6 DNAJC14-KO cell lines by noncp pestiviral infection assay.
Cells transduced with lentiviruses expressing DNAJC14-specific single guide RNAs (sgRNAs) to mediate genomic (micro)deletions in the DNAJC14 gene were functionally screened for their inability to support replication of noncp pestiviral strains after infection. In detail, MDBK DNAJC14-KO cell lines were tested by infecting 3 × 104 cells with noncp BVDV-1 strain NCP7 or NCP8 at an MOI of 3, and SK6 DNAJC14-KO cell lines were analyzed by infecting 3 × 104 cells with noncp CSFV Alfort-Tübingen at an MOI of 3. Virus detection occurred after 72 h by indirect immunofluorescence (IF) analysis using MAb 8.12.7 directed against NS3 of BVDV (90) or MAb C16 against CSFV NS2-3/NS3 (104). Only cell clones which did not support noncp pestiviral replication were further analyzed for DNAJC14-specific genomic (micro)deletions.
Detection of Jiv90-specific genome alterations.
Targeted DNAJC14-specific genome modifications were detected by Sanger sequencing. Genomic DNA was isolated from either MDBK DNAJC14-KO or SK6 DNAJC14-KO cells using the Quick-DNA miniprep kit (Zymo Research Europe GmbH, Freiburg, Germany) and served as the template to amplify bovine- or porcine-specific DNAJC14-specific sequences by DNAJC14 intron-specific primers (for MDBK genomic DNA amplification, Bov-intron se [5′-CTCCCTTTGTCCCTCACTCAATATCCTGAC-3′] and Bov-intron ase [5′-CAGTTCCTTAATAATACGTTAAGACTACC-3′]; for SK6 genomic DNA amplification, Pig-intron se [5′-GCAGAGCCAGAGCAATTAG-3′] and Pig-intron ase [5′-CACTTGTTACATACGCCTAC-3′]) using a Taq DNA polymerase kit (Segenetic, Borken Germany). Amplicons were subcloned into pGEM-T vector (Promega), and individual colonies were sequenced to reveal the clonal genotype. Genome alterations were identified by analyzing the sequences using Vector NTI software (Invitrogen, Karlsruhe, Germany).
The gel-purified intron-specific amplicons were used as templates for seminested PCR using DNAJC14 exon- and intron-specific primers (for MDBK with DNAJC14-KO-specific oligonucleotide, Bov-Exon se [5′-GCAATGAATACGATGATGTGCAGCCGATGCCA-3′] and Bov-intron ase [5′-CAGTTCCTTAATAATACGTTAAGACTACC-3′]; for SK6 with DNAJC14-KO-specific oligonucleotide, Pig-Exon se [5′-CTATGATGCAGCCGATGCCAG-3′] and Pig-intron ase [5′-GCAGAGCCAGAGCAATTAG-3′]) to further verify KO cell clones.
In vitro transcription and RNA electroporation.
In vitro transcription of Bici-388 RLuc-NS3-3′ and Bici-388 RLuc-NS2-3′ replicon RNAs and RNA electroporation into MDBK cells have been described (105). The electroporated cells were resuspended in 0.6 ml medium and seeded immediately postelectroporation. At the indicated time points, cells were processed for Renilla luciferase assay or immunofluorescence assay.
Nucleotide sequencing.
DNA sequencing was performed at LGC Sequencing Services (Berlin, Germany). Sequences were further analyzed by using Vector NTI software (Invitrogen, Karlsruhe, Germany).
SDS-PAGE and immunoblotting.
Proteins were separated in polyacrylamide-Tricine gels (8, 10, or 12% polyacrylamide) (106) and transferred onto nitrocellulose membrane (Pall, Pensacola, FL). The membrane was blocked with 5% (wt/vol) dried skim milk in phosphate-buffered saline (PBS) with 0.05% (vol/vol) Tween 20 (Invitrogen, Karlsruhe, Germany). For antigen detection, the indicated primary antibodies were used and detected by peroxidase-coupled species-specific secondary antibodies. The Western Lightning chemiluminescence reagent (PerkinElmer, Boston, MA) was applied.
DNAJC14 antibody production.
Recombinant His-Ubi-Flag-Jiv90 fusion protein was expressed and purified from Escherichia coli using His-agarose chromatography (Qiagen, Hilden, Germany). The DNAJC14-specific antibody was produced using standard techniques (107) in accordance with the legal regulations of the German Animal Welfare jurisdiction (Tierschutzgesetz). The immunization of mice was subject to authorization and was recorded after approval under reference number Az GZ54-19620/15cGI18/7 JLU 077 ID 266 at the Hessian Ministry for Environment, Climate Protection, Agriculture, and Consumer Protection. Sera were tested for immunoreactivity against GST-Jiv90 expressed from pcite-GST-Jiv90 in BHK21 cells infected with MVA-T7 vaccinia virus as described previously (15). Monoclonal antibodies (MAbs) were purified by standard protein G chromatography (GE Healthcare, Chicago, IL, USA). Monoclonal antibodies were screened for reactivity against Jiv-Flag and GST-Jiv fusion proteins expressed in BHK cells. MAb 9.4E was identified to specifically recognize a linear Jiv epitope in Jiv90.
Virus infection and titration.
Infection of cells with BVDV or BDV strains was performed with either 500 μl of cell culture supernatant derived from approximately 3 × 106 infected cells or at the indicated MOI as described previously (20). To determine virus titers, cell culture supernatants of infected cells were harvested at the indicated time points postinfection and filtered through a 0.2-μm cellulose filter (Sartorius). The 50% tissue culture infection dose (TCID50 per milliliter) of viral supernatants was determined in three replicates by endpoint titration. Viral infection with BVDV or BDV was detected 72 hpi by IF using MAb 8.12.7 directed against NS3/NS2-3, kindly provided by E. J. Dubovi (Cornell University, Ithaca, NY) (90). Infection of SK6 cells with CSFV isolates was detected by indirect IF with MAb C16 directed against CSFV NS2-3/NS3 (104, 108).
Luciferase assay.
Bicistronic reporter constructs encoding Npro-Renilla luciferase in the 5′ ORF and one monomer of ubiquitin followed by either NS2-5B or by NS3-5B in the 3′ ORF were used, and RNA replication efficiencies were determined at the indicated time points (2 h, 24 h, and 48 h) as described previously (33). Luciferase activities were measured with a Junior LB 9509 portable tube luminometer (Junior LB9509; Berthold, Bad Wildbad, Germany).
Immunofluorescence assay.
Immunofluorescence assays were performed as described previously (33). Briefly, cells were fixed with 4% paraformaldehyde for 20 min at 4°C, permeabilized with 0.5% (wt/vol) N-octylglucopyranoside in PBS, and stained with mouse anti-BVDV NS2-3/NS3 MAb 8.12.7 antibody, kindly provided by E. J. Dubovi (Cornell University, Ithaca, NY), to detect BVDV-1 infection. Infection with CSFV was detected by indirect IF with MAb C16 directed against NS2-3/NS3 (104, 108), while infection with Bungowannah virus was detected by a virus-specific porcine antiserum kindly provided by P. Kirkland (Elizabeth Macarthur Agricultural Institute, Department of Primary Industries, Menangle, New South Wales, Australia). For detection of GST-Jiv90 in rescue cell lines, mouse monoclonal antibodies directed against GST (anti-GST; GE Healthcare) or against the Jiv90 domain (anti-Jiv90; this study) were used. Goat anti-mouse IgGs conjugated with either Cy3 or Alexa488 served as the secondary antibodies. Images were obtained with a Zeiss Axio Observer.Z1 fluorescence microscope (Zeiss, Göttingen, Germany).
Infections of MDBK and SK6 cells with selected pestivirus isolates were performed at an MOI of 1 on 24-well plates, which showed approximately 70% to 80% confluent monolayers. Viral infection was performed for 1 h at 37°C with gentle shaking. Subsequently, virus inocula were removed, and infected cells were washed three times with PBS and then incubated in 2 ml medium. After 72 h, medium was removed from infected cell cultures, and cells were washed three times with PBS, dried (5 min, room temperature), and subsequently heat fixed (3 h, 80°C). The MAb C16 (Institute of Virology, University of Veterinary Medicine, Hannover, Germany) served as the primary antibody (diluted 1:50 in 1× PBS, 0.05% Tween 20) to detect CSFV, BDV, BVDV-2, giraffe pestivirus, and PG-2 (2 h, 37°C). Staining (1 h, 37°C) was performed with a Cy3-labeled goat anti-mouse IgG antibody (diluted 1:800; Dianova). Infection with Bungowannah virus was detected with a virus-specific porcine antiserum (1:12,000, 2 h, 37°C) kindly provided by P. Kirkland (Elizabeth Macarthur Agricultural Institute, Department of Primary Industries, Menangle, New South Wales, Australia) (64, 85). A Cy3-labeled goat anti-swine IgG conjugate (diluted 1:250; Dianova) was used as a secondary antibody (1 h, 37°C). Nuclei of cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml; Cell Signaling) for 10 min at room temperature.
ACKNOWLEDGMENTS
We especially thank J. Larsen for excellent technical assistance. We are grateful to E. J. Dubovi (Cornell University, Ithaca, NY) for antibody 8.12.7. The pWPI msc GUN plasmid was a generous gift from Thomas Pietschmann (Twincore, Hannover, Germany). We are grateful to Didier Trono (University of Geneva, Switzerland) for providing us with the packaging vector (pCMVΔR8.91) and the vector expressing VSV-G (pMD.G). We also thank Danilo Dubrau and members of the Tautz laboratory for helpful discussions.
This work was supported by intramural funding from the University of Luebeck (N.T.) and the University of Veterinary Medicine Hannover (P.B.).
Authors made the following contributions: conceived and designed the experiments, O.I., A.P., P.B., and N.T.; generated reagents, E.L.; performed the experiments, O.I., B.B., and A.P.; analyzed the data, O.I., A.P., P.B., and N.T.; wrote the paper, O.I. and N.T.
REFERENCES
- 1.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT, ICTV Report Committee . 2017. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tautz N, Tews BA, Meyers G. 2015. The molecular biology of pestiviruses. Adv Virus Res 93:47–160. doi: 10.1016/bs.aivir.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Collett MS, Larson R, Gold C, Strick D, Anderson DK, Purchio AF. 1988. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology 165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Thiel HJ, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Stark R, Meyers G, Rümenapf T, Thiel H-J. 1993. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol 67:7088–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callens N, Brügger B, Bonnafous P, Drobecq H, Gerl MJ, Krey T, Roman-Sosa G, Rümenapf T, Lambert O, Dubuisson J, Rouillé Y. 2016. Morphology and molecular composition of purified bovine viral diarrhea virus envelope. PLoS Pathog 12:e1005476. doi: 10.1371/journal.ppat.1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiel H-J, Stark R, Weiland E, Rümenapf T, Meyers G. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J Virol 65:4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H-J. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169–1171. [DOI] [PubMed] [Google Scholar]
- 9.Elbers K, Tautz N, Becher P, Stoll D, Rümenapf T, Thiel HJ. 1996. Processing in the pestivirus E2-NS2 region: identification of the nonstructural proteins p7 and E2p7. J Virol 70:4131–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada T, Tautz N, Thiel H-J. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J Virol 74:9498–9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Largo E, Gladue DP, Huarte N, Borca MV, Nieva JL. 2014. Pore-forming activity of pestivirus p7 in a minimal model system supports genus-specific viroporin function. Antiviral Res 101:30–36. doi: 10.1016/j.antiviral.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 12.de Moerlooze L, Desport M, Renard A, Lecomte C, Brownlie J, Martial JA. 1990. The coding region for the 54-kDa protein of several pestiviruses lacks host insertions but reveals a “zinc finger-like” domain. Virology 177:812–815. [DOI] [PubMed] [Google Scholar]
- 13.Desport M, Collins ME, Brownlie J. 1998. Genomic instability in BVDV: an examination of the sequence and structural influences on RNA recombination. Virology 246:352–361. doi: 10.1006/viro.1998.9219. [DOI] [PubMed] [Google Scholar]
- 14.Lackner T, Müller A, Pankraz A, Becher P, Thiel H-J, Gorbalenya AE, Tautz N. 2004. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J Virol 78:10765–10775. doi: 10.1128/JVI.78.19.10765-10775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackner T, Müller A, König M, Thiel H-J, Tautz N. 2005. Persistence of bovine viral diarrhea virus is determined by a cellular cofactor of a viral autoprotease. J Virol 79:9746–9755. doi: 10.1128/JVI.79.15.9746-9755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulin HR, Seuberlich T, Bauhofer O, Bennett LC, Tratschin JD, Hofmann MA, Ruggli N. 2007. Nonstructural proteins NS2-3 and NS4A of classical swine fever virus: essential features for infectious particle formation. Virology 365:376–389. doi: 10.1016/j.virol.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 17.Tautz N, Meyers G, Thiel H-J. 1993. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology 197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- 18.Agapov EV, Murray CL, Frolov I, Qu L, Myers TM, Rice CM. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J Virol 78:2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lattwein E, Klemens O, Schwindt S, Becher P, Tautz N. 2012. Pestivirus virion morphogenesis in the absence of uncleaved nonstructural protein 2-3. J Virol 86:427–437. doi: 10.1128/JVI.06133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemens O, Dubrau D, Tautz N. 2015. Characterization of the determinants of NS2-3-independent virion morphogenesis of pestiviruses. J Virol 89:11668–11680. doi: 10.1128/JVI.01646-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubrau D, Tortorici MA, Rey FA, Tautz N. 2017. A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis. PLoS Pathog 13:e1006134. doi: 10.1371/journal.ppat.1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bazan JF, Fletterick RJ. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A 85:7872–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM. 1989. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res 17:3889–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17:4713–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrens SE, Grassmann CW, Thiel HJ, Meyers G, Tautz N. 1998. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol 72:2364–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grassmann CW, Isken O, Behrens SE. 1999. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol 73:9196–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tautz N, Kaiser A, Thiel H-J. 2000. NS3 serine protease of bovine viral diarrhea virus: characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology 273:351–363. doi: 10.1006/viro.2000.0425. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Mendez E, Caron PR, Lin C, Murcko MA, Collett MS, Rice CM. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol 71:5312–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiskerchen M, Collett MS. 1991. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 184:341–350. [DOI] [PubMed] [Google Scholar]
- 30.Liang D, Chen L, Ansari IH, Gil LH, Topliff CL, Kelling CL, Donis RO. 2009. A replicon trans-packaging system reveals the requirement of nonstructural proteins for the assembly of bovine viral diarrhea virus (BVDV) virion. Virology 387:331–340. doi: 10.1016/j.virol.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Weiskircher E, Aligo J, Ning G, Konan KV. 2009. Bovine viral diarrhea virus NS4B protein is an integral membrane protein associated with Golgi markers and rearranged host membranes. Virol J 6:185. doi: 10.1186/1743-422X-6-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grassmann CW, Isken O, Tautz N, Behrens SE. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J Virol 75:7791–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isken O, Langerwisch U, Schonherr R, Lamp B, Schroder K, Duden R, Rümenapf TH, Tautz N. 2014. Functional characterization of bovine viral diarrhea virus nonstructural protein 5A by reverse genetic analysis and live cell imaging. J Virol 88:82–98. doi: 10.1128/JVI.01957-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed KE, Gorbalenya AE, Rice CM. 1998. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol 72:6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffens S, Thiel H-J, Behrens S-E. 1999. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J Gen Virol 80:2583–2590. doi: 10.1099/0022-1317-80-10-2583. [DOI] [PubMed] [Google Scholar]
- 36.Collett MS, Larson R, Belzer S, Retzel E. 1988. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology 165:200–208. [DOI] [PubMed] [Google Scholar]
- 37.Lai VC, Kao CC, Ferrari E, Park J, Uss AS, Wright-Minogue J, Hong Z, Lau JY. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol 73:10129–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao CC, Del Vecchio AM, Zhong W. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1–7. [DOI] [PubMed] [Google Scholar]
- 39.Zhong W, Gutshall LL, Del Vecchio AM. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol 72:9365–9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moennig V, Frey H-R, Liebler E, Polenz P, Liess B. 1990. Reproduction of mucosal disease with cytopathogenic bovine viral diarrhoea virus selected in vitro. Vet Rec 127:200–203. [PubMed] [Google Scholar]
- 41.Brownlie J, Clarke MC, Howard CJ. 1984. Experimental production of fatal mucosal disease in cattle. Vet Rec 114:535–536. [DOI] [PubMed] [Google Scholar]
- 42.Bolin SR, McClurkin AW, Cutlip RC, Coria MF. 1985. Severe clinical disease induced in cattle persistently infected with noncytopathogenic bovine viral diarrhea virus by superinfection with cytopathogenic bovine viral diarrhea virus. Am J Vet Res 46:573–576. [PubMed] [Google Scholar]
- 43.Hoff HS, Donis RO. 1997. Induction of apoptosis and cleavage of poly(ADP-ribose) polymerase by cytopathic bovine viral diarrhea virus infection. Virus Res 49:101–113. [DOI] [PubMed] [Google Scholar]
- 44.Jordan R, Wang L, Graczyk TM, Block TM, Romano PR. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J Virol 76:9588–9599. doi: 10.1128/JVI.76.19.9588-9599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweizer M, Peterhans E. 1999. Oxidative stress in cells infected with bovine viral diarrhoea virus: a crucial step in the induction of apoptosis. J Gen Virol 80:1147–1155. doi: 10.1099/0022-1317-80-5-1147. [DOI] [PubMed] [Google Scholar]
- 46.Brownlie J, Clarke MC, Howard CJ. 1989. Experimental infection of cattle in early pregnancy with a cytopathic strain of bovine virus diarrhoea virus. Res Vet Sci 46:307–311. [PubMed] [Google Scholar]
- 47.Tautz N, Harada T, Kaiser A, Rinck G, Behrens SE, Thiel H-J. 1999. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J Virol 73:9422–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B. 1996. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol 70:8606–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendez E, Ruggli N, Collett MS, Rice CM. 1998. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol 72:4737–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinck G, Birghan C, Harada T, Meyers G, Thiel H-J, Tautz N. 2001. A cellular J-domain protein modulates polyprotein processing and cytopathogenicity of a pestivirus. J Virol 75:9470–9482. doi: 10.1128/JVI.75.19.9470-9482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller A, Rinck G, Thiel H-J, Tautz N. 2003. Cell-derived sequences located in the structural genes of a cytopathogenic pestivirus. J Virol 77:10663–10669. doi: 10.1128/JVI.77.19.10663-10669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becher P, Meyers G, Shannon AD, Thiel H-J. 1996. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J Virol 70:2992–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lackner T, Thiel H-J, Tautz N. 2006. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc Natl Acad Sci U S A 103:1510–1515. doi: 10.1073/pnas.0508247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallei A, Blome S, Gilgenbach S, Tautz N, Moennig V, Becher P. 2008. Cytopathogenicity of classical swine fever virus correlates with attenuation in the natural host. J Virol 82:9717–9729. doi: 10.1128/JVI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi Z, Sperzel L, Nurnberger C, Bredenbeek PJ, Lubick KJ, Best SM, Stoyanov CT, Law LM, Yuan Z, Rice CM, MacDonald MR. 2011. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog 7:e1001255. doi: 10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi Z, Yuan Z, Rice CM, MacDonald MR. 2012. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J Virol 86:11815–11832. doi: 10.1128/JVI.01022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bozzacco L, Yi Z, Andreo U, Conklin CR, Li MM, Rice CM, MacDonald MR. 2016. Chaperone-assisted protein folding is critical for yellow fever virus NS3/4A cleavage and replication. J Virol 90:3212–3228. doi: 10.1128/JVI.03077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382. doi: 10.1016/j.virol.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 59.Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 60.Kim MY, Shu Y, Carsillo T, Zhang J, Yu L, Peterson C, Longhi S, Girod S, Niewiesk S, Oglesbee M. 2013. hsp70 and a novel axis of type I interferon-dependent antiviral immunity in the measles virus-infected brain. J Virol 87:998–1009. doi: 10.1128/JVI.02710-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung J, Kim J, Roh SH, Jun I, Sampson RD, Gee HY, Choi JY, Lee MG. 2016. The HSP70 co-chaperone DNAJC14 targets misfolded pendrin for unconventional protein secretion. Nat Commun 7:11386. doi: 10.1038/ncomms11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamp B, Riedel C, Roman-Sosa G, Heimann M, Jacobi S, Becher P, Thiel H-J, Rümenapf T. 2011. Biosynthesis of classical swine fever virus nonstructural proteins. J Virol 85:3607–3620. doi: 10.1128/JVI.02206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Postel A, Hansmann F, Baechlein C, Fischer N, Alawi M, Grundhoff A, Derking S, Tenhundfeld J, Pfankuche VM, Herder V, Baumgartner W, Wendt M, Becher P. 2016. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci Rep 6:27735. doi: 10.1038/srep27735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirkland PD, Frost MJ, Finlaison DS, King KR, Ridpath JF, Gu X. 2007. Identification of a novel virus in pigs–Bungowannah virus: a possible new species of pestivirus. Virus Res 129:26–34. doi: 10.1016/j.virusres.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Hause BM, Collin EA, Peddireddi L, Yuan F, Chen Z, Hesse RA, Gauger PC, Clement T, Fang Y, Anderson G. 2015. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J Gen Virol 96:2994–2998. doi: 10.1099/jgv.0.000251. [DOI] [PubMed] [Google Scholar]
- 66.Liu L, Kampa J, Belak S, Baule C. 2009. Virus recovery and full-length sequence analysis of atypical bovine pestivirus Th/04_KhonKaen. Vet Microbiol 138:62–68. doi: 10.1016/j.vetmic.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 67.Schirrmeier H, Strebelow G, Depner K, Hoffmann B, Beer M. 2004. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J Gen Virol 85:3647–3652. doi: 10.1099/vir.0.80238-0. [DOI] [PubMed] [Google Scholar]
- 68.Avalos-Ramirez R, Orlich M, Thiel H-J, Becher P. 2001. Evidence for the presence of two novel pestivirus species. Virology 286:456–465. doi: 10.1006/viro.2001.1001. [DOI] [PubMed] [Google Scholar]
- 69.Vilcek S, Ridpath JF, Van Campen H, Cavender JL, Warg J. 2005. Characterization of a novel pestivirus originating from a pronghorn antelope. Virus Res 108:187–193. doi: 10.1016/j.virusres.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, Lee B, Che X, Quan PL, Lipkin WI. 2014. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 5:e01933-14. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P, Becher P. 2017. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol 98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richter M, Reimann I, Schirrmeier H, Kirkland PD, Beer M. 2014. The viral envelope is not sufficient to transfer the unique broad cell tropism of Bungowannah virus to a related pestivirus. J Gen Virol 95:2216–2222. doi: 10.1099/vir.0.065995-0. [DOI] [PubMed] [Google Scholar]
- 73.Mali P, Esvelt KM, Church GM. 2013. Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirkland PD, Read AJ, Frost MJ, Finlaison DS. 2015. Bungowannah virus–a probable new species of pestivirus–what have we found in the last 10 years? Anim Health Res Rev 16:60–63. doi: 10.1017/S1466252315000031. [DOI] [PubMed] [Google Scholar]
- 75.Tautz N, Meyers G, Stark R, Dubovi EJ, Thiel H-J. 1996. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol 70:7851–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Becher P, Tautz N. 2011. RNA recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol 8:216–224. [DOI] [PubMed] [Google Scholar]
- 77.Meyers G, Tautz N, Dubovi EJ, Thiel H-J. 1991. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology 180:602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyers G, Rümenapf T, Thiel H-J. 1989. Ubiquitin in a togavirus. Nature 341:491. doi: 10.1038/341491a0. [DOI] [PubMed] [Google Scholar]
- 79.Kümmerer BM, Meyers G. 2000. Correlation between point mutations in NS2 and the viability and cytopathogenicity of bovine viral diarrhea virus strain Oregon analyzed with an infectious cDNA clone. J Virol 74:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kümmerer B, Stoll D, Meyers G. 1998. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations. J Virol 72:4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagy PD, Pogany J. 2011. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- 83.Dong D, Lei M, Hua P, Pan YH, Mu S, Zheng G, Pang E, Lin K, Zhang S. 2017. The genomes of two bat species with long constant frequency echolocation calls. Mol Biol Evol 34:20–34. doi: 10.1093/molbev/msw231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schregel V, Jacobi S, Penin F, Tautz N. 2009. Hepatitis C virus NS2 is a protease stimulated by cofactor domains in NS3. Proc Natl Acad Sci U S A 106:5342–5347. doi: 10.1073/pnas.0810950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finlaison DS, Cook RW, Srivastava M, Frost MJ, King KR, Kirkland PD. 2010. Experimental infections of the porcine foetus with Bungowannah virus, a novel pestivirus. Vet Microbiol 144:32–40. doi: 10.1016/j.vetmic.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 86.Geller R, Taguwa S, Frydman J. 2012. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta 1823:698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J. 2015. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163:1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yost SA, Marcotrigiano J. 2013. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr Opin Virol 3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kasza L, Shadduck JA, Christofinis GJ. 1972. Establishment, viral susceptibility and biological characteristics of a swine kidney cell line SK-6. Res Vet Sci 13:46–51. [PubMed] [Google Scholar]
- 90.Corapi WV, Donis RO, Dubovi EJ. 1988. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea infections. J Virol 62:2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Becher P, Orlich M, Thiel H-J. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J Virol 75:6256–6264. doi: 10.1128/JVI.75.14.6256-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng R, Brock KV. 1992. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathogenic bovine viral diarrhea virus strain SD-1. Virology 191:867–879. doi: 10.1016/0042-6822(92)90262-N. [DOI] [PubMed] [Google Scholar]
- 93.Meyers G, Rümenapf T, Thiel H-J. 1989. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology 171:555–567. [DOI] [PubMed] [Google Scholar]
- 94.Postel A, Schmeiser S, Bernau J, Meindl-Boehmer A, Pridotkas G, Dirbakova Z, Mojzis M, Becher P. 2012. Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet Res 43:50. doi: 10.1186/1297-9716-43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Becher P, Shannon AD, Tautz N, Thiel H-J. 1994. Molecular characterization of border disease virus, a pestivirus from sheep. Virology 198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 96.Becher P, Avalos Ramirez R, Orlich M, Cedillo Rosales S, König M, Schweizer M, Stalder H, Schirrmeier H, Thiel H-J. 2003. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology 311:96–104. doi: 10.1016/S0042-6822(03)00192-2. [DOI] [PubMed] [Google Scholar]
- 97.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. 2015. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pham HM, Arganaraz ER, Groschel B, Trono D, Lama J. 2004. Lentiviral vectors interfering with virus-induced CD4 down-modulation potently block human immunodeficiency virus type 1 replication in primary lymphocytes. J Virol 78:13072–13081. doi: 10.1128/JVI.78.23.13072-13081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 103.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. [DOI] [PubMed] [Google Scholar]
- 104.Peters W, Greiser-Wilke I, Moennig V, Liess B. 1986. Preliminary serological characterization of bovine viral diarrhoea virus strains using monoclonal antibodies. Vet Microbiol 12:195–200. [DOI] [PubMed] [Google Scholar]
- 105.Isken O, Langerwisch U, Jirasko V, Rehders D, Redecke L, Ramanathan H, Lindenbach BD, Bartenschlager R, Tautz N. 2015. A conserved NS3 surface patch orchestrates NS2 protease stimulation, NS5A hyperphosphorylation and HCV genome replication. PLoS Pathog 11:e1004736. doi: 10.1371/journal.ppat.1004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. [DOI] [PubMed] [Google Scholar]
- 107.Harlow E, Lane D. 1998. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 108.Greiser-Wilke I, Dittmar KE, Liess B, Moennig V. 1992. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J Gen Virol 73:47–52. doi: 10.1099/0022-1317-73-1-47. [DOI] [PubMed] [Google Scholar]
- 109.Postel A, Becher P, Haas L, Wendt M. 2018. The newly discovered “atypical porcine pestivirus” (APPV): an old player in the “shaking piglets” disease complex? Tierarztl Prax Ausg G Grosstiere Nutztiere 46:261–270. doi: 10.15653/TPG-180053. [DOI] [PubMed] [Google Scholar]