Bacillus velezensis is a Gram-positive plant-beneficial bacterium which is widely used in agriculture. Additionally, Bacillus spp. are some of the model organisms used in the study of biofilms, and as such, the molecular networks and regulation systems of biofilm formation are well characterized. However, the molecular processes involved in root colonization by plant-beneficial Bacillus strains remain largely unknown. Here, we showed that WTAs play important roles in the plant root colonization process. The loss of the gtaB gene affects the ability of B. velezensis SQR9 to sense plant polysaccharides, which are important environmental cues that trigger biofilm formation and colonization in the rhizosphere. This knowledge provides new insights into the Bacillus root colonization process and can help improve our understanding of plant-rhizobacterium interactions.
KEYWORDS: Bacillus velezensis SQR9, UDP-glucose, biofilm formation, root colonization, wall teichoic acids
ABSTRACT
Rhizosphere colonization by plant growth-promoting rhizobacteria (PGPR) along plant roots facilitates the ability of PGPR to promote plant growth and health. Thus, an understanding of the molecular mechanisms of the root colonization process by plant-beneficial Bacillus strains is essential for the use of these strains in agriculture. Here, we observed that an sfp gene mutant of the plant growth-promoting rhizobacterium Bacillus velezensis SQR9 was unable to form normal biofilm architecture, and differential protein expression was observed by proteomic analysis. A minor wall teichoic acid (WTA) biosynthetic protein, GgaA, was decreased over 4-fold in the Δsfp mutant, and impairment of the ggaA gene postponed biofilm formation and decreased cucumber root colonization capabilities. In addition, we provide evidence that the major WTA biosynthetic enzyme GtaB is involved in both biofilm formation and root colonization. The deficiency in biofilm formation of the ΔgtaB mutant may be due to an absence of UDP-glucose, which is necessary for the synthesis of biofilm matrix exopolysaccharides (EPS). These observations provide insights into the root colonization process by a plant-beneficial Bacillus strain, which will help improve its application as a biofertilizer.
IMPORTANCE Bacillus velezensis is a Gram-positive plant-beneficial bacterium which is widely used in agriculture. Additionally, Bacillus spp. are some of the model organisms used in the study of biofilms, and as such, the molecular networks and regulation systems of biofilm formation are well characterized. However, the molecular processes involved in root colonization by plant-beneficial Bacillus strains remain largely unknown. Here, we showed that WTAs play important roles in the plant root colonization process. The loss of the gtaB gene affects the ability of B. velezensis SQR9 to sense plant polysaccharides, which are important environmental cues that trigger biofilm formation and colonization in the rhizosphere. This knowledge provides new insights into the Bacillus root colonization process and can help improve our understanding of plant-rhizobacterium interactions.
INTRODUCTION
Rhizobacteria associated with plant root may provide beneficial effects to their host plants and have been widely used in agriculture (1, 2). Bacillus spp. are typical biocontrol agents that can suppress soilborne pathogens and promote plant growth (3). Their biological control of soilborne pathogens involves various mechanisms, which include antibiosis, competition for ecological niches or substrates, production of inhibitory allelochemicals, and the induction of systemic resistance (ISR) (4–8). Bacillus velezensis SQR9 (formerly Bacillus amyloliquefaciens SQR9) is a well-investigated plant growth-promoting rhizobacterial (PGPR) strain with strong root colonization capabilities and is commercially used as a biocontrol bacterium, being especially efficient against soilborne pathogens (9, 10).
Microbial colonization on the plant roots is a crucial step in the ability of rhizobacteria to exert their beneficial effects on plants, which mainly depend on their ability to form biofilms (11, 12). Biofilms are the conglomeration of multicellular community attached to a surface and held together in a self-produced polymeric materials (13). The biofilm polymeric matrix is extremely important, which provides complex architectural structure for multicellular community (14, 15). The major components produced by Bacillus species in biofilm are exopolysaccharides (EPS) and amyloid-forming protein TasA, encoded by the epsA-epsO operon (epsA-O) and tapA-sipW-tasA operon, respectively (15). In Bacillus subtilis, biofilm formation is triggered by several environmental cues (16, 17). It has been reported that plant polysaccharides (arabinogalactan, pectin, and xylan) served as signals and as the substrates for B. subtilis matrix synthesis, which then stimulate root colonization (11, 18).
Despite intensive studies of the molecular mechanisms of B. subtilis biofilm formation, its role in plant root colonization has not been fully characterized (11). Detection and identification of key proteins or genes involved in in situ root colonization can improve our knowledge of rhizobacterial behavior on the root. Previous studies show the important role for wall teichoic acids (WTAs) in adherence of the Gram-positive bacteria to host cells in vitro (19). Hussain et al. (20) discovered that teichoic acids strengthen the adhesion of Staphylococcus epidermidis to immobilized fibronectin. Moreover, it has been reported that WTAs play a key role in the early stage of biofilm development and that d-alanine-modified teichoic acids are necessary for adherence to polar and nonpolar surfaces (21). In B. subtilis, WTAs are composed of two kinds of forms, major and minor WTAs (22). The biosynthetic genes of the major WTA have been identified, including tagABDEFOP and gtaB (23, 24). The minor WTA form is synthesized by GgaA and GgaB, which consists of glucosyl-N-acetylgalactosamine 1-phosphate (GlcGalNAcP) (23). The role of WTAs in biofilm formation and root colonization, however, has remained elusive.
The attachment of rhizobacteria to the roots of their host plants is considered the initial step in the rhizosphere colonization process. Several WTA polymers and surface-exposed proteins have been previously implicated in Staphylococcus aureus attachment to nasal epithelial cells (23, 25). In this study, we compare the proteomes of the wild-type and sfp mutant strains using the high-throughput isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomics approach. We identified two WTAs involved in in situ root attachment. We present evidence that two WTA biosynthetic enzymes, GgaA and GtaB, are involved in root colonization and biofilm formation. This research enhances the understanding of PGPR root colonization mechanisms and may help improve the application of Bacillus rhizobacteria as a biofertilizer.
RESULTS
Mutations in the sfp gene affect biofilm architecture and protein expression.
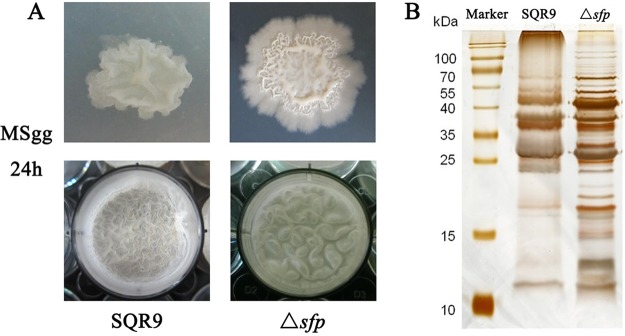
The sfp gene encodes a 4′-phosphopantetheinyl transferase, an enzyme that catalyzes a necessary processing step in the biosynthesis of nonribosomal synthesized peptides (26, 27). Previous studies demonstrated that Sfp is crucially involved in the production of cyclic lipopeptides and polyketides in Bacillus spp. (2, 28); laboratory strain PY79 carries a mutation in the sfp gene and was shown to form atypical surface-associated biofilm structures (15, 29). In addition, the sfp mutant strains of B. velezensis FZB42 were simultaneously affected in their ability to form biofilm and to colonize plant roots (30). Here, we also observed that an sfp gene mutant of SQR9 was unable to form biofilm with normal architecture (Fig. 1A). The sfp deletion mutant exhibited significantly lower levels of wrinkling on both the colony surface and pellicles (see Fig. S1 in the supplemental material). Since several studies indicated that the onset of protein production was correlated with the architecture of biofilms in different developmental stages, we hypothesized that the protein composition would be different between the biofilms of the wild-type and sfp mutant strains. Therefore, we investigated the profiles of biofilm matrix proteins from the wild-type and sfp mutant strains by SDS-PAGE analysis. Interestingly, the protein profiles in the biofilm matrix were distinct between the sfp mutant and wild-type strains (Fig. 1B). These results showed that the effect of the sfp deletion on the biofilm and protein composition in the biofilm matrix may be linked.
FIG 1.
The biofilm architecture and protein profiles were significantly different between the sfp mutant and wild-type strains. (A) The complex colony morphology and microtiter plate assay of biofilm formation by the sfp mutant and wild-type strains in MSgg medium at 24 h. (B) Biofilm matrix protein profiles were distinct between the sfp mutant and wild-type strains. SDS-PAGE gel was stained with sliver staining. Lane Marker, protein molecular weight markers in kilodaltons; lane SQR9, protein extraction from biofilm formed by wild-type SQR9; lane Δsfp, protein extraction from biofilm formed by sfp mutant.
SQR9 wild-type and sfp mutant cells showed different proteomic profiles.
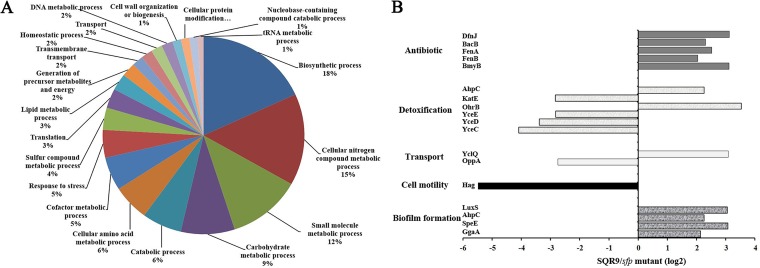
To further understand the physiological changes that resulted in the change in biofilm architecture in the sfp mutant, we compared the complete proteomes of wild-type and mutant strains by using an iTRAQ analysis. A total of 765 common proteins were identified, of which 763 were quantified (see the supplemental material). In total, 87 and 107 proteins were upregulated (SQR9 sfp mutant, >4) and downregulated (SQR9 sfp mutant, <4) with treatment (P < 0.05), and all proteins were classified in clusters of orthologous groups (COGs) (Tables S1 and S2 in the supplemental material). We next performed cluster analysis by combining the protein abundance data using a hierarchical clustering algorithm according to the program instructions. The distribution of these 194 proteins in different functional categories (e.g., cell envelope and cellular processes, intermediary metabolism, information pathways, and other functions) for these 194 proteins is shown in Fig. 2A. Previous studies have revealed the genetic basis of rhizosphere adaptation and the plant-beneficial effects of strain SQR9, and we compared proteins related to biocontrol (antibiotic production), detoxification, transporters, cell motility, and biofilm formation (Fig. 2B). Some proteins involved in antibiotic production were decreased in the sfp mutant, including DfnJ, BacB, FenA, FenB, and BmyB. However, six proteins with higher fold changes were associated with detoxification (KatE, YceD, YceE, and YceC), transporters (OppA), and cell motility (Hag). For biofilm formation, four proteins (LuxS, AhpC, SpeE, and GgaA) were decreased in the sfp mutant. We noted that GgaA is a WTA biosynthetic enzyme, and WTA is involved in cell division, host cell adhesion, and colonization in many Gram-positive bacteria; GgaA showed a 4-fold decrease in expression in the sfp mutant, which implies that GgaA is possibly involved in biofilm development in SQR9.
FIG 2.
The sfp mutant and wild-type strains showed different proteomic profiles by iTRAQ analysis. (A) Distribution in various functional categories of proteins altered in the sfp mutant strain. (B) Distribution of the rhizosphere proteins in the presence of various treatments. The abscissa shows the fold change in the protein ratio of two treatments (SQR9/sfp mutant), and the ordinate represents various indicated proteins.
WTA biosynthetic enzyme GgaA is necessary for biofilm formation and root colonization.
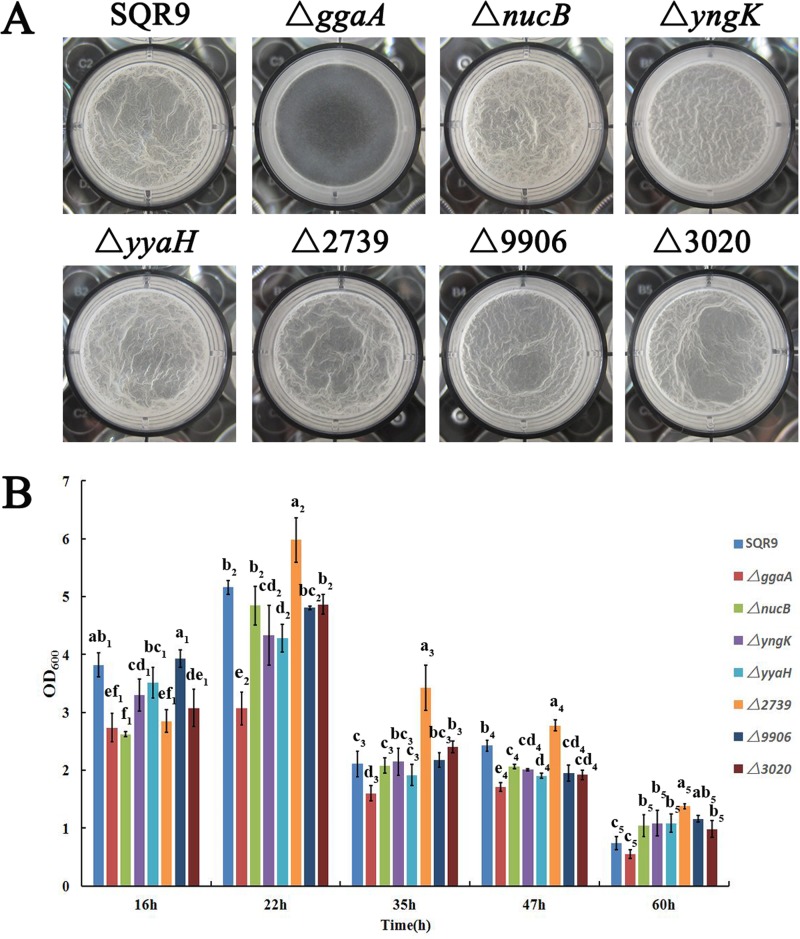
Because the differences in proteome composition of the wild-type and sfp mutant strains may affect biofilm development, we were interested in whether distinct biofilm-specific proteins produced in wild-type biofilms might have important implications for biofilm development. We therefore tested the biofilm formation of seven B. velezensis mutant strains with mutations in proteins that displayed a >4-fold increase in expression compared to the sfp mutant (Fig. 3A and Table S1). Biofilm formation was indicated by the presence of pellicles (floating pellicles at the medium-air interface). During this process, we observed a ggaA mutant that formed thin and flat pellicles, while other mutants showed a biofilm architecture indistinct from that of the wild type (Fig. 3A). Complementation of the ggaA gene in the ΔggaA mutant restored its ability to form a normal biofilm (Fig. S2). At the 22-h time point, this pattern correlated well with the measured values of pellicle biomass (Fig. 3B), thus reinforcing the observation that the ggaA gene is needed for biofilm development by the SQR9 strain.
FIG 3.
Impairment of the ggaA gene postponed the formation of cellular biofilms and decreased the root colonization capability of the B. velezensis SQR9 strain. (A) Microtiter plate assay of biofilm formation by the wild-type and mutant strains. (B) OD600 of solubilized crystal violet from the microtiter plate assay over time for the wild-type and mutant strains. Error bars indicate the standard deviations based on three different replicated experimental values. Different letters above the bars indicate significant differences (P < 0.05), and the subscript numbers differentiate the different time points. Mutant designations Δ2739, Δ9906, and Δ3020 represent mutants with deletions of the gene with the last four numbers of the GI accession number of each gene in the SQR9 genome in the NCBI database (accession no. CP006890), i.e., V529_33780 (https://www.ncbi.nlm.nih.gov/protein/631802739), V529_05450 (https://www.ncbi.nlm.nih.gov/protein/631799906), and V529_36590 (https://www.ncbi.nlm.nih.gov/protein/631803020), respectively.
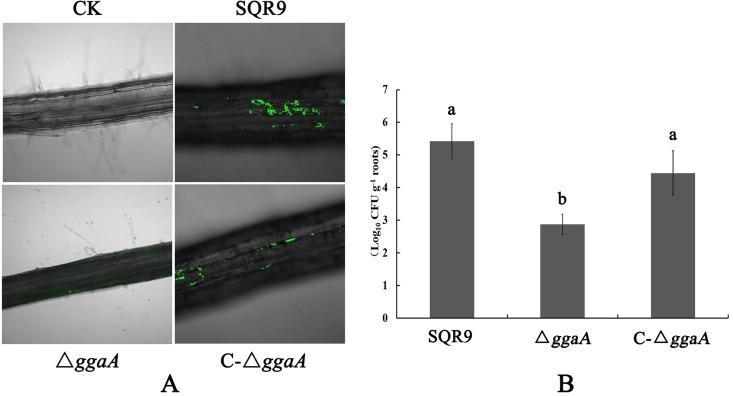
To further investigate whether SQR9 and the ggaA mutant differ in root colonization, we monitored the populations of wild-type and mutant strains colonized on the cucumber roots. After 2 days of incubation in a hydroponic system, green fluorescent protein (GFP)-labeled B. velezensis mutant strains (SQR9-gfp, ΔggaA-gfp, and C-ΔggaA-gfp) were investigated using confocal laser scanning microscopy (CLSM). The formation of biofilms consisting of wild-type GFP-labeled cells on the root surface could be easily observed. Comparatively, only a few small regions of the roots were colonized by ggaA mutant cells, and complementation of the ggaA gene in the ΔggaA mutant restored almost all root colonization ability (Fig. 4A). This was further confirmed by quantitative measurement of the bacterial population that colonized the plant roots. The results showed that approximately 105 CFU g−1 root of SQR9 cells were detected, but only 103 CFU g−1 root of ggaA mutant cells were colonized on the root. Similarly, there were no significant differences in the populations of wild-type SQR9 and the C-ΔggaA-gfp complemented strain that colonized on the roots (Fig. 4B).
FIG 4.
The ggaA mutant strain is deficient in cucumber root colonization. (A) CLSM micrographs of cucumber roots colonization by GFP-tagged wild-type and ggaA mutant strains. Ck is a control which was not inoculated with GFP-tagged SQR9. (B) The populations of wild-type and ggaA mutant strains colonizing cucumber seeding roots. Error bars indicate the standard deviations from the results from three independent experiments. Different letters above the bars indicate significant differences (P < 0.01).
SQR9 lacking gtaB has a significant defect in biofilm formation.
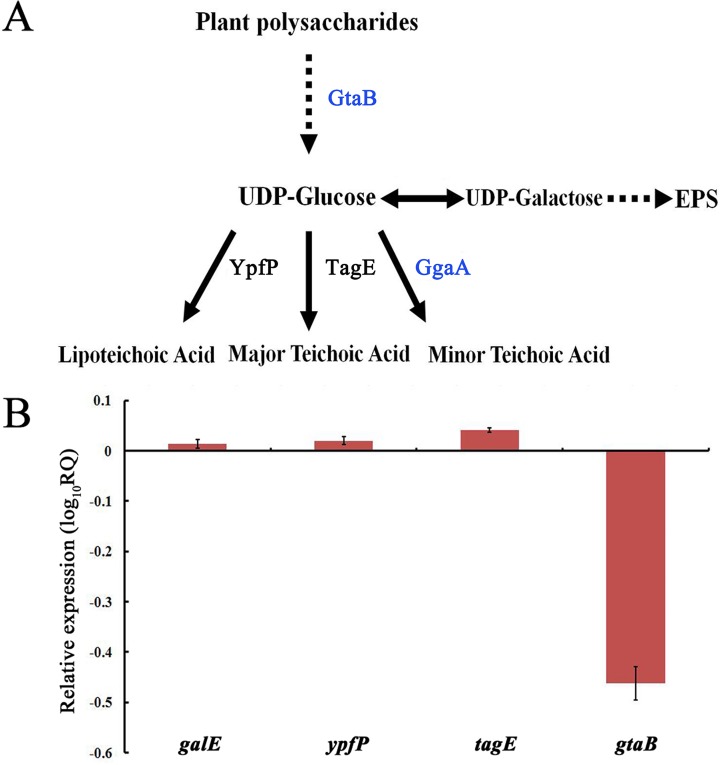
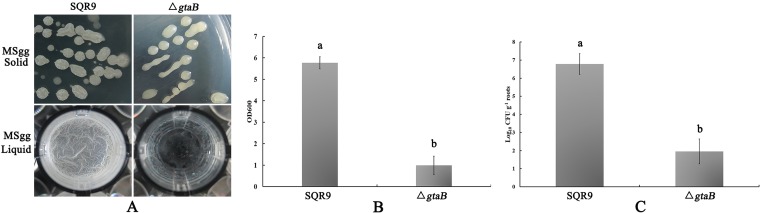
According to the accepted knowledge of WTA synthesis in B. subtilis (11, 22), several genes (galE, ypfP, tagE, and gtaB) are involved in the synthesis of WTAs and lipoteichoic acid (Fig. 5A), and UDP-glucose (UDP-Glc) is a key precursor for WTA polymers. Reverse transcription-quantitative PCR (RT-qPCR) results showed that the transcripts of gtaB were significantly decreased (4-fold) in the ggaA mutant (Fig. 5B). GtaB is an α-glucose-1-phosphate uridylyltransferase that catalyzes the formation of UDP-Glc and is involved in the glucosylation of the major and minor WTAs (22, 31). We hypothesized that gtaB may be involved in biofilm formation in SQR9. We constructed ΔgtaB and ΔgtaB-gfp mutant strains and evaluated their biofilm formation and root colonization abilities compared to those of the wild-type strain SQR9. On MSgg solid medium (see Materials and Methods for ingredients), gtaB mutant colonies were smooth and lacked aerial structures. Moreover, in MSgg liquid medium, the gtaB mutant formed very fragile pellicles that usually split and sank to the bottom of the culture vessel (Fig. 6A). Quantitative analysis of the biofilm biomass showed that the gtaB mutant exhibited significantly lower levels of biofilm biomass (Fig. 6B). In addition, the gtaB mutant population was significantly decreased compared to that of the wild-type strain (Fig. 6C). Overall, these results suggested that the gtaB gene is involved in biofilm formation and root colonization in SQR9.
FIG 5.
(A) Involvement of UDP-glucose in synthesis of WTAs and lipoteichoic acid. (B) Transcriptional levels of galE, ypfP, tagE, and gtaB in the ggaA mutant strain relative to those in wild-type strain SQR9. The experiments were carried out with pellicle obtained from microtiter plates when cells were grown in MSgg medium for 24 h. The B. velezensis SQR9 recA gene was used as an internal reference gene. RQ represents the relative expression level (relative quantification) compared to the wild-type strain. Bars represent standard deviations of data from three biological replicates.
FIG 6.
GtaB is involved in biofilm formation and cucumber root colonization by B. velezensis SQR9. (A) The complex colony morphology and microtiter plate assay of biofilm formation by the gtaB mutant and wild-type strains at 24 h. (B) OD600 of solubilized crystal violet from the microtiter plate assay for the wild-type and gtaB mutant strains at 24 h. (C) The populations of wild-type and gtaB mutant strains colonizing cucumber seeding roots. Error bars indicate the standard deviations from three independent experiments. Different letters above the bars indicate significant differences (P < 0.05).
Response to plant polysaccharides depends on the UDP-glucose pyrophosphorylase GtaB.
Beauregard et al. reported that plant polysaccharides (arabinogalactan, pectin, and xylan) can be used as a carbohydrate source for building the biofilm matrix in Bacillus subtilis and that the production of the EPS requires the synthesis of UDP-galactose (11, 22). We therefore anticipated that a mutation in gtaB would block the successive catalytic steps in UDP-Glc synthesis, which consequently may have been responsible for the defect in biofilm development. To investigate this possibility, we examined the capacity of plant polysaccharides to induce B. velezensis SQR9 and the mutant strains to form biofilms in MSNc medium (see Materials and Methods for ingredients). First, we tested whether the transcripts of ggaA or gtaB could be induced by plant polysaccharides. Our results showed that the expression of gtaB can be induced in the wild-type strain and that xylan had the strongest effect (5-fold) (Fig. S4). Next, the results from our biofilm assay showed that the wild-type SQR9 and ΔggaA mutant strains were able to form biofilm in the presence of plant polysaccharides. These results suggest that plant polysaccharides can serve as an environmental cue for biofilm formation, as has been reported in B. subtilis (Fig. 7). However, the ΔgtaB mutant formed much weaker pellicles under the same conditions. Additionally, we compared the EPS production of the wild-type and mutant strains. Our results showed that both the ggaA and gtaB mutant strains were deficient in EPS production (Fig. S5). Finally, we tested the attachment abilities of ggaA mutant, gtaB mutant, and wild-type strains on the cucumber root surfaces. The results showed that wild-type strain SQR9 exhibited significantly higher cell attachment than did the ggaA and gtaB mutant cells (Fig. S6). These observations suggest that plant polysaccharides can be incorporated into the EPS production by gtaB and that EPS is an important biofilm matrix for root attachment.
FIG 7.
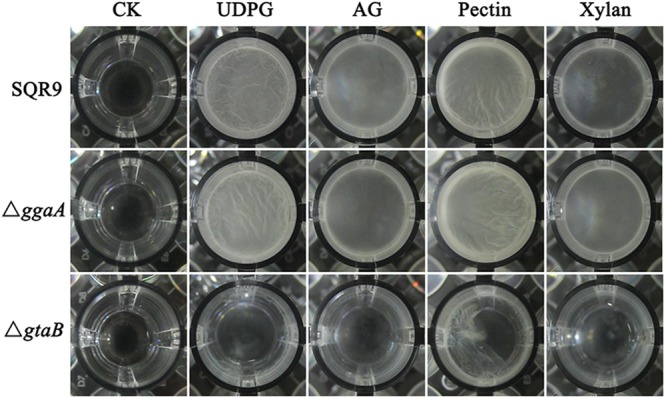
The response to plant polysaccharides depends on the UDP-glucose pyrophosphorylase, GtaB. Top-down view of biofilm assay in which the indicated mutant cells were incubated for 24 h in the presence of UDP-glucose (UDPG), arabinogalactan (AG), pectin, or xylan in MSNc medium. Biofilm formation of wild-type and mutant cells in MSNc without supplements were used as a control (CK). Results are representative of three experiments.
DISCUSSION
Gram-positive Bacillus species are attractive PGPR that are widely used as a biofertilizers (32, 33). The colonization of PGPR strains in the rhizosphere is a prerequisite for them to execute their specific functions (34). In this study, we have shown that the ggaA and gtaB genes, which are required for the biosynthesis of WTAs in B. velezensis SQR9, are essential for biofilm formation and root colonization. By using a proteomic approach to compare the proteomes of wild-type and sfp mutant strains, we noted that the GgaA protein is much more abundant in wild-type cells than in sfp mutant cells. Moreover, we showed that plant polysaccharides are used as a source for the synthesis of biofilm matrix exopolysaccharides by GtaB.
The sfp gene encodes a phosphopantetheinyl transferase that is required for Bacillus cells to produce lipopeptide antibiotics (35). In B. velezensis FZB42, an sfp gene mutant was found to be impaired in biofilm formation and root colonization (28). Here, we focused on the proteome of the biofilm produced by the sfp mutant compared to that of the wild-type strain. First, we found that the expression of BmyB, FenA, FenB, DfnJ, and BacB was indeed lower in the sfp mutant, and Sfp is crucially involved in the biosynthesis of lipopeptides. Then, we found that four proteins (LuxS, AhpC, SpeE, and GgaA) that influence biofilm formation were also decreased in the sfp mutant. Lombardia et al. (36) reported that LuxS activity was required for the formation of sophisticated aerial colonies and biofilm formation. Zwick et al. (37) demonstrated that AhpC is highly expressed in both stationary-phase planktonic cells and biofilms and that strains lacking ahpC do not form a biofilm. In addition, SpeE is a spermidine synthase, and Hobley et al. (38) found that spermidine promotes B. subtilis biofilm formation by activating the expression of the matrix regulator slrR. Overall, it is likely that the lower expression of these proteins affects the biofilm development of the sfp mutant. The defect in surfactant activity of the sfp mutant might also influence the biofilm formation, since both bacillomycin D and fengycin have surfactant properties. However, the addition of the lipopeptide surfactin, which is a more powerful surfactant than those two lipopeptides (Fig. S1), did not restore the biofilm-defective phenotype of sfp mutant. These results indicated that a more intricate mechanism was involved in this biofilm-defective mutant.
The ggaA gene is required for biosynthesis of the galactosamine-containing minor WTA (39). However, relatively little is known regarding the biological function of this gene in plant-microorganism interactions. As major components of the cell envelope, one of the main roles of WTAs is the maintenance of the global negative charge of the cell surface (40). Here, we present results demonstrating that the GgaA protein is involved in the rhizobacterium SQR9 colonization of cucumber roots and that this protein may also be responsible for the sfp mutant defect in biofilm development. We also constructed two mutants lacking YpfP and TagE, which are required for the synthesis of major teichoic acid and lipoteichoic acid, that still formed normal architecture pellicles similar to the wild-type strain in biofilm formation (data not shown). These results suggest a specificity role of GgaA in biofilm formation by B. velezensis SQR9. Several studies have revealed that bacterium-environment interactions can be influenced by WTAs (41, 42). In Staphylococcus aureus, WTA mutants affected in the early stage of cell attachment to artificial surfaces. Similarly, WTA null mutants are defective both in biofilm development and colonization of nasal passages of mice (41). Taken together, these findings highlight the phenomenon that WTAs are involved in biofilm formation and the colonization process.
In Gram-positive bacteria, the gtaB gene encodes UTP glucose-1-phosphate-uridilyl-transferase, which is partly controlled by sigma B (43). In addition, GtaB is needed for nucleotide sugar phosphates involved in EPS biosynthesis (44). In our study, we showed that gtaB null mutants are impaired in biofilm formation and root colonization and that wild-type and gtaB mutant strains showed indistinguishable growth curves (Fig. S3). In accordance with these results, Lazarevic et al. (22) discovered that gtaB-deficient mutants were also deficient in biofilm formation. UDP-Glc serves as a glucosyl donor for the synthesis of all phosphate-containing anionic envelope polymers, including lipoteichoic acid, major teichoic acid, and minor teichoic acid (22). The absence of UDP-Glc in the gtaB mutant might explain the deficiency in biofilm formation, since UDP-Glc is an important precursor for the extracellular matrix required for biofilm development. In many environmental bacteria, UDP-Glc is necessary for the synthesis of exopolysaccharides (EPS) (44, 45). For example, a Bradyrhizobium japonicum mutant that lacks UDP-Glc-4′-epimerase activity produces less extracellular EPS than does the wild-type strain (46). Additionally, in another report, UDP-Glc and UDP-galactose were shown to be involved as signaling molecules or as precursors for the synthesis of EPS (44). In our study, EPS production was indeed lower in gtaB-deficient mutants than in the wild-type strain. In the rhizosphere, the production of extracellular matrix, including both EPS and protein components, is essential for root colonization, as Bacillus strains carrying knockout mutations in either component failed to colonize on the plant root.
Rhizobacteria in the rhizosphere sense plant extracts and root exudate components released by plants (47, 48). For instance, organic acids, such as malic acid and citric acid, in root exudates recruit Bacillus spp. in the rhizosphere (49). After that, other molecules that PGPR encounter on the plant root include the polysaccharides from the plant cell wall. Beauregard et al. reported that plant polysaccharides (arabinogalactan, pectin, and xylan) act both as an environmental cue and as a substrate for the synthesis of the B. subtilis 3610 biofilm matrix and that these plant polysaccharides promote biofilm development in various plant growth-promoting Bacillus strains, including B. subtilis GB03 and B. amyloliquefaciens FZB42 (11). In this study, we provide evidence that loss of the gtaB gene affects the ability of SQR9 to sense plant polysaccharides and form a biofilm, which can partially explain the defect of the gtaB strain in cucumber root colonization.
Over the past few decades, research on the process of Bacillus biofilm formation has been well studied under laboratory conditions (13, 14, 18). However, relatively little is known about how this bacterium colonizes plant roots. We showed that WTAs may play important roles in the root colonization process. Although there is evidence that two WTA biosynthetic enzymes, GgaA and GtaB, positively influence biofilm formation and root colonization, determining exactly how WTAs affect the root colonization process will be an important challenge for the future.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. B. velezensis strain SQR9 (CGMCC accession no. 5808; China General Microbiology Culture Collection Center) was used throughout this study. Escherichia coli DH5α was used as the host strain for all plasmids. B. velezensis strains were grown in Luria-Bertani medium and, where appropriate, in minimal medium (MSgg; 5 mM potassium phosphate, 100 mM morpholinepropanesulfonic acid [MOPS] [pH 7], 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 mM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg ml−1 tryptophan, 50 μg ml−1 phenylalanine, and 50 μg ml−1 threonine) (15). For transformant selection, colonies were selected on MGY-Cl agar plates (minimal medium-glucose-yeast extract [MGY] medium contained glucose [5 g/liter], yeast extract [4 g/liter], NH4NO3 [1 g/liter], NaCl [0.5 g/liter], K2HPO4 [1.5 g/liter], KH2PO4 [0.5 g/liter], and MgSO4 [0.2 g/liter], and MGY-Cl medium is an MGY-based medium supplemented with 5 mM p-Cl-Phe [Sigma]). Biofilm assays were performed in 48-well plates with 1 ml of MSNc (5 mM potassium phosphate buffer [pH 7], 0.1 M MOPS [pH 7], 2 mM MgCl2, 0.05 mM MnCl2, 1 μM ZnCl2, 2 μM thiamine, 700 μM CaCl2, 0.2% NH4Cl, and 0.5% cellobiose) medium supplemented or not with the purified plant polysaccharides (11). B. velezensis strains were incubated at 37°C. Antibiotics were added as required at the following concentrations: 20 μg ml−1 zeocin, 5 μg ml−1 chloramphenicol, and 100 μg ml−1 ampicillin for E. coli strains.
TABLE 1.
Microorganisms and plasmids used in this study
| Strain or plasmid | Description or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Φ80dlacZΔDM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 phoA | Invitrogen (Shanghai, China) |
| E. coli BL21 | ||
| Bacillus velezensis SQR9 | Wild type | CGMCC no. 5808 |
| B. velezensis SQR9-gfp | GFP-labeled SQR9 (Kanr) | |
| B. velezensis SQR9M6 | SQR9 Δsfp::cm | 60 |
| SQR9 ΔggaA | This study | |
| GFP-labeled SQR9 ΔggaA (Cmr) | This study | |
| SQR9 Δ3020 | This study | |
| GFP-labeled SQR9 Δ3020 (Cmr) | This study | |
| SQR9 ΔgtaB | This study | |
| GFP-labeled SQR9 ΔgtaB (Cmr) | This study | |
| Plasmids | ||
| pTPC | pMD19-T harboring the Pbc-pheS*-cat(PC) cassette | 54 |
| pNW33n | Cmr; MCS | BGSC |
| pNW33n-GFP | This study |
Kanr, kanamycin resistance; Cmr, chloramphenicol resistance; MCS, multiple-cloning site; Δ3020, mutant with deletion of V529_36590 (https://www.ncbi.nlm.nih.gov/protein/631803020).
Protein extraction analysis.
Biofilms were incubated in MSgg medium and harvested at the 24-h time point. Protein extraction was carried out as described by Kierul et al. (50). The cells were removed by centrifugation (11,000 × g, 4°C, 20 min) and filtered through a 0.22-µm-pore-size membrane (Millipore). Trichloroacetic acid was added to the filtered exudates to a final concentration of 10%, and the solution was stored at 4°C overnight. Subsequently, extracellular proteins were collected by centrifugation (15,000 × g, 4°C, 30 min). The protein pellet was washed three times with 1 ml of ice-cold acetone and centrifuged (15,000 rpm, 4°C, 20 min). Similarly, two washing steps with 96% ice-cold ethanol were performed. The extracellular proteins obtained were stored in 96% ethanol at −20°C.
iTRAQ labeling and automated two-dimensional liquid chromatography with tandem mass spectrometry protein identification.
The protein samples were resuspended in lysis buffer (7 M urea, 2 M thiourea, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]), thoroughly sonicated, and then incubated at 37°C for 30 min. After centrifugation at 20,000 × g at 4°C for 30 min, the supernatant was collected. Protein concentration and quality were determined using a Bradford assay. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the method described by Laemmli (51), and the SDS-PAGE gel was stained with sliver staining (52).
Protein digestion and labeling were carried out by the method of Qiu et al. (53), and the peptide samples were labeled using the iTRAQ reagent multiplex kit (Applied Biosystems, Foster City, CA). The automated two-dimensional liquid chromatography with tandem mass spectrometry (2D LC-MS/MS) analysis was similar to what has been previously described (53). Peptides were desalted using ZipTip C18 reverse resin (Millipore Corporation, Billerica, MA) and separated via C18 reversed-phase column by high-performance liquid chromatography (HPLC; Agilent Technologies, Santa Clara, CA). MS was performed using an LTQ Orbitrap mass spectrometer (Thermo Electron Corp.), and data were collected using the Xcalibur software (Thermo Electron).
Protein identification and quantification were performed with the ProteinPilot 4.0 software (Applied Biosystems, USA). The database used for searching was the B. velezensis SQR9 (GenBank accession no. CP006890) entry in the National Center for Biotechnology Information (NCBI) database. Proteins with fold changes significantly >4.0 or <0.25 were considered differentially expressed (P < 0.05).
B. velezensis SQR9 mutant construction.
The marker-free deletion strains of targeted genes were constructed using the Pbc-pheS*-cat (PC) cassette and an overlap PCR-based strategy, essentially as has been previously described (54). To delete the targeted genes in B. velezensis SQR9, 1-kb fragments located upstream and downstream of the target gene were amplified. The 1.1-kb chloramphenicol resistance gene (Cmr) was amplified from pNW33N, and the PC cassette was amplified from pTPC as a template. These four fragments were fused using overlap PCR and directly transformed into SQR9. The transformants were selected on LB plates containing Cm. Cmr colonies were cultivated to an optical density at 600 nm (OD600) of 1.0 without Cm, and a 100-μl aliquot of a 10-fold dilution of the cultures (approximately 105 cells) was plated on MGY-Cl medium (54). Mutants growing on MGY-Cl were further confirmed by PCR and DNA sequencing. The primers used in this experiment are listed in Table 2.
TABLE 2.
Primers used in this study
| Primera | Oligonucleotide sequence (5′–3′) |
|---|---|
| PggaALF-F | TTTTGTATCAAAGCCTATCGT |
| PggaALF-R | GCTAGACAATAAGAGGAGAATTTAAAGTGATGATTGTGATTGGATATG |
| PggaADR-F | CGTATAGCATACATTATACGAACGGCGGTTCATCGCCATCTTCTC |
| PggaADR-R | TAGCGGTAGTCGTTCCTGTT |
| PggaARF-F | CATATCCAATCACAATCATCACTTTAAATTCTCCTCTTATTGTCTAGC |
| PggaARF-R | CGCAGTTTAAACAGGCGTAATAAAGTTCAACCTGCAAAAACAAAAAAAGG |
| PggaAPS-F | CCTTTTTTTGTTTTTGCAGGTTGAACTTTATTACGCCTGTTTAAACTGCG |
| PggaAPS-R | GAGAAGATGGCGATGAACCGCCGTTCGTATAATGTATGCTATACG |
| PgtaBLF-F | TCAAACTCTTCAACGACTCT |
| PgtaBLF-R | GGATTCGGCAGCTTTTTTGAATAACACCATCCTTGT |
| PgtaBDR-F | CGTATAGCATACATTATACGAACGGTGCTTCCAATCGTTGATAAG |
| PgtaBDR-R | GGCGTCCGTTAATTGAATT |
| PgtaBRF-F | ACAAGGATGGTGTTATTCAAAAAAGCTGCCGAATCC |
| PgtaBRF-R | CGCAGTTTAAACAGGCGTAATAAGACGGATGTTCAGCTTAATG |
| PgtaBPS-F | CATTAAGCTGAACATCCGTCTTATTACGCCTGTTTAAACTGCG |
| PgtaBPS-R | CTTATCAACGATTGGAAGCACCGTTCGTATAATGTATGCTATACG |
| PnucBLF-F | ATCAGGGTCTGCTTTTGTTT |
| PnucBLF-R | AAACTCCGGCGAACCGGAGAAGCTATCACATCCTCCTTAAGAAGT |
| PnucBDR-F | CGTATAGCATACATTATACGAACGGTTGGTGATCTCTCTTCAGTTCG |
| PnucBDR-R | CTCTCGTGCCGTCGGAATA |
| PnucBRF-F | ACTTCTTAAGGAGGATGTGATAGCTTCTCCGGTTCGCCGGAGTTT |
| PnucBRF-R | CGCAGTTTAAACAGGCGTAATAAGTTACCGCAGGGGGCCAGAATGATT |
| PnucBPS-F | AATCATTCTGGCCCCCTGCGGTAACTTATTACGCCTGTTTAAACTGCG |
| PnucBPS-R | CGAACTGAAGAGAGATCACCAACCGTTCGTATAATGTATGCTATACG |
| PyngKLF-F | TCCGCCCGCCGTCTAATGTATATAAAAATT |
| PyngKLF-R | AAGCAGAGAGGGGGAAAAACTAGCATTATTAATAAAACTGACAGTTGCGGCGC |
| PyngKDR-F | CGTATAGCATACATTATACGAACGGGAAGCCTGTCAACGGAAGTAACGATAT |
| PyngKDR-R | GGGTGCGGAACTACGATGATTTATACG |
| PyngKRF-F | GCGCCGCAACTGTCAGTTTTATTAATAATGCTAGTTTTTCCCCCTCTCTGCTT |
| PyngKRF-R | CGCAGTTTAAACAGGCGTAATAAGATCATTCTTGCCAGCTACC |
| PyngKPS-F | GGTAGCTGGCAAGAATGATCTTATTACGCCTGTTTAAACTGCG |
| PyngKPS-R | ATATCGTTACTTCCGTTGACAGGCTTCCCGTTCGTATAATGTATGCTATACG |
| PyyaHLF-F | GCAGGCTACTTAGGGCT |
| PyyaHLF-R | AGAAAAAAAGGGAGGCGGACATTAACAGTGCGGGGGAAC |
| PyyaHDR-F | CGTATAGCATACATTATACGAACGGCCATCCTCTATCACGCTCTCAT |
| PyyaHDR-R | GCAGCGATATGGGTGAAAGATT |
| PyyaHRF-F | GTTCCCCCGCACTGTTAATGTCCGCCTCCCTTTTTTTCT |
| PyyaHRF-R | CGCAGTTTAAACAGGCGTAATAATTCTGGTCGGATGCCATGATAT |
| PyyaHPS-F | ATATCATGGCATCCGACCAGAATTATTACGCCTGTTTAAACTGCG |
| PyyaHPS-R | ATGAGAGCGTGATAGAGGATGGCCGTTCGTATAATGTATGCTATACG |
| P2739LF-F | TTACGACGAAAAAGTATCCGAC |
| P2739LF-R | AGGAACATGGTGACTGTCCTTGATATGACATATCGAACACCG |
| P2739DR-F | CGTATAGCATACATTATACGAACGGACCATTCCAGCGTTCTCCAT |
| P2739DR-R | CCGACAGATCATGCCGATTATG |
| P2739RF-F | CGGTGTTCGATATGTCATATCAAGGACAGTCACCATGTTCCT |
| P2739RF-R | CGCAGTTTAAACAGGCGTAATAATTGACCACTCCGTCGTATTC |
| P2739PS-F | GAATACGACGGAGTGGTCAATTATTACGCCTGTTTAAACTGCG |
| P2739PS-R | ATGGAGAACGCTGGAATGGTCCGTTCGTATAATGTATGCTATACG |
| P9906LF-F | AGATATCGATCCTGCGATTT |
| P9906LF-R | CGGCTTTTGCGGTCTTTTTATTCAAGAACTCCTTTGCCTT |
| P9906DR-F | CGTATAGCATACATTATACGAACGGAACAGCCAATCTTCAGGAG |
| P9906DR-R | CGTCAGTCGGTGTATAGTC |
| P9906RF-F | AAGGCAAAGGAGTTCTTGAATAAAAAGACCGCAAAAGCCG |
| P9906RF-R | CGCAGTTTAAACAGGCGTAATAACTACTTTGCGCCTTTTTTGG |
| P9906PS-F | CCAAAAAAGGCGCAAAGTAGTTATTACGCCTGTTTAAACTGCG |
| P9906PS-R | CTCCTGAAGATTGGCTGTTCCGTTCGTATAATGTATGCTATACG |
| P3020LF-F | CGTATAACGTTTCTGTCGCATTAC |
| P3020LF-R | CTATTGAAAAGGGGAAAAAGCATAAGAAAAGGTGCCGTTTATCG |
| P3020DR-F | CGTATAGCATACATTATACGAACGGATGAACTCGCTGCCATTGTAA |
| P3020DR-R | GCTCGGATTGATGCTGACTG |
| P3020RF-F | CGATAAACGGCACCTTTTCTTATGCTTTTTCCCCTTTTCAATAG |
| P3020RF-R | CGCAGTTTAAACAGGCGTAATAAGACTTTGAATGGAAACGAAACG |
| P3020PS-F | CGTTTCGTTTCCATTCAAAGTCTTATTACGCCTGTTTAAACTGCG |
| P3020PS-R | TTACAATGGCAGCGAGTTCATCCGTTCGTATAATGTATGCTATACG |
| ggaA-qF | GTCCAAGCCGTCCGTAGAAG |
| ggaA-qR | ACAGGAACGACTACCGCTACCT |
| gtaB-qF | CGTTGATAAGCCGACAATTCAG |
| gtaB-qR | CATTGTCGAAATGGTCTTCTATTG |
| ggaALF-F | GAGAAGGCGTCGTAAAC |
| ggaALF-R | TAACGGCAGTAAAGAGGT |
| ggaACm-F | ATTCAAAACCTCTTTACTGCCGTTAGCATAAAGTGTAAAGCCTGGGG |
| ggaACm-R | CCTTTCGCTGAAAGTAACAAAATGAAATGTGGAATTGGGAACGGAAA |
| ggaACG-F | TCATTTTGTTACTTTCAGCG |
| ggaACG-R | ATACAAACCATTCTGCACATAATCAGAACACAGAAAGCTGATTCT |
| ggaARF-F | TGATTATGTGCAGAATGGT |
| ggaARF-R | TCTCGATAATATGGTAGGC |
| gtaBLF-F | GAGAAGGCGTCGTAAAC |
| gtaBLF-R | TAACGGCAGTAAAGAGGT |
| gtaBCm-F | ATTCAAAACCTCTTTACTGCCGTTAGCATAAAGTGTAAAGCCTGGGG |
| gtaBCm-R | AAAACCAGATATCGTTTTGGTCCACAATGTGGAATTGGGAACGGAAA |
| gtaBCG-F | GTGGACCAAAACGATATCT |
| gtaBCG-R | ATACAAACCATTCTGCACATAATCACTATTCGTCAGCTTCTTCTT |
| gtaBRF-F | TGATTATGTGCAGAATGGT |
| gtaBRF-R | TCTCGATAATATGGTAGGC |
Designations 2739, 9906, and 3020 refer to the genes with the last four numbers of the GI accession number in the SQR9 genome in the NCBI database (accession no. CP006890), i.e., V529_33780 (https://www.ncbi.nlm.nih.gov/protein/631802739), V529_05450 (https://www.ncbi.nlm.nih.gov/protein/631799906), and V529_36590 (https://www.ncbi.nlm.nih.gov/protein/631803020), respectively.
Complementation of disrupted ggaA and gtaB genes.
The ggaA and gtaB gene fragments were complemented at the amyE locus in each mutant strain. The left flanking (LF) region (∼1,000 bp), complement gene (CG) sequences, and right flanking (RF) region (∼1,000 bp) were amplified from the SQR9 strain using the primer pairs ggaALF-F/ggaALF-R and gtaBLF-F/gtaBLF-R, ggaACG-F/ggaACG-R and gtaBCG-F/gtaBCG-R, and ggaARF-F/ggaARF-R and gtaBRF-F/gtaBRF-R, respectively. The Cm region (∼1,000 bp) was amplified with the primer pairs ggaACm-F/ggaACm-R and gtaBCm-F/gtaBCm-R using pNW33N as the template. These four fragments were fused and directly transformed into the ΔggaA and ΔgtaB mutant strains. The transformants were selected on LB plates containing Cm.
Biofilm assays of B. velezensis SQR9 and mutants.
The biofilm assay was carried out in 48-well microtiter plates in MSgg medium, as described by Hamon and Lazazzera (55). Biofilm formation was quantified by staining with crystal violet (CV). Cells of a biofilm were stained with CV, and then unbound CV was removed with distilled water. The remaining CV was solubilized with 1 ml of 80% ethanol–20% acetone. The absorbance of CV at 570 nm was measured using the SpectraMax i3x analysis system (Molecular Devices Corporation, CA).
Root colonization assay.
The bacterial suspensions of SQR9 and mutants were inoculated into sterile cucumber seedlings in 1/4 Murashige and Skoog (MS) culture medium. After 4 days, the cells colonized on the cucumber roots were collected and quantified using the method described by Qiu et al. (53).
Congo red binding assay.
Strains were incubated at 37°C overnight in Luria-Bertani medium, and 1 ml of cells was collected by centrifugation (10,000 × g, 4°C, 2 min). The supernatant was removed, the pellet was resuspended in 1 ml of tryptone broth (10 g/liter tryptone), and 500 μl of cells was transferred to a new tube. A total of 5 μl of Congo red (CR) stock solution (4 mg/ml CR, filtered) was added to 500 μl cells and 500 μl T-broth as a blank. Subsequently, the tubes were mixed well and shaken vigorously at 30°C for 2 h. The OD600 value was measured using the remaining cells (10× dilution, 100 μl cells plus 900 μl medium). After 2 h of incubation, samples were centrifuged at 15,000 rpm for 5 min, and the OD490 value of the supernatant was measured. The amount of CR bound to the cells was calculated as follows: CR (μg/OD600) = [(OD490-blank – OD490-sample) × 44.676/OD600] (56).
Bacterial attachment assay.
The B. velezensis attachment assay was carried out using the methods from Smit et al. (57) and Dardanelli et al. (58). Strains were incubated at an OD600 of 1.0, pelleted by centrifugation (6,000 × g, 4°C, 10 min), and then washed three times with phosphate-buffered saline (PBS [pH 7.4]). The pellet was suspended in the same solution to a final OD600 of 0.1. Three sterile cucumber roots were immersed in 50 ml of bacterial suspension for 2 h at room temperature under gentle agitation. Then, the roots were washed 10 times with sterile distilled water and ground up in a sterilized mortar. The number of bound bacteria was quantified by dilution plate counting.
Microscopy.
To determine if B. velezensis colonized the cucumber root surface, SQR9-gfp-, ΔggaA-gfp mutant-, and ΔgtaB-gfp mutant-colonized roots were investigated at 2 days after inoculation in hydroponics. The roots were washed with distilled water and viewed by a confocal laser scanning microscope (CLSM; model TCS SP2; Leica, Heidelberg, Germany). Images were taken using Leica confocal software, version 2.61 (12).
Quantification of gene transcription by real-time PCR.
Total RNA was obtained from biofilms formed by B. velezensis using the E.Z.N.A. bacterial RNA kit (Toyobo, Japan), according to the instructions. The RNA was detected on a 1% agarose gel, and a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) was used to check the quality and concentration. RNA was reverse transcribed using the PrimeScript RT reagent kit with a genomic DNA (gDNA) eraser (Toyobo). Transcript levels of ggaA and gtaB were measured by reverse transcription-quantitative PCR (RT-qPCR) using a SYBR Premix Ex Taq (perfect real time) kit (TaKaRa, Dalian, China). The recA gene was used as an internal control. Reactions were carried out on an ABI 7500 system (Applied Biosystems, USA) under the following conditions: cDNA was denatured for 10 s at 95°C, followed by 40 cycles consisting of 5 s at 95°C and 34 s at 60°C. The 2−ΔΔCT method was used to analyze the RT-qPCR data (59).
Statistical analysis.
Differences among the treatments were calculated and statistically analyzed with a one-way analysis of variance (ANOVA). Duncan’s multiple-range test was used when one-way ANOVA indicated a significant difference (P < 0.05). All statistical analyses were performed with IBM SPSS Statistics 20. To identify and classify differentially expressed proteins, the Gene Ontology (GO) (www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) databases were used.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Nature Science Foundation of China (grants 31501833 and 31330069), the National Key Research and Development Program (grants 2016YFD0200305 and 2017YFD0200805), the China Science and Technology Ministry (973 Program, grant 2015CB150500), the China Postdoctoral Science Foundation (grant 2015M581813), and the Innovative Research Team Development Plan of the Ministry of Education of China (grant IRT_17R56).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02116-18.
REFERENCES
- 1.Lugtenberg BJJ, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 2.Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R. 2007. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 3.Chet I, Chernin L. 2003. Biocontrol, microbial agents in soil, p 450–465. In Bitton G. (ed), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, NY. [Google Scholar]
- 4.Abed H, Rouag N, Mouatassem D, Rouabhi A. 2016. Screening for Pseudomonas and Bacillus antagonistic rhizobacteria strains for the biocontrol of Fusarium wilt of chickpea. Eurasian J Soil Sci 5:182–191. doi: 10.18393/ejss.2016.3.182-191. [DOI] [Google Scholar]
- 5.Kloepper JW, Ryu CM, Zhang S. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathol 94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 6.Lugtenberg BJJ, Chin-A-Woeng TFC, Bloemberg GV. 2002. Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81:373–383. doi: 10.1023/A:1020596903142. [DOI] [PubMed] [Google Scholar]
- 7.Moeinzadeh A, Sharifzadeh F, Ahmadzadeh M, Tajabadi FH. 2010. Biopriming of sunflower (Helianthus annuus L.) seed with Pseudomonas fluorescens for improvement of seed invigoration and seedling growth. Aust J Crop Sci 4:564–570. [Google Scholar]
- 8.Ramey BE, Koutsoudis M, Von Bodman SB, Fuqua C. 2004. Biofilm formation in plant-microbe associations. Curr Opin Microbiol 7:602–609. doi: 10.1016/j.mib.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, Shen Q. 2011. Bacillus subtilis SQR9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47:495–506. doi: 10.1007/s00374-011-0556-2. [DOI] [Google Scholar]
- 10.Xu Z, Shao J, Li B, Yan X, Shen Q, Zhang R. 2013. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl Environ Microbiol 79:808–815. doi: 10.1128/AEM.02645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:1621–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Zhang R, Wang D, Qiu M, Feng H, Zhang N, Shen Q. 2014. Enhanced control of cucumber wilt disease by Bacillus amyloliquefaciens SQR9 by altering the regulation of its DegU phosphorylation. Appl Environ Microbiol 80:2941–2950. doi: 10.1128/AEM.03943-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–268. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudrappa T, Czymmek KJ, Paré PW, Bais HP. 2008. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 18.Mielich-Süss B, Lopez D. 2015. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. doi: 10.1111/1462-2920.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain M, Heilmann C, Peters G, Herrmann M. 2001. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb Pathog 31:261–270. doi: 10.1006/mpat.2001.0469. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama K, Mizuguchi H, Araki Y, Kaya S, Ito E. 1989. Biosynthesis of linkage units for teichoic acids in Gram positive bacteria distribution of related enzymes and their specificities for UDP-sugars and lipid-linked intermediates. J Bacteriol 171:940–946. doi: 10.1128/jb.171.2.940-946.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarevic V, Soldo B, Medico N, Pooley H, Bron S, Karamata D. 2005. Bacillus subtilis α-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71:39–45. doi: 10.1128/AEM.71.1.39-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. 2008. The major and minor wall teichoic acids prevent the sidewall localization of vegetative dl-endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- 25.Winstel V, Kühner P, Salomon F, Larsen J, Skov R, Hoffmann W, Peschel A, Weidenmaier C. 2015. Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 6:e00632-15. doi: 10.1128/mBio.00632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julkowska D, Obuchowski M, Holland IB, Seror SJ. 2005. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J Bacteriol 187:65–76. doi: 10.1128/JB.187.1.65-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Yan F, Chai Y, Liu H, Kolter R, Losick R, Guo J. 2013. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol 15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter A, Gottschalk G, Sussmuth R, Borriss R. 2006. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB42. J Bacteriol 188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davey ME, Caiazza NC, O’Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietel K, Beator B, Budiharjo A, Fan B, Borriss R. 2013. Bacterial traits involved in colonization of Arabidopsis thaliana roots by Bacillus amyloliquefaciens FZB42. Plant Pathol J 29:59–66. doi: 10.5423/PPJ.OA.10.2012.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross M, Cramton SE, Gotz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chowdhury SP, Hartmann A, Gao XW, Borriss R. 2015. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—a review. Front Microbiol 6:780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Compant S, Duffy B, Nowak J, Clément C, Barka EA. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cascioferro S, Cusimano M, Schillaci D. 2014. Antiadhesion agents against Gram-positive pathogens. Future Microbiol 9:1209–1220. doi: 10.2217/fmb.14.56. [DOI] [PubMed] [Google Scholar]
- 35.Nakano M, Corbell N, Besson J, Zuber P. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol Gen Genet 232:313–321. [DOI] [PubMed] [Google Scholar]
- 36.Lombardia E, Rovetto A, Arabolaza A, Grau R. 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J Bacteriol 188:4442–4452. doi: 10.1128/JB.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwick J, Noble S, Ellaicy Y, Coe G, Hakey D, King A, Sadauskas A, Faulkner M. 2017. AhpA is a peroxidase expressed during biofilm formation in Bacillus subtilis. Microbiologyopen 6:e00403. doi: 10.1002/mbo3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobley L, Li B, Wood J, Kim S, Naidoo J, Ferreira A, Khomutov M, Khomutov A, Stanley-Wall N, Michael A. 2017. Spermidine promotes Bacillus subtilis biofilm formation by activating expression of the matrix regulator slrR. J Biol Chem 292:12041–12053. doi: 10.1074/jbc.M117.789644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, Mcguinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of novel wall teichoic acid inhibitors as effective anti-MRSA β-lactam combination agents. Chem Biol 20:272–284. doi: 10.1016/j.chembiol.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, Götz F, Peschel A. 2008. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol 298:505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Weidenmaier C, Goerke C, Wolz C. 2012. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol 20:243–250. doi: 10.1016/j.tim.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Soldo B, Lazarevic V, Margot P, Karamata D. 1993. Sequencing and analysis of the divergon comprising gtaB, the structural gene of UDP-glucosepyrophosphorylase of Bacillus subtilis 168. J Gen Microbiol 139:3185–3195. doi: 10.1099/00221287-139-12-3185. [DOI] [PubMed] [Google Scholar]
- 43.Soldo B, Lazarevic V, Pooley HM, Karamata D. 2002. Characterization of a Bacillus subtilis thermosensitive teichoic acid-deficient mutant: gene mnaA (yvyH) encodes the UDP-N- acetylglucosamine 2-epimerase. J Bacteriol 184:4316–4320. doi: 10.1128/JB.184.15.4316-4320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. 2012. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3:e00184-12. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law A, Gu Y, Marshall V. 2001. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol Adv 19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 46.Quelas JI, López-García SL, Casabuono A, Althabegoiti MJ, Mongiardini EJ, Pérez-Giménez J, Couto A, Lodeiro AR. 2006. Effects of N-starvation and C-source on Bradyrhizobium japonicum exopolysaccharide production and composition, and bacterial infectivity to soybean roots. Arch Microbiol 186:119–128. doi: 10.1007/s00203-006-0127-3. [DOI] [PubMed] [Google Scholar]
- 47.Badri DV, Weir TL, van der Lelie D, Vivanco JM. 2009. Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol 20:642–650. doi: 10.1016/j.copbio.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Mark GL, Dow JM, Kiely PD, Higgins H, Haynes J, Baysse C, Abbas A, Foley T, Franks A, Morrissey J, O’Gara F. 2005. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc Natl Acad Sci U S A 102:17454–17459. doi: 10.1073/pnas.0506407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang N, Yang D, Wang D, Miao YZ, Shao JH, Zhou X, Xu ZH, Li Q, Feng HC, Li SQ, Shen QR, Zhang RF. 2015. Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16:685. doi: 10.1186/s12864-015-1825-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kierul K, Voigt B, Albrecht D, Chen XH, Carvalhais LC, Borriss R. 2015. Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Microbiology 161:131–147. doi: 10.1099/mic.0.083576-0. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 53.Qiu M, Xu Z, Li X, Li Q, Zhang N, Shen Q, Zhang R. 2014. Comparative proteomics analysis of Bacillus amyloliquefaciens SQR9 revealed the key proteins involved in in situ root colonization. J Proteome Res 13:5581–5591. doi: 10.1021/pr500565m. [DOI] [PubMed] [Google Scholar]
- 54.Zhou C, Shi L, Ye B, Feng H, Zhang J, Zhang R, Yan X. 2017. phe*, an effective host-genotype-independent counter-selectable marker for marker-free chromosome deletion in Bacillus amyloliquefaciens. Appl Microbiol Biotechnol 101:217–227. doi: 10.1007/s00253-016-7906-9. [DOI] [PubMed] [Google Scholar]
- 55.Hamon M, Lazazzera B. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. [DOI] [PubMed] [Google Scholar]
- 56.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smit G, Kijne JW, Lugtenberg BJJ. 1986. Correlation between extracellular fibrils and attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol 168:821–827. doi: 10.1128/jb.168.2.821-827.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dardanelli M, Angelini J, Fabra A. 2003. A calcium-dependent bacterial surface protein is involved in the attachment of rhizobia to peanut roots. Can J Microbiol 49:399–405. doi: 10.1139/w03-054. [DOI] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Li B, Li Q, Xu Z, Zhang N, Shen Q, Zhang R. 2014. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front Microbiol 5:636. doi: 10.3389/fmicb.2014.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.