Pterostilbene, an analog of resveratrol, is a stilbene allelochemical produced by plants to inhibit microbial infection. As a potent antioxidant, pterostilbene acts more effectively than resveratrol as an antifungal agent. Bacterial degradation of this plant natural product would affect the allelopathic efficacy and fate of pterostilbene and thus its ecological role. This study explores the isolation and abundance of bacteria that degrade resveratrol and pterostilbene in peanut tissues and rhizosphere, the catabolic pathway for pterostilbene, and the molecular basis for the initial cleavage of pterostilbene. If plant allelopathy is an important process in agriculture and management of invasive plants, the ecological role of bacteria that degrade the allelopathic chemicals must be equally important.
KEYWORDS: Sphingobium, allelopathy, carotenoid cleavage oxygenase, degradation pathway, pterostilbene
ABSTRACT
Many plants produce allelopathic chemicals, such as stilbenes, to inhibit pathogenic fungi. The degradation of allelopathic compounds by bacteria associated with the plants would limit their effectiveness, but little is known about the extent of biodegradation or the bacteria involved. Screening of tissues and rhizosphere of peanut (Arachis hypogaea) plants revealed substantial enrichment of bacteria able to grow on resveratrol and pterostilbene, the most common stilbenes produced by the plants. Investigation of the catabolic pathway in Sphingobium sp. strain JS1018, isolated from the rhizosphere, indicated that the initial cleavage of pterostilbene was catalyzed by a carotenoid cleavage oxygenase (CCO), which led to the transient accumulation of 4-hydroxybenzaldehyde and 3,5-dimethoxybenzaldehyde. 4-Hydroxybenzaldehyde was subsequently used for the growth of the isolate, while 3,5-dimethoxybenzaldehyde was further converted to a dead-end metabolite with a molecular weight of 414 (C24H31O6). The gene that encodes the initial oxygenase was identified in the genome of strain JS1018, and its function was confirmed by heterologous expression in Escherichia coli. This study reveals the biodegradation pathway of pterostilbene by plant-associated bacteria. The prevalence of such bacteria in the rhizosphere and plant tissues suggests a potential role of bacterial interference in plant allelopathy.
IMPORTANCE Pterostilbene, an analog of resveratrol, is a stilbene allelochemical produced by plants to inhibit microbial infection. As a potent antioxidant, pterostilbene acts more effectively than resveratrol as an antifungal agent. Bacterial degradation of this plant natural product would affect the allelopathic efficacy and fate of pterostilbene and thus its ecological role. This study explores the isolation and abundance of bacteria that degrade resveratrol and pterostilbene in peanut tissues and rhizosphere, the catabolic pathway for pterostilbene, and the molecular basis for the initial cleavage of pterostilbene. If plant allelopathy is an important process in agriculture and management of invasive plants, the ecological role of bacteria that degrade the allelopathic chemicals must be equally important.
INTRODUCTION
Allelopathic chemicals produced by plants can affect seed germination and growth of other nearby plants (1, 2), symbiotic plant associations (3), aggregation of microbial communities (4), and interactions with pathogens (5). Plants can release diverse organic exudates that contain allelochemicals into the rhizosphere in substantial quantities (6–8). The exudates greatly increase microbial activity in the rhizosphere, resulting in bacterial population densities one to two orders of magnitude higher than those in in bulk soil (9). Microbial diversity in the rhizosphere, however, is significantly lower than that in bulk soil, suggesting that strong selective forces are at play (7). Through the exudation of specific compounds, plants can select for specific microbial partners and thus regulate the soil microbial community in their immediate vicinity (6, 7). These interactions between plant roots and rhizosphere bacteria have important ecological and evolutionary significance and can lead to beneficial associations that foster plant growth or to associations that negatively impact plant health (6, 7, 10). One such negative association is the interference of soil bacteria in allelochemical-mediated plant-plant or plant-fungus competition (11, 12). Although there is a growing awareness of the phenomenon, the microbes involved and their ecological roles are poorly understood.
Stilbenes are bioactive, polyphenolic plant compounds with promising chemotherapeutic properties related to anti-inflammation, cancer, diabetes, obesity, and dyslipidemia (13–16). In vitro studies on cancer cells showed that stilbene compounds, especially pterostilbene, act as inducers of multiple cell-death pathways, including apoptosis, cell cycle arrest, and autophagy (13, 17, 18). Trans-resveratrol, pterostilbene, and other stilbenoid compounds are considered to be allelochemicals due to their antimicrobial activity and proposed role in resistance to fungal infection (19, 20). They are produced by a wide range of spermatophytes, and potential biomedical applications of the compounds from members of the family Vitaceae (grapes) and from Arachis hypogaea L. (peanut) have received wide attention (21–23). Production of the various stilbenoids represents a major factor in the resistance of peanut plants to agriculturally significant fungal pathogens, including Aspergillus flavus, Aspergillus caelatus, Aspergillus niger, Botrytis cinerea, and other species (23, 24). Natural stilbenes are likely to be quickly biodegraded by soil bacteria in situ, but the identities and ecological roles of such bacteria are unknown. Recently, we reported the isolation and characterization of an Acinetobacter sp. strain (JS678) that grows on resveratrol (25). Pterostilbene is a methylated derivative of resveratrol. The dimethyl ether structure of pterostilbene is believed to be more stable and more lipophilic than that of resveratrol, which leads to reduced metabolism and greater bioavailability (13, 15, 26). Whether and how it is biodegraded by bacteria have not been reported.
The enzymes involved in initial cleavage of the central double bond of stilbenes in bacteria have not been widely studied. The physiological roles of such enzymes have been established in only two isolates (25, 27). The enzyme responsible for initial cleavage of lignostilbene as part of a productive catabolic pathway was first identified in Sphingomonas paucimobilis TMY1009 and was designated lignostilbene-α,β-dioxygenase (27–29). Recently, we identified a homologous oxygenase that initiates the biodegradation of resveratrol in an Acinetobacter strain isolated from the rhizosphere of peanuts (25). The most extensive biochemical characterization of related oxygenases has been done with the homologues of carotenoid cleavage oxygenases (CCOs), NOV1 and NOV2, in Novosphingobium aromaticivorans DSM 12444 (30, 31). Both enzymes are iron-containing oxygenases that cleave the interpenyl α,β double bond of resveratrol and several related substrates, but which did not attack carotenoids and apocarotenoids (30).
Here, we report the isolation of stilbenoid-degrading bacteria from peanut plants and the initial steps in the degradation pathway of pterostilbene. The results reveal a surprising abundance and diversity of plant-associated stilbene-degrading bacteria and provide the basis for evaluating the ecological role of the bacteria in plant allelopathy.
RESULTS
Abundance of stilbenoid-degrading bacteria associated with peanut plants.
Previous enrichments with resveratrol (25) yielded six bacterial isolates (4 species of Pseudomonas, 1 species of Acinetobacter, and 1 species of Burkholderia) from peanut rhizosphere soil samples. In the present study, 25 strains were further isolated from rhizosphere soils, shells, and seeds of the peanut plants by using resveratrol or pterostilbene as the sole carbon source in minimal media (Table S1).
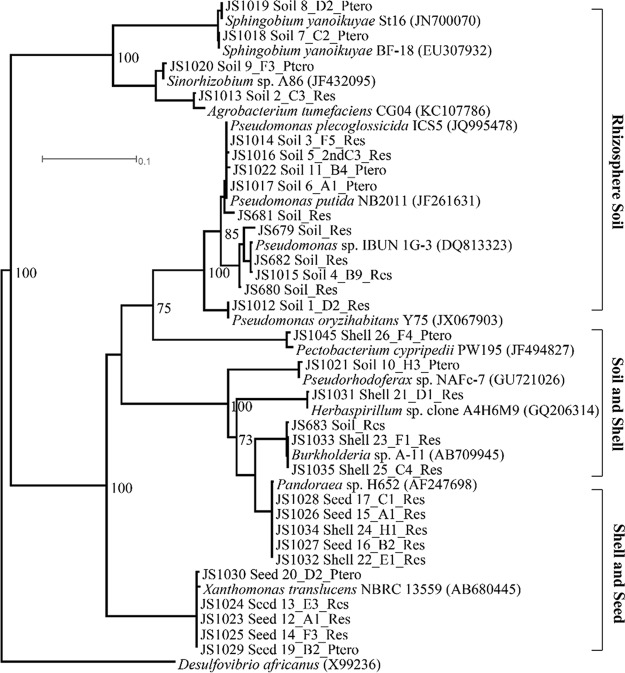
Phylogenetic analyses of the 16S rRNA genes indicated that most of the isolates from the rhizosphere soil were clustered into two clades, the Sphingobium-Sinorhizobium and Pseudomonas groups, and were clearly distinct from strains isolated from peanut seeds and shells (Fig. 1). Isolates from peanut shells and seeds tended to group together, indicating that their phylogenies and niches were similar. The analyses suggest that, although the three types of samples were located in close proximity, stilbenoid-degrading bacteria displayed distinct community clusters in the different microhabitats.
FIG 1.
Phylogenetic maximum-likelihood analyses of 16S rRNA genes of bacterial isolates from peanut plants. Isolate designations identify the strain number (the initial two letters followed by three or four numbers), isolation source (plant tissues or rhizosphere soil), plate codes (numbers with letters), and enrichment substrates (Res, resveratrol; Ptero, pterostilbene). Numbers at branching points denote bootstrap values as a percentage of successful resamplings out of 100.
The most probable number (MPN) results indicated that resveratrol- and pterostilbene-degrading bacteria were enriched in both peanut rhizosphere soil and tissues compared to those in nonrhizosphere soil (Table 1). Of the cultivable heterotrophs associated with peanut plants, bacteria that could degrade stilbenes (both resveratrol and pterostilbene) represented 39.2% in seeds, 4.6% in shells, and 0.2% in rhizosphere soil (Table 1). In addition, all of the strains isolated on pterostilbene could grow on resveratrol, whereas only 11 out of 16 strains isolated on resveratrol could grow on pterostilbene (Table S1).
TABLE 1.
Enumeration of stilbenoid-degrading bacteria from the peanut field in Dawson, Georgia (October 2012)
| Substrate for enrichments | Cell counts (95% confidence limits) froma: |
|||
|---|---|---|---|---|
| Peanut |
Open field bulk soil | |||
| Seed | Shell | Rhizosphere soil | ||
| Resveratrol | 486 (189–1,249) | 1.6 × 105 (6.1 × 104–4.0 × 105) | 9.2 × 105 (3.6 × 105–2.4 × 106) | 1.2 × 104 (4.8 × 103–3.2 × 104) |
| Pterostilbene | 265 (103–681) | 4.0 × 105 (1.6 × 105–1.0 × 106) | 1.0 × 105 (4.0 × 104–2.6 × 105) | 547 (212–1,406) |
| 1/4 TSAb | 1,916 (745–4,924) | 1.2 × 107 (4.7 × 106–3.1 × 107) | 4.6 × 108 (1.8 × 108–1.2 × 109) | 5.5 × 105 (2.1 × 105–1.4 × 106) |
Cell counts (viable cells · g−1 wet weight) and 95% confidence limit (lower and upper, in parentheses) by eight-well microMPN methods.
TSA, tryptic soy agar.
Pterostilbene biodegradation by strain JS1018.
Sphingobium sp. strain JS1018 was used for subsequent experiments to investigate growth on pterostilbene. The strain was grown on pterostilbene in short-term (24 h; Fig. 2A1 and A2) and midterm incubations (7 days; Fig. 2B) in order to analyze the occurrence and consumption of intermediate metabolites. In the short-term test, metabolites that accumulated transiently were identified by high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) analyses as 4-hydroxybenzaldehyde (Fig. 2A1) and 3,5-dimethoxybenzaldehyde (Fig. 2A2). A novel product with a retention time (RT) of 6.23 min, presumably derived from 3,5-dimethoxybenzaldehyde, accumulated as a dead-end product (Fig. 2A2 and B).
FIG 2.
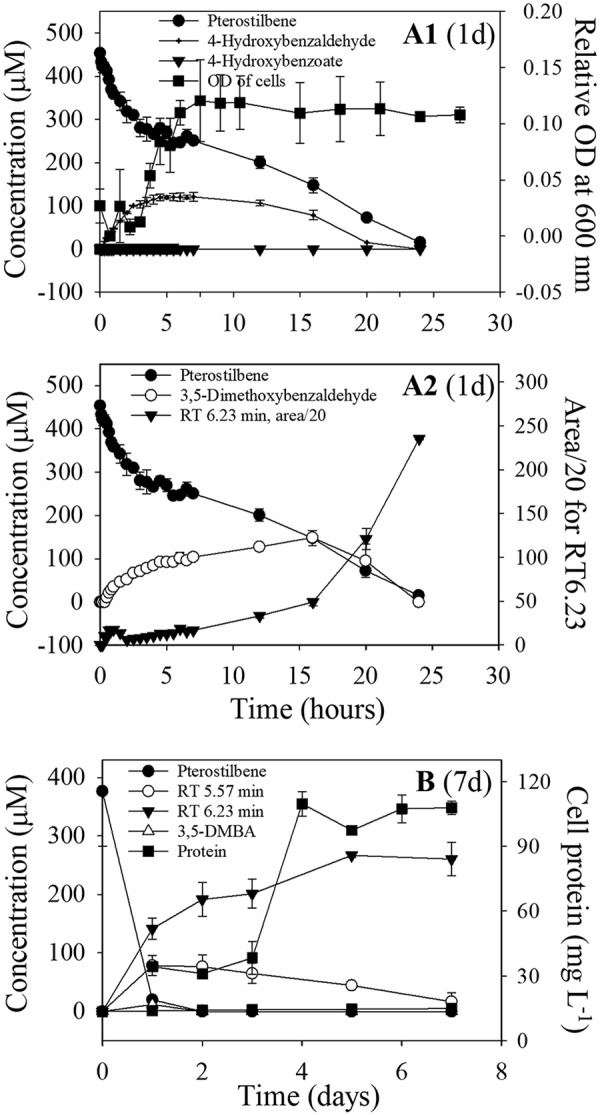
Degradation kinetics of pterostilbene by succinate-grown cells of Sphingobium sp. strain JS1018 over 24 h (A1 and A2) and over 7 days (B). Two novel metabolites with HPLC retention times of 5.57 and 6.23 min are labeled (area/20) in panels A2 and B. 3,5-DMBA, 3,5-dimethoxybenzaldehyde.
In the 7-day test (Fig. 2B), 4-hydroxybenzaldehyde and 3,5-dimethoxybenzaldehyde were not detected, but a derivative of 3,5-dimethoxybenzaldehyde with an RT of 6.23 min (major) and another with an RT of 5.57 min (minor) accumulated. The growth yield of cells grown on resveratrol (Table 2) was substantially higher than that on pterostilbene, which, taken with accumulation of the metabolites from pterostilbene, indicates that the pterostilbene-degrading bacteria only used half of the molecule for growth.
TABLE 2.
Growth yields of JS1018 with various substrates
| Substrate | Yield (g protein·g substrate−1) |
|---|---|
| Pterostilbene | 0.146 ± 0.011 |
| Resveratrol | 0.217 ± 0.009 |
| 4-Hydroxybenzaldehyde | 0.218 ± 0.013 |
| 3,5-Dihydroxybenzaldehyde | 0.203 ± 0.006 |
| 3.5-Dimethoxybenzaldehyde | No growth |
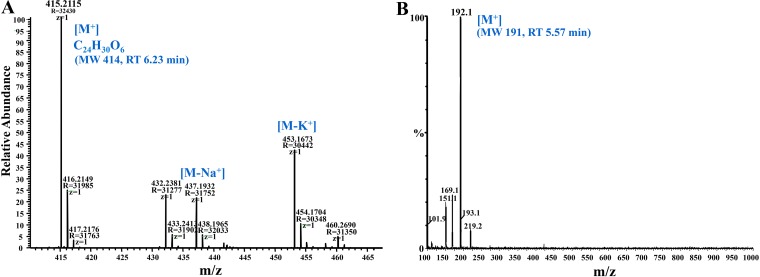
All reference compounds and intermediate products analyzed by HPLC were further confirmed by LC-MS analysis. The unknown metabolites (RTs of 6.23 and 5.57 min in HPLC) were collected and analyzed by LC-MS. The mass fragmentation pattern for the product that eluted at an RT of 6.23 indicated major ions at m/z (mass-to-charge ratio) values of 415 (M+), 437 (M-Na+), and 453 (M-K+), respectively (Fig. 3A and Fig. S1A). The results are consistent with a molecular weight of 414.2 and a molecular formula of C24H31O6 (Fig. 3A) and suggest a condensation product from 3,5-dimethoxybenzaldehyde. The metabolite eluting from HPLC at an RT of 5.57 gave a conjugate ion with an m/z value of 192, indicating a molecular weight of 191 (Fig. 3B). The two dead-end products at an RT of 6.23 and 5.57 min also accumulated during pterostilbene degradation by two other isolates, Agrobacterium tumefaciens JS1013 (Fig. S1) and an Acinetobacter sp. isolate (data not shown).
FIG 3.
Mass fragmentation patterns of pterostilbene metabolites identified by LC-MS from Sphingobium sp. JS1018. (A) Metabolite (MW 414) with an HPLC RT of 6.23 min (RT of 28.18 min on LC-MS). (B) Metabolite (MW 191) with an HPLC RT of 5.57 min (or an RT of 20.28 min on LC-MS).
Enzyme assays with cell extracts from strain JS1018.
To determine the initial steps in the catabolic pathway of stilbene, assays of enzymes from crude cell extracts were conducted with resveratrol or pterostilbene as the substrate (30, 32). Enzymes in lysates from pterostilbene-grown cells converted resveratrol to 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde (Fig. 4A), with a specific activity of 454 ± 87 nmol·mg protein−1 · min−1. After addition of NAD+ at 40 min, the two intermediates were converted to 4-hydroxybenzoate and 3,5-dihydroxybenzoate. No activity was detected in cells grown on succinate.
FIG 4.
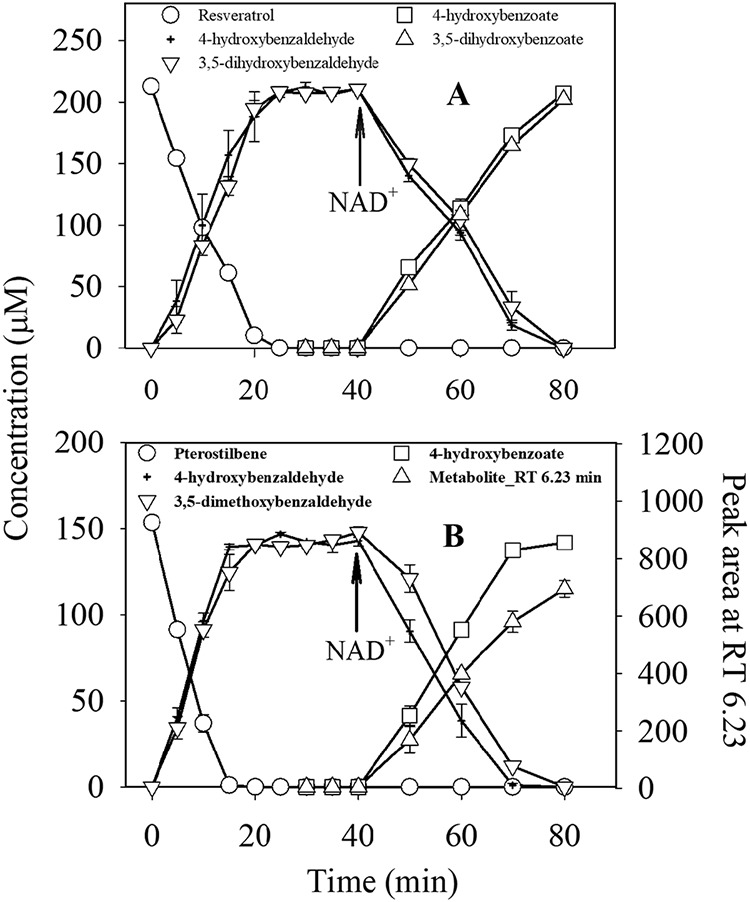
Transformation of resveratrol (A) or pterostilbene (B) by dialyzed crude extract of JS1018 (26 ± 5 μg of protein·ml−1). NAD (3 mM final) was added at 40 min during the incubation period.
When pterostilbene was the substrate of the assay, the specific activity (485 ± 93 nmol·mg protein−1 · min−1) was similar to the activity with resveratrol. 4-Hydroxybenzaldehyde accumulated and was oxidized to 4-hydroxybenzoate upon addition of NAD+ (Fig. 4B). 3,5-Dimethoxybenzaldehyde accumulated and was slowly converted to the unidentified dead-end metabolite with an RT of 6.23 after addition of NAD+. The results were consistent with the pattern of degradation by the intact cells (Fig. 2A2 and B). There was no accumulation of a metabolite consistent with 3,5-dimethoxybenzoate. When the above-described experiments were repeated with 3,5-dimethoxybenzaldehyde as the substrate, the metabolite with an RT of 6.23 accumulated (Fig. S2), which indicated clearly that the metabolite was synthesized from 3,5-dimethoxybenzaldehyde. Extracts prepared from resveratrol-grown JS1018 cells gave very similar results to those from pterostilbene-grown cells (Fig. S3), which indicates that different preexposures to the two compounds did not change the end metabolites in the degradation.
Gene identification and overexpression of CCO from JS1018 in Escherichia coli.
The draft genome sequence of strain JS1018 revealed that two putative CCO homologs (1,479 bp for cco1 and 1,500 bp for cco2) might encode CCO1 (492 amino acids [aa]) and CCO2 (499 aa), with 54% and 32% amino acid similarity to resveratrol cleavage oxygenase (RCO) from Acinetobacter sp. strain JS678 (GenBank accession number KY888940). The next most closely related biochemically characterized homolog to CCO1 and CCO2 was lignostilbene-α,β-dioxygenase isozyme I from Sphingomonas paucimobilis (GenPept accession number Q53353), with 49.4% and 31.5% amino acid similarity.
To establish the gene function, the putative cco genes from JS1018 were cloned into Escherichia coli, and the enzyme activities were tested by whole-cell biotransformation assay. The heterologously expressed cco2 gene gave a band of the appropriate size (55.4 kDa) on polyacrylamide gel electrophoresis, but it was not active in degrading pterostilbene or resveratrol (with a specific activity of 0 μmol·mg protein−1 · min−1). E. coli cells expressing cco1 (E. coli pJS801) rapidly transformed pterostilbene (Fig. 5A), with a specific activity of 0.22 ± 0.02 μmol·mg protein−1 · min−1. 4-Hydroxybenzaldehyde and 3,5-dimethoxybenzaldehyde accumulated with the disappearance of pterostilbene. E. coli pJS801 could also transform resveratrol under the same conditions (Fig. 5B), with a specific activity (0.18 ± 0.04 μmol·mg protein−1 · min−1) that was consistent with the ratios of activities in cell extracts from JS1018. The results clearly indicate that CCO1 is a stilbene oxygenase, and also that an aldehyde oxidase expressed in JS1018 is required for subsequent conversion of the aldehydes to acids that can enter central metabolism.
FIG 5.
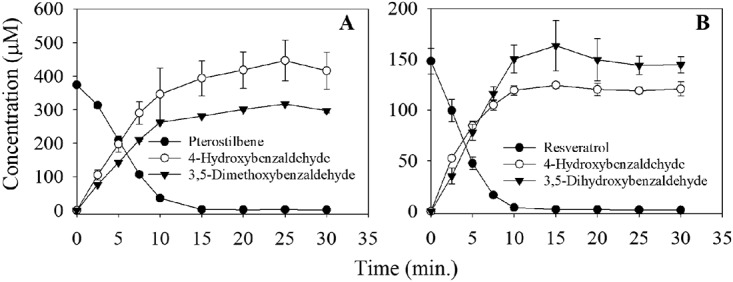
Cleavage of pterostilbene (A) and resveratrol (B) by E. coli cells expressing recombinant CCO1 enzyme from strain JS1018. Rosetta 2 (DE3) cells (without insert) as a negative control did not transform the two stilbenes (data not shown).
From the phylogenetic analysis, a subset of the CCOs form a cluster with enzymes originally identified as lignostilbene dioxygenases (Fig. S4). Alignment of the amino acid sequence of CCO1 with those of previously characterized carotenoid dioxygenases revealed a conserved region of CCO1 that contains four histidines and three glutamic acids (Fig. S5), indicating that CCO1 has an iron binding site with tetradentate iron coordination. The results were further confirmed by modeling the CCO1, using the crystal structure of stilbene cleavage oxygenase from Novosphingobium aromaticivorans DSM 12444 (Nov1 enzyme) as a template (Fig. S6). The predicted structure revealed the iron binding site of CCO1 and justified the addition of iron to the enzyme assays.
DISCUSSION
Stilbenoid-degrading bacteria in peanut allelopathy.
The fact that peanut seeds also contained stilbenoid degraders extends the findings of Sobolev and colleagues (33), which revealed the presence of bacterial endophytes in the seeds but not their catabolic capabilities. Although stilbenoid-degrading bacteria in the rhizosphere outnumbered those in control soil from a nearby open field, the overall community of heterotrophs was much larger in the rhizosphere. This finding suggests an important ecological role of stilbene catabolic capability in the peanut-associated microbiome. The phylogenetic analyses of 16S rRNA genes showed that stilbene-degrading bacteria were quite diverse. The biodegradation of resveratrol or pterostilbene is probably not genus- or species-specific and is presumably strain-specific, which suggests that the catabolic trait enables diverse bacteria to opportunistically degrade pterostilbene and its analogs. Because growth-dependent enumeration methods might underestimate microbial populations, molecular methods based on our identification of the key genes will be required for more accurate evaluation of the in situ stilbenoid-degrading guilds.
Consistent with previous work by Inderjit and colleagues (1) showing that soil bacteria can diminish or eliminate allelopathic effects on target organisms, our results suggest that the bacteria degrade stilbenes as a source of carbon and energy and thus are likely to reduce their allelopathic effects. The high phylogenetic diversity among the isolates supports the hypothesis that the bacteria are opportunistic. The alternative hypothesis is that the bacteria are involved in a symbiosis with the plants and provide benefits to the plants that offset the costs of elimination of the allelopathic chemicals. The fact that many of the isolates are related to plant growth-promoting bacteria identified in previous studies (Table S1) supports the second hypothesis (34, 35). The two hypotheses are not mutually exclusive and the bacteria could opportunistically utilize stilbenoids released from plants as carbon and energy sources while enhancing plant growth by producing plant hormones and/or improving plant nutrition. Such complex interactions seem especially likely for the endophytic bacteria in seeds and shells, but the potential for specific plant-microbe interactions was not evaluated here.
Catabolic pathways for resveratrol and pterostilbene.
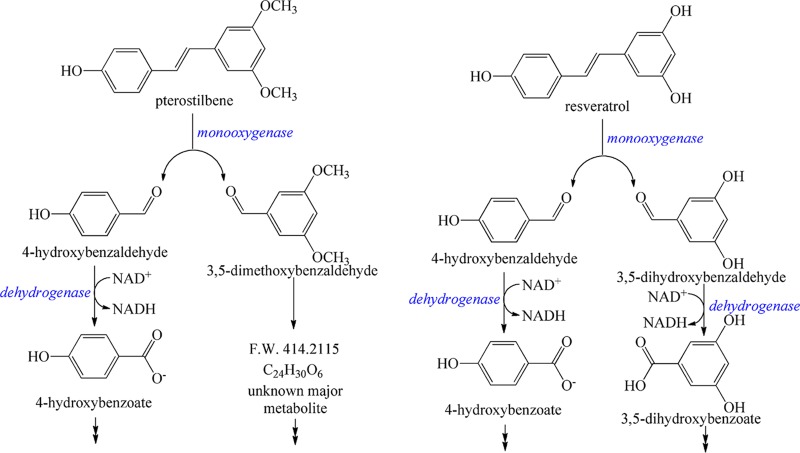
Our results indicate that enzymes in strain JS1018 catalyze cleavage of resveratrol at the interphenyl α-β double bond to produce 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde (Fig. 6). The reaction is consistent with cleavage of resveratrol by enzymes in Novosphingobium aromaticivorans DSM 13444 (30) (which was not reported to grow on resveratrol) and Acinetobacter sp. strain JS678 (25) (grown on resveratrol). The conversion of the two resveratrol pathway intermediates, 4-hydroxybenzaldehyde and 3,5-dihydroxybenzaldehyde, into 4-hydroxybenzoate and 3,5-dihydroxybenzoate was consistent with the resveratrol catabolic pathway in Acinetobacter sp. strain JS678 (25). The results presented here establish that the initial reactions in pterostilbene biodegradation and subsequent assimilation of 4-hydroxybenzaldehyde are the same as with resveratrol (Fig. 6). The results are consistent with the hypothesis that the ability to degrade pterostilbene arose as an extension of the ability to degrade resveratrol, and the pterostilbene catabolic pathway is likely in the early stages of evolution. It is likely that 3,5-dimethoxybenzaldehyde formed in the initial oxidation of pterostilbene was subsequently converted to the unidentified major product with an HPLC RT of 6.23 min (Fig. S2) and a molecular weight of 414. The synthesis, identities, and biological functions of the novel metabolites are under investigation.
FIG 6.
Proposed degradation pathways for pterostilbene and resveratrol in Sphingobium sp. JS1018.
CCOs are nonheme iron-type enzymes that are widely distributed in plants, animals, bacteria, and fungi, where they catalyze cleavage of carotenoids. The products are often aldehydes or ketones, essential precursors of vitamins, plant hormones (abscisic acid), visual pigments, signaling molecules, flavors, aromas, and defense compounds (36–38). Several CCOs have been biochemically characterized (36). The targeting substrates in degradation of CCOs may vary. For example, CCOs from Novosphingobium aromaticivorans (NOV1 and NOV2) catalyzed cleavage of resveratrol, but not that of carotenoids, whereas CCOs from Bradyrhizobium spp. catalyzed only the cleavage of farnesol (30). However, the above studies did not establish the physiological roles of the CCO homologs. The CCOs from Acinetobacter sp. strain JS678, reported previously as a resveratrol degrader, could not degrade pterostilbene (25). The results suggest that pterostilbene has selected for the evolution of a CCO with a broader substrate range. Here, the ability of Sphingobium sp. strain JS1018 to grow on resveratrol and pterostilbene, cleavage of the compounds in cell extracts, heterologous expression in E. coli, and analogy with the resveratrol cleavage oxygenase from Acinetobacter sp. JS678, the function of which was established by knockouts (25), provide strong evidence that CCO1 catalyzes cleavage of resveratrol and pterostilbene and initiates the stilbenoid degradation pathway.
Evolution of stilbenoid diversity in plant allelopathy.
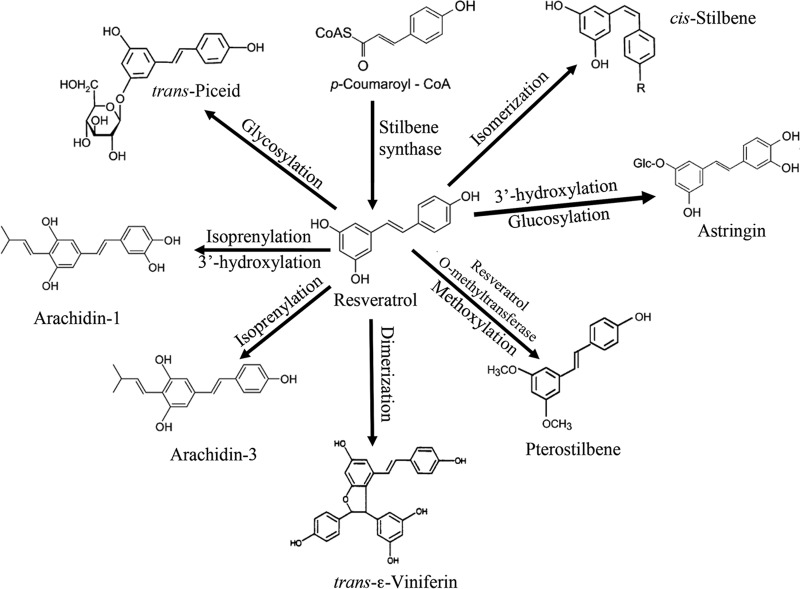
As a family of plant secondary metabolites, stilbenoids are actively involved in both constitutive (phytoanticipins) and inducible (phytoalexins) defense mechanisms in plants (22). Sobolev (23) established that when peanut seeds were challenged by microbial pathogens, resveratrol was initially detected at high concentrations, followed by its exponential decline and the appearance of multiple stilbenoid derivatives. Resveratrol seems to be the precursor of a variety of analogs synthesized in plants in response to pathogen attack (22, 23) (Fig. 7). Most analogs are simple derivatives of resveratrol that contain additional functional groups. The two methyl ether groups in pterostilbene enable it to be more effective than resveratrol as an antifungal agent (24, 39) and seem to make it less readily biodegradable. It seems reasonable to suggest that the biosynthetic pathway of resveratrol evolved first, and that subsequent modifications emerged when the original molecule lost its effectiveness toward pathogens. Loss of effectiveness could result from two different mechanisms. The first would be the emergence of resistance mechanisms in target organisms, which is well supported by the existence of resveratrol-cleaving enzymes in fungi (Fig. S4). The second, less obvious mechanism would be the evolution of catabolic pathways in soil bacteria, leading to the rapid biodegradation of resveratrol or other allelopathic chemicals. Modifications of the molecules by plants (Fig. 7) would circumvent both mechanisms that cause loss of effectiveness. The two models would both lead to an arms race, selecting for a variety of resveratrol derivatives and resistance mechanisms. This exploration of the isolation and abundance of stilbenoid-degrading bacteria and the identification of the key enzyme will provide the basis for further evaluation of the ecological roles of the bacteria in plant-fungus interactions and plant allelopathy in general.
FIG 7.
Analogs of resveratrol synthesized by plants as antifungal allelochemicals (modified from reference 22).
MATERIALS AND METHODS
Isolation and growth of stilbenoid-degrading bacteria.
Rhizosphere soil and plant tissues (seeds and shells) of peanut plants (Arachis hypogaea L.) were collected at a USDA experimental farm near Dawson, Georgia, USA (N 31°47.088′, W 84°29.238′). Bacteria were isolated by spread-plating serial dilutions of soil or homogenized samples of surface-sterilized plant tissue on agar plates with one-quarter-strength minimal salt base (MSB) (40) containing resveratrol or pterostilbene (400 μM) as the sole carbon source. The seeds were surface sterilized by immersion in 2% bleach for 2 min, following a previously described method (41). Isolates that grew on pterostilbene or resveratrol medium were purified by three transfers on agar plates with the pterostilbene or resveratrol medium.
Identification and phylogenetic analysis of the bacterial strains.
The genomic DNA of the isolates was extracted with a PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). The bacterial 16S rRNA genes were amplified by PCR with 8F (42) and 1489R primers (43), purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA), and sequenced using both the forward and reverse primer for each strain by Genewiz, Inc. (Piscataway, NJ). The final sequences were obtained by merging the forward and reverse sequences after base error correction. The sequences (1,361 bp) were analyzed by using BLASTn in GenBank (https://www.ncbi.nlm.nih.gov/) (44). Phylogenetic analyses of the sequences were performed by using PhyML (45) and the Phylogeny Inference Package (PHYLIP) version 3.695 (46) with LG and JTT models, respectively.
Quantification of stilbenoid-degrading bacteria in peanut plants.
Samples of peanut rhizosphere, seeds, and shells were collected in August and October 2012 from a peanut field as described above, with a composited bulk soil sample taken from a nearby unplanted soil field as the control for the cell count assay. Stilbene-degrading bacteria were enumerated in 96-well microplates (eight replicates) by most probable number (MPN) assays through serial dilutions of homogenized samples in a minimal medium (47) containing resveratrol or pterostilbene (400 μM) as the sole carbon source. The optical density at 600 nm (OD600) was monitored with a microplate reader (48).
Pterostilbene degradation by intact cells of stilbenoid-degrading bacteria.
Sphingobium sp. strain JS1018 was used for determination of the molecular basis for the initial reaction in the pterostilbene catabolic pathway. The strain was grown initially in one-quarter MSB with succinate (1 mM), harvested by centrifugation, washed twice with one-quarter MSB, suspended in one-quarter MSB containing 400 to 500 μM pterostilbene at an initial OD600 of 0.05, and incubated at 22°C with shaking (125 rpm). At appropriate intervals, samples were analyzed by HPLC for quantification of substrates and metabolites. Cell protein was assayed with the bicinchoninic acid (BCA) method.
Enzyme assays.
JS1018 cells were grown for 4 days in one-quarter MSB with resveratrol (Fig. S3) or pterostilbene (Fig. 4) at 0.5 to 1.0 mM added repeatedly every 12 h from concentrated stocks (0.5 M in dimethyl sulfoxide). Cells were harvested by centrifugation, washed twice with ice-cold potassium phosphate buffer (pH 7.2, 50 mM), and suspended in the same buffer. Cells were lysed by three passages through a French pressure cell at 20,000 lb/in2, followed by centrifugation at 20,000 × g (4°C, 30 min). The cell extract was subjected to ultrafiltration with an Amicon Ultra 0.5-ml centrifugal filter device (10 kDa molecular weight cutoff), and the retentate was washed twice with phosphate buffer to remove soluble cofactors.
CCO activity was assayed at 30°C by monitoring the disappearance of resveratrol or pterostilbene (400 μM) and formation of metabolites (30, 49). Specific enzyme activity is defined as the conversion rate in μmol of substrate per mg cell protein per min. Briefly, reaction mixtures in a final volume of 500 μl included cell extracts (0.3 mg protein), resveratrol or pterostilbene (400 μM), and assay buffer. The assay buffer contained phosphate buffer (50 mM, pH 7.2), NaCl (300 mM), sodium ascorbate (10 mM), and FeSO4 (0.5 mM). The FeSO4 was added from a freshly prepared 0.5 M stock acidified with H2SO4. At appropriate intervals, samples were removed, trifluoroacetic acid (TFA) was added at a ratio of 1:100 to stop the reaction, an equal volume of acetonitrile was added, and the sample was centrifuged prior to HPLC analysis.
Genome sequencing for identifying putative CCO genes in strain JS1018.
Genomic DNA from strain JS1018 was extracted and purified according to the standard Illumina protocols. Genome sequencing was performed with an Illumina HiSeq 2000 sequencer at the Center for Genome Research and Biocomputing at Oregon State University (Corvallis, OR). The draft genome was assembled using the Velvet sequence assembler (50), and assembled genes were curated by GeneMark (51, 52) and annotated using UniProt (53).
Cloning and expression of cco genes in E. coli and stilbenoid transformation assays.
Two primer sets were designed based on the putative carotenoid cleavage oxygenase genes cco1 and cco2 that were identified by BLAST in the genome of JS1018 (Table S2). The cco1 gene was amplified by PCR using the primer set 5′-TTTCATATGATGACATCGTGCTTTCCTGACGATCC-3′ and 5′-TTTAAGCTTTCAAGCCGGCAATGGCGA-3′. The cco2 gene was amplified with the primer set 5′-TTT CATATGATGGCAACTTCGTTACGGCACC and 3′-TTTAAGCTTTCAATTTCCCGTCGCATAAGCAGC. After purification with a QIAquick PCR purification kit, PCR products were digested with NdeI and HindIII and ligated into a pET-21a vector (Invitrogen Corp., Carlsbad, CA). The recombinant plasmids were transformed into Escherichia coli DH5α (New England BioLabs, Ipswich, MA) for screening and maintenance and then subsequently transformed into E. coli Rosetta 2(DE3) competent cells (Novagen) for overexpression according to the manufacturer’s protocol.
CCO activity was tested by growing E. coli (pJS801) cells in LB with ampicillin (50 mg · liter−1) and chloramphenicol (30 mg · liter−1) at 37 °C until the OD600 reached 0.6 to 0.8. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at a final concentration of 0.4 mM. After 6 h of incubation at 30 °C, E. coli pJS801 cells were harvested by centrifugation, washed twice with 40 mM phosphate buffer, suspended in one-quarter MSB (110 mg protein · liter−1) containing resveratrol (200 μM) or pterostilbene (400 μM), and incubated at 30°C with shaking. Samples were collected and analyzed at appropriate intervals, following the methods described above for strain JS1018. Rosetta 2(DE3) cells with pET21a containing no insert served as a negative control for the biotransformation assays.
Analytical methods.
High-performance liquid chromatography (HPLC) analyses of stilbenes and metabolites were performed on an Agilent 1100 system equipped with a diode array detector and a Merck Chromolith column (100 mm by 4.6 mm). The mobile phase consisted of 0.1% trifluoroacetic acid (TFA) in water (part A) and 0.05% TFA in acetonitrile (part B). The gradient consisted of 98% A/2% B, increased linearly to 50% A/50% B over 7 min and then held isocratically for 1 min. The flow rate was 1 ml · min−1. Concentrations of trans-resveratrol and pterostilbene were measured at 305 and 318 nm, respectively. 3,5-Dihydroxybenzaldehyde, 3,5-dimethoxybenzaldehyde, 4-hydroxybenzaldehyde, 4-hydroxybenzoate, and 3,5-dihydroxybenzoate were analyzed under the same HPLC conditions at 284, 212, 284, 284, and 318 nm, respectively. The above compounds were identified and quantified by comparison with standards. Cell cultures or crude extracts were mixed with equal volumes of acetonitrile and centrifuged prior to analysis by HPLC. Extraction efficiencies ranged from 85% to 99% based on spiked standards.
Stilbene metabolites were further analyzed for chemical identification by LC-MS in the Bioanalytical Mass Spectrometry Facility at Georgia Institute of Technology. Mass spectrometry analysis was conducted using a Micromass Quattro liquid chromatograph (Waters Corp., Milford, MA) with a Phenomenex Gemini C18 column (2.1 × 150 mm) and with 5-μm particles. The MS analysis was done in positive mode electrospray ionization (ESI), with a capillary voltage of 3.5 kV and a cone voltage of 25 V. The instrument was scanned from 100 to 1,000 Da at 2.5 s/scan. The LC system was an Agilent 1100. Solvent A was 5% acetonitrile in water, and solvent B was 95% acetonitrile in water. Both solvents contained 0.1% (vol/vol) formic acid. The solvent gradient started at 100% A and was held there for 8 min. The gradient was ramped to 100% B at 45 min and held there until 53 min, when it was reset to 100% A until the end of the run at 62 min. The flow rate was 200 μl · min−1. Twenty microliters of each solution was injected for each sample.
Chemicals.
Resveratrol was purchased from AK Scientific, Inc.; pterostilbene was from Biotang, Inc.; 4-hydroxybenzaldehyde, 4-hydroxybenzoate, and 3,5-dimethoxybenzaldehyde were from Sigma-Aldrich; 3,5-dihydroxybenzaldehyde was from Acros Organics; and 3,5-dihydroxybenzoic acid was from Alfa Aesar.
Data availability.
The 16S rRNA gene sequences of the isolates sequenced here were archived in GenBank under the accession numbers KX507136 to KX507160. The sequences of the two candidate cco genes, cco1 and cco2, were deposited in GenBank under the accession numbers KX523172 and KX523173.
Supplementary Material
ACKNOWLEDGMENTS
We thank Renee S. Arias, Victor S. Sobolev, and Phat M. Dang for help in field sampling and constructive suggestions for isolating peanut seed endophytes.
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011-67019-30184 from the USDA National Institute of Food and Agriculture.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02154-18.
REFERENCES
- 1.Inderjit, Wardle DA, Karban R, Callaway RM. 2011. The ecosystem and evolutionary contexts of allelopathy. Trends Ecol Evol 26:655–662. doi: 10.1016/j.tree.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Latif S, Chiapusio G, Weston LA. 2017. Allelopathy and the role of allelochemicals in plant defenc, p 19–54. In Becard G. (ed), Advances in botanical research: how plants communicate with their biotic environment, vol 82 Academic Press, Cambridge, MA. [Google Scholar]
- 3.Hale AN, Kalisz S. 2012. Perspectives on allelopathic disruption of plant mutualisms: a framework for individual- and population-level fitness consequences. Plant Ecol 213:1991–2006. doi: 10.1007/s11258-012-0128-z. [DOI] [Google Scholar]
- 4.Dias F, Antunes JT, Ribeiro T, Azevedo J, Vasconcelos V, Leao PN. 2017. Cyanobacterial allelochemicals but not cyanobacterial cells markedly reduce microbial community diversity. Front Microbiol 8:1495. doi: 10.3389/fmicb.2017.01495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adrian M, Jeandet P. 2012. Effects of resveratrol on the ultrastructure of Botrytis cinerea conidia and biological significance in plant/pathogen interactions. Fitoterapia 83:1345–1350. doi: 10.1016/j.fitote.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Mhlongo MI, Piater LA, Madala NE, Labuschagne N, Dubery IA. 2018. The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front Plant Sci 9:112. doi: 10.3389/fpls.2018.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Massalha H, Korenblum E, Tholl D, Aharoni A. 2017. Small molecules below-ground: the role of specialized metabolites in the rhizosphere. Plant J 90:788–807. doi: 10.1111/tpj.13543. [DOI] [PubMed] [Google Scholar]
- 9.Morgan JAW, Bending GD, White PJ. 2005. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot 56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- 10.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 11.Mishra S, Upadhyay RS, Nautiyal CS. 2013. Unravelling the beneficial role of microbial contributors in reducing the allelopathic effects of weeds. Appl Microbiol Biotechnol 97:5659–5668. doi: 10.1007/s00253-013-4885-y. [DOI] [PubMed] [Google Scholar]
- 12.Inderjit, Evans H, Crocoll C, Bajpai D, Kaur R, Feng YL, Silva C, Carreon JT, Valiente-Banuet A, Gershenzon J, Callaway RM. 2011. Volatile chemicals from leaf litter are associated with invasiveness of a neotropical weed in Asia. Ecology 92:316–324. doi: 10.1890/10-0400.1. [DOI] [PubMed] [Google Scholar]
- 13.Tsai HY, Ho CT, Chen YK. 2017. Biological actions and molecular effects of resveratrol, pterostilbene, and 3′-hydroxypterostilbene. J Food Drug Anal 25:134–147. doi: 10.1016/j.jfda.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen RJ, Kuo HC, Cheng LH, Lee YH, Chang WT, Wang BJ, Wang YJ, Cheng HC. 2018. Apoptotic and nonapoptotic activities of pterostilbene against cancer. Int J Mol Sci 19:287. doi: 10.3390/ijms19010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrela JM, Ortega A, Mena S, Rodriguez ML, Asensi M. 2013. Pterostilbene: biomedical applications. Crit Rev Clin Lab Sci 50:65–78. doi: 10.3109/10408363.2013.805182. [DOI] [PubMed] [Google Scholar]
- 16.Pan MH, Wu JC, Ho CT, Lai CS. 2018. Antiobesity molecular mechanisms of action: Resveratrol and pterostilbene. Biofactors 44:50–60. doi: 10.1002/biof.1409. [DOI] [PubMed] [Google Scholar]
- 17.Chang HP, Lu CC, Chiang JH, Tsai FJ, Juan YN, Tsao JW, Chiu HY, Yang JS. 2018. Pterostilbene modulates the suppression of multidrug resistance protein 1 and triggers autophagic and apoptotic mechanisms in cisplatin-resistant human oral cancer CAR cells via AKT signaling. Int J Oncol 52:1504–1514. doi: 10.3892/ijo.2018.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Ding L, Wang X, Zhang J, Han W, Feng L, Sun J, Jin H, Wang XJ. 2012. Pterostilbene simultaneously induces apoptosis, cell cycle arrest and cyto-protective autophagy in breast cancer cells. Am J Transl Res 4:44–51. [PMC free article] [PubMed] [Google Scholar]
- 19.Chalal M, Klinguer A, Echairi A, Meunier P, Vervandier-Fasseur D, Adrian M. 2014. Antimicrobial activity of resveratrol analogues. Molecules 19:7679–7688. doi: 10.3390/molecules19067679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert C, Bisson J, Waffo-Téguo P, Papastamoulis Y, Richard T, Corio-Costet M-F, Mérillon J-M, Cluzet S. 2012. Phenolics and their antifungal role in grapevine wood decay: focus on the Botryosphaeriaceae family. J Agric Food Chem 60:11859–11868. doi: 10.1021/jf303290g. [DOI] [PubMed] [Google Scholar]
- 21.Jeandet P, Douillet-Breuil A-C, Bessis R, Debord S, Sbaghi M, Adrian M. 2002. Phytoalexins from the Vitaceae: biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J Agric Food Chem 50:2731–2741. doi: 10.1021/jf011429s. [DOI] [PubMed] [Google Scholar]
- 22.Chong J, Poutaraud A, Hugueney P. 2009. Metabolism and roles of stilbenes in plants. Plant Science 177:143–155. doi: 10.1016/j.plantsci.2009.05.012. [DOI] [Google Scholar]
- 23.Sobolev VS. 2013. Production of phytoalexins in peanut (Arachis hypogaea) seed elicited by selected microorganisms. J Agric Food Chem 61:1850–1858. doi: 10.1021/jf3054752. [DOI] [PubMed] [Google Scholar]
- 24.Sobolev VS, Khan SI, Tabanca N, Wedge DE, Manly SP, Cutler SJ, Coy MR, Becnel JJ, Neff SA, Gloer JB. 2011. Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J Agric Food Chem 59:1673–1682. doi: 10.1021/jf104742n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurt Z, Minoia M, Spain JC. 2018. Resveratrol as a growth substrate for bacteria from the rhizosphere. Appl Environ Microbiol 84:e00104-18. doi: 10.1128/AEM.00104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin HS, Yue BD, Ho PC. 2009. Determination of pterostilbene in rat plasma by a simple HPLC-UV method and its application in pre-clinical pharmacokinetic study. Biomed Chromatogr 23:1308–1315. doi: 10.1002/bmc.1254. [DOI] [PubMed] [Google Scholar]
- 27.Kamoda S, Habu N, Samejima M, Yoshimoto T. 1989. Purification and some properties of lignostilbene-alpha-beta-dioxygenase responsible for the C-alpha-C-beta cleavage of a diarylpropane type lignin model-compound from Pseudomonas sp. TMY1009. Agric Biol Chemy 53:2757–2761. doi: 10.1271/bbb1961.53.2757. [DOI] [Google Scholar]
- 28.Kamoda S, Saburi Y. 1993. Cloning, expression, and sequence analysis of a lignostilbene-alpha, beta-dioxygenase gene from Pseudomonas paucimobilis TMY1009. Biosci Biotechnol Biochem 57:926–930. doi: 10.1271/bbb.57.926. [DOI] [PubMed] [Google Scholar]
- 29.Kamoda S, Terada T, Saburi Y. 2003. A common structure of substrate shared by lignostilbenedioxygenase isozymes from Sphingomonas paucimobilis TMY1009. Biosci Biotechnol Biochem 67:1394–1396. doi: 10.1271/bbb.67.1394. [DOI] [PubMed] [Google Scholar]
- 30.Marasco EK, Schmidt-Dannert C. 2008. Identification of bacterial carotenoid cleavage dioxygenase homologues that cleave the interphenyl alpha, beta double bond of stilbene derivatives via a monooxygenase reaction. Chembiochem 9:1450–1461. doi: 10.1002/cbic.200700724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAndrew RP, Sathitsuksanoh N, Mbughuni MM, Heins RA, Pereira JH, George A, Sale KL, Fox BG, Simmons BA, Adams PD. 2016. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc Natl Acad Sci U S A 113:14324–14329. doi: 10.1073/pnas.1608917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt H, Kurtzer R, Eisenreich W, Schwab W. 2006. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J Biol Chem 281:9845–9851. doi: 10.1074/jbc.M511668200. [DOI] [PubMed] [Google Scholar]
- 33.Sobolev VS, Orner VA, Arias RS. 2013. Distribution of bacterial endophytes in peanut seeds obtained from axenic and control plant material under field conditions. Plant Soil 371:367–376. doi: 10.1007/s11104-013-1692-2. [DOI] [Google Scholar]
- 34.Anandham R, Gandhi PI, Madhaiyan M, Sa T. 2008. Potential plant growth promoting traits and bioacidulation of rock phosphate by thiosulfate oxidizing bacteria isolated from crop plants. J Basic Microbiol 48:439–447. doi: 10.1002/jobm.200700380. [DOI] [PubMed] [Google Scholar]
- 35.Lucy M, Reed E, Glick BR. 2004. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek 86:1–25. doi: 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- 36.Harrison PJ, Bugg TD. 2014. Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles. Arch Biochem Biophys 544:105–111. doi: 10.1016/j.abb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Kloer DP, Schulz GE. 2006. Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci 63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. 2005. The structure of a retinal-forming carotenoid oxygenase. Science 308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 39.Schmidlin L, Poutaraud A, Claudel P, Mestre P, Prado E, Santos-Rosa M, Wiedemann-Merdinoglu S, Karst F, Merdinoglu D, Hugueney P. 2008. A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol 148:1630–1639. doi: 10.1104/pp.108.126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 41.Horn BW. 2005. Colonization of wounded peanut seeds by soil fungi: selectivity for species from Aspergillus section Flavi. Mycologia 97:202–217. doi: 10.1080/15572536.2006.11832854. [DOI] [PubMed] [Google Scholar]
- 42.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY. [Google Scholar]
- 43.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Felsenstein J. 2013. PHYLIP (Phylogeny Inference Package) version 3.695. Department of Genome Sciences, University of Washington, Seattle, WA. [Google Scholar]
- 47.Cohen-Bazire G, Sistrom WR, Stanier RY. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol 49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 48.Wallenius K, Lappi K, Mikkonen A, Wickstrom A, Vaalama A, Lehtinen T, Suominen L. 2012. Simplified MPN method for enumeration of soil naphthalene degraders using gaseous substrate. Biodegradation 23:47–55. doi: 10.1007/s10532-011-9485-x. [DOI] [PubMed] [Google Scholar]
- 49.Marasco EK, Vay K, Schmidt-Dannert C. 2006. Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J Biol Chem 281:31583–31593. doi: 10.1074/jbc.M606299200. [DOI] [PubMed] [Google Scholar]
- 50.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu WH, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.UniProt Consortium 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences of the isolates sequenced here were archived in GenBank under the accession numbers KX507136 to KX507160. The sequences of the two candidate cco genes, cco1 and cco2, were deposited in GenBank under the accession numbers KX523172 and KX523173.