Tooth decay, a costly and painful disease affecting the vast majority of people worldwide, is caused by the bacterium Streptococcus mutans. The bacteria utilize dietary sugars to build and strengthen biofilms, trapping acids onto the tooth’s surface and causing demineralization and decay of teeth. As knowledge of our body’s microbiomes increases, the need for developing therapeutics targeted to disease-causing bacteria has arisen. The significance of our research is in studying and identifying a novel therapeutic target, a dynamic biofilm matrix that is mediated by a new virulence factor and membrane vesicles. The study increases our understanding of S. mutans virulence and also offers a new opportunity to develop effective therapeutics targeting S. mutans. In addition, the mechanisms of membrane vesicle-mediated biofilm matrix dynamics are also applicable to other biofilm-driven infectious diseases.
KEYWORDS: Streptococcus mutans, biofilms, eDNA, extracellular matrix, glucans, glycosyltransferases
ABSTRACT
Streptococcus mutans is a key cariogenic bacterium responsible for the initiation of tooth decay. Biofilm formation is a crucial virulence property. We discovered a putative glycosyltransferase, SMU_833, in S. mutans capable of modulating dynamic interactions between two key biofilm matrix components, glucan and extracellular DNA (eDNA). The deletion of smu_833 decreases glucan and increases eDNA but maintains the overall biofilm biomass. The decrease in glucan is caused by a reduction in GtfB and GtfC, two key enzymes responsible for the synthesis of glucan. The increase in eDNA was accompanied by an elevated production of membrane vesicles, suggesting that SMU_833 modulates the release of eDNA via the membrane vesicles, thereby altering biofilm matrix constituents. Furthermore, glucan and eDNA were colocalized. The complete deletion of gtfBC from the smu_833 mutant significantly reduced the biofilm biomass despite the elevated eDNA, suggesting the requirement of minimal glucans as a binding substrate for eDNA within the biofilm. Despite no changes in overall biofilm biomass, the mutant biofilm was altered in biofilm architecture and was less acidic in vitro. Concurrently, the mutant was less virulent in an in vivo rat model of dental caries, demonstrating that SMU_833 is a new virulence factor. Taken together, we conclude that SMU_833 is required for optimal biofilm development and virulence of S. mutans by modulating extracellular matrix components. Our study of SMU_833-modulated biofilm matrix dynamics uncovered a new target that can be used to develop potential therapeutics that prevent and treat dental caries.
IMPORTANCE Tooth decay, a costly and painful disease affecting the vast majority of people worldwide, is caused by the bacterium Streptococcus mutans. The bacteria utilize dietary sugars to build and strengthen biofilms, trapping acids onto the tooth’s surface and causing demineralization and decay of teeth. As knowledge of our body’s microbiomes increases, the need for developing therapeutics targeted to disease-causing bacteria has arisen. The significance of our research is in studying and identifying a novel therapeutic target, a dynamic biofilm matrix that is mediated by a new virulence factor and membrane vesicles. The study increases our understanding of S. mutans virulence and also offers a new opportunity to develop effective therapeutics targeting S. mutans. In addition, the mechanisms of membrane vesicle-mediated biofilm matrix dynamics are also applicable to other biofilm-driven infectious diseases.
INTRODUCTION
Dental caries, commonly known as tooth decay or cavity, is characterized by the demineralization and subsequent destruction of tooth enamel, resulting in the formation of cavities. Streptococcus mutans is considered a major etiological agent because it can produce acids from the catabolism of various sugars that cause dissolution of the tooth enamel, survive the low pH niche created largely by itself, and colonize and accumulate on the tooth surface in a robust acidic biofilm community (1, 2). The biofilm also protects the bacteria housed within it and traps acids close to the tooth surface, facilitating the development of carious lesions. The extracellular biofilm matrix of S. mutans consists of glucan polysaccharides, extracellular DNA (eDNA), and proteins. The most important and well-studied component of the biofilm is the glucan matrix, which provides a platform for S. mutans to adhere to the tooth surface, mechanical stability, and acidic microenvironments (3). The glucan matrix is synthesized and organized by extracellular glucosyltransferases (GtfB, GtfC, and GtfD). Under high-sucrose conditions, Gtfs break down sucrose and polymerize the glucose moiety into sticky glucose polymers. GtfB synthesizes water-insoluble glucans, GtfD synthesizes water-soluble glucans, and GtfC synthesizes a mixture of both (4, 5). Deletion of GtfB has been shown to be sufficient to abolish microcolony formation within the biofilm, and deletion of both GtfB and GtfC results in a severe defect in S. mutans biofilm formation (6) and bacterial cariogenicity (7).
Extracellular DNA (eDNA) is a major component of the biofilm extracellular matrix of many bacterial species. eDNA plays an important role in bacterial adhesion and accumulation, as well as the architecture and stability of biofilms (8–11). Additionally, eDNA is crucial for the formation of dental plaque biofilms, especially in the early stages (12). Programmed cell death and lysis are considered the major source of eDNA (13, 14); however, there is also growing evidence that DNA can be actively released via the type IV secretion system, membrane vesicles, and probably other lysis-independent mechanisms (11, 15–19). In S. mutans, it has been shown that DNA release can occur through cell death induced by the competence-stimulating peptides CSP/XIP (20, 21) and via secreted membrane vesicles in a lysis-independent manner (11). eDNA has been shown to be important in early S. mutans biofilm development, as pretreatment of S. mutans biofilms with DNase reduces the biofilm formation (11, 22). In addition, recent studies have suggested that in the presence of glucans, eDNA can further enhance S. mutans adherence (11). However, little is known regarding how various matrix components interact and contribute to the cariogenic biofilm formation. Understanding this new activity may reveal new targets that are amenable to the design of selective antibiofilm inhibitors.
The genome of S. mutans UA159 encodes several uncharacterized glycosyltransferases, enzymes responsible for the transfer of a sugar moiety from an activated nucleotide-sugar to an acceptor, which may be a lipid, protein, or growing oligosaccharide. Glycosyltransferases in S. mutans, including the aforementioned and well-characterized glucosyltransferases (GtfB, GtfC, and GtfD), have been shown to be vitally important for the organism's fitness and virulence and play important roles in cell wall biogenesis, biofilm formation, and protein homeostasis (4–6, 23–26). Our lab has been interested in uncovering the role of the putative glycosyltransferases with no known function. The smu_833 gene is predicted to encode a hypothetical glycosyltransferase highly conserved among mutans group streptococci but absent among oral commensals. The SMU_833 protein is predicted to have two transmembrane helices and shares high homology with proteins involved in cell wall biogenesis and cell stress response, suggesting this protein may be important for the establishment, survival, or virulence of S. mutans.
In this study, we further investigated the function of SMU_833 and showed that the loss of SMU_833 resulted in a significant decrease in amounts of Gtfs and a concurrent reduction in glucan matrix but without any major effect on total biofilm biomass. Further analysis revealed an increase in extracellular DNA and an increased production of membrane vesicles. It appears that the increased eDNA and membrane vesicle production were able to compensate the loss of glucans to maintain the biofilm biomass but were not sufficient to replace the loss of glucans to generate a cariogenic biofilm. Despite no apparent loss of in vitro biofilm biomass, the lack of SMU_833 led to defects in bacterial colonization and caries formation in a rat caries model, demonstrating that SMU_833 is an important S. mutans virulence factor.
RESULTS
Deletion of smu_833 affects production of high-molecular-weight proteins.
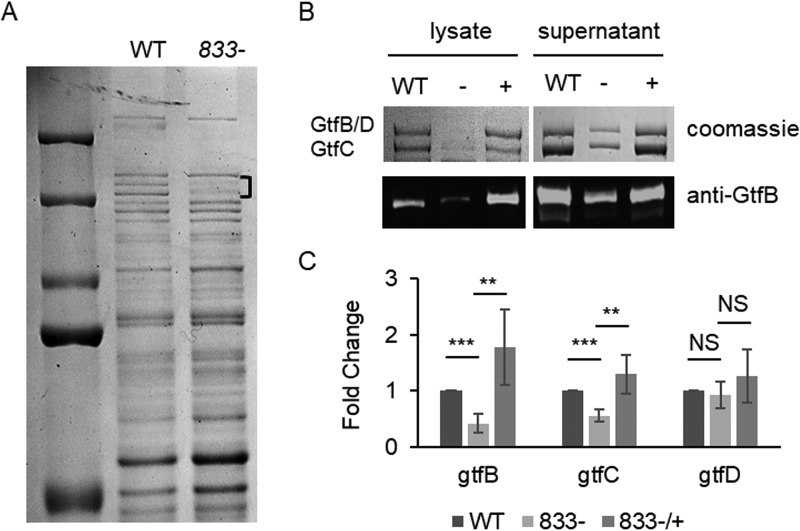
SMU_833 is a putative glycosyltransferase whose function has yet to be elucidated. To gain insight into the function of SMU_833 in S. mutans, an in-frame deletion mutant for smu_833 was constructed, confirmed, and characterized. The protein profiles of cell lysates from the parent strain UA159 and the smu_833 deletion mutant were examined by SDS-PAGE analysis. Although the banding patterns of most proteins were similar between the wild-type strain and the mutant, there were reduced amounts of two protein bands just above 150 kDa in the mutant lysate (Fig. 1A). Mass spectrometry analysis of the protein bands revealed they are GtfB, GtfC, and GtfD (data not shown). These proteins are known to be secreted and are needed for the formation of the biofilm glucan matrix. Because the majority of these proteins are found in the culture supernatants (4), levels of Gtfs were further analyzed in both the cell lysates and cell supernatant fractions (Fig. 1B, top panel). A decrease in Gtfs in both fractions of the mutant was evident. In addition, Western blot analysis confirmed the decrease in GtfB in the smu_833 mutant (Fig. 1B, bottom panel). Furthermore, complementation restored the amount of Gtfs in the mutant to the level found in the parent strain. Together, these data demonstrate that deletion of smu_833 decreases overall Gtf levels and that the decrease is not due to the redistribution of protein localization.
FIG 1.
Effect of smu_833 deletion on protein expression. (A) Coomassie-stained SDS-PAGE gel comparing total protein profiles of the smu_833 mutant and its parent strain, UA159. Bracket indicates bands with decreased expression. (B) SDS-PAGE and Western blot of Gtf proteins in both the whole-cell lysate and cell supernatant from wild-type (WT), smu_833 mutant (−), and complementation (+) strains. A GtfB-specific antibody was used in the Western blot. (C) qRT-PCR analysis of gtfB, gtfC, and gtfD gene expression levels in WT, smu_833 mutant, and complement strains. Data presented here represent the mean values from four independent experiments. ***, P < 0.001; **, P < 0.01; NS, not significant compared with the wild type. Statistical analysis was done using Student’s t test.
Next, we determined whether the decrease in Gtf protein levels in the mutant was related to a decrease in transcript levels of the gtf genes. Reverse transcription-quantitative PCR (qRT-PCR) analysis revealed that the loss of SMU_833 resulted in a decrease in the transcript levels of gtfB and gtfC but not gtfD (Fig. 1C). This result suggests that SMU_833 affects the transcription of gtfB and gtfC and that the decrease in protein levels of the Gtfs is at least partially due to the decrease in transcript levels. GtfB and GtfD are not able to be separated by SDS-PAGE and, therefore, run as one band (Fig. 1B, the top band). It is possible that the decrease in the protein band for GtfB and GtfD is just a decrease in the levels of GtfB and the protein levels of GtfD have remained unchanged.
Loss of SMU_833 results in a large decrease in glucans within the bacterial biofilm but retains comparable total biofilm biomass.
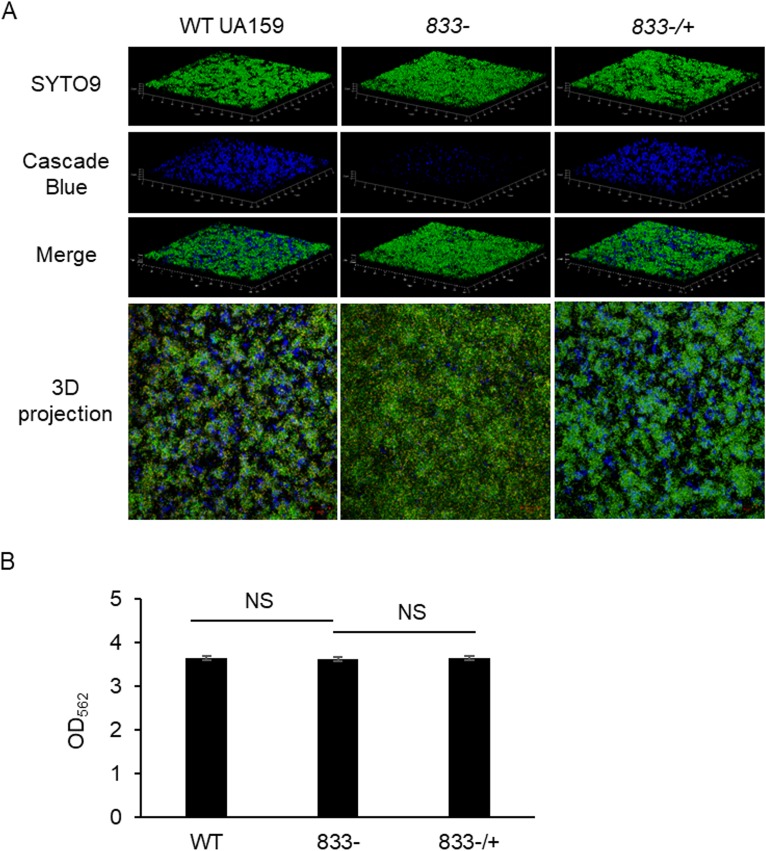
GtfB and GtfC are key enzymes needed to produce the glucan matrix component of the S. mutans biofilm. Because there was a decrease in the Gtfs in the smu_833 deletion mutant, we hypothesized the smu_833 mutant would yield less glucan matrix and have a reduced total biofilm biomass. To determine whether a decrease in Gtfs translated to the production of less glucan matrix, we used confocal laser scanning microscopy (CLSM) to evaluate the S. mutans biofilm. The parent strain formed distinct microcolonies with an abundant glucan matrix to support the biofilm structure (Fig. 2A, left panel). However, the mutant biofilm had greatly reduced glucan matrix formation and showed an altered structure with less-defined microcolonies (Fig. 2A, middle panel). The biofilm formed by the complement strain displays a phenotype similar to that of the parent strain (Fig. 2A, right panel). Despite the large decrease in the glucan matrix, the mutant appears to yield a large biofilm, comparable in overall mass with the wild type (Fig. 2A, top panel; SYTO9). To quantitatively determine if the total biofilm biomass was altered by the loss of smu_833, crystal violet staining of biofilms was performed. Surprisingly, despite the decrease in the major structural component of the biofilm, the smu_833 mutant was still capable of producing a total biomass that was comparable to the parent strain (Fig. 2B), suggesting other components of the S. mutans biofilm matrix may be increased to compensate for the glucan defect. Nevertheless, these data illustrate that SMU_833 is important for the glucan matrix formation and the biofilm structure.
FIG 2.
Visualization of bacterial biofilms and measurement of total biofilm biomass. (A) Confocal laser scanning microscopy of wild-type, mutant, and complement biofilms. Bacterial cells were stained with SYTO9 green, and the glucan matrix was stained with Cascade blue conjugated to dextran. Images are representative of at least three independent experiments. (B) Comparison of total biofilm biomasses produced by wild-type, smu_833 mutant, and complementation strains. Biofilms were grown on 96-well polystyrene plates in TYE medium supplemented with 1% sucrose for 18 h. Biofilms were stained with crystal violet, and OD562 was read to ascertain total biofilm biomass. Data are representative of those from at least three independent experiments for which there was no significant difference (NS; P > 0.05). Statistical analysis was done using Student’s t test.
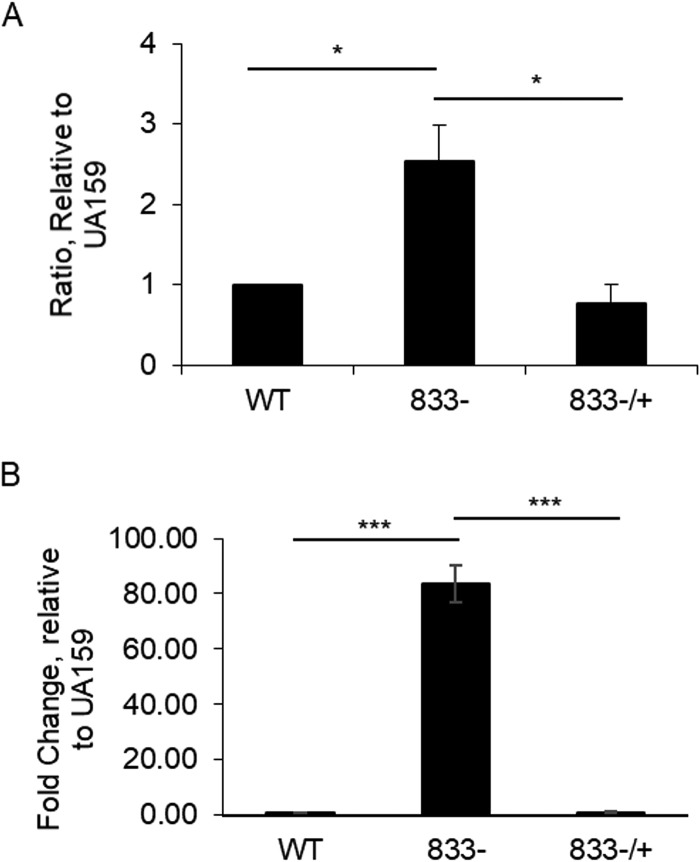
Loss of SMU_833 increases extracellular DNA but does not significantly change cell viability.
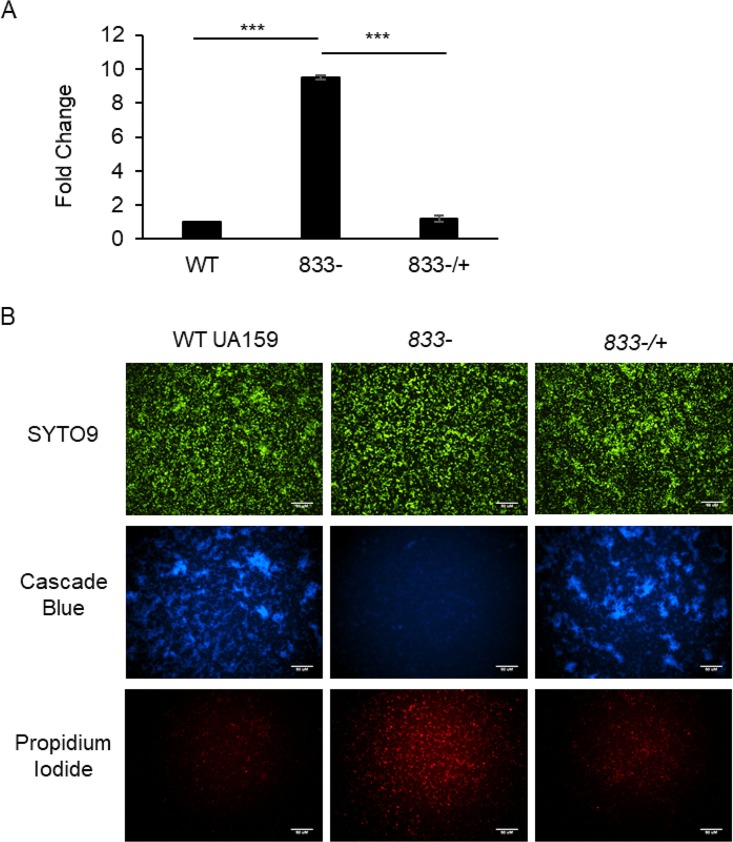
Extracellular DNA (eDNA) is another component of the S. mutans biofilm which is known to play an imperative role in the biofilm formation of many other bacterial species, as well as early biofilm formation in S. mutans (11, 22, 27). To determine if the smu_833 mutant produces more eDNA than its parent strain, supernatants of planktonic bacterial cultures were first analyzed for relative eDNA concentration via quantitative PCR (qPCR) using primers specific to S. mutans 16S rRNA. The supernatant of the mutant contained almost 10-fold more eDNA than that of UA159 (Fig. 3A). The DNA levels in the complementation strain were similar to those in the wild type, indicating that the increase in eDNA in the smu_833 mutant was due to the deletion of smu_833 (Fig. 3A). To determine if the smu_833 deletion also caused an increase in eDNA within the biofilm, levels of eDNA were visualized with fluorescence microscopy. Early biofilms (5 h) were used for easier visualization of eDNA and to avoid the use of mature biofilms, which often contain too many dead cells. The biofilm of wild-type (WT) UA159 contained a large amount of glucan matrix but only a small amount of eDNA/dead cells (Fig. 3B, left panel). In contrast, the smu_833 mutant biofilm contained very little glucan matrix but a larger quantity of eDNA/dead cells than the WT UA159 (Fig. 3B, middle panel). Complementation of smu_833 again restored levels of glucan matrix and eDNA/dead cells to wild-type levels (Fig. 3B, right panel). Again, the overall biomass produced by each strain appeared equal to the others (Fig. 3B, top panel; SYTO9). Together, these results suggest that the eDNA levels increased within the smu_833 mutant and that the increase in eDNA helps to compensate for the lack of glucan matrix.
FIG 3.
Measurement and visualization of extracellular DNA in planktonic cultures and biofilm. Comparison of extracellular (eDNA) levels in wild-type, smu_833 deletion mutant, and complemented bacterial strains. (A) Real-time PCR using 16S rRNA specific primers was used to analyze the eDNA concentration in the supernatants of planktonic cultures grown to an OD470 of 1.0. Data presented here represent the average of values from three independent experiments. ***, P < 0.001. Statistical analysis was done using Student’s t test. (B) Fluorescence microscopy was used to visually determine the relative amount of eDNA and cell death within the bacterial biofilms grown for 5 h. The biofilm glucan matrix was stained using Cascade blue conjugated to dextran, SYTO9 green for DNA of living cells, and propidium iodide red for DNA in dead cells/eDNA within the biofilm. Images are representative of at least three independent experiments.
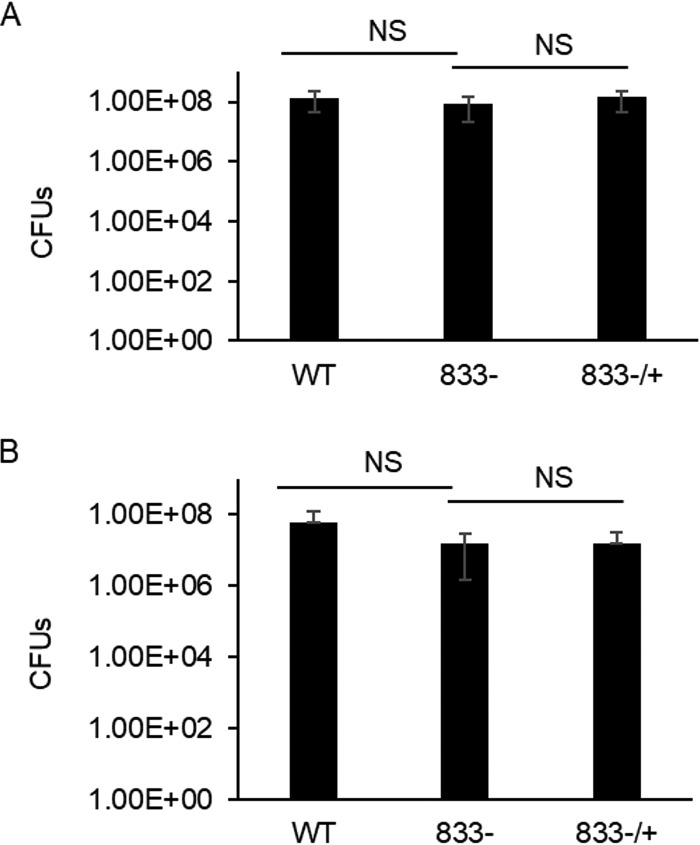
One possible reason for an increase in eDNA is an increase in cell death and lysis. To determine if deletion of smu_833 reduces cell viability, cultures from wild-type UA159, the smu_833 mutant, and its complement were grown to exponential phase, and their viability was measured by counting colony forming units (CFU) (Fig. 4A). Exponential-phase cultures were used to match the time point used in Fig. 3. There was no significant difference in the cell viability between the strains, indicating that the large increase in eDNA in the mutant is not due to an increase in cell lysis. A similar assay was performed to determine if the loss of smu_833 caused an increase in cell lysis within the bacterial biofilm. Again, no significant change in cell viability was observed with the deletion of smu_833 (Fig. 4B). To corroborate these data, alamarBlue was used as another means to assess cell viability within the biofilm. alamarBlue staining also indicated that there is no significant difference in cell viability between these strains (Fig. S1). Taken together, these results indicate that smu_833 deletion does not cause a discernible decrease in cell viability. It is likely that a cell lysis-independent mechanism accounts for the increase in eDNA.
FIG 4.
Viability of wild-type UA159, smu_833 mutant, and complement strains in planktonic and biofilm cultures. Bacterial cultures were either grown planktonically in THB to an OD470 of 1.0 (A) or allowed to form a biofilm for 5 h in TYE with 1% sucrose (B). Appropriate dilutions of the cultures were made and plated onto THB plates, and CFU were enumerated. Data are averages of those from at least five independent experiments. NS, no significant difference (P > 0.05). Statistical analysis was done using Student’s t test.
Deletion of smu_833 results in an increase in production of membrane vesicles and eDNA.
Membrane vesicles are known to be a source of eDNA in Gram-negative bacteria and similar results have also been recently reported in Gram-positive bacteria (15–17). S. mutans has been shown to be capable of producing DNA containing membrane vesicles in a lysis-independent manner (11). To determine if the smu_833 deletion mutant produces more membrane vesicles, membrane vesicles were prepared from cell-free supernatants by ultracentrifugation and estimated using the bicinchoninic acid (BCA) protein assay. The results showed that the mutant contained 2.54-fold more membrane vesicle proteins, indicative of an increase in vesicle production (Fig. 5A). In addition, bacterial DNA concentrations of vesicle preparations were measured by real-time quantitative PCR. The SMU_833 membrane vesicles contained at least 50-fold-higher concentrations of DNA than the wild-type and complemented strains (Fig. 5B). These data support the notion that eDNA released by membrane vesicles in the smu_833 mutant contributes to the biofilm matrix.
FIG 5.
Analysis of membrane vesicle production and DNA content. Membrane vesicle yield was estimated by measuring protein concentration using BCA assay. (A) Ratio of membrane vesicle production relative to S. mutans UA159. Data represent the average (±SD in error bars) values from three independent sets of experiments. *, P < 0.05 versus UA159. (B) DNA content of membrane vesicles was measured with real-time PCR using 16S specific primers. Results presented are representative of those from at least three independent experiments. ***, P < 0.001. Statistical analysis was done using Student’s t test.
Glucan interacts with eDNA and is required to form a biofilm.
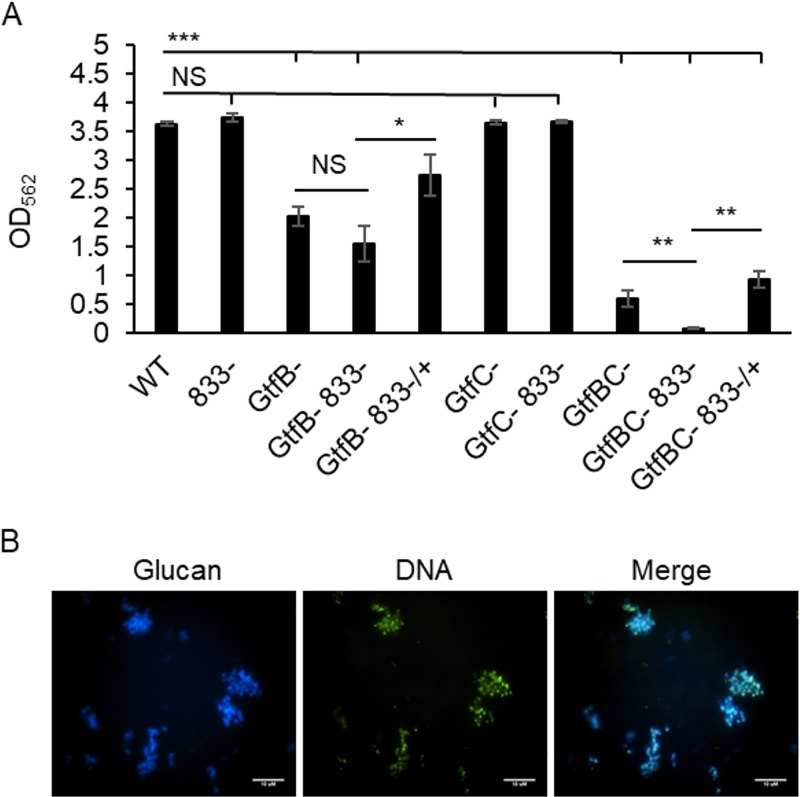
Glucans are an important component of the S. mutans biofilm matrix. We have found that the SMU_833-deficient mutant is capable of forming a biofilm with a biomass similar to that of the wild type but with a greatly reduced glucan matrix. However, it remains to be determined if this mutant can form a biofilm in the complete absence of Gtfs. To determine this, each gtf gene was deleted from the smu_833 mutant, and the total biofilm biomasses of different deletion variant strains were compared. A deletion of gtfC did not affect the total biomass of the biofilm formed in a rich medium, tryptone-yeast extract (TYE) (Fig. 6A). However, a deletion of gtfB caused a large reduction in biofilm formation. The double deletion of gtfB and smu_833 did not further reduce biofilm formation compared with that of the gtfB mutant (Fig. 6A), while double deletion of gtfB and gtfC resulted in an even greater reduction. In addition, the triple deletion of gtfB, gtfC, and smu_833 led to the greatest reduction in the biofilm biomass (Fig. 6A). Complementation of smu_833 in both the gtfB smu_833 double mutant and the gtfBC smu_833 triple mutant increased biofilm formation beyond the levels of either the double or triple mutant. These results suggest that despite the increase in eDNA, a certain amount of glucan matrix is still required for the smu_833 mutant to support biofilm formation.
FIG 6.
Glucan contribution to biofilm formation. (A) Strains containing a Gtf deletion with and without a deletion of smu_833 were allowed to form a biofilm for 18 h in TYE medium supplemented with 1% sucrose. Biomass was quantified with crystal violet staining, and the optical density at 562 nm was read. Results are representative of those from at least three independent experiments. NS, P > 0.05; *, P < 0.05, **, P < 0.01; ***, P < 0.001. Statistical analysis was done using Student’s t test. (B) Filtered culture supernatants containing eDNA and Gtf proteins were incubated with sucrose overnight and allowed to form cell-free biofilms, which were visualized with fluorescence microscopy. Cascade blue conjugated to dextran was used to visualize glucan, and SYTO9 green was used for visualization of eDNA.
Because our data suggest that some glucans are needed for the smu_833 mutant to form a biofilm despite an increase in eDNA, we hypothesize that glucan may prime the binding of eDNA, and, thus, the two components directly interact within the S. mutans biofilm. In a cell-free matrix formation system, Gtf proteins and eDNA prepared from the supernatant of wild-type S. mutans were allowed to form glucans in TYE medium supplemented with 1% sucrose. This cell-free system was used to simplify and better visualize possible interactions between matrix components. Fluorescence microscopy revealed that eDNA and glucan colocalized with one another (Fig. 6B). This interaction and the need for the presence of some glucans together suggest that glucans and eDNA are integral components of the biofilm matrix.
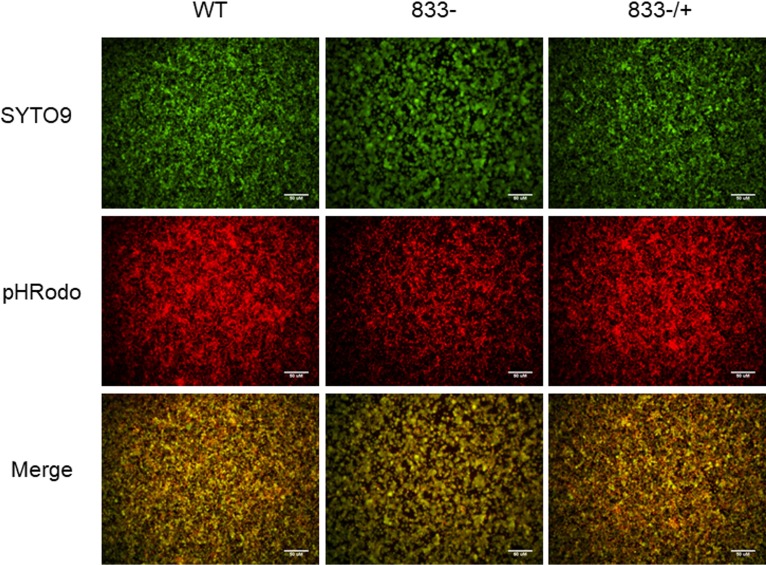
Loss of SMU_833 results in an increased biofilm pH.
As the S. mutans biofilm develops, more bacteria accumulate and ferment more sugars, which cause the pH of the biofilm to drop and demineralization of tooth enamel to occur. The biofilm matrix facilitates the formation of punctate areas of low pH next to the surface on which the biofilm is attached (28). Because the matrix and the overall structure of the mutant biofilm differ from those of wild-type UA159, we evaluated the pH of the biofilms. In the wild-type biofilm (Fig. 7, left panel), there were many areas with an intense red coloring signifying many areas of low pH. However, in the mutant biofilm (Fig. 7, middle panel), the majority of the fluorescence was of a low intensity, indicative of a less acidic pH. The complemented strain regained wild-type levels of red fluorescence intensity and area coverage (Fig. 7, right panel). These results indicate that the smu_833 deletion leads to the formation of a less acidic biofilm.
FIG 7.
Effect of smu_833 deletion on biofilm pH. Wild-type, smu_833 mutant, and complemented bacterial strains were allowed to form a biofilm on polystyrene plates for 16 h. pHRodo red conjugated to dextran was added after the formation of the biofilm. An increase in red is indicative of a decrease in pH. SYTO9 green was used to stain bacterial cells. Images are representative of those from three independent experiments.
SMU_833 mutant exhibits reduced bacterial colonization and cariogenicity in vivo.
The smu_833 mutant forms a biofilm that exhibits an alternate structure and a higher pH environment, which may become less cariogenic. An established rat caries model (29–33) was used to determine whether the loss of smu_833 had any impact on S. mutans colonization and cariogenicity in vivo. Rats were infected with either the wild-type S. mutans or the smu_833 mutant and fed a 5% sucrose diet to aid in the formation of S. mutans biofilms. The results showed that there were significantly fewer carious lesions in rats infected with the mutant (Table 1). Additionally, mutant-infected rats yielded fewer recoverable S. mutans CFU (Table 1). Taken together, these data indicate that SMU_833 is a virulence factor that contributes to bacterial colonization and cariogenicity.
TABLE 1.
Colonization and cariogenicity of S. mutans wild type and smu_833 mutant in ratsa
| Group | Mean ± SEM caries score by location and severityb |
S. mutans (CFU/ml [× 106])c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buccal |
Sulcal |
Proximal |
|||||||||
| E | DS | DM | DX | E | DS | DM | DX | E | DS | ||
| UA159 | 13.0 ± 0.7 | 9.4 ± 0.7 | 7.4 ± 0.5 | 5.0 ± 0.5 | 20.2 ± 0.9 | 15.0 ± 0.3 | 8.6 ± 0.5 | 4.4 ± 0.4 | 5.2 ± 0.8 | 2.8 ± 0.5 | 11.0 ± 0.3 |
| smu_833 mutant | 8.8 ± 0.7 | 6.2 ± 0.4 | 4.4 ± 0.6 | 1.8 ± 0.4 | 16.2 ± 0.4 | 11.2 ± 0.2 | 4.8 ± 0.6 | 1.2 ± 0.5 | 2.8 ± 0.8 | 0 ± 0 | 4.3 ± 0.6 |
| Statistical significance | ** | ** | ** | *** | ** | *** | *** | *** | NS | *** | *** |
n = 5 per group. Weights (g): wild type, 136 ± 11; smu_833 mutant, 128 ± 10. There was no significant difference between the weights of the groups. **, P < 0.01; ***, P < 0.001; NS, no significance.
E, enamel; DS, dentinal slight; DM, dentinal moderate; DX, dentinal extensive.
Values were determined by plating plaque samples on mitis salivarius (MS) agar.
DISCUSSION
The biofilm matrix produced by cariogenic bacteria is the scaffold that shapes and strengthens the biofilm, and it enables the formation of acidic microenvironments, which facilitate demineralization of tooth enamel and caries formation (34). One of the most important components of the biofilm matrix is the glucan polymers produced by the glucosyltransferases GtfB and GtfC secreted by S. mutans. In this study, we characterized a putative glycosyltransferase, SMU_833, and determined that deletion of smu_833 resulted in an unexpected decrease in GtfB and GtfC levels and, subsequently, reduced glucan production within the biofilm. A point mutation of the catalytic DXD motif of SMU_833 results in the same reduced Gtf and glucan phenotype (Fig. S2). Additional studies are required to determine the exact mechanism by which SMU_833 causes a reduction in Gtfs. Protein glycosyltransferase of streptococci (PgfS) modifies and prevents degradation of the collagen binding protein Cnm in S. mutans (25). As SMU_833 is a putative glycosyltransferase, it is possible that SMU_833 functions in a similar manner and is needed for glycosylation and stability of the Gtfs.
Gtfs are the enzymes responsible for the building of the sticky glucan matrix, which allows S. mutans to adhere rigorously to the tooth surface (4, 23, 35). Therefore, it was not surprising to observe a decrease in glucan matrix production in the smu_833 mutant biofilm. Interestingly, the mutant still maintained a total biofilm biomass comparable with that of the parent strain. This result was unexpected, as it has been well documented that the glucan matrix is key to the formation of robust S. mutans biofilms and that the loss of Gtfs often results in little to no biofilm being formed (6, 23, 35). This finding may reveal a new dynamic in the biofilm matrix. Indeed, we discovered that the amount of extracellular DNA was elevated in the smu_833 mutant, which likely compensated for the loss of the glucan matrix. It is unclear how S. mutans may be able to coordinate this dynamic modulation. It has been shown that the exopolysaccharide Pel can compensate for the lack of a different exopolysaccharide, Psl, in Pseudomonas aeruginosa (36, 37). However, the underlying mechanism for this remains unknown. It was speculated that the presence of the Psl polysaccharide can act as an environmental signal that downregulates Pel production (36). In a similar manner, S. mutans may be able to sense the reduction in glucans and, in response, upregulate DNA release. In addition, it was shown that polysaccharide biofilm matrix components can be upregulated in response to the loss of surface adhesin proteins LapA and LapF in Pseudomonas putida, and it was suggested that the second messenger molecule cyclic di-GMP may play a role in the balance of matrix components (38). In S. mutans, altered levels of the second messenger c-di-AMP can affect biofilm formation (32, 39), which may be a possible means by which the smu_833 mutant can alter its biofilm.
eDNA plays an important role in biofilm formation in many bacterial species and has been shown to play a role in early biofilm formation in S. mutans (11, 22, 27). Both cell lysis-dependent and -independent mechanisms of DNA release have been proposed in S. mutans (11, 20, 21). Our data suggest that the increased eDNA present in the smu_833 mutant is a result of the active release of membrane vesicles and not due to an increase in cell lysis. Emerging studies provide evidence of the presence of eDNA and increased secretion of nucleic acid-containing membrane vesicles in Gram-positive bacteria, including S. mutans (11, 16, 17, 40); however, how they contribute to the biofilm matrix or how they are regulated is not defined. It is possible that the cargo for the membrane vesicles packaged in S. mutans is linked to a regulatory network by which the bacteria may be able to sense the decrease in glucans and, thereby, upregulate eDNA release into the biofilm via vesicles. Furthermore, a decrease in the production of glucans led to an increase in pH. Acidic pH can become a signal sensed by some bacteria, including S. mutans, to induce the gene expression controlled by bacterial two-component regulatory systems (41, 42). Changes in this gene expression pattern may lead to the activated secretion of outer membrane vesicles and eDNA. Further investigation into the regulation of alternate matrix components would provide new insights into our understanding of S. mutans matrix biology.
A recent study of S. mutans biofilms using field emission scanning electron microscopy analysis indicates interactions between nanofibrous eDNA and wool-like glucans (11), similar to what has been reported in Myxococcus spp. (8). The results from our current study demonstrate the dependence of the biofilm formation on the interaction between glucan matrix and eDNA. Elevated eDNA can compensate for the partial loss of glucans but not the complete loss of glucans. The biofilm formation supported by increased eDNA only occurs when small amounts of glucans exist. Furthermore, the glucan and eDNA of S. mutans colocalize, suggesting that the glucan acts as a scaffold for eDNA to bind. This further demonstrates the importance of glucans in biofilm formation and reveals the dynamic interactions between two matrix components. It has been reported that eDNA and glucan matrix components of biofilms interact with one another in P. aeruginosa (43) and the negative charge of eDNA allows for binding to occur with the positively charged polysaccharides of P. aeruginosa (36). Furthermore, eDNA can acidify the biofilm and trigger the formation of acidic biofilms in P. aeruginosa (41). This is different from S. mutans where, despite an increase in eDNA, the net pH increases upon the deletion of smu_833 and the reduction of Gtfs. In S. aureus, eDNA was shown to facilitate the formation of functional amyloids in biofilms (44). S. mutans is also shown to form amyloids, structured protein fibrils which, too, contribute to biofilm formation (45). However, the exact mechanism for how glucans, eDNA, and amyloids of the major biofilm matrices interact in S. mutans is unclear and needs further investigation. The DNABII family of DNA-binding proteins have also been shown to play a role in the structure and stability of DNA within biofilms (46–49), and it is possible that proteins such as these also play a role in the matrix interactions in S. mutans.
Despite the formation of a comparable in vitro biofilm biomass by the smu_833 mutant, the biofilm structure and composition were altered, resulting in a higher biofilm pH suggestive of a reduced cariogenic potential. Our studies revealed that SMU_833 is an important virulence factor in vivo. The smu_833 mutant reduced bacterial colonization and attenuated bacterial virulence in a rat model of dental caries. Because both the wild-type and mutant strains were able to colonize the rat initially, reduced colonization of the mutant at the end of the experiment suggests either the altered matrix was unable to facilitate bacterial adherence to the tooth surface as well or the bacteria themselves were unable to compete with the other bacteria present in the oral cavity. Further studies are ongoing to determine the fitness and competitiveness of the smu_833 mutant. The complex biofilm architecture supported by the glucan matrix is required for the creation of acidic, cariogenic microenvironments (28), which is essential for bacterial virulence. Our findings reveal a new virulence factor, SMU_833, whose deficiency alters the dynamic of the glucan and eDNA matrices of S. mutans biofilm and, thus, modulates bacterial virulence. It is still unclear precisely how SMU_833 affects the production of Gtfs and the release of eDNA by membrane vesicles.
In Enterococcus faecalis, eDNA is localized at the septum and the release may occur from metabolically active, living cells with a membrane potential (15). This may be the mechanism responsible for the active secretion of eDNA by membrane vesicles reported for S. mutans. In this regard, the smu_833 gene is located downstream of and adjacent to the gene cluster (smu_825 to smu_831) that encodes the biosynthetic pathway for the rhamnose-glucose polysaccharides (RGPs) located on the surface of S. mutans (24, 50). However, neither the mutant unable to produce any surface RGPs (ΔrgpG) nor the mutant that produces the rhamnose backbone without the glucose side chains (ΔrgpE) showed any decrease in glucan matrix (Fig. S3). This result indicates that the smu_833 biofilm phenotype is independent of the RGPs. However, further studies are ongoing to determine if SMU_833 affects cell envelope biogenesis and membrane vesicle release by another means. Figure S4 shows that the smu_833 mutant is more susceptible to autolysis by Triton X. Therefore, although the mutant is viable and metabolically active, the deletion of smu_833 may affect the cell wall or membrane in some manner, decreasing its stability under certain stressors. This could be a potential explanation for an increase in membrane vesicles or possible evidence that an alteration in the cell wall or membrane has made the bacterial cells more “leaky” and able to release DNA or other cell materials more readily. More studies are underway to further elucidate the role of SMU_833 in S. mutans. Understanding the underlying mechanisms may reveal new targets that can be used to develop potential therapeutics that prevent and treat dental caries.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptococcus mutans UA159 (51) and Escherichia coli Top 10 (Invitrogen) were used in this study. S. mutans strains were maintained in either Todd-Hewitt broth (THB) or tryptone-yeast extract (TYE) medium and grown statically at 37°C with 5% CO2. E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates and grown aerobically at 37°C. Antibiotics were used at the following concentrations: 1 mg ml−1 kanamycin and 10 µg ml−1 erythromycin for S. mutans, and 50 µg ml−1 kanamycin, 50 µg ml−1 ampicillin, and 300 µg ml−1 erythromycin for E. coli.
Construction and complementation of mutants.
Mutant strains were derived from S. mutans UA159 using the allelic replacement mutagenesis strategy by inserting a nonpolar kanamycin resistance cassette in place of smu_833. The smu_833 open reading frame plus its 1-kb flanking regions was amplified from S. mutans UA159 genomic DNA using PCR. The fragment was ligated into a pGEM-T plasmid, transformed into E. coli Top10, and selected for using plates containing ampicillin, isopropyl-β-d-thiogalactopyranoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The plasmid was isolated, and inverse PCR adding XbaI restriction sites was used to amplify everything except smu_833. The PCR product and the kanamycin cassette were digested with XbaI, ligated, and transformed into E. coli using ampicillin and kanamycin for selection. The resulting plasmid was transformed into S. mutans UA159 and selected for using kanamycin (1 mg ml−1). The replacement of smu_833 with a kanamycin resistance cassette was verified by PCR. The complete open reading frame of smu_833 was amplified and cloned into the E. coli-Streptococcus shuttle vector pVPT (57) to generate the corresponding complementation plasmid pVPT-smu_833. The plasmid was then transformed into the smu_833 mutant strain to generate a complemented strain. The gtfB, gtfC single mutant and gtfBC double mutant in which gtf was replaced with a kanamycin resistance cassette, aphA3 (encoding an aminoglycoside phosphotransferase), are gifts from Robert Burne’s laboratory, University of Florida, Gainesville, FL. The pGEM-T plasmid containing the kanamycin resistance cassette and the regions flanking smu_833 which was used to delete smu_833 from WT S. mutans (described above) was used to create the smu_833 gtfB and smu_833 gtfC double mutants and the smu_833 gtfB gtfC triple mutant. Mutants were selected for using kanamycin (1 mg ml−1) and verified with PCR.
SDS-PAGE and Western blotting.
The same cell numbers of each bacterial sample were spun down and resuspended in lysis buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, and 0.5% [vol/vol] NP-40). Glass beads were added, and the samples were vortexed for 60 min. Cell debris was separated from the protein extracts by centrifugation at 9,279 × g for 10 min. Cell supernatants were harvested and precipitated with 30% ethanol at −80°C for at least 1 h. Supernatant proteins were harvested by centrifugation at 15,682 × g for 15 min at 4°C. Protein samples were then dissolved into SDS loading buffer and boiled for 10 min. Proteins were separated by SDS-PAGE, blotted onto polyvinyl difluoride membranes, and incubated with a GtfB-specific antibody followed by goat anti-rabbit IgG. Signals were detected using Li-COR Biosciences Odyssey infrared imaging system.
Biofilm formation assay.
S. mutans biofilms were grown in TYE medium (2.5% tryptone, 1.5% yeast extract) containing 1% sucrose. Overnight cultures were subcultured into fresh THB, grown to an optical density at 470 nm (OD470) of 0.6, diluted to an OD470 of 0.01, subsequently aliquoted in a 96-well microtiter plate, and grown at 5% CO2 at 37°C under static conditions. Biofilm samples were collected after 16 h and stained with crystal violet. Crystal violet staining at OD562 was used to monitor biofilm formation as described previously (52). Each assay was carried out with at least triplicate samples and replicated at least three times.
Confocal laser scanning microscopy analysis.
Biofilms were formed as stated above in TYE medium with 1 μM dextran-conjugated Cascade blue (Molecular Probes, Invitrogen) to label exopolysaccharide (EPS) on 8-well ibiTreat slides (catalog number 80826; ibidi) under 5% CO2 conditions at 37°C for 16 h. The biofilm samples were gently washed with phosphate-buffered saline (PBS) three times to remove any nonadherent cells and then stained with SYTO9 green (Molecular Probes, Invitrogen). The stained samples were then examined by CLSM (Zeiss LSM 710 laser confocal microscope) with a 63× oil immersion objective. Images were obtained from serial optical sections. At least three independent experiments were performed on different days, and the images displayed are representative.
Biofilm eDNA fluorescence.
Biofilms were formed as stated above in TYE medium supplemented with 1% sucrose with the addition of Cascade blue conjugated to dextran. Diluted cultures were plated onto a 24-well plate, and biofilms were grown statically for 5 h at 37°C in 5% CO2. Biofilms were washed 3 times with PBS, and then SYTO9 and propidium iodide diluted in PBS were added to the wells. Fluorescence microscopy was performed using the 10× objective magnification on the Nikon Eclipse TE2000-E inverted fluorescence microscope. At least three independent experiments were performed on different days, and the images displayed are representative.
Visualization of biofilm pH with fluorescence microscopy.
Biofilms were grown as stated above. After incubating for 16 h in 24-well plates, 1 μM dextran-conjugated pHRodo red (Molecular Probes, Invitrogen) was added to the wells and the mixture was incubated for 3 h at room temperature. The biofilms were then washed with PBS three times to remove any nonadherent cells and stained with SYTO9 (Molecular Probes, Invitrogen) diluted in PBS. Fluorescence microscopy was performed using the 10× objective magnification on the Nikon Eclipse TE2000-E inverted fluorescence microscope. At least three independent experiments were performed on different days, and the images displayed are representative.
Planktonic eDNA quantification.
Overnight cultures were diluted into fresh THB and grown at 37°C in 5% CO2 to an OD470 of 1.0. Cultures were centrifuged, and the supernatant was filtered with a 0.22-µm filter (Millex-GS). Quantitative PCR was performed using SYBR green master mix on an iCycler iQ5 machine (Bio-Rad). The S. mutans 16S rRNA gene was used to quantify the amount of DNA present in the supernatant and the delta CT method was used to calculate fold change.
Preparation of membrane vesicles and DNA measurement.
Membrane vesicles (MVs) were prepared by following the protocol of Rivera et al. (11, 53, 54). S. mutans strains were grown in FMC medium with glucose (55) overnight (16 h) or in THB to an OD470 of 1.0 for DNA quantification. Following centrifugation at 6,000 × g at 4°C for 10 min, the cell-free culture supernatants were filtered through 0.22-µm syringe filters to remove residual cells. MVs in the cell-free supernatants were collected and washed once with sterile PBS after centrifugation at 140,153 × g at 4°C for 2 h. After being washed, the pellets were resuspended in 200 µl PBS. MV yield was estimated by measuring protein concentration using the BCA assay (Pierce) and normalized by bacterial cell numbers in CFU. DNA measurement was obtained with qPCR using S. mutans-specific 16S rRNA primers (Table 2).
TABLE 2.
Primers used in this study
| Primera | Sequence (5′–3′) | Description |
|---|---|---|
| Smu_833 pGEM F | CTTCCTGAAAAGGGTTATCTATATG | smu_833 deletion |
| Smu_833 pGEM R | ACCTAACGCAAAAGACAAGG | smu_833 deletion |
| Smu_833 Xba1 Inv F | CGCGTCTAGAAGGCTATAGGCATTTATTTAGAA | smu_833 deletion |
| Smu_833 Xba1 Inv R | CGCGTCTAGATTTTCCCCCTTGAAAATTAATAATTGG | smu_833 deletion |
| Smu_833 Kpn1 F | CGCACGGGTACCATGAAAAAACTTTCAATAG | smu_833 complement |
| Smu_833 BspH1 R | GGTCAGTCATGAGTCCTTTTTCCTTAAC | smu_833 complement |
| 16S rRNA F | CTTACCAGGTCTTGACATCCC | qRT-PCR |
| 16S rRNA R | CCAACATCTCACGACACGAG | qRT-PCR |
| gtfB F | ACACTTTCGGGTGGCTTG | qRT-PCR |
| gtfB R | GCTTAGATGTCACTTCGGTTG | qRT-PCR |
| gtfC F | CAAAATGGTATTATGGCTGTCG | qRT-PCR |
| gtfC R | TGAGTCTCTATCAAAGTAACGCAG | qRT-PCR |
| gtfD F | CGGTATTCAGGTTATTGCG | qRT-PCR |
| gtfD R | CTCACCATAATCGTTGACACG | qRT-PCR |
F, forward; R, reverse.
Colocalization of glucan-eDNA in cell-free matrix formation system.
S. mutans was grown in THB to an OD470 of 1.0 and centrifuged at 1,950 × g at 4°C for 10 min, and the cell-free culture supernatants were filtered through 0.22-µm syringe filters to remove residual cells. The cell-free supernatants were then concentrated with the addition of 100% ethanol (300 µl ml−1 supernatant) and frozen at −80°C for 1 h. Supernatant proteins and eDNA were harvested by centrifugation at 15,682 × g for 10 min at 4°C, resuspended in TYE medium supplemented with 1% sucrose and Cascade blue conjugated to dextran, added to the wells of a 96-well plate, and incubated at 37°C in 5% CO2 for 16 h. SYTO9 was added to the wells, and the mixture was incubated for 20 min. A total of 10 µl was taken from the well and placed on a microscope slide for imaging using the 60× objective magnification on the Nikon Eclipse TE2000-E inverted fluorescence microscope.
Rat caries model.
The virulence of the S. mutans smu_833 mutant was compared with that of the WT using a rat caries model of infection (29–33, 56). Briefly, male and female rat pups (Fischer 344), 18 days of age, were weaned and divided into 2 groups of 5 animals each. Rats were challenged at 22 days of age with either wild-type S. mutans or the smu_833 mutant by oral swabbing with a fresh overnight culture. Rats were provided a caries promoting diet (MIT 305) containing 5% sucrose and sterile drinking water ad libitum. Oral swabs were taken 5 days postinfection to confirm colonization. Rats were weighed at weaning and at the termination of the experiment. The rats were sacrificed at 60 days of age (day 38) and their mandibles excised for plaque analysis and scoring for caries by the method of Keyes (58). Bacterial CFU were enumerated from mitis salivarius (MS) agar. Statistical analysis was done using Student’s t test. All animal studies were done according to National Institutes of Health guidelines, using protocols approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Dental and Craniofacial Research, National Institutes of Health (grants F31DE025805 to K.R., R01DE022350 to H.W., R01DE017954 to H.W., and R01DE019452 and R21 DE25348 to Z.T.W.).
We thank Ashton Jorgenson for her assistance with the membrane vesicle assays and S. Stephanie Garcia for her help with CLSM. We also thank Robert Burne at the University of Florida for the gtfB, gtfC, and gtfBC deletion mutant strains and Noel Childers for the GtfB antibody.
K.R. and H.W. designed experiments and wrote the manuscript. Z.T.W. conducted membrane vesicle quantification experiments. S.M.M. performed the rat caries study. K.R. conducted the remaining experiments. All authors reviewed the final draft of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02247-18.
REFERENCES
- 1.Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B, Skalniak A. 2014. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis 33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schilling KM, Bowen WH. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun 60:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada S, Slade HD. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev 44:331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkitaraman AR, Vacca-Smith AM, Kopec LK, Bowen WH. 1995. Characterization of glucosyltransferaseB, GtfC, and GtfD in solution and on the surface of hydroxyapatite. J Dent Res 74:1695–1701. doi: 10.1177/00220345950740101101. [DOI] [PubMed] [Google Scholar]
- 6.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun 61:3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, Li L, Sharma S, Wang J, McHardy I, Lux R, Yang Z, He X, Gimzewski JK, Li Y, Shi W. 2012. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One 7:e51905. doi: 10.1371/journal.pone.0051905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 10.Dominiak DM, Nielsen JL, Nielsen PH. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol 13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 11.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rostami N, Shields RC, Yassin SA, Hawkins AR, Bowen L, Luo TL, Rickard AH, Holliday R, Preshaw PM, Jakubovics NS. 2017. A critical role for extracellular DNA in dental plaque formation. J Dent Res 96:208–216. doi: 10.1177/0022034516675849. [DOI] [PubMed] [Google Scholar]
- 13.Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, Heuser J, Dodson KW, Caparon MG, Hultgren SJ. 2009. Contribution of autolysin and sortase A during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun 77:3626–3638. doi: 10.1128/IAI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Burne RA. 2011. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol 193:2826–2837. doi: 10.1128/JB.00056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnes AMT, Ballering KS, Leibman RS, Wells CL, Dunny GM. 2012. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio 3:e00193-12. doi: 10.1128/mBio.00193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grande R, Celia C, Mincione G, Stringaro A, Di Marzio L, Colone M, Di Marcantonio MC, Savino L, Puca V, Santoliquido R, Locatelli M, Muraro R, Hall-Stoodley L, Stoodley P. 2017. Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front Microbiol 8:1040. doi: 10.3389/fmicb.2017.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Kong Q, Roland KL, Curtiss R III.. 2014. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 304:431–443. doi: 10.1016/j.ijmm.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreth J, Vu H, Zhang Y, Herzberg MC. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol 191:6281–6291. doi: 10.1128/JB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 20.Perry JA, Cvitkovitch DG, Levesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336:104–112. doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol 187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata Y, Ozaki K, Seki M, Kawato T, Tanaka H, Nakano Y, Yamashita Y. 2003. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J Clin Microbiol 41:4107–4112. doi: 10.1128/JCM.41.9.4107-4112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avilés-Reyes A, Miller JH, Simpson-Haidaris PJ, Hagen FK, Abranches J, Lemos JA. 2014. Modification of Streptococcus mutans Cnm by PgfS contributes to adhesion, endothelial cell invasion, and virulence. J Bacteriol 196:2789–2797. doi: 10.1128/JB.01783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano K, Koga T. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol 180:5803–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okshevsky M, Regina VR, Meyer RL. 2015. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol 33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barletta RG, Michalek SM, Curtiss R III. 1988. Analysis of the virulence of Streptococcus mutans serotype c gtfA mutants in the rat model system. Infect Immun 56:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowley PJ, Brady J, Michalek SM, Bleiweis AS. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun 67:1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, Jespersgaard C, Lamberty-Mallory L, Katz J, Huang Y, Hajishengallis G, Michalek SM. 2002. Enhanced immunogenicity of a genetic chimeric protein consisting of two virulence antigens of Streptococcus mutans and protection against infection. Infect Immun 70:6779–6787. doi: 10.1128/IAI.70.12.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng X, Michalek S, Wu H. 2016. Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of Streptococcus mutans revisit. Environ Microbiol 18:3612–3619. doi: 10.1111/1462-2920.13440. [DOI] [PubMed] [Google Scholar]
- 33.Garcia SS, Blackledge MS, Michalek S, Su L, Ptacek T, Eipers P, Morrow C, Lefkowitz EJ, Melander C, Wu H. 2017. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J Dent Res 96:807–814. doi: 10.1177/0022034517698096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koo H, Falsetta ML, Klein MI. 2013. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent Res 92:1065–1073. doi: 10.1177/0022034513504218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooshima T, Matsumura M, Hoshino T, Kawabata S, Sobue S, Fujiwara T. 2001. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J Dent Res 80:1672–1677. doi: 10.1177/00220345010800071401. [DOI] [PubMed] [Google Scholar]
- 36.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghafoor A, Hay ID, Rehm BH. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez-Gil M, Quesada JM, Ramos-González MI, Soriano MI, de Cristóbal RE, Espinosa-Urgel M. 2013. Interplay between extracellular matrix components of Pseudomonas putida biofilms. Res Microbiol 164:382–389. doi: 10.1016/j.resmic.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 39.Peng X, Zhang Y, Bai G, Zhou X, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol 99:945–959. doi: 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resch U, Tsatsaronis JAL, Rhun A, Stubiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Wai SN, Charpentier E. 2016. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 7:e00207-16. doi: 10.1128/mBio.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilton M, Charron-Mazenod L, Moore R, Lewenza S. 2016. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:544–553. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li YH, Lau PCY, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol 184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Liu X, Liu H, Zhang L, Guo Y, Yu S, Wozniak DJ, Ma LZ. 2015. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep 7:330–340. doi: 10.1111/1758-2229.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. 2016. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol 99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besingi RN, Wenderska IB, Senadheera DB, Cvitkovitch DG, Long JR, Wen ZT, Brady LJ. 2017. Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology 163:488–501. doi: 10.1099/mic.0.000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. 2013. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J Cyst Fibros 12:384–389. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandstetter KA, Jurcisek JA, Goodman SD, Bakaletz LO, Das S. 2013. Antibodies directed against integration host factor mediate biofilm clearance from Nasopore. Laryngoscope 123:2626–2632. doi: 10.1002/lary.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devaraj A, Justice SS, Bakaletz LO, Goodman SD. 2015. DNABII proteins play a central role in UPEC biofilm structure. Mol Microbiol 96:1119–1135. doi: 10.1111/mmi.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. 2011. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol 4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 50.Ozaki K, Shibata Y, Yamashita Y, Nakano Y, Tsuda H, Koga T. 2002. A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS Lett 532:159–163. doi: 10.1016/S0014-5793(02)03661-X. [DOI] [PubMed] [Google Scholar]
- 51.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H, Zeng M, Fives-Taylor P. 2007. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun 75:2181–2188. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. 2013. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol 195:213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Nijampatnam B, Hua Z, Nguyen T, Zou J, Cai X, Michalek SM, Velu SE, Wu H. 2017. Structure-based discovery of small molecule inhibitors of cariogenic virulence. Sci Rep 7:5974. doi: 10.1038/s41598-017-06168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou M, Fives-Taylor P, Wu H. 2008. The utility of affinity-tags for detection of a streptococcal protein from a variety of streptococcal species. J Microbiol Methods 72:249–256. doi: 10.1016/j.mimet.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keyes PH. 1958. Dental caries in the molar teeth of rats. II. A method for diagnosing and scoring several types of lesions simultaneously. J Dent Res 37:1088–1099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.