High-throughput-sequencing-based marine microbiome profiling is rapidly expanding and changing how we study the oceans. Although powerful, the technique is not fully quantitative; it provides taxon counts only in relative abundances. In order to address this issue, we present a method to quantitatively estimate microbial abundances per unit volume of seawater filtered by spiking known amounts of internal DNA standards into each sample. We validated this method by comparing the calculated abundances to other independent estimates, including chemical markers (pigments) and total bacterial cell counts by flow cytometry. The internal standard approach allows us to quantitatively estimate and compare marine microbial community profiles, with important implications for linking environmental microbiomes to quantitative processes such as metabolic and biogeochemical rates.
KEYWORDS: amplicon sequencing, community profiling, internal standard, marine microbiome
ABSTRACT
An inherent issue in high-throughput rRNA gene tag sequencing microbiome surveys is that they provide compositional data in relative abundances. This often leads to spurious correlations, making the interpretation of relationships to biogeochemical rates challenging. To overcome this issue, we quantitatively estimated the abundance of microorganisms by spiking in known amounts of internal DNA standards. Using a 3-year sample set of diverse microbial communities from the Western Antarctica Peninsula, we demonstrated that the internal standard method yielded community profiles and taxon cooccurrence patterns substantially different from those derived using relative abundances. We found that the method provided results consistent with the traditional CHEMTAX analysis of pigments and total bacterial counts by flow cytometry. Using the internal standard method, we also showed that chloroplast 16S rRNA gene data in microbial surveys can be used to estimate abundances of certain eukaryotic phototrophs such as cryptophytes and diatoms. In Phaeocystis, scatter in the 16S/18S rRNA gene ratio may be explained by physiological adaptation to environmental conditions. We conclude that the internal standard method, when applied to rRNA gene microbial community profiling, is quantitative and that its application will substantially improve our understanding of microbial ecosystems.
IMPORTANCE High-throughput-sequencing-based marine microbiome profiling is rapidly expanding and changing how we study the oceans. Although powerful, the technique is not fully quantitative; it provides taxon counts only in relative abundances. In order to address this issue, we present a method to quantitatively estimate microbial abundances per unit volume of seawater filtered by spiking known amounts of internal DNA standards into each sample. We validated this method by comparing the calculated abundances to other independent estimates, including chemical markers (pigments) and total bacterial cell counts by flow cytometry. The internal standard approach allows us to quantitatively estimate and compare marine microbial community profiles, with important implications for linking environmental microbiomes to quantitative processes such as metabolic and biogeochemical rates.
INTRODUCTION
Since the first application of Roche 454 pyrosequencing to marine 16S rRNA gene amplicon samples (1), high-throughput sequencing of environmental PCR-amplified marker genes has transformed the study of marine microbiomes. It has been at the core of multiple recent programs varying in scale and breadth, including the International Census of Marine Microbes (2), TARA expeditions (3, 4), the Malaspina 2010 Expedition (5), Ocean Sampling Day (6) and the Long Term Ecological Research (LTER) sites (Palmer, HOT, Tahiti, and other sites). These studies and other programs have revealed unprecedented microbial diversity and biogeographic patterns and have advanced our understanding of marine microbial ecology (7) and biogeochemistry (4, 8).
An important limitation of the rRNA gene tag-based DNA sequencing approach is that it provides only compositional data, i.e., taxonomical profiles in relative proportions. While useful, compositional data are incomplete. As an example, should species A be equally abundant in two samples, its relative abundance in the first sample will be double that in the second sample if the total cell concentration is twice as high in the second sample. More broadly, compositional data can lead to various statistical issues, mainly due to two geometric features (9). First, the distance between two points has no absolute scale (e.g., counts of 1 and 2 have the same information as counts of 100 and 200) (10), and thus, the counts from different samples could have different uncertainties, making it difficult to identify statistically significant differences by standard tests (11). Second, compositional data are constrained by the “sum of 1,” and its projection in space is restricted to a simplex for which common statistical analyses based on Euclidean space may not be applicable (12–15). For example, it has long been realized that correlation analyses on compositional data can yield spurious correlations (16). This problem is particularly severe when communities have dominant taxa (14), as is commonly observed in some environmental samples (1, 17). These issues hinder cross-study comparisons of the rapidly expanding communal rRNA gene data sets. Various transformation (e.g., centered log-ratio transformation) and specialized data analysis routines have been developed to overcome these issues (e.g., programs such as DESeq2 consider the weighting of each taxon [18, 19]). However, such routines make it difficult to interpret the underlying biological and ecological mechanisms.
To palliate the artifacts associated with relative microbiome profiling (RMP), two approaches have recently been developed for quantitative microbiome profiling (QMP). The first approach is to normalize the 16S rRNA gene operational taxonomic unit (OTU) counts to total bacterial counts estimated by flow cytometry (FCM) (20, 21). The second approach, internal standard normalization (ISN), consists of spiking known concentrations of internal standards (DNA or cells) into samples before DNA extraction (22). This approach was adapted from the use of internal RNA standards in metatranscriptomics (23). ISN has recently been applied to study prokaryotic community composition in soils (24) and in the human gastrointestinal tract (25). In this study, as a proof of concept, we estimated the QMP of oceanic prokaryotes and eukaryotic plankton sampled from the Western Antarctica Peninsula (WAP) (Fig. 1A) using 16S and 18S rRNA gene amplicon sequencing combined with internal DNA standards. The large environmental gradients (e.g., coast versus open-ocean and open-water ice-covered regions) at the WAP lead to diverse and highly variable microbial communities (8, 26), thereby providing an ideal stage to test the ISN. Below, we present the ISN method as applied to marine samples. In order to validate the method, we (i) assessed the precision of ISN by spiking in various amounts of standards, (ii) compared phytoplankton abundances based on ISN to those based on estimates obtained with CHEMTAX, a program to calculate phytoplankton abundances based on pigment analyses (27), and (iii) compared total bacterial counts estimated by the 16S rRNA gene ISN to direct measurements by cell-counting flow cytometry. As an example of the numerous applications of this new approach, we demonstrated how the QMP and the relation of phytoplankton chloroplast 16S to genomic 18S rRNA gene abundances may provide insight into plankton ecology and photophysiology.
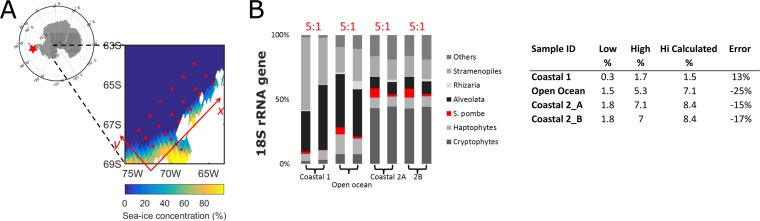
FIG 1.
(A) Sampling location at the WAP across the gradients of coast to open ocean (y axis) and open water to ice edge (x axis). (B) Relative abundances of 18S rRNA gene from a test run with a 5:1-diluted internal standard using representative samples from the open ocean and the coast. Coastal 2A and 2B are duplicate samples from the same location. The internal standard 18S rRNA gene reads (in red) are a small portion of the total reads and are proportional to the dilution factor.
RESULTS AND DISCUSSION
Brief description of the method.
A thorough description of the method is presented in Materials and Methods. Briefly, known amounts of genomic DNA (gDNA) from organisms not expected to be present in the natural seawater samples, i.e., Schizosaccharomyces pombe for 18S rRNA genes and Thermus thermophilus for 16S rRNA genes, were added to each sample before DNA extraction. The abundance of OTUi (in 16S or 18S rRNA gene copies per ml seawater) in sample j was calculated as
where Ri,j is the number of reads of OTUi in sample j, Rs,j is the number of 16S or 18S rRNA gene standard reads sequenced in sample j, Cs is the total number of 16S or 18S rRNA gene copies spiked into each sample, and Vj is the filtered seawater volume in milliliters. For double-stranded DNA, assuming that the average weight of a base pair is 650 daltons (650 g per mol), Cs can be calculated as
where rrns is the 16S or 18S rRNA gene copy number per cell. In our study, the spiked 16S rRNA gene standard was 14.85 ng of T. thermophilus gDNA, with rrn = 2 and genome size = 2.13 Mb (28); thus, Cs = 1.29 × 107 rRNA gene copies per sample. For the 18S rRNA gene standard S. pombe, the rrn could vary from 100 to 120 copies per cell (29). For our 18S rRNA gene calculation, we used a median number rrn = 110, which may introduce up to a 10% bias. However, this bias should be the same across all the samples and thus should not influence the comparison between samples. With 16.1 ng of spiked 18S rRNA standard per sample and the genome size of 13.8 Mb (29), Cs = 1.19 × 108 copies per sample. With a known number of rRNA gene copy number per cell rrni for OTUi (e.g., 1 copy per cell for SAR11), the cell abundance in sample j (in cells milliliter−1) can be calculated as Ai,j/rrni. We note that this is only possible when the rrni is known and assuming a single genome per cell.
Validation of the method.
To validate the method, 56 samples were collected at the WAP on three Palmer LTER annual cruises (years 2012, 2013, and 2015) (Fig. 1A). Internal standard recoveries averaged 0.8% (0.2% to 2.9%) of total prokaryotic 16S rRNA gene reads and 2.4% (0.7% to 5.7%) of total eukaryotic 18S rRNA gene reads, well within the range appropriate for detection (i.e., ≥0.1%) without overwhelming the environmental reads. Based on ISN, the abundance of rRNA genes between stations varied by 16- and 27-fold for eukaryotes and prokaryotes, respectively (Fig. 2). Using rrn from the rrnDB database (30), we converted OTU2 (SAR11) and OTU5 (Polaribacter) rRNA gene counts to cell abundances (see Fig. S1 in the supplemental material). The average cell abundance of the SAR11 OTU in our samples was 2.0 × 105 cells ml−1, in line with SAR11 estimates reported by other studies in the Southern Ocean (31–33). As described below, we assessed the precision of the ISN by spiking in two different amounts of internal standards. We also corroborated our results with abundance estimates using two independent methods, CHEMTAX pigment analyses for the 18S rRNA gene and FCM for the 16S rRNA gene abundances.
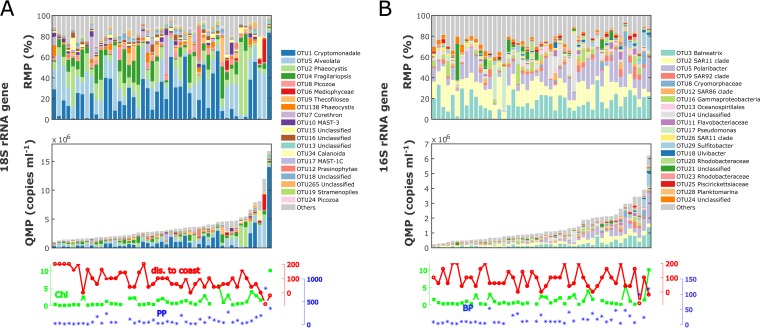
FIG 2.
A 3-year data set of WAP community OTU counts (97% similarity) in RMP (upper panels) and QMP in rRNA gene copies milliliter−1 (lower panels) for eukaryotes (A) and prokaryotes (B). The bar plots present QMP and RMP, with the top 20 OTUs associated with their taxonomic identifications (down to the finest identified level in the SILVA 128 database) and all other OTUs combined into one bin labeled “others” in gray. Stations are ranked in ascending total 16S or 18S rRNA gene counts. Environmental variables are plotted at the bottom: grid station or the approximate distance to shore (kilometers) in red, chlorophyll a concentration (milligrams meter−3) in green, and primary production (milligrams meter−3 day−1) or bacterial production (leucine incorporation [picomoles liter−1 hour−1]) in blue. QMP captures significant variations across samples and is correlated with environmental variables.
(i) Precision of ISN.
In a test sequencing run to optimize the standard amount, we added the eukaryotic internal standards at two different concentrations (16.1 and 3.22 ng) into representative samples (see details in Materials and Methods). The response was proportional to the spiked-in level (Fig. 1B), with a maximum deviation estimated at 25% (average, 18%) across the various communities sampled at the coastal and open-ocean sites. For comparison, the traditional quantitative PCR (qPCR) methods can yield errors as large as the signal (34), with typical coefficients of variation (CVs) ranging from 15% to 50% (35, 36). This comparison should be interpreted with caution because the precision of qPCR has been verified over a wider range of concentrations (i.e., 7 to 9 orders of magnitude) (37, 38) than for most internal standard studies (39). To test the reproducibility of the sequencing technique, we also barcoded and sequenced a coastal sample in duplicates (Coastal 2A and 2B), and the resulting community profiles are highly similar (Fig. 1B). The CV for estimated taxon abundance was 2.8% on average and 12.3% at maximum (see Table S1 in the supplemental material), with higher uncertainties for rarer taxa.
(ii) Method comparison.
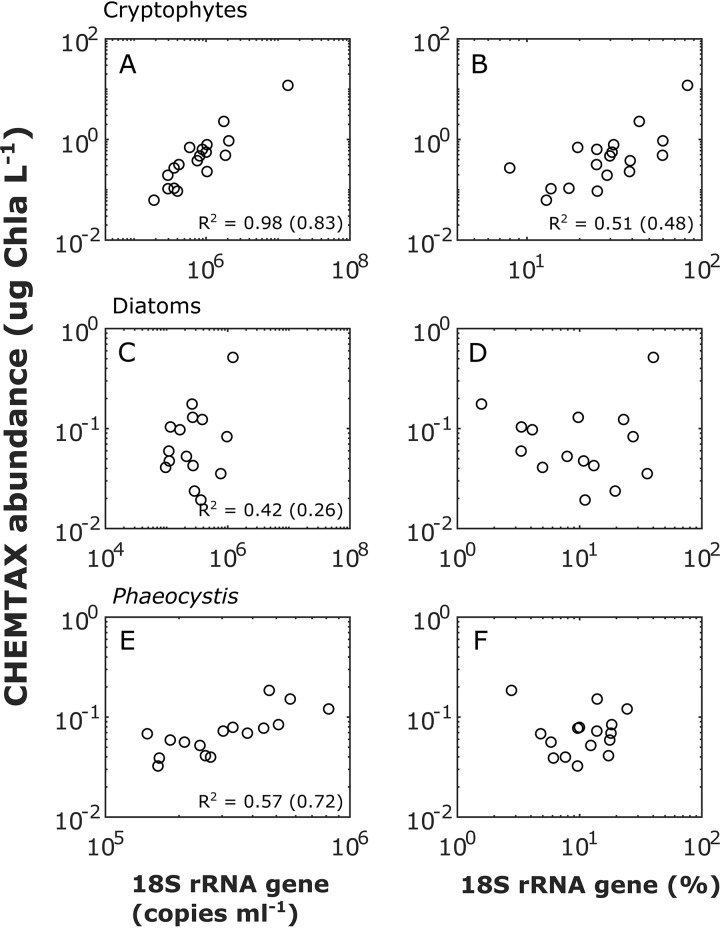
(a) Phytoplankton 18S rRNA gene ISN versus CHEMTAX abundance. We compared the phytoplankton QMP estimated by ISN with the traditional CHEMTAX analysis of high-performance liquid chromatography (HPLC) pigment profiles (26, 40) for three phytoplankton groups commonly observed at the WAP, i.e., cryptophytes, diatoms, and Phaeocystis. The cryptophyte abundances calculated by using the 18S rRNA gene and CHEMTAX were highly correlated (Pearson’s r2 = 0.98, P < 0.0001) (Fig. 3A). Significant correlations were also observed for diatoms (r2 = 0.42, P < 0.0001) (Fig. 3C) and Phaeocystis (r2 = 0.57, P < 0.0001) (Fig. 3E), although the relationships were weaker. Because alloxanthin is present only in cryptophytes, their CHEMTAX estimates are likely more robust than the ones for diatoms and Phaeocystis. In addition, alloxanthin was the most abundant pigment in our sample set, with an average concentration of 0.61 μg/liter. In comparison, the other accessory pigments were substantially less abundant (19′-butanoyloxyfucoxanthin, 0.01 μg/liter; chlorophyll c2 [Chl c2], 0.18 μg/liter; chlorophyll c3, 0.02 μg/liter; chlorophyll b, 0.01 μg/liter; fucoxanthin, 0.13 μg/liter; and hexanoyloxyfucoxanthin, 0.13 μg/liter). Low concentrations of accessory pigments could introduce errors in CHEMTAX estimates of diatoms and Phaeocystis. Using RMP, a significant but weaker correlation was observed for cryptophytes (r2 = 0.51, P < 0.001) (Fig. 3B). No significant correlation between RMP and CHEMTAX estimates was observed for diatoms (Fig. 3D) and Phaeocystis (Fig. 3F).
FIG 3.
Abundances of cryptophytes (A and B), diatoms (C and D), and Phaeocystis (E and F) estimated by 18S rRNA gene QMP (left panels) and RMP (right panels) compared to abundances estimated by CHEMTAX-HPLC pigment analyses. For significant correlations (P < 0.05), Pearson’s r2 values for original data and log10-transformed data (in parentheses) are shown at the bottom of the plot.
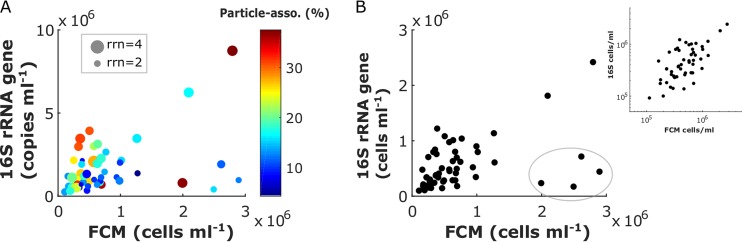
(b) Bacterial 16S rRNA gene ISN versus FCM bacterial abundance. The total prokaryotic 16S rRNA gene abundances were significantly correlated with the bacterial FCM counts, albeit with a small correlation coefficient (Pearson’s r2 = 0.19, P < 0.001; or r2 = 0.20, P < 0.001 after log transformation) (Fig. 4A). In general, rRNA gene copy numbers were much higher than the FCM cell counts. A variety of factors may explain this. First, for the four points circled in gray in Fig. 4B, the FCM estimates of ≥2.0 × 106 cells ml−1 were anomalously high compared to the corresponding leucine incorporation rates or Chl a concentrations. Second, while bacteria associated with particles were efficiently captured by DNA sequencing, they may have been missed by FCM counts if the vortex step did not break down the particle-bacterium associations. In polar and coastal regions, a significant proportion of bacteria could be attached to particles (41). Corroborating this hypothesis, we found that samples where ISN predicted a higher abundance of bacteria than FCM tended to have a higher percentage of particle-associated OTUs (Fig. 4A). Finally, the difference in rrn for different OTUs could also explain the discrepancy between the ISN and FCM bacterial abundances. For example, the rrns in SAR11 and Marinomonas sp. strain MWYL1 are 1 and 8, respectively (30). Populations with larger rrn should have higher 16S rRNA gene/FCM count ratios. In addition, the fact that multiple genomes may exist within a single cell (42) could also contribute to the discrepancy. To estimate cell abundances, the top 20 classified OTU QMPs were divided by their rrn estimated by rrnDB (30), and the resulting OTU cell abundances were summed for each sample. Taxa previously identified as particle-associated bacteria through size-fractionated filtration (41) were then excluded. After discarding the four potential outliers and correcting for rrn and particle association effects, cell abundances estimated by rRNA gene and FCM counts displayed a substantially higher correlation coefficient (r2 = 0.61, P < 0.001; or r2 = 0.44, P < 0.001 after log transformation) and were close to the 1:1 line (Fig. 4B).
FIG 4.
Comparison of total bacterial abundances estimated by FCM and 16S rRNA gene QMP. (A) The size of each data point represents the rrn effect, calculated as the averaged rrn of the top 20 classified OTUs in each sample. The color coding represents the particle association effect, calculated as the cumulative cell percentage (after rrn correction) of the top 10 most abundant particle-associated OTUs, identified as the >3-µm fraction bacteria reported by Delmont et al. (41). An exhaustive survey of the particle-associated OTUs is not feasible considering that significant portions of the prokaryotic OTUs are of unknown physiology or are even unclassified in the current rRNA database (SILVA 128). (B) FCM versus rrn and particle association effect-corrected 16S rRNA QMP in cells milliliter−1. The four points inside the gray circle are likely outliers. After excluding these four points, data points are plotted on both linear and log scales.
We note that the rrn correction is important not only for ISN but also for the normalization of FCM (see, e.g., reference 20). The absolute cell abundance of OTU x in a particular sample should be calculated as , where Cx is the rRNA gene counts for OTU x. Should be constant for a particular taxon, a change in the numerator introduces a systematic bias when comparing relative changes in absolute abundances between samples. However, because , the denominator may lead to uneven biases across samples. A simple example using two OTUs commonly found in the WAP is presented in Table S2 in the supplemental material. Without taking into account the rrn, the estimates of absolute OTU abundances based on FCM normalization could be off by 5-fold, and the estimated abundance variation between two samples could be off by 3.6-fold in this particular example. Caution should therefore be taken in applying the FCM normalization method without resolving the community rrn profile.
One approach to estimating the rrn profile is to use the phylogenetic information to predict the rrns of OTUs based on existing rrn databases, such as rrnDB (30, 43). A recent human microbiome study corrected the 16S rRNA gene matrix using rrnDB (21). However, substantial uncertainties associated with the rrn correction remain, as (i) a significant portion of the OTUs are unclassified and (ii) the limited number of known rrns from sequenced genomes likely does not reflect the natural variability in rrn.
When applying the FCM normalization method to marine samples, the difference in sampling volume for DNA and FCM should be considered. Cells for DNA analyses are generally filtered from liters of seawater, while FCM samples are generally estimated from less than 1 ml of seawater. In patchy environments, these two volumes may reflect different communities.
Application: case study at the WAP.
In our WAP case study, the estimated total eukaryotic rRNA gene abundance was significantly correlated with environmental variables, including the distance to shore (Pearson’s r = −0.6, P < 0.001; Spearman’s ρ = −0.6, P < 0.001), Chl a concentration (r = 0.8, P < 0.001; ρ = 0.7, P < 0.001), and primary production rate (r = 0.7, P < 0.001; ρ = 0.5, P < 0.001). Conversely, the estimated total prokaryotic rRNA gene abundance was not significantly correlated with distance to shore (r = −0.3, P > 0.05; ρ = −0.2, P > 0.1) but was significantly correlated with Chl a (r = 0.6, P < 0.001; not significant by Spearman test [ρ = 0.3, P > 0.05]) and significantly correlated with bacterial production measured by [3H]leucine incorporation (r = 0.7, P < 0.001; ρ = 0.6, P < 0.001). Looking at specific taxa, the abundance of Polaribacter OTU5 increased significantly with increasing Chl a (r = 0.8, P < 0.001; ρ = 0.5, P < 0.001) (Fig. S1), which is consistent with the observations that Polaribacter thrives during phytoplankton blooms (44, 45). The SAR11 OTU2 cell abundances did not show a clear trend across Chl a gradients (r = −0.02, P = 0.9; ρ = −0.01, P = 0.9). This could be a result of patterns at finer taxonomic scales, e.g., amplicon sequence variants resolved down to the single-nucleotide level (46). The relative abundance of SAR11 OTU decreased with increasing Chl a (r = −0.5, P < 0.001; ρ = −0.5, P < 0.001), but this could be a spurious correlation stemming from an increase in the total bacterial abundance.
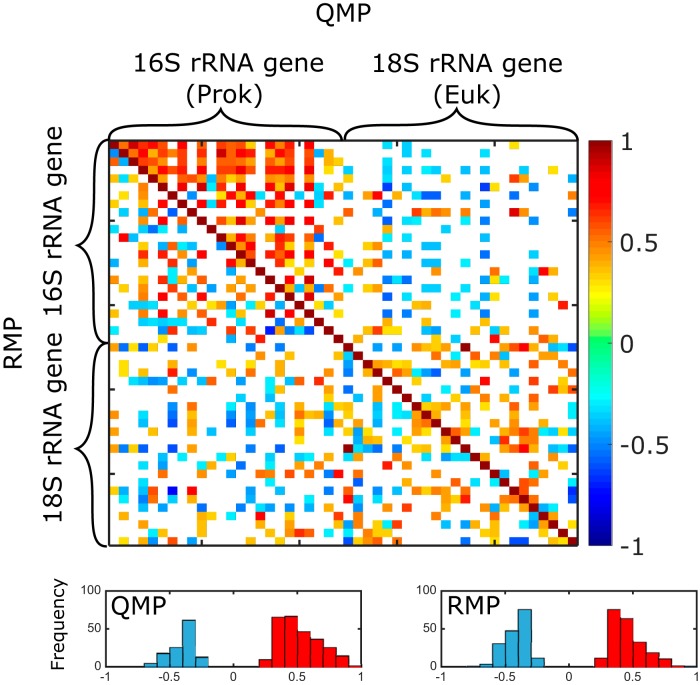
Community cooccurrence matrices based on Spearman’s correlation coefficients (Fig. 5) showed that QMP and RMP matrices were significantly different (P < 0.001) by the Jennrich test (47) and by the Steiger test (48). QMP resulted in more positive correlations (270, versus 218 for RMP), mostly appearing within the prokaryotic communities, and fewer negative correlations overall (124, versus 172 for RMP). Interestingly, similar differences in cooccurrence patterns based on RMP and QMP have also been observed in human gut microbiome studies using the FCM normalization method (21).
FIG 5.
Taxon cooccurrence matrices based on QMP (rRNA gene copies per milliliter) versus RMP (rRNA gene copy percentage). The top 24 most abundant prokaryotic (16S rRNA gene) OTUs and eukaryotic (18S rRNA gene) OTUs were used to construct the pairwise correlation matrices based on QMP (upper triangle) and RMP (lower triangle). Spearman’s ρ values for significant correlations (P < 0.05) are presented as squares in the heat map. The bottom histograms display the distribution of negative (blue) and positive (red) ρ values for matrices based on QMP and RMP.
(i) Quantitatively estimating eukaryotic phytoplankton abundances using chloroplast 16S rRNA gene abundances.
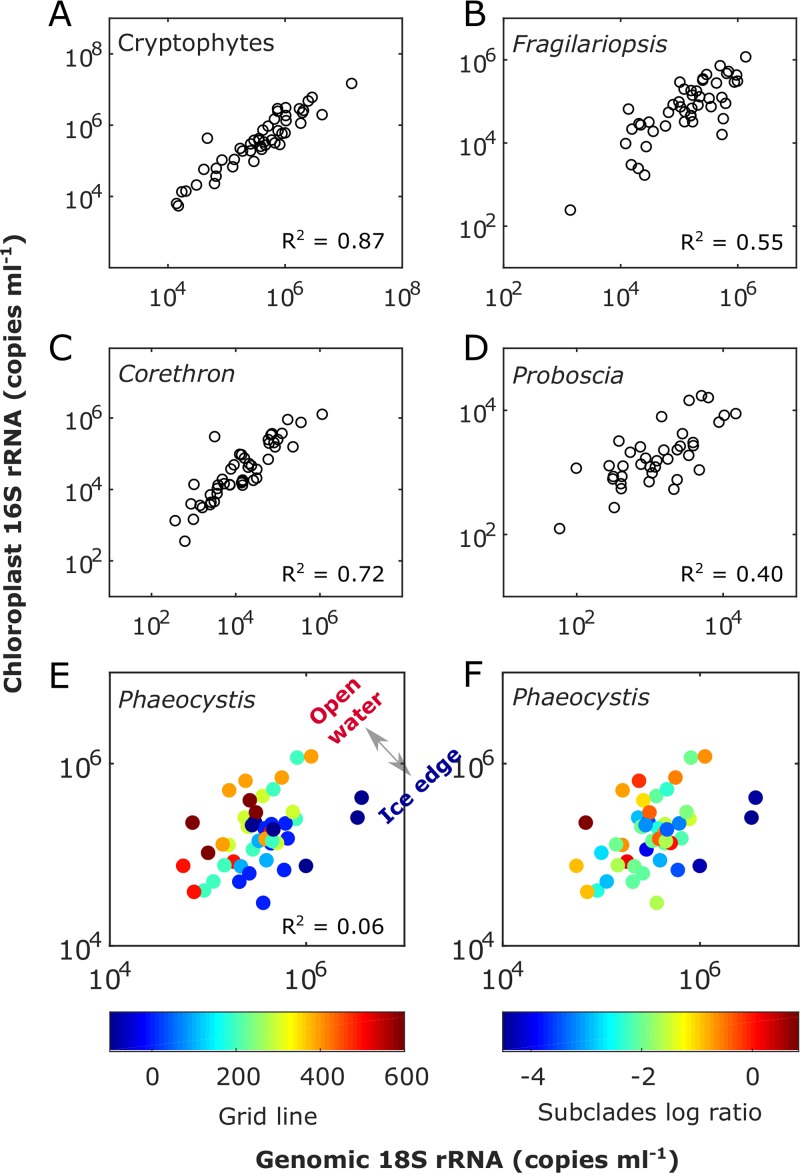
The QMPs of five eukaryotic phytoplankton groups calculated from internal standard-normalized 18S rRNA gene abundances and the corresponding chloroplast 16S rRNA gene counts were compared (Fig. 6). Strong linear correlations using the type II least-square fit were observed between the chloroplast 16S rRNA gene counts and genomic 18S rRNA gene counts for cryptophytes (r2 = 0.87, P < 0.0001) and diatoms, including Fragilariopsis (r2 = 0.55, P < 0.0001), Corethron (r2 = 0.72, P < 0.0001), and Proboscia (r2 = 0.40, P < 0.0001). A weak correlation was observed for Phaeocystis using the type II least-square fit (r2 = 0.06, P < 0.0001) but not with a Pearson coefficient (r2 = 0.06, P = 0.09). These results show that eukaryotic autotroph abundances can be reliably estimated from their corresponding chloroplast 16S rRNA gene abundances for the three phytoplankton groups examined, i.e., cryptophytes, diatoms, and Phaeocystis.
FIG 6.
Comparison of phytoplankton abundances based on normalized genomic 18S and chloroplast 16S rRNA genes for cryptophytes (A), Fragilariopsis (diatom) (B), Corethron (diatom) (C), and Proboscia (diatom) (D) and for Phaeocystis in relationship to the north-south geographic gradient (color-coded by Palmer LTER line number as an index for open water to ice edge gradient) (E) and Phaeocystis subclade ratios calculated as ln(OTU138/OTU2) (F). OTU2 and OTU138 are the top two Phaeocystis OTUs, comprising 87.27% and 12.70% of the total Phaeocystis 18S rRNA gene counts, respectively. The slope and intercept values are presented in Table S3 in the supplemental material.
Chloroplast 16S rRNA genes can represent a large fraction of total community 16S rRNA gene library reads, especially in productive oceanic regions where phototrophic eukaryotes tend to dominate. For example, 52% of the total 16S rRNA gene reads were annotated as chloroplasts at our study sites (averaged over all sampled stations). While these chloroplast reads are generally discarded, they may provide valuable information about the phototrophic eukaryote abundance without incurring the additional cost of 18S rRNA gene amplicon sequencing. Several recent studies inferred eukaryotic phytoplankton relative abundances from the chloroplast 16S rRNA gene reads (41, 49). The method described here may allow for estimating the host phytoplankton abundances from the ISN chloroplast sequences (Fig. 6).
(ii) 18S/16S rRNA gene ratios as a measure of phytoplankton ecophysiology.
ISN can also be used to quantify variability in the ratio of the chloroplast 16S rRNA gene to the genomic 18S rRNA gene and thus to gain insight into phytoplankton ecophysiology. Compared to diatoms and cryptophytes, laboratory data suggest that Phaeocystis is well adapted to variability in light availability (50). This photoacclimation capacity could result from a greater plasticity in pigments per chloroplast (51) or chloroplasts per cell under different light regimes. The latter strategy could explain the variability in chloroplast 16S versus genomic 18S rRNA gene reads in Phaeocystis observed in our study. As shown in Fig. 6E, the ratios of Phaeocystis chloroplast 16S to genomic 18S rRNA genes generally decreased from north to south. Phytoplankton physiology is influenced by sea ice dynamics at the WAP (52, 53). Considering that the ice generally retreated from north to south, the southern communities closer to the ice edge might have been more recently exposed to higher light levels. The northern communities, on the other hand, had been in open water for a longer period of time, being exposed to stronger wind-induced vertical mixing, and were therefore more likely to be light limited. This may explain the higher chloroplast 16S/genomic 18S rRNA gene ratios in the south. These geographic variations were consistent with changes in the relative abundances of two Phaeocystis subclades (Fig. 6F) which may be adapted to different light conditions. The correlation to mixed-layer depth was not as strong as that to the geographic gradients (see Fig. S2 in the supplemental material). Overall, the chloroplast 16S/genomic 18S rRNA gene ratio could prove to be a valuable indicator of in situ algal photophysiology adaptations when combined with laboratory experiments for further validation.
Limitations of ISN.
There are several limitations to ISN. The first issue is associated with the extraction efficiency. Since the extraction efficiency is never 100%, the calculated rRNA gene abundance represents a lower bound on the true abundance. This could partially be addressed by spiking in cells instead of genomic DNA, although cell standards could also introduce biases due to (i) differences in extraction efficiency between the standard cells and the natural cells and (ii) variability in the number of genomes per cell (42). A second issue is the high uncertainty in rrn correction (54), which is relevant only when converting rRNA gene copy numbers to cell numbers or when combining groups with mixed rrn. For example, large eukaryotes, such as some dinoflagellates, could have high rrns (>1,000 copies per cell) (55), and thus their 18S rRNA gene abundances could be orders of magnitude higher than their cell numbers. However, should a specific OTU have a constant rrn, the relative changes in absolute abundances across samples will still be captured because the copy numbers are proportional to the cell density. As the rrn is more comparable at finer taxonomic levels (56), it is best to apply the rrn normalization down to single genotypes. Defining OTUs at coarse taxonomic levels may combine groups with differing rrns. In this case, the rRNA gene copy numbers are no longer proportional to the true cell numbers, thus complicating the interpretation of the rRNA gene counts. Finally, a third issue is that some eukaryotic species have high plasticity in rrn (57). Variability in their 18S rRNA gene counts may not reflect variability in their cell numbers. On the other hand, positive correlations of rrn versus cell biovolume have been reported across different eukaryotic plankton taxon, including diatoms and dinoflagellates (52, 54). If this relation is valid, groups with different rrns could be combined, and the rRNA gene copy numbers could be used as an index for group-specific biomass. This is important because biomass is often of more relevance to biogeochemical budgets (e.g., carbon and nitrogen) than cell numbers.
PCR bias could skew the relative abundances of mixed-community members estimated from the PCR products (58, 59). One main concern specific to our approach is the biased PCR amplification caused by the varying template GC contents. Due to the triple hydrogen bonds between G and G, templates with higher GC contents have higher melting temperatures and are less efficiently amplified (59, 60). T. thermophilus, the 16S rRNA internal standard used in our study, has a high GC content (69% for the whole genome [61] and 65% for the amplified V4 region). A high GC content can cause underestimation of the internal standard abundance and overestimation of the natural community member abundance. A second concern is the amplification bias introduced by the degenerate primers. DNA sequences with G/C at the degenerate position can be overamplified compared to sequences with A/T. The deviation in PCR product due to a single base difference at the priming site could be over 100% after 35 PCR cycles (58). Various methods have been developed to reduce PCR biases: combining PCR replicates (combined triplicates were used in this study), minimizing PCR cycle numbers and the degeneracy of primers, and reconditioning PCR (62). On the other hand, despite the significant PCR biases, intersample variability could still be precisely captured by PCR method (58). A time series study reported that PCR primer selection affects the estimated population abundances but not the community dynamic patterns (63). Although the abundance estimates by PCR-based ISN may deviate from the absolute cell numbers due to PCR bias and rrn issues, the estimated intersample variability is less affected. Hence, this may not be as much of an issue for correlation analyses, e.g., time series community dynamics, community cooccurrence, and correlations to environmental variables.
Conclusions.
Addition of internal standards to the amplicon rRNA gene sequencing approach allowed us to quantitatively compare microbial communities across different samples, as well as phytoplankton chloroplast 16S and genomic 18S rRNA gene abundances. Conceptually, the ISN could provide information equivalent to qPCR measurements targeting rRNA genes but with the advantage of examining a diverse community in a single assay. In our case study at the WAP, significant correlations observed in phytoplankton abundances based on 18S rRNA gene versus CHEMTAX abundances and in total bacterial abundances based on 16S rRNA gene versus FCM counts confirm that the ISN is quantitative. Our study also shows that chloroplast 16S rRNA gene sequences could be used to estimate phytoplankton abundances and that the ratio of chloroplast 16S to genomic 18S rRNA genes may be an insightful indicator of phytoplankton in situ photophysiology. The ISN comes at a minimal cost of implementation and could be applied in conjunction with metagenomics (64). Overall, the ISN allows for an improved statistical, and ultimately ecological, interpretation of the rich and rapidly expanding marine microbiome data sets. More broadly, this approach could be valuable to researchers interested in relating microbial ecology to quantitative processes such as microbial interactions, metabolic rates, energy and material fluxes, and eventually quantitative ecosystem modeling.
MATERIALS AND METHODS
DNA extraction with internal standard DNA addition.
Samples for DNA extraction were collected by seawater filtration (for details, see the supplemental material). Each filter with a recorded filtration volume (4 liters for most samples) was split into two, with one half used for DNA extraction and the other half stored for later RNA work. We note that this step could introduce errors due to an uneven cell distribution on the filter. Just prior to DNA extraction, gDNAs from two organisms representing eukaryotic and prokaryotic taxa not expected to be present in marine surface water samples were added to the tube containing the sample filter and lysis buffer (see below for optimization of internal standard addition). For the 18S rRNA gene internal standard, 50 μl of Schizosaccharomyces pombe gDNA (ATCC [Manassas, VA, USA] 24843D-5) at 0.322 ng/μl was spiked into each sample. For the 16S rRNA gene internal standard, 50 μl of Thermus thermophilus gDNA (ATCC 27634D-5) at 0.297 ng/μl was added to each sample. The internal standard working solutions were made in single-use aliquots to avoid DNA being lost during freeze-thaw cycles. gDNA standard stock solutions and dilution concentrations were measured using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). After spiking in internal standards, DNA extraction was performed as described previously (8).
Optimizing the amount of internal standard added to a sample.
In order to get enough standard signal without overwhelming the environmental signal, we added the internal gDNA standards targeting a final concentration of around 1% of the total 16S and 18S rRNA gene reads. The amount of the prokaryotic genomic internal standard to spike in was based on the anticipated total extracted DNA mass as estimated with trial samples (22). For example, if we expected 10 µg of total genomic DNA in the sample, we added 100 ng of prokaryotic gDNA internal standard. Because the fraction of eukaryotic gDNA in total community DNA and the eukaryotic rRNA gene copy numbers per unit weight of gDNA are highly variable in different marine environments, a test sequencing run was conducted to optimize the internal standard amount to be spiked in. Test libraries were constructed with representative samples spiked with different amounts of internal eukaryotic genomic standard (16.1 ng or 3.22 ng) Schizosaccharomyces pombe gDNA (Fig. 1B). The test amplicon libraries were subsequently sequenced using the Illumina MiSeq platform (nano format) as a customized run at the Duke Institute for Genomic Sciences and Policy (IGSP) with 300-bp single-coverage forward reads and 10-bp reverse reads to read the reverse barcodes. The averaged read count per sample was 50,661 after demultiplexing (see Table S4 in the supplemental material).
Amplicon library construction.
16S rRNA genes were amplified by PCR using V4 primer set 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) (65) and 805R (5′-GACTACNVGGGTATCTAAT-3′), modified from references 66 and 67). 18S rRNA genes were amplified by PCR using V4 primer set EukF (5′-CCAGCASCYGCGGTAATTCC-3′) (68) and EukR (5′-ACTTTCGTTCTTGAT-3′), modified from reference 68 as described in reference 8, to increase coverage for haptophytes.
Dual-indexed fusion primers had 6-bp barcodes at each end, which were constructed using error-proof Hamming codes (69). In order to improve the “low sequence diversity” issue of the rRNA amplicon library, 0- to 5-bp heterogeneity spacers were added to each primer (70). PCRs were performed in triplicates for each sample. 18S rRNA gene PCR and library pooling were performed as described previously (8). 16S rRNA gene library construction was similar to that for 18S rRNA gene except that 2 U of Platinum Taq DNA high-fidelity polymerase (Invitrogen) was added to each reaction mixture, and the PCR annealing temperature was 60°C.
Amplicon libraries were sequenced at the Duke IGSP using the Illumina MiSeq 250PE platform for 16S rRNA amplicons and the MiSeq 300PE platform for 18S rRNA amplicons. For each library, reads per sample after multiplexing are reported in Table S4.
Bioinformatic analyses.
For each library, paired-end reads were assembled using VSEARCH v2.3.4 (71) with quality scores of the merged bases calculated as described previously (72). Assembled reads were further processed using USEARCH (73) and QIIME (74) as described previously (8). In brief, 16S or 18S rRNA gene reads were quality controlled, including quality filtering and chimera checking, and then were trimmed for barcodes and primer sequences. Singletons were discarded. OTUs (97% similarity) were then clustered using USEARCH, and the representative sequences were assigned taxonomy based on the SILVA small-subunit (SSU) database 128 (76) using QIIME.
For the 16S rRNA gene library, sequences identified in SILVA as mitochondria were removed. Sequences classified as chloroplasts were filtered out as a separate data set. In order to further identify the phytoplankton host taxonomy from the chloroplast sequences, representative chloroplast sequences were subjected to a BLAST search against the NCBI nucleotide collection database using BLAST+ 2.6.0 (75). The top three hits for each sequence are reported in Table S5 in the supplemental material.
Accession number(s).
Sequence data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database under accession numbers PRJNA508517 and PRJNA508514.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Laboratoire d'Excellence LabexMER (ANR-10-LABX-19) in France and U.S. NSF grant OPP-1043339 to N.C. This research was undertaken during the January 2012, 2013, and 2015 cruises by the Palmer Antarctica Long Term Ecological Research (PAL-LTER) project, supported by NSF awards OPP-0823101 and 1440435 to J.D. and NASA ROSES award NNX14AL86G to O.S.
We thank Bruce Barnett, Rachel Eveleth, and Naomi Shelton for sampling and all the scientists and the crew on R/V L. M. Gould for shipboard assistance. We are also grateful to Damien Eveillard and Samuel Chaffron for insightful discussion and comments.
Y.L. and N.C. conceived and designed the study. Y.L., H.D., and O.S. collected field samples and data during the cruises. Y.L. processed the DNA samples and analyzed the data. Y.L., N.C., and S.G. wrote the manuscript with contributions from the other authors.
We declare that we have no competing financial interests in regard to the work described.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02634-18.
REFERENCES
- 1.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere. Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinger L, Amaral-Zettler LA, Fuhrman JA, Horner-Devine MC, Huse SM, Welch DBM, Martiny JBH, Sogin M, Boetius A, Ramette A. 2011. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vargas C, Audic S, Henry N, Decelle J, Mahe F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury J-M, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horak A, Jaillon O, Lima-Mendez G, Luke J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Acinas SG, Bork P, Bowler C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki ME, Speich S, Stemmann L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E, Boss E, Follows M, Karp-Boss L, Krzic U, Reynaud EG, Sardet C, Sullivan MB, Velayoudon D. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 4.Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, Darzi Y, Audic S, Berline L, Brum JR. 2016. Plankton networks driving carbon export in the oligotrophic ocean. Nature 532:465–470. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pernice MC, Giner CR, Logares R, Perera-Bel J, Acinas SG, Duarte CM, Gasol JM, Massana R. 2016. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J 10:945–958. doi: 10.1038/ismej.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopf A, Bicak M, Kottmann R, Schnetzer J, Kostadinov I, Lehmann K, Fernandez-Guerra A, Jeanthon C, Rahav E, Ullrich M, Wichels A, Gerdts G, Polymenakou P, Kotoulas G, Siam R, Abdallah RZ, Sonnenschein EC, Cariou T, O'Gara F, Jackson S, Orlic S, Steinke M, Busch J, Duarte B, Caçador I, Canning-Clode J, Bobrova O, Marteinsson V, Reynisson E, Loureiro CM, Luna GM, Quero GM, Löscher CR, Kremp A, DeLorenzo ME, Øvreås L, Tolman J, LaRoche J, Penna A, Frischer M, Davis T, Katherine B, Meyer CP, Ramos S, Magalhães C, Jude-Lemeilleur F, Aguirre-Macedo ML, Wang S, et al. 2015. The ocean sampling day consortium. Gigascience 4:27. doi: 10.1186/s13742-015-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhrman JA, Cram JA, Needham DM. 2015. Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13:133–146. doi: 10.1038/nrmicro3417. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, Cassar N, Marchetti A, Moreno C, Ducklow H, Li Z. 2017. Specific eukaryotic plankton are good predictors of net community production in the Western Antarctic Peninsula. Sci Rep 7:14845. doi: 10.1038/s41598-017-14109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn TP, Richardson MF, Lovell D, Crowley TM. 2017. propr: an R-package for identifying proportionally abundant features using compositional data analysis. Sci Rep 7:16252. doi: 10.1038/s41598-017-16520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Boogaart KG, Tolosana-Delgado R. 2008. “compositions”: a unified R package to analyze compositional data. Comput Geosci 34:320–338. doi: 10.1016/j.cageo.2006.11.017. [DOI] [Google Scholar]
- 11.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aitchison J. 1982. The statistical analysis of compositional data. J R Stat Soc Ser B 44:139–177. doi: 10.1111/j.2517-6161.1982.tb01195.x. [DOI] [Google Scholar]
- 13.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R. 2016. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 10:1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H. 2015. Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu Rev Stat Appl 2:73–94. doi: 10.1146/annurev-statistics-010814-020351. [DOI] [Google Scholar]
- 15.Lin W, Shi P, Feng R, Li H. 2014. Variable selection in regression with compositional covariates. Biometrika 101:785–797. doi: 10.1093/biomet/asu031. [DOI] [Google Scholar]
- 16.Pearson K. 1896. Mathematical contributions to the theory of evolution. III. Regression, heredity, and panmixia. Philos Trans R Soc London Ser A 187:253–318. doi: 10.1098/rsta.1896.0007. [DOI] [Google Scholar]
- 17.Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R, Claverie J-M, Decelle J, Dolan JR, Dunthorn M, Edvardsen B, Gobet A, Kooistra WHCF, Mahé F, Not F, Ogata H, Pawlowski J, Pernice MC, Romac S, Shalchian-Tabrizi K, Simon N, Stoeck T, Santini S, Siano R, Wincker P, Zingone A, Richards TA, de Vargas C, Massana R. 2014. Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24:813–821. doi: 10.1016/j.cub.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Props R, Kerckhof F-M, Rubbens P, De Vrieze J, Sanabria EH, Waegeman W, Monsieurs P, Hammes F, Boon N. 2017. Absolute quantification of microbial taxon abundances. ISME J 11:584. doi: 10.1038/ismej.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. [DOI] [PubMed] [Google Scholar]
- 22.Satinsky BM, Gifford SM, Crump BC, Moran MA. 2013. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol 531:237–250. doi: 10.1016/B978-0-12-407863-5.00012-5. [DOI] [PubMed] [Google Scholar]
- 23.Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. 2011. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J 5:461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smets W, Leff JW, Bradford MA, McCulley RL, Lebeer S, Fierer N. 2016. A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol Biochem 96:145–151. doi: 10.1016/j.soilbio.2016.02.003. [DOI] [Google Scholar]
- 25.Stämmler F, Gläsner J, Hiergeist A, Holler E, Weber D, Oefner PJ, Gessner A, Spang R. 2016. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 4:28. doi: 10.1186/s40168-016-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang K, Ducklow H, Vernet M, Cassar N, Bender ML. 2012. Export production and its regulating factors in the West Antarctica Peninsula region of the Southern Ocean. Global Biogeochem Cycles 26:GB2005. doi: 10.1029/2010GB004028. [DOI] [Google Scholar]
- 27.Mackey MD, Mackey DJ, Higgins HW, Wright SW. 1996. CHEMTAX—a program for estimating class abundances from chemical markers: application to HPLC measurements of phytoplankton. Mar Ecol Prog Ser 144:265–283. doi: 10.3354/meps144265. [DOI] [Google Scholar]
- 28.Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk H-P, Kramer W, Merkl R, Gottschalk G, Fritz H-J. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22:547. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- 29.Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 30.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straza TRA, Ducklow HW, Murray AE, Kirchman DL. 2010. Abundance and single-cell activity of bacterial groups in Antarctic coastal waters. Limnol Oceanogr 55:2526–2536. doi: 10.4319/lo.2010.55.6.2526. [DOI] [Google Scholar]
- 32.Wietz M, Gram L, Jørgensen B, Schramm A. 2010. Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microb Ecol 61:179–189. doi: 10.3354/ame01443. [DOI] [Google Scholar]
- 33.Thiele S, Fuchs BM, Ramaiah N, Amann R. 2012. Microbial community response during the iron fertilization experiment LOHAFEX. Appl Environ Microbiol 78:8803–8812. doi: 10.1128/AEM.01814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith CJ, Osborn AM. 2009. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 35.Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. 2007. Statistical significance of quantitative PCR. BMC Bioinformatics 8:131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison TB, Weis JJ, Wittwer CT. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24:954–958. [PubMed] [Google Scholar]
- 37.Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ, Chisholm SW. 2006. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol 72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labrenz M, Brettar I, Christen R, Flavier S, Bötel J, Höfle MG. 2004. Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea. Appl Environ Microbiol 70:4971–4979. doi: 10.1128/AEM.70.8.4971-4979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gifford SM, Becker JW, Sosa OA, Repeta DJ, DeLong EF. 2016. Quantitative transcriptomics reveals the growth-and nutrient-dependent response of a streamlined marine methylotroph to methanol and naturally occurring dissolved organic matter. mBio 7:e01279-16. doi: 10.1128/mBio.01279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schofield O, Saba G, Coleman K, Carvalho F, Couto N, Ducklow H, Finkel Z, Irwin A, Kahl A, Miles T, Montes-Hugo M, Stammerjohn S, Waite N. 2017. Decadal variability in coastal phytoplankton community composition in a changing West Antarctic Peninsula. Deep Sea Res Part I Oceanogr Res Pap 124:42–54. doi: 10.1016/j.dsr.2017.04.014. [DOI] [Google Scholar]
- 41.Delmont TO, Hammar KM, Ducklow HW, Yager PL, Post AF. 2014. Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front Microbiol 5:646. doi: 10.3389/fmicb.2014.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sargent EC, Hitchcock A, Johansson SA, Langlois R, Moore CM, LaRoche J, Poulton AJ, Bibby TS. 2016. Evidence for polyploidy in the globally important diazotroph Trichodesmium. FEMS Microbiol Lett 363:fnw244. doi: 10.1093/femsle/fnw244. [DOI] [PubMed] [Google Scholar]
- 43.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. 2015. rrn DB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 45.Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. 2013. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol 15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- 46.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jennrich RI. 1970. An asymptotic χ2 test for the equality of two correlation matrices. J Am Stat Assoc 65:904–912. [Google Scholar]
- 48.Steiger JH. 1980. Tests for comparing elements of a correlation matrix. Psychol Bull 87:245. doi: 10.1037//0033-2909.87.2.245. [DOI] [Google Scholar]
- 49.Needham DM, Sachdeva R, Fuhrman JA. 2017. Ecological dynamics and co-occurrence among marine phytoplankton, bacteria and myoviruses shows microdiversity matters. ISME J 11:1614–1629. doi: 10.1038/ismej.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arrigo KR, Mills MM, Kropuenske LR, van Dijken GL, Alderkamp A-C, Robinson DH. 2010. Photophysiology in two major Southern Ocean phytoplankton taxa: photosynthesis and growth of Phaeocystis antarctica and Fragilariopsis cylindrus under different irradiance levels. Integr Comp Biol 50:950–966. doi: 10.1093/icb/icq021. [DOI] [PubMed] [Google Scholar]
- 51.Moisan TA, Ellisman MH, Buitenhuys CW, Sosinsky GE. 2006. Differences in chloroplast ultrastructure of Phaeocystis antarctica in low and high light. Mar Biol 149:1281–1290. doi: 10.1007/s00227-006-0321-5. [DOI] [Google Scholar]
- 52.Montes-Hugo M, Doney SC, Ducklow HW, Fraser W, Martinson D, Stammerjohn SE, Schofield O. 2009. Recent changes in phytoplankton communities associated with rapid regional climate change along the western Antarctic Peninsula. Science 323:1470–1473. doi: 10.1126/science.1164533. [DOI] [PubMed] [Google Scholar]
- 53.Obryk MK, Doran PT, Friedlaender AS, Gooseff MN, Li W, Morgan-Kiss RM, Priscu JC, Schofield O, Stammerjohn SE, Steinberg DK, Ducklow HW. 2016. Responses of Antarctic marine and freshwater ecosystems to changing ice conditions. Bioscience 66:864–879. doi: 10.1093/biosci/biw109. [DOI] [Google Scholar]
- 54.Louca S, Doebeli M, Parfrey LW. 2018. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6:41. doi: 10.1186/s40168-018-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godhe A, Asplund ME, Härnström K, Saravanan V, Tyagi A, Karunasagar I. 2008. Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl Environ Microbiol 74:7174–7182. doi: 10.1128/AEM.01298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. 2014. CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. doi: 10.1186/2049-2618-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. 2015. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci U S A 112:2485–2490. doi: 10.1073/pnas.1416878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reysenbach A-L, Giver LJ, Wickham GS, Pace NR. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol 58:3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oshima T, Imahori K. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Evol Microbiol 24:102–112. doi: 10.1099/00207713-24-1-102. [DOI] [Google Scholar]
- 62.Thompson JR, Marcelino LA, Polz MF. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res 30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wear EK, Wilbanks EG, Nelson CE, Carlson CA. 2018. Primer selection impacts specific population abundances but not community dynamics in a monthly time-series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environ Microbiol 20:2709–2726. doi: 10.1111/1462-2920.14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satinsky BM, Crump BC, Smith CB, Sharma S, Zielinski BL, Doherty M, Meng J, Sun S, Medeiros PM, Paul JH, Coles VJ, Yager PL, Moran MA. 2014. Microspatial gene expression patterns in the Amazon River Plume. Proc Natl Acad Sci U S A 111:11085–11090. doi: 10.1073/pnas.1402782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 66.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 68.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 69.Bystrykh LV. 2012. Generalized DNA barcode design based on Hamming codes. PLoS One 7:e36852. doi: 10.1371/journal.pone.0036852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:67. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC, Flyvbjerg H. 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 73.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 74.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.