The gut microbiota affects many important host functions, including the immune response and the nervous system1. However, while substantial progress has been made in growing diverse microorganisms of the microbiota2, 23–65% of species residing in the human gut remain uncultured3,4, which is an obstacle for understanding their biological roles. A likely reason for this unculturability is the absence of key growth factors in artificial media, which are provided by neighboring bacteria in situ5,6. In the present study, we used co-culture to isolate KLE1738, which required the presence of Bacteroides fragilis to grow. Bio-assay driven purification of B. fragilis supernatant led to the isolation of the growth factor, which, surprisingly, is the major inhibitory neurotransmitter γ-aminobutyric acid (GABA). GABA was the only tested nutrient that supported growth of KLE1738, and genome analysis supported a GABA-dependent metabolism. Using growth of KLE1738 as an indicator, we isolated a variety of GABA-producing bacteria, and found that Bacteroides ssp. produced large quantities of GABA. Genome based metabolic modeling of the human gut microbiota revealed multiple genera with the predicted capability to produce or consume GABA. Transcriptome analysis of human stool from healthy individuals showed that GABA producing pathways are actively expressed by Bacteroides, Parabacteroides, and Escherichia species. By coupling 16S rRNA sequencing with fMRI imaging in patients with Major Depressive Disorder (MDD), a disease associated with an altered GABAergic response, we found that relative abundance levels of fecal Bacteroides are negatively correlated with brain signatures associated with depression.
We previously identified quinones as growth factors for uncultured bacteria of the human microbiome6. In this study, we searched for previously undescribed growth factors. On a densely inoculated plate, some uncultured bacteria may be growing because they are in proximity to cultivable organisms producing growth factors. Using this rationale, a fecal sample from a healthy human donor was plated on Fastidious Anaerobic Agar supplemented with yeast extract (FAAy), and newly formed colonies were noted daily for a week. Late forming colonies (appearing after 4–7 days – “candidate dependent”) were diluted and spread on FAAy, and a heavy inoculum of a neighboring, early forming colony (appearing after1–3 days – “candidate helper”) was spotted (Fig. 1A). Given that E. coli induced the growth of all quinone-dependent organisms in our previous study6, candidate dependents were counter-screened against E. coli (Fig. 1B).
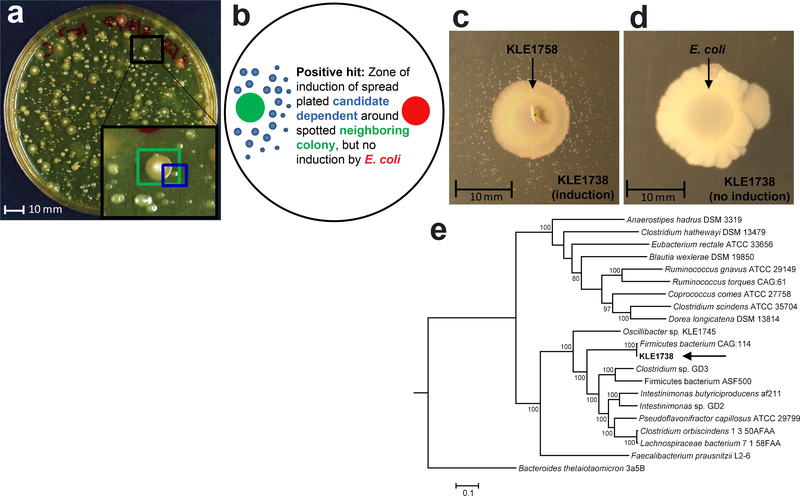
Fig. 1. Co-culture assay to isolate KLE1738.
(A) Diluted human fecal sample was plated on Fastidious Anaerobe Agar with 0.5% yeast extract (FAAy), and slower growing, smaller colonies (formed after 4–7 days -- “candidate dependent”, example in blue box) were tested for dependence on their faster growing, larger neighboring colonies (formed after 1–3 days -- “candidate helper”, example in green box) by co-culture. (B) To identify growth factors, the candidate dependent was also tested for growth promotion with Escherichia coli BW25113. Using this method we isolated KLE1738, which grew around the helper (C) Bacteroides fragilis KLE1758, but not Escherichia coli (D). Experiments describing dependency phenotypes were repeated in triplicate. (E) Phylogenetic tree of closely related type and representative genomes belonging to the Ruminococcaceae family. Tree assembled using Randomized Axelerated Maximum Likelihood in PATRIC. Parts of Figure modified from Fenn, 20176; the culture plate and colonies highlighted in (A) is used for an illustration, and is not the source plate for KLE1738. A single stool sample yielded the KLE1738-KLE1758 helper-dependent pair.
After screening approximately 200 colonies for the desired dependency phenotype, we identified a single isolate that failed to grow in the presence of E. coli, but grew on a plate with B. fragilis. This isolate, KLE1738, required the presence of Bacteroides fragilis KLE1758 (100% similar by 16S rRNA sequence to Bacteroides fragilis ATCC 25852), for growth (Fig. 1C). KLE1738 is a Gram-positive bacterium of the Ruminococcaceae family, and is 93.4% similar to Flavonifractor plautii VPI 0310 [S1]7 as well as Intestinimonas butyriciproducens SRB-521–5-I(T)8 by 16S rRNA gene sequence, with its closest relative by genome sequence similarity being the uncultured Firmicutes bacterium CAG:114 (Fig. 1D). Its dissimilarity by 16S rRNA gene sequence to existing type strains suggests KLE1738 is the first representative of an unreported genus of bacteria, using recent guidelines for assigning taxonomy9. KLE1738 is found on the NIH Most Wanted list10, indicating it has not been cultured, although it is fairly prevalent in the human gut microbiome, being detectable in 6,001 of 31,451 (19.08%) of human gut metagenomes available in the Integrated Microbial Next Generation Sequencing database (16S similarity threshold >99%, with a minimum sequence length of 200bp)11. However, KLE1738 appears to be a relatively minor constituent of the microbiome, detectable at a relative abundance >1% in only 60/31,451 human gut metagenomes (0.19%).
The supernatant of a 48-hour culture of B. fragilis KLE1758 grown in rich medium induced growth of KLE1738 (Supplemental Information Fig. 1A, control in 1B), enabling bioassay-driven purification of the growth factor. The supernatant was solvent-partitioned with ethyl acetate, and the aqueous fraction induced growth of KLE1738. The aqueous fraction was then separated on an HP-20 column, and the most polar fraction induced growth of KLE1738 (Supplemental Information Fig. 1C, non-inducing fraction in 1D). This active fraction was then further fractionated by preparative HPLC, yielding a single active fraction. NMR analysis revealed that it contained 10 compounds including 6 common amino acids, 3 carboxylic acids and gamma-aminobutyric acid (GABA) (Supplemental Information Fig. 2). While all compounds identified in the active fraction were tested, only GABA induced growth of KLE1738 (Supplemental Information Fig. 1E, medium without GABA in 1F).
The growth requirement of GABA a was surprising, since this suggested that KLE1738 was unable to grow on glucose or the numerous amino acids and other nutrients present in the rich medium. Precursors and breakdown products of GABA, and over 100 common carbon or nitrogen sources (including all standard amino acids) were tested for their ability to induce growth of KLE1738, but none were active (Supplemental Information Table 1). In addition to the rich FAAy medium, multiple commercially available and published growth media were tested -- including those shown to enable recovery of diverse gut bacteria, YCFAg2 and GMM12 -- but none were found to promote the growth of KLE1738 in the absence of exogenous GABA. Systematically excluding components of FAAy medium showed that KLE1738 grows on solid agar with only peptone and GABA (Supplemental Information Table 2). These results indicate that GABA is a required nutrient for KLE1738 in the tested experimental conditions.
To gain insight into the unusual growth requirements of KLE1738, the genome was sequenced and annotated. The annotated draft genome of KLE1738 revealed an unusual profile – there were no apparent entry points into central metabolism for common sugars, amino acids, or other carbon sources. Bacteria use phosphotransferases to uptake a variety of sugars, such as glucose, fructose, and mannose. Uptake is coupled to phosphorylation by the membrane transporter Enzyme II, the last component of a phosphorylation pathway: phosphoenolpyruvate – Enzyme I – Hpr – Enzyme II13. KLE1738 appears to lack Enzyme I. The fact that only one component of the phosphorylation pathway is missing suggests a recent loss of function. Similarly, KLE1738 is predicted to have a limited set of ABC transporters (Supplemental Information Table 3). This agrees with the inability of KLE1738 to grow on the tested nutrients. The utilization of GABA for carbon and nitrogen by bacteria has been reported before, although not as a required nutrient. The GABA shunt serves as a pathway for its conversion into succinate, which then enters into the TCA cycle14. However, KLE1738 appears to lack succinate semialdehyde dehydrogenase, an essential enzyme of this pathway. An alternative ATP-generating pathway for GABA consumption was described for the environmental anaerobe Clostridium aminobutyricum15. KLE1738 has homologs of all enzymes of this pathway (Supplemental Information Fig. 3), with some enzymes present in multiple copies (Supplemental Information Table 4). Feeding KLE1738 with 13C, 15N labeled GABA allowed us to detect its metabolites, butyrate and acetate, as well as hexanoic acid, an intermediate of fatty-acid biosynthesis (Supplemental Information Fig. 4 and Fig. 5A-D, 4E-H). No incorporation of the 15N label was found. These results suggest that GABA is a carbon and energy source for KLE1738.
For some species, including E. coli, Lactobacillus ssp., and Bifidobacterium ssp., GABA secretion has been reported to serve as an acid-resistance mechanism. Decarboxylation of glutamate is induced at a low pH and produces GABA, which is then exported from the cell in a protonated form, alkalinizing the cytoplasm14. While E. coli does not induce growth of KLE1738, a strain overexpressing glutamate decarboxylase in E. coli K12 does, comparably to B. fragilis KLE1758 (Supplemental Information Fig. 6A-D).
Bacteroides fragilis, the helper of KLE1738, is a common gut bacterium, but we found that similarly to E. coli, GABA production by Bacteroides fragilis is only observed at a low pH (≤5.5) (Supplemental Information Fig. 7). Apparently, Bacteroides fragilis growing on a Petri dish acidifies the medium enough to induce GABA production. We therefore sought to find gut microorganisms capable of producing GABA at a physiologically relevant pH for the human large intestine (pH 5.7 – 7.4)16.
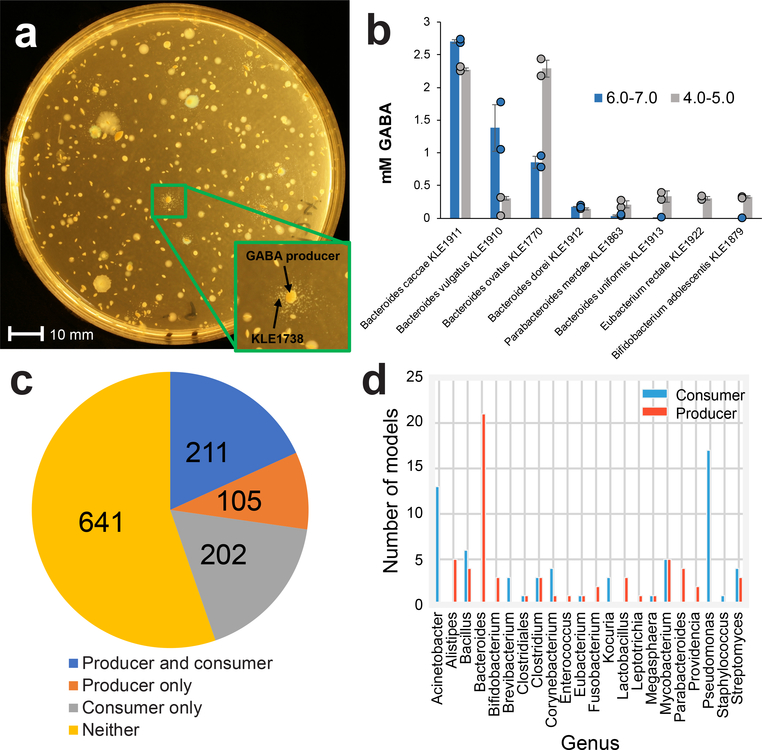
The unique GABA-dependence of KLE1738 was exploited to identify additional GABA producers in a co-culture assay. Stool samples were mixed with molten FAAy agar, allowed to solidify, and KLE1738 was then spread on top of the medium. By looking for zones of KLE1738 growth, we were able to identify gut bacteria that produce GABA (Fig. 2A, B). In addition to Bacteroides species, representatives from the Parabacteroides, Eubacterium, and Bifidobacterium genera were identified as GABA producers in this assay (Fig. 2B). Of those, Bacteroides ssp. (and to a lesser extent Parabacteroides sp.) were found to produce GABA within the pH range of the human large intestines (Fig. 2B).
Fig. 2. In vitro and in silico identification of GABA modulating bacteria.
(A) To screen for GABA-producing bacteria, homogenized human stool sample was diluted and mixed with molten FAAy, with or without pH 7.0 MOPS buffer. KLE1738 was then spread on top of the agar and plates were incubated anaerobically for a week. Colonies that KLE1738 grew around were GABA producers (Inset). (B) Identified GABA producers were grown in liquid medium buffered at a pH of 5.0 and 7.0, and GABA levels of the spent medium was quantified using LC/MS and final pH of the medium was tested with pH strips. N=2 independent samples, and error is based on standard error. (C) 1,159 available gut genomes (consisting of 919 species) were analyzed for the genetic potential to produce and/or consume GABA (pathways associated with production or consumption highlighted in Supplemental Information Table 5). (D). The biochemical potential of 533 available gut-related metabolic models in KBase were examined for the capability to produce GABA or consume GABA. Shown are genera that represent at least 0.5% of the 533 models and contain either GABA producers or consumers.
We next sought to profile the GABA modulating potential of the human gut microbiome in silico. All available bacterial genes encoding for GABA consumption and production were obtained from RAST17 and/or UniProt18. These sequences were then used as input for a bidirectional best hit analysis against genomes of known members of the gut microbiota, as evident by their presence in the fecal 16S rRNA data set of the American Gut Project – a crowd sourced microbiome sequencing project with over 10,000 participants19. Genomes were scored as “producer” if they had at least one complete metabolic route for GABA production or a “consumer” if they had the GABA shunt or GABA transaminase and at least 7/11 of the remaining enzymes of the KLE1738 pathway (the presence of 7/11 enzymes was chosen due to poor annotation of the remaining enzymes). Of 1,159 genomes analyzed (consisting of 919 species), we identified 105, 205, and 211 species harboring the genetic potential to only produce, only consume, or both produce and consume GABA, respectively (Fig. 2C; more detail in Supplemental Information Table 5). Due to its dissimilarly to type strains and unique growth requirements, we propose the name “Evtepia gabavorous” for KLE1738. It will be described in a future manuscript, using traditional description methods for bacteria20.
To complement this approach, we applied a more rigorous in silico constraint-based modeling method to survey the potential GABA consumption and/or production of the gut microbiota. This was performed using KBase, a database that contains metabolic models computationally derived from genome sequences of many microorganisms21. Of the 919 species used in the above bi-directional analysis, 533 had models represented in KBase, and these were examined for their ability to produce GABA from known precursors – glutamate, arginine, putrescine, and ornithine – or consume GABA via the KLE1738 pathway or the GABA shunt. The analysis predicted that 97 organisms had the capability to produce GABA, mostly via glutamate decarboxylase (Supplemental Information Fig. 7). Of these, >25% belong either to the genera Bacteroides or Parabacteroides (Fig. 2D). For consumption, we identified 102 potential consumers of this neurotransmitter, with the majority belonging to the Pseudomonas, Acinetobacter, and Mycobacterium genera (Fig. 2D). The number of GABA consumers is likely an underestimate, since the KLE1738 GABA consumption is poorly annotated and thus not captured in KBase.
To validate activity of the genes of interest in humans, we surveyed an existing human transcriptome stool dataset22 for active expression of transcripts associated with the bottlenecks of bacterial GABA metabolism (glutamate decarboxylase, gamma-aminobutyrate:alpha-ketoglutarate aminotransferase and succinate semialdehyde dehydrogenase). The RNAseq data was assembled using the Trinity platform23, and retrieved coding sequences were subjected to BLASTN (90% cutoff at 1e−5). No transcripts were identified for enzymes involved in GABA consumption (perhaps attributable to the depth of sequencing), but multiple hits were found for glutamate decarboxylase (Supplemental Information Table 6). These transcripts were successfully mapped to the nearest type strains of some of our GABA producing panel, suggesting Bacteroides and Parabacteroides species produce GABA in humans (Supplemental Information Table 7). Surprisingly, a set of transcripts also mapped to Escherichia coli (Supplemental Information Table 6). Given the pH restrictions for GABA production by E. coli in vitro14, this may indicate the presence of acidic microenvironments in the large intestine.
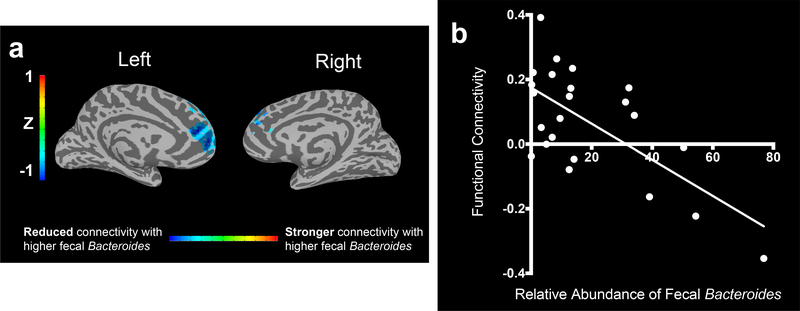
In germ-free mice, GABA levels have been reported to be significantly decreased both in the stool and the blood24 (but an older report showed no difference in GABA blood levels in germ-free and specific pathogen free rats25). Similarly, in specific pathogen free mice, fecal GABA levels can be modifiable by antibiotics26, suggesting that the microbiota may contribute to circulating levels of GABA. This is of interest, because multiple diseases are associated with an altered GABAergic profile, such as depression27. Accordingly, we sought to explore whether Bacteroides, perhaps the major bacterial producers of GABA in the human gut, were associated with clinically diagnosed Major Depressive Disorder (MDD). To do this, stool from 23 patients with MDD was collected and analyzed by 16S rRNA sequencing using the American Gut protocols. Resting state fMRI was acquired within 3 days of stool sample collection and scans were co-registered to MNI-space. To investigate the relationship between neural circuitry important in depression and fecal Bacteroides abundance we focused, a priori, on the left dorsolateral prefrontal cortex (DLPFC) and default mode network (DMN). Our choice of the left DLPFC was guided by the highly replicated finding that the left DLPFC is hypoactive in depression28. Similarly, the DMN is involved in self-referential processing29 and negative rumination in depression30. Functional connectivity is elevated both within the DMN and with other networks in depression31 and normalizes with treatment response32. We found an expansive region of the default mode network spanning the left anterior medial frontal cortex in which functional connectivity with the left DLPFC was inversely correlated with the relative abundance of fecal Bacteroides (Fig. 3A,B). This region of significance overlapped extensively with both the left medial prefrontal and left frontopolar cortex, regions highly interconnected to the limbic system and thought to be important in emotional reappraisal and reward processing33,34. This cluster was unique, and we found no clusters in which functional connectivity was positively correlated with the relative abundance of Bacteroides. We found no associations with KLE1738, perhaps due to its low abundance. Interestingly, a recent study of 40 healthy women found that levels of Bacteroides were associated with increased gray matter in the cerebellum, hippocampus, and frontal regions of the brain, and exhibited reduced levels of anxiety, distress, and irritability after looking at photos to evoke an emotional response35. Furthermore, a high fat diet has been shown to reduce GABA levels in the rat prefrontal cortex by about 40%, which was associated with reduced levels Bacteroides and depressive-like behavior36. Nonetheless, our pilot cohort has limitations – the sample size was small and consequently we did not correct for medications (listed in Supplemental Information Table 8), and the sequencing data was not quantitative. As such, this cohort should be expanded, to further profile whether Bacteroides, or the GABA they may produce, are involved in affecting behaviors.
Fig. 3. Fecal Bacteroides relative abundance inversely correlates with functional connectivity between left DLPFC and DMN structures in patients with Major Depressive Disorder (MDD).
(A) 3-Dimensional plots of the medial surface of the left and right hemispheres in patients with MDD (n=23). Significant cluster of 387 voxels in which fecal Bacteroides relative abundance correlated inversely with functional connectivity between this cluster and the left DLPFC. Colorbar shows Z scores of beta weights of the Bacteroides relative abundance covariate of a multiple linear regression with functional connectivity as the dependent variable. (B) Scatter plot of the average functional connectivity (Z score) over a sphere of radius 5mm centered at the voxel of peak significance (+12, −57, 0 in MNI coordinates) and abundance of fecal Bacteroides (Pearson r = −0.67, p=.0005). n=23.
Notably, it has been reported that treatment of mice with the probiotic bacterium Lactobacillus rhamnosus JB-1 reduces stress and depression-like behavior in a vagus nerve-dependent manner37. This was accompanied by changes in the expression of GABA receptors in several areas of the brain, including the amygdala and hippocampus37, and a later study showed elevated brain GABA in mice post-supplementation with JB-1 after four weeks38. While JB-1 was not explicitly tested for its ability to produce GABA, other L. rhamnosus strains have been shown to produce GABA around a pH of 3.6–5.239. Some Bifidobacterium have been shown to produce GABA40, and introduction of a GABA producing Bifidobacterium strain resulted in an improved visceral sensitivity in a rat model of pain41. Importantly, a recent human study found that fecal microbiome transplant from lean to obese individuals resulted in increased levels of GABA in the plasma42, showing manipulating the microbiome may alter GABA levels. Our findings, combined with these reports, suggest microbial-derived GABA may influence the host, and are the first step in understanding the biology of this intriguing connection.
Methods
Human stool collection
For cultivation experiments, stool samples from an adult healthy human donor were collected using a stool collection vessel (Medline Industries). Within five minutes of collection, 1 gram of stool was resuspended in 9 mL of sterile 20% glycerol in PBS and homogenized for 30 seconds using a vortex. 1 mL aliquots of this mixture were loaded in cryotubes and stored at −80 °C. Stool samples for cultivation experiments were collected with informed consent following an IRB approved protocol at Northeastern University (IRB# 08–11-16).
For the Major Depressive Disorder cohort, study participants were provided with sterile plastic 4 oz specimen collection cups at their first visit. They were instructed to collect stool the day of or the night before their second visit depending on their ability to produce a sample to make sure no more than 24 hours pass between stool sample collection and processing. Study participants were instructed to keep their stool samples at room temperature until they bring it to their second visit. Once a sample was received by study personnel, it was processed within an hour. American Gut sample kits were shipped at room temperature the day of sample processing (the standard shipping protocol used for the American Gut). Two 1.5 ml screw top plastic tubes per sample were filled with stool and immediately frozen at −80C for future studies.
Cultivation of helper-uncultured pairs from human stool samples
All cultivation was performed in a Coy Anaerobic Vinyl chamber with an atmosphere of 5% hydrogen, 10% CO2, 85% nitrogen. Stool samples were thawed and serially diluted in PBS under anaerobic conditions and bead-spread (7–10 beads/plate) on 1X Fastidious Anaerobic Agar (Accumedia) plates with 2.5% yeast extract (FAAy). Plates were incubated at 37 °C anaerobically for one week, and each day appearance of colonies was tracked by spotting the outside of the plates with different color markers. At the end of the week, serial dilutions of late forming colonies (appearance after 4–7 days) were prepared in PBS and bead spread on FAAy plates. Nearby (< 2 cm), early forming colonies (appearance after 1–3 days) were then resuspended in PBS at a high density. Five μL of this suspension was spotted on plates with their respective spread-plated candidate dependent, incubated for up to one week in the chamber, and observed daily. Growth induction of the dependent organism around the spotted helper indicated a positive hit. Strain nomenclature – KLE = Kim Lewis; # = strain number)
Taxonomic assignment
PCR was performed for candidate dependents, helpers, and other isolates using the general bacterial primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) to amplify the 16S rRNA gene. The PCR reaction mixture was 12.5 μL GoTaq Master Mix (Promega), 1 μL 10 μM 27F and 1492R primers (Operon), 9.5 μL Nuclease Free Water (Promega), and 1 μL of a colony resuspended in 100 μL sterilized distilled water. The amplification conditions were one cycle of 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s; and finally one cycle of 72 °C for 7 min. Amplification of PCR reactions were confirmed using gel electrophoresis on a 0.8% agarose gel containing ethidium bromide. Successful PCRs were sequenced by Macrogen Corporation using the 27F and 1492R primers using the Applied Biosystems 3730×l DNA analyzer. Quality control for sequences was performed using DNA Baser (www.DnaBaser.com, version 4.36.0), in which ends were trimmed until there were more than 75% good bases (defined by having a QV score of higher than 25) in an 18 base window. Identification of phylogenetic neighbors and calculation of pairwise sequence similarity were done using the EZTaxon server (http://www.eztaxon.org). The phylogenic tree for KLE1738 was built using the Randomized Axelerated Maximum Likelihood (RAxML) method43 via PATRIC44 (version 3.5.23).
Identification of GABA as a growth factor for KLE1738
A single colony of Bacteroides fragilis KLE1758 was inoculated into Brain Heart Infusion Broth (Becton Dickinson) with 0.1% cysteine-HCL, 5.0 mg/mL yeast extract, and 15 mg/L hemin (BHIych) and incubated in the anaerobic chamber at 37 °C. The 48-hour B. fragilis KLE1758 culture was then filter sterilized using a 0.22 μm syringe filter unit and 200 μL of the supernatant was loaded in a Millicell Single Well Hanging Insert (pore size of 0.4 μm) and placed on top of a BHIych agar plate with bead-spread (7–10 beads/plate) KLE1738. Induction of KLE1738 growth was observed after 48 hours. For all induction experiments, cells of KLE1738 were taken from 48-hour cultures on solid BHIych plates with 1 mg/mL GABA, or from plates spotted with B. fragilis.
The supernatant of B. fragilis KLE1758 (1 L) was solvent-partitioned with ethyl acetate (3 × 500 mL, each 3 hours) to yield ethyl acetate-soluble fraction and water residue, respectively. All extractions and fractionations of the supernatant were tested for KLE1738 induction by loading 200 μL into a Millicell Single Well Hanging Insert, as described above. The water residue part induced growth of KLE1738, which allowed the highly polar water fraction to be applied to a HP-20 column for further fractionation, yielding six fractions [A-F, A: water eluted fraction (2L); B: 20% MeOH eluted fraction (1L); C: 40% MeOH eluted fraction (1L); D: 60% MeOH eluted fraction (1L); E: 80% MeOH eluted fraction (1L); and F: 100% MeOH eluted fraction (2L)]. The most polar fraction (A) turned out to be the active fraction for inducing growth of KLE1738, and was consequently separated by HPLC using an Agilent 1100 series HPLC system (Agilent Technologies) equipped with a photo diode array detector. The active fraction A (3.8 g) was further fractionated by a preparative HPLC (phenyl-hexyl column, Phenomenex Luna, 250 × 21.2 mm, 5 μm) with a flow rate of 10 mL/min using an isocratic solvent system of 1% aqueous acetonitrile for 30 min, then to 100% acetonitrile in the next 6 min, and 100% acetonitrile for the next 7 min to give 14 fractions [A1 – A14, A1 (3–5 min); A2 (5–8 min); A3 (8–10 min); A4 (10–13 min); A5 (13–16 min); A6 (16–19 min); A7 (19–22 min); A8 (22–25 min); A9 (25–29 min); A10 (29–33 min); A11 (33–36 min); A12 (36–38 min); A13 (38–40 min); and A14 (40–43 min)] according to HPLC chromatography analysis. Among these fractions, the fraction A2 induced growth of KLE1738. The active fraction was directly applied to NMR analysis including 1H, 13C, 1H-1H COSY, TOCSY, HSQC, and HMBC experiments to identify its constituents in the fraction. All NMR experiments were carried out on a Varian INOVA 600 MHz NMR spectrometer equipped with an indirect detection probe.
Testing other compounds for induction of KLE1738
Multiple compounds were tested for the ability to induce the growth of KLE1738. Stocks of each compound (purchased from Sigma, excluding the ATCC Mineral and Vitamin mixes purchased from ATCC) were prepared dependent on solubility in water (compounds and tested concentrations are found in Supplemental Information Table 1). 5 μL of the stocks were then spotted on FAAy plates spread with KLE1738, and incubated in the anaerobic chamber at 37 °C for a week. Biolog plates were tested per manufacturer’s instructions (Biolog).
Media exclusion experiments
To facilitate media exclusion experiments, we prepared batches of Fastidious Anaerobic Agar (FAA) using the recipe from Accumedia (Lansing, MI): 23 g/L Bacto proteose peptone no. 3 (BD), 5 g/L sodium chloride (Sigma), 1g/L Difco soluble starch (BD), 0.4 g/L sodium bicarbonate (Sigma), 1 g/L glucose (Sigma), 1 g/L sodium pyruvate (Sigma), 0.5 g/L L-cysteine HCl-H2O (Sigma), 0.25 g/L sodium pyrophosphate (Sigma), 1 g/L L-arginine (Sigma), 0.5 g/L sodium succinate (Sigma), 0.01 g/L hemin (Sigma), 0.001 g/L menadione (Sigma), 12 g/L agar (Sigma), and 1.0 mg/mL GABA (Sigma). We then prepared batches of this “Homemade” Fastidious Anaerobic Agar (HFAA) with individual or several components removed to test the nutritional requirements of KLE1738. All induction experiments were performed using 48-hour cultures of KLE1738 grown on FAAy agar with 1.0 mg/mL GABA, and incubated anaerobically as described above.
Whole genome sequencing and annotation
DNA from cells of KLE1738 grown 48 hours anaerobically on FAAy plates with 1.0 mg/mL GABA was isolated for genome sequencing using the PowerSoil® DNA Isolation Kit (Mo Bio, San Diego, CA) according to manufacturer specifications, yielding ~5.0 μg of high quality DNA. Genomic sequencing and de novo assembly was performed by the Genomic Core at Tufts University in Boston, MA. The genome of KLE1738 was sequenced on an Illumina MiSeq using MiSeq V2 500 cycles chemistry with a paired-end 250 bases format. Briefly, 100 ng of genomic DNA was sheared on a Covaris M220 to an average fragment size of around 600 bases. Using the fragmented DNA as input, the sequencing library was prepared with Illumina TruSeq Nano DNA Sample Preparation Kit per the manufacturer instruction. Base calling and demultiplexing was performed on the raw data from the MiSeq using CASAVA and fastq files were generated. De novo assembly of the genome was performed using Edena V3.131028 with a customized parameter optimization pipeline. The best assembled genome, as assessed by the contig statistic, was reported. Assembly yielded 68 contigs (n), with all contigs having a sequence length longer than 200 bases (n:200). There are 7 contigs with a larger value than the N50 (119748), and the minimal contig length is 355 (min). The N20, N50 and N80 are 33403, 119748 and 204670, respectively. The largest contig length (max) is 344080, and the estimated genome size is 2500009. The draft genome was annotated using the RAST server and the KAAS (KEGG Automatic Annotation Server) analysis tool of the KEGG (Kyoto Encyclopedia of Genes and Genomes) database45, version 2.0.
Preparation of U-13C, 15N-GABA and 13C feeding experiments
Recombinant glutamate decarboxylase (GAD) was purified from an Escherichia coli strain harboring a his-tagged gadB from the ASKA library using the suggested protocol46. The His-tagged GadB enzyme was eluted off of a Ni-NTA column, and was further purified by dialysis in PBS at 4 °C for 5h, with a 2–10K MWCO filter. GadB was quantitated by UV (μ~85000 M−1cm−1) and diluted to give a concentration of 1.7 μg/ μL. The following protocol was followed to convert U-13C, 15N-Glu (Cambridge Isotope Laboratories) to U-13C, 15N-GABA. Stocks of pyridoxal-5’-phosphate (PLP, 10 mM, in water) and dithiothreitol (DTT, 1 mM, in water) were prepared fresh each experiment. In an Eppendorf tube, U-13C,15N-Glu (9.4 mg, 0.06 mmol 60 mM final) is dissolved in 0.2 M NaOAc buffer (750 μL, pH 4.6). To this is added 10 mM PLP (100 μL, 1 mM final), 1 mM DTT (100 μL, 0.1 mM final), and 1.7 μg/ μL GAD (50 μL, 85 μg final). This is incubated in a 37 °C water bath for 18 h. The mixture is passed through a Ni-NTA column to remove protein, adjusted to pH 1 with 2 M HCl and lyophilized. This was repeated 4 times (~ 40 mg scale) to produce enough U-13C, 15N-GABA for analysis.
13C, 15N-GABA was fed to KLE1738 at a final concentration of 100 μg/mL on FAAy plates. After 72 hours of incubation in the anaerobic chamber, cells we resuspended in PBS, washed once, and pelleted. Agar was also collected. 13C metabolites were detected by MS (notably no 15N metabolites were identified). Bacteria were pelleted through centrifugation, the supernatant was discarded, and 2 M HCl (100 μL) was added. The sample was subsequently vortexed, H2O (Mill-Q, 500 μl) was added, the sample was vortexed again, and then was allowed to stand at room temperature for 10 min. Afterwards the sample was extracted with Et2O (3 × 2 mL), and the consolidated organic phase was dried with Na2SO4. The organic phase was then evaporated to dryness and the resulting material was resuspended in Et2O (500 μL). For extraction of the agar on which the bacteria was grown, the agar was sliced into small squares and 2 M HCl (10 mL) was added. The sample was vortexed, H2O (Mill-Q, 50 mL) was then added, and the sample was vortexed again. The mixture was subsequently allowed to stand at room temperature for 10 min, and then was extracted with Et2O (3 × 50 mL). The consolidated organic phase was dried with Na2SO4, evaporated to dryness, and the resulting material was resuspended in Et2O (500 μL). The resuspended material was spun-down to pellet insoluble particulate, and the supernatant was passed through a 0.2 micron syringe filter to afford samples suitable for GC/MS analysis. All samples were analyzed on an Agilent 6890 GC with a Waters Quattro micro GC/MS/MS triple quadrupole mass spectrometer using electron ionization (EI) with a sample injection volume of 1 μL. A fused-silica capillary column of cross-linked DB-624UI (30 m x 0.32 mm ID, 1.80 μm film thickness, Agilent) was used. The GC conditions were as follows: inlet and transfer line temperate, 240 °C and 220 °C respectively; over temperature program, held at 50 °C for 7 min, then increased to 250 °C at 50 °C/min, lastly held for 5 min; inlet helium carrier gas flow rate, 2.3 mL/min; split ratio: 30:1. The electron impact (EI)-MS conditions were as follows: ion source temperature, 200 °C; full scan m/z range 10–640 Da; selected ion recording (SIR) mode, m/z of 60 for monitoring of acetic and butyric acids. Data were acquired and analyzed with Waters MassLynx V4.1 SCN805 software package and the NIST spectral library was used in searches to determine compound identities.
Quantification of GABA production
To measure the impact of pH on GABA production of KLE1758, triplicate cultures of B. fragilis KLE1758 were grown in 3 mL BHIych (pH 5.0, 5.5, 6.0 or 6.5) anaerobically for 48 hours, the cells centrifuged, and the supernatant was filtered through a 0.2 μm filter. Samples were stored at 4°C until analysis (<48 hours). To analyze the samples, an aliquot (2 μL) of each sample was added to AccQ reaction buffer (16 μL), CSA internal standard (2 μL of a 50 μg/mL solution in buffer), followed by the addition of the AccQ reagent (20 μL). These samples were heated to 55 °C for ten minutes, and then transferred directly into an LC/MS vial fitted with a glass insert. An aliquot of each sample (10 μL) was injected onto the LC/MS, and separated following the same injection program as used for the calibration curve. The total EIC area under curves representing GABA, Glu and CSA was determined using ChemStation software (Agilent). Each injection represented 25% of the original media concentration, therefore the total amount of sample determined (in ng) was multiplied by a factor of four to determine the original concentration (in ng/μL = μg/mL). All areas were normalized to the area under the curve of the internal standard (CSA), which was held at constant concentration throughout the experiment.
For quantification of GABA by strains identified in the co-culture screen with KLE1738, the above procedure was employed where one culture per strain was used instead, samples were stored at 4°C until analysis, and each prepared sample from a bacterial culture was run on the LC/MS twice. pH was measured using pH strips.
Co-culture screen for GABA producers using KLE1738
Molten FAAy was loaded into 50 mL centrifuge tubes and cycled into the anaerobic chamber. Diluted stool sample (from frozen stock) was mixed with the molten agar to reach a final dilution of 10−6 – 10−9 and 15 mL was plated in triplicate for each dilution. Once solidified, KLE1738 was resuspended in pre-reduced PBS (stored anaerobically for >24 hours) and bead spread on top of the agar. Inoculated plates were incubated for one week, and growth of KLE1738 around colonies embedded in the agar indicated candidate GABA producing colonies. These candidate GABA producers were then restreaked on fresh FAAy and identified by 16S rRNA gene sequencing. All strains were confirmed to produce GABA on their respective mediums by co-culture with KLE1738 in isolation.
Co-culture of KLE1738 with E. coli overexpressing glutamate decarboxylase.
24 hour cultures of E. coli clones harboring native glutamate decarboxylases (gadA, gadB) or the GABA antiporter (gadC) in the pCA24N IPTG inducible high-copy number vector, were tested for GABA production via co-cultivation assay with KLE1738 on FAAy (described in Fig 1) with the addition of 1 mM IPTG. An E. coli clone harboring the empty vector and a 48 hour culture of B. fragilis KLE1758 growth in BHIYch served as a control. E. coli strains were taken from the ASKA library46.
Metagenomic analysis of potential GABA producing or consuming bacteria
To identify potential GABA-producing and -consuming bacteria, a bidirectional best hit (BBH) nucleotide BLAST was performed. Operon nucleotide fasta sequences were retrieved from KBase (ftp://ftp.kbase.us/assets/KBase_Reference_Data/blast/fasta/kbase.ffn) and filtered for members of the gut microbiota by comparing 16S rRNA sequences that shared >99% sequence similarity to the healthy fecal set of organisms from the American Gut Project19. In total, sequences for 913 different gut bacteria were found in the KBase operon file (some species had multiple strain representatives, bringing the total analyzed genomes to 1,159). To identify potential GABA consumers, genes involved in the KLE1738 GABA consumption pathway were downloaded from RAST45 and UniProt18. To identify organisms utilizing the GABA shunt, the list of GABA consumption genes was completed by adding all bacterial succinate semialdehyde dehydrogenase sequences found in UniProt. To identify potential GABA-producing organisms, nucleotide sequences for genes involved in the different pathways associated with GABA production (decarboxylation of glutamate, degradation of putrescine, or from arginine or ornithine) were downloaded from UniProt. The compiled lists of genes involved in GABA production or consumption were used as input for the BBH analysis against the gut microbe operon nucleotide sequences from KBase.
Metabolic modeling
In March 2016, all of the metabolic models from KBase, which were automatically generated in 2014 or earlier from all publicly available prokaryotic genome sequences, were downloaded. By comparing 16S rRNA sequences that shared >99% sequence similarity to the healthy fecal set of organisms from the American Gut project, 533 models were identified as gut-related (not all 913 microbes identified as gut related in Kbase have associated models). The models were forced to produce GABA (cpd00281) intracellularly ([c]) by introducing the constraint “cpd00281[c] → “ and maximizing it, with all exchange reactions (except for GABA) allowed to be unbounded (−1000 to 1000). Reversing the GABA constraint revealed that no model can consume free GABA. To identify potential GABA consumers, the biochemical reactions involved in the KLE1738 GABA consumption pathway were defined, and models were examined for similar reactions. Specificity of cofactor molecules were not considered, only that GABA was being metabolized.
Transcriptomic analysis
We surveyed a published human stool transcriptome dataset22 to explore the activity of three major genes involved in GABA metabolism i.e. glutamate decarboxylase (gad), gamma-aminobutyrate:alpha-ketoglutarate aminotransferase and succinate semialdehyde dehydrogenase in humans. The transcriptome data was assembled using Trinity23 which was further annotated for CDSs using prodigal47. BLASTN was used at % identity cut-off of 75% and E-value of 1e-5 to retrieve active transcripts from the CDS-annotated transcriptome data using gene sequences. Five active transcripts were obtained for glutamate decarboxylase with lengths 519, 234, 231, 276 and 286 bp. This was additionally confirmed by mapping of raw transcriptome reads data on to the gene sequences which led to reconstruction of a 500 bp long gad transcript. Transcripts for gamma-aminobutyrate:alpha-ketoglutarate aminotransferase and succinate semialdehyde dehydrogenase could not be recovered from this data.
Major Depressive Disorder Cohort -- Subjects
23 currently depressed subjects between the ages of 19 and 65 (15 female) participated in the study. Subjects were recruited through referral from the outpatient clinic in the Department of Psychiatry at Weill Cornell Medical College. Subjects were also self-referred by directly contacting our mood disorders research program or from the local community via flyers, outreach at local events, or direct contact. The recruitment procedure and all other aspects of our experimental protocol were approved by the Institutional Review Board of Weill Cornell Medical College, and all experiments were conducted in accordance with institutional guidelines and regulations. Patients provided written informed consent.
All subjects participated in an initial screening interview to determine eligibility for enrollment in the study. Patients were eligible for inclusion if they met DSM-IV-Text Revision criteria for a major depressive episode with a diagnosis of major depressive disorder or bipolar II disorder and if they also met criteria for treatment resistance, including a failure to respond to at least two previous antidepressant trials at adequate doses for 8 weeks during the current episode. Diagnoses were determined by a Board-Certified psychiatrist (MJD) in an unstructured clinical interview and through consultation with family members and the current treating psychiatrist. Potential subjects were excluded from the study if they presented with a history of claustrophobia, seizure disorder or other neurological disorder, head injury resulting in loss of consciousness, metal implants, pacemakers, intrauterine contraceptive devices, or braces, or if they were currently pregnant or lactating. Potential subjects were also excluded if they had bipolar I disorder or a psychotic disorder, were actively suicidal with plan or intent, had been in their current episode for longer than 3 years, had a history of clinically significant personality disorder as established in the diagnostic interview, or had substance abuse disorder or substance dependence within the past 3 years.
16S rRNA sequencing of the Major Depressive Disorder stool samples via the American Gut
Sequence data for the MDD patients in the American Gut dataset were produced in accordance with the standard Earth Microbiome Project 16S rRNA V4 amplicon sequencing protocol49. The MDD sequences analyzed corresponded to the set of samples represented by the April 26th 2017 processing. The American Gut sequences were processed using Deblur v1.0.250, and sequences corresponding to blooms were removed as previously reported51. Relative abundances of Bacteroides post-bloom filtering were used for the functional connectivity analysis.
Magnetic resonance imaging data acquisition and preprocessing
Magnetic resonance imaging data was collected on a 3.0 Tesla Siemens Trio MRI Scanner that was upgraded to a 3.0 Tesla Prisma MRI Scanner (Siemens, Munich, Germany) after the acquisition of imaging data from the first 10 subjects. Data were obtained from subjects in one session that occurred on or prior to the day of stool sample collection (mean=1.6d; SD=3.6d). Each imaging session included a resting-state fMRI (rsfMRI) sequence (Trio Scanner: repetition time 2.25sec, 161 volumes; Prisma Scanner: repetition time 0.77sec, 425 volumes) and a T1-weighted (MP-RAGE) anatomical scan. Preprocessing of rs-fMRI data was conducted with the AFNI (http://afni.nimh.nih.gov/afni/) and FSL (http://www.fmrib.ox.ac.uk/fsl/) software packages and included motion correction (AFNI), spatial smoothing (6-mm full-width half-maximum Gaussian kernel; FSL), temporal band-pass filtering (0.005–0.1 Hz; AFNI), linear and quadratic detrending (AFNI), and removal of nuisance signals by regression on six motion parameters (roll, pitch, yaw, and translation in three dimensions) and signal time courses for white matter and cerebrospinal fluid (CSF) regions-of-interest (ROIs) determined on an individual basis using an automated segmentation algorithm (FSL). We did not use global signal regression52.
Regions of interest
The region of interest comprising the DMN was defined a priori based on previously published reports53. The left DLPFC seed was an ROI within BA46 (9 mm seed centered on MNI coordinates: –44, 40, 29)32.
Functional connectivity analysis
We first quantified functional connectivity between the left DLPFC and DMN areas by testing for correlations between the BOLD signal time series seeded from the left DLPFC with voxels in the DMN (see above). Next, to identify cortical clusters in the DMN in which left-DLPFC:DMN functional connectivity correlated with Bacteroides relative abundance, we performed an analysis of covariance (ANCOVA; implemented using AFNI’s 3dttest++ function), with age, gender, head motion, scanner (pre vs. post upgrade) and Bacteroides relative abundance as covariates. This generated a statistical map identifying areas of the DMN in which functional connectivity with the left DLPFC varied with Bacteroides relative abundance, controlling for the remaining covariates. Importantly, this analysis showed that the effects of relative Bacteroides abundance on left DLPFC:DMN functional connectivity were not driven by Bacteroides effects on head motion. This analysis also showed that effects of relative Bacteroides abundance on left DLPFC:DMN functional connectivity were indistinguishable in the groups of subjects scanned before and after the scanner upgrade. Next, a Monte Carlo simulation was performed for this map using AFNI’s 3dClustSim function to determine statistical thresholds for voxel cluster size needed to achieve a family-wise α < 0.01 at voxel-wise p < 0.05. This yielded a threshold voxel cluster size of 213 voxels. In addition, the spatial mean connectivity value (Z) of each significant cluster was extracted for each subject and a linear regression was performed for each cluster with Bacteroides relative abundance.
Supplementary Material
Acknowledgements:
The authors would like to thank Sarah Rubin for help with the cultivation of KLE1738, and Jennifer Wang of the Small Molecule Mass Spectrometry Facility, a Harvard Faculty of Arts and Science (FAS) Division of Science Core Facility, for analyzing the GC/MS samples. This work was supported by grants R01HG005824 to KL, R01GM086158 to J.C. and F32GM108415 to T.R.R.
Footnotes
Data Availability: he 16S rRNA sequence and genome for KLE1738 are available from NCBI (MH636586 and PRJNA482656, respectively). The American Gut sequence data is in EBI under accession ERP012803. fMRI data is available at the discretion of author M.D. All other data that support the findings of this study are available from the corresponding author upon request. Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to K.L. (k.lewis@neu.edu).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Competing interests: P.S. and K. L. declare competing financial interests as they are founders of Holobiome, Inc.
References:
- 1.Fung TC, Olson CA & Hsiao EY Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 20, 145–155, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne HP et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier JC et al. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev 28, 237–264, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagkouvardos I, Overmann J & Clavel T Cultured microbes represent a substantial fraction of the human and mouse gut microbiota. Gut Microbes 8, 493–503, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Onofrio A et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17, 254–264, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenn K et al. Quinones are growth factors for the human gut microbiota. Microbiome 5, 161, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlier JP, Bedora-Faure M, K’Ouas G, Alauzet C & Mory F Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Seguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. International journal of systematic and evolutionary microbiology 60, 585–590, (2010). [DOI] [PubMed] [Google Scholar]
- 8.Klaring K et al. Intestinimonas butyriciproducens gen. nov., sp. nov., a butyrate-producing bacterium from the mouse intestine. International journal of systematic and evolutionary microbiology 63, 4606–4612, (2013). [DOI] [PubMed] [Google Scholar]
- 9.Yarza P et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12, 635–645, (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fodor AA et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS One 7, e41294, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagkouvardos I et al. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep 6, 33721, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman AL et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108, 6252–6257, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher J, Francke C & Postma PW How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70, 939–1031, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feehily C & Karatzas KA Role of glutamate metabolism in bacterial responses towards acid and other stresses. Journal of applied microbiology 114, 11–24, (2013). [DOI] [PubMed] [Google Scholar]
- 15.Hardman JK & Stadtman TC Metabolism of omega-amino acids. I. Fermentation of gamma-aminobutyric acid by Clostridium aminobutyricum n. sp. J Bacteriol 79, 544–548, (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fallingborg J Intraluminal pH of the human gastrointestinal tract. Dan Med Bull 46, 183–196, (1999). [PubMed] [Google Scholar]
- 17.Aziz RK et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman A et al. UniProt: a hub for protein information. Nucleic Acids Research 43, D204–D212, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald D, Hyde ER, Debelius JW, Morton JT, Gonzalez A, Ackermann G American gut: an open platform for citizen‐science microbiome research. mSystems, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sneath PH Principles of bacterial taxonomy. Proc R Soc Med 65, 851–852, (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arkin A. P. e. a. The DOE Systems Biology Knowledgebase. bioRxiv, (2016). [Google Scholar]
- 22.Ni Y, Li J & Panagiotou G A Molecular-Level Landscape of Diet-Gut Microbiome Interactions: Toward Dietary Interventions Targeting Bacterial Genes. MBio 6, e01263–01215, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas BJ et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8, 1494–1512, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto M et al. Colonic Absorption of Low-Molecular-Weight Metabolites Influenced by the Intestinal Microbiome: A Pilot Study. PLoS One 12, e0169207, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Berlo CL et al. gamma-Aminobutyric acid production in small and large intestine of normal and germ-free Wistar rats. Influence of food intake and intestinal flora. Gastroenterology 93, 472–479, (1987). [DOI] [PubMed] [Google Scholar]
- 26.Fujisaka S et al. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep 22, 3072–3086, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luscher B, Shen Q & Sahir N The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry 16, 383–406, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson RJ, Pizzagalli D, Nitschke JB & Putnam K Depression: perspectives from affective neuroscience. Annu Rev Psychol 53, 545–574, (2002). [DOI] [PubMed] [Google Scholar]
- 29.Greicius MD, Krasnow B, Reiss AL & Menon V Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A 100, 253–258, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greicius MD et al. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biological psychiatry 62, 429–437, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheline YI et al. The default mode network and self-referential processes in depression. Proc Natl Acad Sci U S A 106, 1942–1947, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liston C et al. Default mode network mechanisms of transcranial magnetic stimulation in depression. Biological psychiatry 76, 517–526, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koechlin E & Hyafil A Anterior prefrontal function and the limits of human decision-making. Science 318, 594–598, (2007). [DOI] [PubMed] [Google Scholar]
- 34.Wager TD, Davidson ML, Hughes BL, Lindquist MA & Ochsner KN Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron 59, 1037–1050, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tillisch K et al. Brain structure and response to emotional stimuli as related to gut microbial profiles in healthy women. Psychosom Med, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan AM et al. High-fat diet induces depression-like behaviour in mice associated with changes in microbiome, neuropeptide Y, and brain metabolome. Nutr Neurosci, 1–17, (2018). [DOI] [PubMed] [Google Scholar]
- 37.Bravo JA et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 108, 16050–16055, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janik R et al. Magnetic resonance spectroscopy reveals oral Lactobacillus promotion of increases in brain GABA, N-acetyl aspartate and glutamate. Neuroimage 125, 988–995, (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lin Q Submerged fermentation of Lactobacillus rhamnosus YS9 for gamma-aminobutyric acid (GABA) production. Braz J Microbiol 44, 183–187, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett E, Ross RP, O’Toole PW, Fitzgerald GF & Stanton C gamma-Aminobutyric acid production by culturable bacteria from the human intestine. Journal of applied microbiology 113, 411–417, (2012). [DOI] [PubMed] [Google Scholar]
- 41.Pokusaeva K et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil 29, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kootte RS et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab 26, 611–619 e616, (2017). [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wattam AR et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45, D535–D542, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wixon J & Kell D The Kyoto encyclopedia of genes and genomes--KEGG. Yeast 17, 48–55, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitagawa M et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12, 291–299, (2005). [DOI] [PubMed] [Google Scholar]
- 47.Hyatt D et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton M A rating scale for depression. J Neurol Neurosurg Psychiatry 23, 56–62, (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amir A et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amir A et al. Correcting for Microbial Blooms in Fecal Samples during Room-Temperature Shipping. mSystems 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang C & Glover GH Effects of model-based physiological noise correction on default mode network anti-correlations and correlations. Neuroimage 47, 1448–1459, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shirer WR, Ryali S, Rykhlevskaia E, Menon V & Greicius MD Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22, 158–165, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.