Abstract
For over 20 years, native chemical ligation (NCL) has played a pivotal role in enabling the total synthesis and semisynthesis of increasingly complex peptide and protein targets. Classical NCL proceeds by the chemoselective reaction of two unprotected polypeptide chains in near-neutral pH aqueous solution and is made possible by the presence of a thioester moiety on the C-terminus of the N-terminal peptide fragment and a natural cysteine residue on the N-terminus of the C-terminal peptide fragment. The reaction yields an amide bond adjacent to cysteine at the ligation site, furnishing the native protein backbone in a traceless manner. This protocol will highlight a number of recent and powerful advances to the methodology and will outline their particular uses, facilitating their continued application in the synthesis of challenging protein targets.
Keywords: native chemical ligation, peptides, proteins, chemoselective
INTRODUCTION
Native chemical ligation (NCL) enables the assembly of long peptides and proteins from their cognate peptide building blocks (Camarero & Muir, 2001; P. Dawson, Muir, Clark-Lewis, & Kent, 1994). The reaction condenses two unprotected peptide fragments, in aqueous media of near-neutral pH and under mild reaction conditions, to generate a native amide linkage in a chemoselective and high-yielding manner. Importantly, the unprotected peptide fragments can be readily prepared by optimized N-Boc- or N-Fmoc-protected solid phase peptide synthesis (SPPS) in high purity and yield and can be readily characterized by reversed-phase high-performance liquid chromatography (RP-HPLC) and electrospray mass spectrometry (ESI-MS). A number of techniques for the solid-phase synthesis of peptide thioester fragments by Boc-SPPS and thioester surrogate fragments by Fmoc-SPPS are available, in addition to a wide variety of ligation methods and conditions (Behrendt, White, & Offer, 2016; Jaradat, 2018; Li & Dong, 2017). However, despite the wealth of variations on the NCL approach, these methodologies have much in common. This unit aims to guide the selection of a synthetic method best suited to the peptide or protein of interest and provide detailed protocols for some approaches that have been developed in our laboratory.
The first section discusses how to select ligation site(s) in the target protein, along with a brief overview of synthetic considerations in accessing peptide thioester and thioester surrogate fragments (see Strategic Planning).
The second section provides the method for classical NCL from an alkylthioester peptide that can be accessed by a number of approaches including Boc-SPPS, Fmoc-SPPS and biological expression (see Basic Protocol 1) (Muir, Sondhi, & Cole, 1998; Vila-Perello et al., 2013). The third section provides the method for classical sequential NCL from three or more peptide fragments (see Alternate Protocol 1). The fourth section provides the method to synthesize alkyl thioester fragments by Boc-SPPS (see Support Protocol 1).
The fifth section provides the method for ligation from 3-(Fmoc-amino)-4-(methylamino) benzoic acid (Fmoc-MeDbz) thioester surrogate fragments (see Basic Protocol 2). The sixth section provides a method to synthesize the MeDbz linker (Support Protocol 2A). The seventh section provides the method to chemically synthesize either of these thioester surrogate fragments (see Support Protocol 2B).
The eighth section provides the method for ligation from Fmoc-protected peptide hydrazides as thioester surrogate fragments through the formation of a unique pyrazole intermediate (see Basic Protocol 3). The final section provides the means to synthesize Fmoc-protected peptide hydrazides (see Support Protocol 3).
STRATEGIC PLANNING
The first step in the chemical synthesis of a protein is selecting the ligation site(s). Ideally, the target protein is split up into synthetically accessible fragments of up to ~50 residues, the approximate limit for efficient production by routine N-Boc- or N-Fmoc-protected SPPS. As the choice of the ligation site defines the polypeptide fragments used in the ligation, a number of considerations must be made with regards to the amino acid sequence, as well as the primary, secondary and tertiary structure of the protein. Additionally, the scope of the synthesis project should be considered: the preparation of a small, highly pure quantity of a difficult target protein, versus using various alternate fragments in parallel ligations to generate library of synthetic proteins. In the latter case, it is important to select a ligation site where the exchangeable fragment would be of the smallest possible size to aid the chemical synthesis effort. As to the ligation site itself, the ideal location is a naturally occurring X-Cys motif, where X can be any residue (Hackeng, Griffin, & Dawson, 1999). Beta-branched amino acids such as Val, Ile, and Thr and also Pro (Pollock & Kent, 2011) ligate more slowly, but will proceed in under 24 hours as pre-formed arylthioesters. Thioester peptides with a C-terminal Asp or Glu can isomerize to facilitate ligation on the side chain, a side reaction that can be eliminated through an orthogonal protection strategy (Dang, Kubota, Mandal, Bezanilla, & Kent, 2013; Villain, Gaertner, & Botti, 2003). On the other hand, certain residues at the X position will often yield rapid ligations, such as Gly or His. If a naturally occurring Cys residue is not available in the primary sequence, it will be necessary to replace another residue with Cys to generate fragments appropriate for ligation. Non-conserved, surface exposed residues, remote from known active sites or binding sites are typically selected, using sequence alignment or protein structures as a guide.
Following the division of the protein of interest into synthetically accessible fragments, the next step is their chemical synthesis or biological expression. For fragments of ~50 amino acids or less, specific approaches for SPPS by both Boc and Fmoc-protected strategies are covered in Support Protocols 1, 2 and 3. Alkyl thioester peptides can be prepared directly by Boc-SPPS and cleaved by anhydrous HF. Alternatively, as the thioester moiety is not stable to the basic N-Fmoc deprotection conditions (20% 4-methylpiperidine in DMF) through each round of SPPS, a thioester surrogate can be prepared via Fmoc-SPPS and exchanged to an arylthioester in situ during the NCL reaction with an aromatic thiol. Standard aromatic thiols used for this purpose include thiophenol, mercaptophenylacetic acid (MPAA), 4-mercaptophenol (MPOH), or mercaptoethylsulfonic acid (MESNa). Thioester peptides can also be obtained through engineered C-terminal intein fusion proteins expressed in an organism of choice, an approach that will not be covered in this protocol. The N-terminal Cys peptide may be prepared by either Boc- or Fmoc-SPPS using standard SPPS synthetic cycles. For fragments of greater than ~50 amino acids, biosynthesis is more often employed, with the N-terminal Cys generated through processing of the terminal N-formyl-Met or through proteolytic cleavage of a fusion protein containing either a factor Xa or TEV recognition site (Erlanson, Chytil, & Verdine, 1996; Gentle, De Souza, & Baca, 2004; Tolbert & Wong, 2002).
BASIC PROTOCOL 1
NATIVE CHEMICAL LIGATION OF TWO POLYPEPTIDES UTILIZING A C-TERMINAL THIOESTER
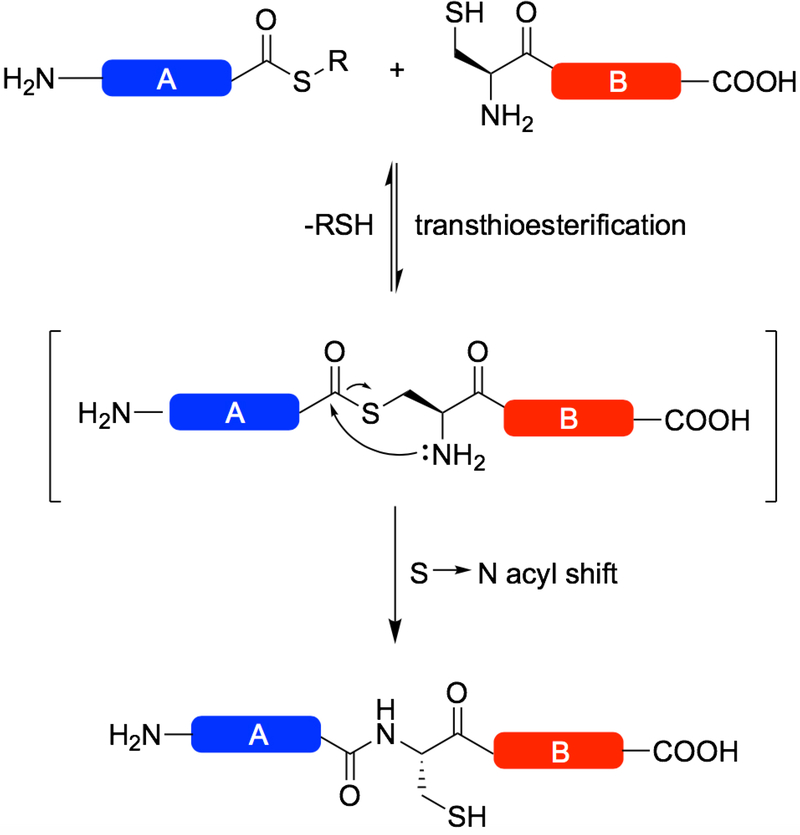
This protocol describes the condensation of two polypeptide chains to afford a single product linked through a native amide bond at the ligation site (Figure 1). The purified peptides are dissolved in aqueous, denaturing buffer in an equimolar fashion at high concentration to a final pH of 7.0–7.5, at which point a thiol catalyst is added to exchange the alkylthioester produced during SPPS to a more labile arylthioester, ready for transthioesterification. The reaction typically proceeds between 4–48 hours; new ligation reactions are usually afforded an overnight reaction time to ensure measurable completion upon taking the first timepoint. Following the reaction, the product is purified by RP-HPLC and folded to its native state.
Figure 1:
Scheme describing Native Chemical Ligation (NCL). An unprotected peptide fragment bearing an N-terminal Cys residue attacks the C-terminal thioester of another unprotected peptide fragment, undergoing transthioesterification and subsequent S to N acyl transfer to form an amide bond at the ligation site.
Note: Peptide purity is paramount to the success of these reactions – see the Critical Parameters section for more details. >95% pure by RP-HPLC peak area integration is suitable.
Materials
Ligation Buffer: 6 M guanidine-HCl, 200 mM Na2HPO4, pH 8.5 (see Reagents and Solutions)
Purified polypeptide with N-terminal Cys residue (lyophilized, >95% pure)
Purified polypeptide with C-terminal thioester functionality (lyophilized, >95% pure)
Thiophenol
Tris(2-carboxyethyl)phosphine HCl (TCEP)
1.5 mL Eppendorf tubes
pH meter with microprobe (Hamilton MiniTrobe pH electrode) or pH indicator strips (pH range 0–14, universal indicator)
1 M NaOH
1 M HCl
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
0.22 μm syringe filters
Analytical RP-HPLC column (such as Phenomenex Jupiter Proteo®: 4 μm, 90 Å, 150 × 4.6 mm)
Preparative RP-HPLC column (such as Phenomenex Jupiter Proteo®: 10 μm, 90 Å, 250 × 21.20 mm)
If the target protein contains tertiary structure, requiring a folding step:
Folding Buffer: 6 M guanidine-HCl, 5 mM DTT, pH 7 (see Reagents and Solutions)
Dialysis Buffer: 6 M guanidine-HCl, 1 mM DTT, pH 7 (see Reagents and Solutions)
Dialysis membrane system with molecular weight cutoff (MWCO) significantly smaller than target protein molecular weight (i.e.; 5,000 MWCO for a target protein of 10,000 Daltons)
If the target protein contains disulfide bonds, requiring an oxidation step:
Disulfide Formation Buffer: 0.5 M guanidine-HCl, 100 mM Tris-HCl, pH 8 (see Reagents and Solutions)
100 mL round-bottom flask
Stir bar
Protocol Steps
-
Prepare 100 mL of ligation buffer as described in Reagents and Solutions.
The ligation buffer is shelf-stable for up to a year once prepared and pH balanced to 8.5.
-
Weigh equimolar amounts of the peptide fragments to be ligated (one containing an N-terminal Cys residue, one containing a C-terminal thioester) in an appropriately sized vessel, such as a 1.5 mL Eppendorf tube.
For example, a test-sized ligation of two 36-mer fragments, both of molecular weight ~4000 Da, will utilize 1 mg of each fragment (0.25 μmol). For a test-sized ligation of two polypeptides of equal length, ~1 mg of each fragment should be targeted, while a preparative-scale ligation will require ~10 mg or more of each fragment. The remainder of this protocol will be written at the scale of the test-sized reaction but can simply be scaled proportionately for preparatory scale. Additionally, if one peptide fragment is more expensive or challenging to prepare than the other, then the less expensive/challenging fragment can be used at 1.5 equivalents to ensure full consumption of the challenging fragment.
-
Dissolve the dry peptides in ligation buffer to a final concentration between 1 mM – 5 mM with respect to each peptide. Ideally, 2 mM or greater concentrations of peptide should be used if the fragments display reasonable solubility. If the peptides have poor solubility, lower concentrations may be used, but the ligation may need a long reaction time to reach completion. Once the buffer has been added to the peptides, vortex to dissolve.
Continuing from above, the test ligation of two equally sized 36-mer fragments would use 1 mg of each fragment (0.25 μmol) dissolved in 100 μL of ligation buffer, which is a concentration of ~10 mg peptide per 1 mL of ligation buffer. A preparative scale ligation of the same polypeptides would use 10 mg of each fragment dissolved together in 1 mL of ligation buffer.
Add thiophenol to the ligation mixture as a 2% volume; i.e., 2 μL of thiophenol to a 100 μL ligation. DO NOT VORTEX TO MIX AT THIS STEP. Instead, emulsify the thiophenol into the ligation by raking the Eppendorf tube across an Eppendorf tube rack 5–6 times. The ligation reaction will appear milky.
-
Confirm that the pH of the solution is now ~7.1 by using a pH meter equipped with a microprobe to verify the pH. Adjust the pH accordingly with 1 M NaOH or 1 M HCl by adding a 2 μL droplet on the interior of the Eppendorf cap and shaking vigorously before taking additional pH readings. Adding the droplet to the cap and shaking rather than into the reaction liquid itself will prevent immediate thioester hydrolysis where the droplet is added.
At this point, the ligation should be near its target pH of ~7.1 for the reaction to take place. LIGATIONS SHOULD NOT BE PERFORMED AT pH 8.5. The buffer pH should drop significantly from 8.5 upon addition of peptides lyophilized as trifluoroacetic acid (TFA) salts, as well as upon addition of mildly acidic thiol additives. If a pH meter with a microprobe is not available, the pH of the ligation can be verified by spotting 3 μL of the ligation on pH indicator strips until a pH of 7.1 is achieved.
-
Allow the ligation reaction to occur by leaving the tube at room temperature for between 4 and 24 hours. The tube can be flipped or raked across the Eppendorf tube rack to mix the components every few hours if desired.
Note that thiophenol disulfide that forms during the course of the ligation is not soluble and will precipitate. This is not a problem and can help to better maintain the reducing environment of the ligation buffer compared to more soluble thiols such as 4-mercaptoacetic acid (MPAA). The precipitate can be spun down, filtered or extracted with organic solvent following ligation.
For new ligation reactions, the first analytical timepoint should be checked at 4 hours. To check the progress of the ligation reaction, take a 2 μL aliquot of the ligation (10–20 μg of peptide) and dilute the aliquot into 100 μL of an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA. After vortexing to mix, add one or two crystals of TCEP-HCl (<0.1 mg) from a spatula tip and shake until completely dissolved. Allow the aliquot to rest for 5 minutes, then filter the sample through a 0.22 μm single-use syringe filter.
-
Analyze the filtered sample by analytical LC-MS on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, to confirm the loss of starting material fragments and appearance of the desired protein product.
For a partially completed ligation, the HPLC trace will often show the starting material alkylthioester fragment, a small amount of thiophenol-exchanged arylthioester fragments nearby in the trace, and the N-terminal Cys fragment. Thiophenol and thiophenol disulfide will be observed as off-scale integrations but can be eluted by running 10–20% acetonitrile (isocratic) through the column for 10 minutes before starting the gradient elution program. The ligated product often appears with an elution time between the elution times of the two starting material fragments, but this is not always the case.
Repeat steps 7–8 until the ligation is complete (>90%). At this point, dilute the full reaction in 10 ligation reaction volumes of 50% MeCN in H2O containing 0.05% TFA, or whichever RP-HPLC buffer is readily employed by the experimenter. For example, a 500 μL ligation will be brought up to 5 mL with RP-HPLC buffer. Add 15 molar equivalents of TCEP (15 times the number of moles of the larger of the two polypeptides used in the ligation reaction), shake to dissolve, and allow to rest for 15 minutes. Filter the crude reaction through a 0.22 μm single-use syringe filter.
-
Purify the crude ligation mixture by preparative HPLC on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, guiding the choice of gradient by the known elution time from the analytical HPLC runs. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
Very high product yields (>90%) are typically obtained from the ligation step. In most cases, for peptides that exhibit complete solubility in the reaction medium, we observe complete conversion to the product (>95%) and achieve about 70% total yield after RP-HPLC purification and lyophilization.
If the target protein contains tertiary structure, fold the target protein by dissolving the lyophilized target protein to a concentration of 100 μM in freshly prepared Folding Buffer (6 M Guanidine-HCl, 5 mM DTT). Using a dialysis membrane system, dialyze the protein solution against freshly prepared Dialysis Buffer (6 M Guanidine-HCl, 1 mM DTT) for 12 hours. Repeat the dialysis step two additional times using Dialysis Buffer containing less Guanidine: first with 4 M Guanidine (Gdn) and 1 mM DTT, then with 2 M Gdn and 1 mM DTT, each for 12 hours. Perform the final dialysis step for 12 hours into the storage buffer of choice, such as standard PBS, pH 7.
If the target protein contains disulfide bonds, an additional oxidation step will have to be performed. Dissolve the protein to a concentration of 100 μM in freshly prepared Disulfide Formation Buffer (0.5 M Guanidine-HCl, 100 mM Tris-HCl, pH 8) in a round-bottom flask equipped with a stir bar. Stir overnight (12+ hours) open to atmosphere and check the progress of the oxidation reaction by taking an aliquot as described in Steps 7 and 8 of this protocol.
Divide the folded protein into suitable aliquots and freeze at −70°C for storage.
ALTERNATE PROTOCOL 1
SEQUENTIAL LIGATION OF THREE OR MORE POLYPEPTIDES
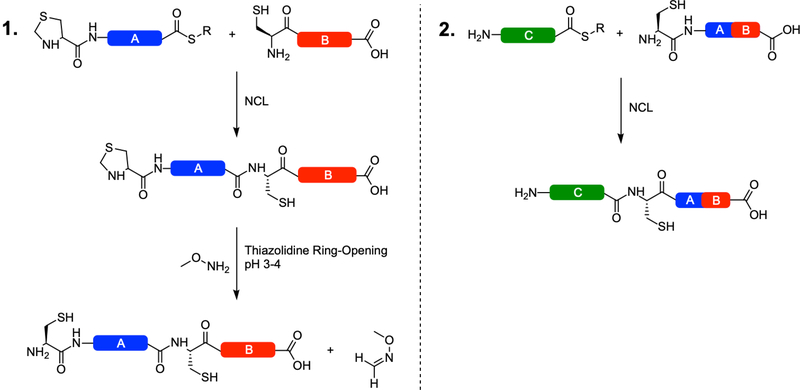
This protocol describes the assembly of a protein of interest from three or more polypeptide fragments by the use of N-terminally protected Cys residues as a thiazolidine (Thz) moiety at the N-terminus of the intermediate fragment(s). Following the successful ligation of two fragments as described in Basic Protocol 1, the N-terminal Thz moiety is unmasked to produce a native Cys residue by treatment with methoxylamine-HCl in aqueous acidic solution. The resulting product is then purified by RP-HPLC, lyophilized, and used as the C-terminal fragment in subsequent ligations, thereby assembling the protein from the C-terminus through the N-terminus as additional fragments are ligated, unmasked and purified (Figure 2). In particular, this protocol is useful when the target protein would require two fragments to be greater than ~50 amino acids each in length, as chemical synthesis of these fragments would be challenging,
Figure 2:
Thiazolidine decryption via methoxyamine at the N-terminus of a ligated peptide and subsequent sequential ligation(s). Following the first NCL reaction, the thiazolidine ring is opened in acidic conditions, freeing formaldehyde (Panel 1). The formaldehyde must be scavenged by a methoxy compound to prevent ring reformation. The resulting fragment AB is then purified by RP-HPLC and lyophilized for use as the C-terminal fragment in the next NCL reaction (Panel 2). In this manner, the protein is constructed in the C-terminal to N-terminal direction. Note that Fragment C could also function as an intermediate fragment, bearing a thiazolidine group at its N-terminus and be ring-opened in the same manner as in Panel 1. This approach is repeated for as many fragments as necessary to complete the protein.
Materials
Ligation Buffer: 6 M guanidine-HCl, 200 mM Na2HPO4, pH 8.5 (see Reagents and Solutions)
Thz Deprotection Buffer: 6 M guanidine-HCl, 500 mM methoxylamine-HCl (CH3ONH2-HCl), pH 1.9 (see Reagents and Solutions)
Fragment A: purified polypeptide with C-terminal thioester functionality and N-terminal Thz moiety (lyophilized, >95% pure)
Fragment B: purified polypeptide with N-terminal Cys residue (lyophilized, >95% pure)
Fragment C: purified polypeptide with C-terminal thioester functionality (lyophilized, >95% pure)
Thiophenol
Tris(2-carboxyethyl)phosphine HCl (TCEP)
1.5 mL Eppendorf tubes
pH meter with microprobe (Hamilton MiniTrobe pH electrode) or pH indicator strips (pH range 0–14, universal indicator)
1 M NaOH
1 M HCl
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
0.22 μm syringe filters
Analytical RP-HPLC column (such as Phenomenex Jupiter Proteo®: 4 μm, 90 Å, 150 × 4.6 mm)
Preparative RP-HPLC column (such as Phenomenex Jupiter Proteo®: 10 μm, 90 Å, 250 × 21.20 mm)
Protocol Steps
Perform the NCL reaction between Fragments A and B exactly as described in Basic Protocol 1 (steps 1–8), through to identifying the ligated product, Fragment AB, by RP-HPLC. The ligated product contains an N-terminal Thz group which will then be unmasked to Cys for the subsequent ligation to Fragment C, following purification and lyophilization of unmasked Fragment AB.
-
Freshly prepare the Thz deprotection buffer as described in Reagents and Solutions.
Unlike the standard ligation buffer, which is shelf stable, the Thz deprotection buffer should be freshly prepared on the day it is to be used.
-
Add 2.5 reaction volumes of Thz deprotection buffer to the crude ligation reaction of Fragments A and B. In this instance, the ligation is 100 μL in size, so 250 μL of the Thz deprotection buffer is added.
Typically, the mixture of 100 μL of ligation buffer of pH ~7.1 and 250 μL of Thz deprotection buffer of pH ~1.9 yields a final pH of ~3.7 for the Thz unmasking reaction without the need for further pH adjustment. If necessary, add more Thz deprotection buffer in 25 μL increments until a pH of ~3.7 is reached, rather than using 1 M HCl to lower the pH.
-
Allow the Thz deprotection reaction to occur by leaving the tube at room temperature for between 4 and 24 hours. The tube can be flipped or raked across the Eppendorf tube rack to mix the components every few hours if desired.
Typically, the reaction is performed overnight (12+ hrs) to ensure complete Thz unmasking for convenience. However, we have observed that this step can be completed in as little as 4 hours at room temperature.
Check the progress of the Thz unmasking reaction by taking a 10 μL aliquot of the reaction (10–20 μg of peptide) and dilute the aliquot into 100 μL of an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA. After vortexing to mix, add one or two crystals of TCEP-HCl (<0.1 mg) from a spatula tip and shake until completely dissolved. Allow the aliquot to rest for 5 minutes, then filter the sample through a 0.22 μm single-use syringe filter.
Analyze the filtered sample by analytical LC-MS on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, to confirm the appearance of the native Cys peptide and disappearance of the Thz-containing peptide, as evidenced by a mass shift of −12 Da.
When the reaction is complete, dilute the full reaction in 5 to 10 reaction volumes of 50% MeCN in H2O containing 0.05% TFA, or whichever HPLC buffer is readily employed by the experimenter. Add 15 molar equivalents of TCEP (15 times the number of moles of the larger of the two polypeptides used in the ligation reaction), shake to dissolve, and allow to rest for 15 minutes. Filter the crude reaction through a 0.22 μm single-use syringe filter.
Purify the crude reaction mixture by preparative HPLC on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, guiding the choice of gradient by the known elution time from the analytical HPLC runs. Pool the fractions containing the target product mass (Fragment AB) at the highest purity (>95%) and proceed to lyophilization.
-
Ligated, purified and lyophilized Fragment AB, now bearing an N-terminal Cys, may now be ligated with Fragment C by following the full Basic Protocol 1 (steps 1–10).
Very high product yields (>90%) are typically obtained from the ligation step. Quantitative yields are typically obtained from the Thz unmasking step (>99%). In most cases, for peptides that exhibit complete solubility in the reaction medium, we observe complete conversion to the product (>95%) and achieve about 70% total yield after RP-HPLC purification and lyophilization. If the target protein requires more than 3 fragments, additional middle fragment ligations may be performed as described in this protocol, with the final ligation taking place as described in Basic Protocol 1. If the target protein contains tertiary structure and/or disulfide bonds in the final product, follow Steps 11–13 of Basic Protocol 1.
SUPPORT PROTOCOL 1
CHEMICAL SYNTHESIS OF C-TERMINAL ALKYLTHIOESTER PEPTIDES BY BOC-SPPS
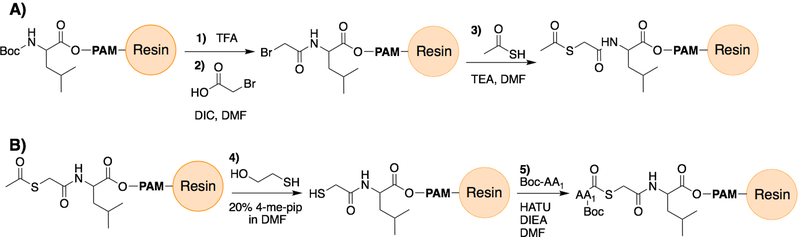
This protocol describes a method for the chain assembly of a peptide containing a C-terminal thioester moiety by Boc-SPPS using leucine-preloaded PAM resin or MBHA-resin (Figure 3). Chain elongation must be performed using the in situ neutralization protocols for Boc-SPPS developed by Alewood and Kent to avoid diketopiperazine formation, and to facilitate the generation of high quality crude synthetic products (Schnolzer, Alewood, Jones, Alewood, & Kent, 1992; Schnölzer, Alewood, Jones, Alewood, & Kent, 2007). For thioester peptides, the C-terminus of the peptide incorporates a commercially available mercaptoproprionic acid linker joined with a leucine residue (MPAL) or mercaptoacetic acid (MAAL), forming the alkylthioester which exchanges with thiophenol during the ligation reaction to form the more reactive arylthioester in situ. In the MPAL linker strategy, the removal of the trityl protecting group is afforded with two 1-min treatments with 2.5% triisopropylsilane and 2.5% H2O in TFA prior to peptide chain elongation. For both linkers, the Leu residue serves to ensure that the highly optimized sensitivity to acidolysis of the Pam- or MBHA-resin linker is maintained (Hackeng et al., 1999; Hojo et al., 1993). Following SPPS, side chain deprotection and cleavage from resin is afforded by treatment with anhydrous HF, which has been recently reviewed in detail (Muttenthaler, Albericio, & Dawson, 2015).
Figure 3:
Scheme describing the on-resin assembly of the mercaptoacetic acid leucine thioester (MAAL) moiety for use in Boc-SPPS. Panel A shows (1) Boc deprotection by TFA and (2) diisopropylcarbodiamide (DIC)-promoted acylation by bromoacetic acid in DMF, followed by (3) displacement through treatment with thioacetic acid in triethylamine (TEA). Panel B described the (4) unmasking of the thioacetic thioester to a free sulfhydryl group by treatment with 2-mercaptoethanol in 20% 4-methylpiperidine solution, which is then (5) acetylated with the C-terminal Boc-protected amino acid of the desired peptide fragment by routine SPPS coupling conditions (HATU, DIEA, DMF).
Materials
Boc-Leu-PAM resin (BACHEM; 200–400 mesh, 0.4–0.7 mmol/g loading)
Dimethylformamide (DMF)
Diisopropylcarbodiamide (DIC)
Bromoacetic acid
Trifluoroacetic acid (TFA)
Diisopropylethylamine (DIEA)
Thioacetic acid
Triethylamine (TEA)
β-mercaptoethanol
4-methyl-piperidine
N-Boc-protected amino acid building blocks
0.5 M solution of HATU in DMF
Anhydrous HF
Thioanisole
Diethyl ether
Reaction vessel for Solid Phase Peptide Synthesis (fritted glass resin support, PTFE stopcock)
Glass stir rod
Diaphragm vacuum pump
Erlenmeyer filter flask (2 liter capacity)
Black rubber vacuum tubing
Protocol Steps
Swell a 0.25 mmol scale quantity of Boc-Leu-PAM resin in 10 mL DMF for 1 hour.
-
Preactivate 520 mg of bromoacetic acid (15 equivalents to resin, 3.75 mmol) in 2.5 mL DMF with 525 μL of DIC (15 equivalents to resin, 3.75 mmol) for 30 min to obtain bromoacetic anhydride (1.87 mmol).
A white precipitate corresponding to the diisopropyl urea will appear during the activation reaction. If the precipitate does not form, most likely due to old DIC (>2 years) being used, the reaction has not taken place and should be repeated with fresh reagents.
With 15 minutes of the preactivation period remaining, deprotect the Boc-Leu-PAM resin with 2 × 1 min treatments of 10 mL TFA, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total). The resin is well-washed when absolutely no TFA fuming occurs upon the addition of flow wash DMF.
In a separate container, combine 1 mL of DIEA with 9 mL of DMF to make 10 mL of a 10% DIEA solution. Add the solution to the resin following the DMF flow wash step to neutralize the resin for 10 minutes and follow with a DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Add the preactivated bromoacetic anhydride mixture to the resin and couple for 5 minutes, follow by a DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Combine 450 μL thioacetic acid (25 equivalents to resin, 6.25 mmol), 144 μL triethylamine (4 equivalents to resin, 1 mmol) and 3 mL of DMF in a separate container and mix well. Add this mixture to the resin and allow to couple for 75 minutes (the resin may appear dark brown during this step). Follow with a DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Combine 1 mL β-mercaptoethanol with 9 mL of 20% 4-methyl-piperidine in DMF; treat the resin with 5 mL of this solution for 5 minutes, 2 times. The dark brown color of the resin will be removed during this step. Flow wash the resin with DMF for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Couple the first amino acid of the peptide of interest under standard Boc-SPPS conditions. Preactivate 5 equivalents of N-Boc-protected amino acid in 5 equivalents of 0.5 M HATU in DMF for 5 minutes. Add 8 equivalents of DIEA to the activated amino acid mixture and add the total mixture to the resin for coupling for 1 hour. Follow with a standard DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
-
Continue to chain assemble the peptide using in situ neutralization cycles for Boc-SPPS through the remainder of the fragment sequence. A standard cycle consists of 2 × 1 min treatments of 10 mL TFA, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total). The resin is well-washed when absolutely no TFA fuming occurs upon the addition of flow wash DMF. During the TFA deprotection periods, preactivate 5 equivalents of N-Boc-protected amino acid in 5 equivalents of 0.5 M HATU in DMF for 5 minutes. Following the DMF flow wash after the TFA deprotection, add 8 equivalents of DIEA to the activated amino acid mixture and add the total mixture to the resin for coupling for 20 minutes. Follow with a standard DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Typically, a peptide of ~50 residues will be broken up into 3 full days of manual couplings, as the total time for one cycle will be approximately 30 minutes. The use of an automated peptide synthesizer at this step will reduce the chain assembly time to approximately 1 day, depending on the length of the peptide sequence.
-
Proceed to HF cleavage, which has been covered in detail in accompanying protocols (Muttenthaler, Albericio, & Dawson, 2015). Great caution should be used when handling anhydrous HF.
During chain assembly, the choice of certain side-chain protected Boc amino acid building blocks should be avoided as they require nucleophilic removal before HF cleavage which is incompatible with the thioester linkage. If Histidine is present in the sequence, Boc-His(Dnp)-OH is recommended but is not removed before cleavage and remains protected. The Dnp group will be removed rapidly by thiol additives during ligation. If Tryptophan is present in the sequence, Bos-Trp(formyl)-OH should be avoided in favor of unprotected Boc-Trp-OH. While the acid labile His(Bom) and Trp(Hoc) protecting groups are compatible with the approach, associated side reactions limit their utility.
Lyophilize the crude peptide from 50% MeCN in H2O containing 0.05% TFA, or whichever HPLC buffer is readily employed by the experimenter. Proceed to analytical RP-HPLC, product identification by ESI-MS, and purification by preparatory RP-HPLC. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
BASIC PROTOCOL 2
NATIVE CHEMICAL LIGATION FROM POLYPEPTIDES CONTAINING N-METHYL-DBZ THIOESTER SURROGATES
This protocol describes the modifications to be applied to Basic Protocol 1 for usage with polypeptide fragments prepared by Fmoc-SPPS using Dbz/Nbz as thioester surrogates. The Dbz method for native chemical ligation was originally developed to allow for simple and efficacious access to the peptide-aryl-thioester intermediate used in native chemical ligation (Blanco-Canosa & Dawson, 2008). The Dbz linker has also been adapted for in situ activation through a benzotriazole intermediate (Wang et al., 2015), but this protocol will focus on the use of N-methyl-Dbz (MeDbz) via on-resin conversion to N-methyl-Nbz, a method that has been optimized to minimize the risk of acylation of the second Dbz amine during peptide elongation (Blanco-Canosa, Nardone, Albericio, & Dawson, 2015). This protocol is broadly similar to Basic Protocol 1, and synthesis of the activated Nbz peptides is described in Support Protocol 2.
Note: This basic protocol is fully compatible with sequential ligation through the post-ligation thiazolidine ring-opening approach as described in Alternate Protocol 1.
Materials
Ligation Buffer: 6 M guanidine-HCl, 200 mM Na2HPO4, pH 8.5 (see Reagents and Solutions)
Purified polypeptide with N-terminal Cys residue (lyophilized, >95% pure)
Purified polypeptide with C-terminal C-terminal MeNbz functionality (lyophilized, >95% pure)
Thiophenol or 4-mercaptophenylacetic acid (MPAA)
Tris(2-carboxyethyl)phosphine HCl (TCEP)
1.5 mL Eppendorf tubes
pH meter with microprobe (Hamilton MiniTrobe pH electrode) or pH indicator strips (pH range 0–14, universal indicator)
1 M NaOH
1 M HCl
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
0.22 μm syringe filters
Analytical RP-HPLC column (such as Phenomenex Jupiter Proteo®: 4 μm, 90 Å, 150 × 4.6 mm)
Preparative RP-HPLC column (such as Phenomenex Jupiter Proteo®: 10 μm, 90 Å, 250 × 21.20 mm)
Protocol Steps
Prepare 100 mL of ligation buffer as described in Reagents and Solutions.
-
Weigh equimolar amounts of the peptide fragments to be ligated (one containing an N-terminal Cys residue, one containing a C-terminal MeNbz functionality) in an appropriately sized vessel, such as a 1.5 mL Eppendorf tube. For example, a test-sized ligation of two 36-mer fragments, both of molecular weight ~4000 Da, will utilize 1 mg of each fragment (0.25 μmol).
For a test-sized ligation of two polypeptides of equal length, ~1 mg of each fragment should be targeted, while a preparative-scale ligation will require ~10 mg or more of each fragment. The remainder of this protocol will be written at the scale of the test-sized reaction but can simply be scaled proportionately for preparatory scale.
Additionally, if one peptide fragment is more expensive or challenging to prepare than the other, then the less expensive/challenging fragment can be used at 1.5 equivalents to ensure full consumption of the challenging fragment.
-
Dissolve the dry peptides in ligation buffer to a final concentration between 1 mM – 5 mM with respect to each peptide. Ideally, 2 mM or greater concentrations of peptide should be used if the fragments display reasonable solubility. If the peptides have poor solubility, lower concentrations may be used, but the ligation may need a long reaction time to reach completion. Once the buffer has been added to the peptides, vortex to dissolve. MOVE TO STEP 4 WITHOUT TIME DELAY.
Continuing from above, the test ligation of two equally sized 36-mer fragments would use 1 mg of each fragment (0.25 μmol) dissolved in 100 μL of ligation buffer, which is a concentration of ~10 mg peptide per 1 mL of ligation buffer. A preparative scale ligation of the same polypeptides would use 10 mg of each fragment dissolved together in 1 mL of ligation buffer.
-
Add thiophenol to the ligation mixture as a 2% volume; i.e., 2 μL of thiophenol to a 100 μL ligation. DO NOT VORTEX TO MIX AT THIS STEP. Instead, emulsify the thiophenol into the ligation by raking the Eppendorf tube across an Eppendorf tube rack 5–6 times. The ligation will appear milky.
Some laboratories prefer not to use thiophenol due to concerns over odor or solubility, despite its utility as an extremely reliable and efficient thiol source for the reaction. For these groups, a ligation buffer containing 200 mM 4-mercaptophenylacetic acid (MPAA) and 20 mM Tris(2-carboxyethyl)phosphine HCl (TCEP) in addition to the guanidine and phosphate can be employed. Note that this buffer should be prepared the day it will be used, and that if it is prepared from separate stock solutions of 6 M Guanidine-HCl and 200 mM Na2HPO4, the acid additions will lower the pH significantly, requiring readjustment to pH 8.5.
-
Confirm that the pH of the solution is now ~7.1 by using a pH meter equipped with a microprobe to verify the pH. Adjust the pH accordingly with 1 M NaOH or 1 M HCl by adding a 2 μL droplet on the interior of the Eppendorf cap and shaking vigorously before taking additional pH readings. Adding the droplet to the cap and shaking rather than into the reaction liquid itself will prevent immediate thioester hydrolysis where the droplet is added.
At this point, the ligation should be near its target pH of ~7.1 for the reaction to take place. LIGATIONS SHOULD NOT BE PERFORMED AT pH 8.5. The buffer pH should drop significantly from 8.5 upon addition of peptides lyophilized as trifluoroacetic acid (TFA) salts, as well as upon addition of mildly acidic thiol additives. If a pH meter with a microprobe is not available, the pH of the ligation can be verified by spotting 3 μL of the ligation on pH indicator strips until a pH of 7.1 is achieved.
-
Allow the ligation reaction to occur by leaving the tube at room temperature for between 4 and 24 hours. The tube can be flipped or raked across the Eppendorf tube rack to mix the components every few hours if desired.
Note that thiophenol disulfide that forms during the course of the ligation is not soluble and will precipitate. This is not a problem and can help to better maintain the reducing environment of the ligation buffer compared to more soluble thiols such as 4-mercaptoacetic acid (MPAA). The precipitate can be spun down, filtered or extracted with organic solvent following ligation.
-
For new ligation reactions, the first analytical timepoint should be checked at 4 hours. To check the progress of the ligation reaction, take a 2 μL aliquot of the ligation (10–20 μg of peptide) and dilute the aliquot into 100 μL of an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA. After vortexing to mix, add one or two crystals of TCEP-HCl (<0.1 mg) from a spatula tip and shake until completely dissolved. Allow the aliquot to rest for 5 minutes, then filter the sample through a 0.22 μm single-use syringe filter.
The time of the ligation will depend on the specific peptides involved. Notably, peptides with a hindered residue following the C-terminal MeNbz, such as valine or proline, may take longer to react. By contrast, peptides with glycine at that position tend to react more rapidly. Taking timepoints is recommended to ensure the reaction is complete.
-
Analyze the filtered sample by analytical LC-MS on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, to confirm the loss of starting material fragments and appearance of the desired protein product.
For a partially completed ligation, the HPLC trace will often show the starting material MeNbz fragment, a small amount of thiophenol-exchanged arylthioester fragments nearby in the trace, and the N-terminal Cys fragment. Thiophenol and thiophenol disulfide will be observed as off-scale integrations but can be eluted by running 10–20% acetonitrile (isocratic) through the column for 10 minutes before starting the gradient elution program. The ligated product often appears with an elution time between the elution times of the two starting material fragments, but this is not always the case.
Repeat steps 7–8 until the ligation is complete (>90%). At this point, dilute the full reaction in 10 ligation reaction volumes of 50% MeCN in H2O containing 0.05% TFA, or whichever RP-HPLC buffer is readily employed by the experimenter. For example, a 500 μL ligation will be brought up to 5 mL with RP-HPLC buffer. Add 15 molar equivalents of TCEP (15 times the number of moles of the larger of the two polypeptides used in the ligation reaction), shake to dissolve, and allow to rest for 15 minutes. Filter the crude reaction through a 0.22 μm single-use syringe filter.
-
Purify the crude ligation mixture by preparative HPLC on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, guiding the choice of gradient by the known elution time from the analytical HPLC runs. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
Very high product yields (>90%) are typically obtained from the ligation step. In most cases, for peptides that exhibit complete solubility in the reaction medium, we observe complete conversion to the product (>95%) and achieve about 70% total yield after RP-HPLC purification and lyophilization.
If the target protein contains tertiary structure and/or disulfide bonds in the final product, follow Steps 11–13 of Basic Protocol 1.
SUPPORT PROTOCOL 2A
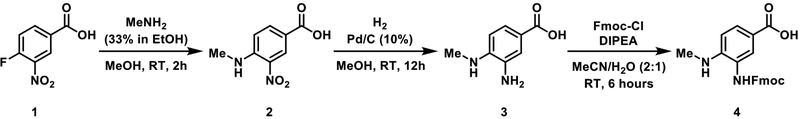
CHEMICAL SYNTHESIS OF FMOC-3-AMINO-4-(METHYLAMINO)BENZOIC ACID (Fmoc-MeDbz)
This protocol describes the solution-phase synthesis of Fmoc-MeDbz, Scheme 1, for use as described in Basic Protocol 2 after the desired peptide chain is assembled according to Support Protocol 2B. This procedure is adapted from (Kuzniewski, Gertsch, Wartmann, & Altmann, 2008) and (Blanco-Canosa et al., 2015). Fmoc-MeDbz (CAS: 1788861–35-7) can also be purchased from a number of suppliers, such as Peptide Institute, Iris Biotech, and Peptides International.
Materials
Water
Acetonitrile (MeCN)
Methanol (MeOH)
250-mL round-bottom flask
500-mL round-bottom flask
3 M HCl solution
MeNH2 (33% solution in EtOH)
3-nitro-4-fluorobenzoic acid
Celite
H2 gas
N2 gas
Rotary evaporator
Stir bar
Magnetic stir plate
Buchner funnel
Fritted glass Buchner funnel
High vacuum apparatus
Rubber septa
Needles
Balloons
Syringes
Protocol Steps
Add 7.405 g (40 mmol) 3-nitro-4-fluorobenzoic acid, 1, to a 250-mL round-bottom flask equipped with a large stir bar.
Add 100 mL MeOH and stir until the suspension is evenly dispersed.
Add 45 mL MeNH2 (33% solution in EtOH) and stir vigorously for 2 hours at room temperature.
Add 100 mL distilled water.
Acidify the solution by the slow addition of 3 M HCl to pH 5, at which point a yellow precipitate will form from the red suspension.
Collect this yellow precipitate via filtration and wash the filter cake with 2 × 50 mL distilled water.
-
Dry the yellow precipitate under high vacuum for 18 hours.
7.753 g (99% yield) of 3-nitro-4-(methylamino)benzoic acid 2 were obtained. Spectral properties matched those reported previously (Kuzniewski et al., 2008).
Add 7.74 grams of 3-nitro-4-(methylamino)benzoic acid 2 to a 500-mL round-bottom flask equipped with a large stir bar.
Add 160 mL MeOH, then 774 mg Pd/C (10%).
Seal the round-bottom using a rubber septum and puncture with an exit needle to allow for H2 gas exit flow.
Introduce a balloon of H2 gas to the reaction vessel using a long needle and submerge the needle tip into the stirring suspension to saturate the solution with H2 gas and initiate the reaction. Keep H2 gas flowing through the suspension with vigorous stirring for 5 minutes. Make sure the exit needle is open to air to allow the H2 to flow from the balloon, through the reaction mixture, and out of the vessel. Retract the H2 gas needle to the headspace above the suspension and remove the exit needle from the septum.
Stir the reaction mixture vigorously for 12 hours while maintaining a constant supply of H2 gas. Often, multiple H2 balloons will be depleted during the course of this reaction and new ones must be introduced to maintain a reducing environment.
When no yellow solid remains (after 12 hours), remove the H2 balloon, open the flask to atmosphere using an exit needle, and sparge the suspension with N2 for 10 minutes to displace the H2 gas.
Filter this suspension through a MeOH wetted pad of Celite on a fritted glass Buchner funnel, eluting with 4 × 100 mL MeOH. DO NOT PULL AIR THROUGH THE Pd/C SOLID THAT YOU ARE FILTERING OFF, as this may result in ignition of residual H2 bound to the Pd/C. Make sure there is always MeOH above the level of the filter cake.
Dispose of the Celite/Pd/C filter cake in a dedicated waste receptacle, using water to rinse. Keep a layer of water above the solid in the waste container at all times.
-
Concentrate the methanolic filtrate using a rotary evaporator and high vacuum to remove residual solvent.
6.033 g (92% yield) of 3-amino-4-(methylamino)benzoic acid 3 were obtained. Spectral properties matched those reported previously (Kuzniewski et al., 2008).
Add 6.00 g of 3-amino-4-(methylamino)benzoic acid 3 into a 250-mL round-bottom flask equipped with a stir bar.
Add 40 mL water and 40 mL MeCN.
Add 7.23 mL N,N-diisopropylethylamine (DIEA).
Add a solution of 9.81 g fluorenylmethyloxycarbonyl chloride (Fmoc-Cl) in 40 mL MeCN via syringe with vigorous stirring and stir for 6 hours.
Remove MeCN on a rotary evaporator and filter the resultant suspension. Wash the filter cake with 3 × 25 mL MeCN.
-
Dry the filter cake under high vacuum.
10.939 g (78% yield) of Fmoc-3-amino-4-(methylamino)benzoic acid 4 were obtained. Spectral properties matched those reported previously (Blanco-Canosa et al., 2015).
SUPPORT PROTOCOL 2B
CHEMICAL SYNTHESIS OF C-TERMINAL NBZ PEPTIDES BY FMOC-SPPS
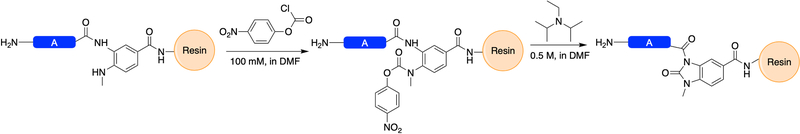
The synthesis of peptides containing an Nbz ester as the activated thioester surrogate primarily proceeds under standard conditions for Fmoc SPPS. While first generation Dbz linkers were susceptible to secondary acylation during synthesis in the presence of excess base or microwaves, the second generation MeDbz linker has significantly reduced susceptibility to this undesired side reaction. Following synthesis of the MeDbz-containing peptide, activation to MeNbz is carried out on-resin by acylation of the second aryl amine by a chloroformate, followed by cyclization to the N-acyl urea by attack of the amide nitrogen on the Dbz ring on the carbamate (Figure 5). The resulting N-acyl urea is highly susceptible to thiolysis, and rapidly converts to arylthioester under ligation conditions (Basic Protocol 2).
Figure 5:
On-resin acylation with 4-nitrophenylchloroformate and subsequent formation of the N-acyl benzimidazolinone species by treatment with base (DIEA). The activated MeDbz peptide is then ready for standard TFA cleavage, lyophilization, purification and NCL.
Materials
Rink Amide resin (BACHEM; 200–400 mesh, 0.4–0.8 mmol/g loading)
Dimethylformamide (DMF)
Dichloromethane (DCM)
Deprotection solution: 20% 4-methylpiperidine in DMF
MeDbz Coupling Solution: 0.4 M HATU, 0.4 M HOAt, in DMF
Diisopropylethylamine (DIEA)
N-Fmoc-protected amino acid building blocks
Fmoc-MeDbz-OH
100 mM 4-nitrophenyl chloroformate in DCM
0.5 M DIEA in DMF
0.4 M solution of HATU in DMF
Trifluoroacetic acid (TFA)
Triisopropylsilane (TIPS)
Milli-Q purified H2O
N2 gas
Diethyl ether
20 mL Disposable filter columns (such as Bio-Rad® Econo-Pac® Chromatography columns)
50 mL Falcon® centrifuge tubes
Benchtop centrifuge (Sorvall XTR or equivalent)
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
Reaction vessel for Solid Phase Peptide Synthesis (fritted glass resin support, PTFE stopcock)
Glass stir rod
Diaphragm vacuum pump
Erlenmeyer filter flask (2 L capacity)
Black rubber vacuum tubing
Protocol Steps
-
Swell a 0.20 mmol scale quantity of Rink Amide resin in 10 mL DMF for 1 hour.
The choice of resin should be made based on the specific requirements of the peptide being synthesized, as the Dbz method does not typically require special resins. As the C-terminus of the synthesized peptide is exchanged with thiol during the ligation (Basic Protocol 2) the C-terminal functionality is not important. Therefore, amide resin is selected as all couplings can be performed under normal conditions, unlike the higher base first coupling onto an alcohol-bearing Wang resin. Standard polystyrene resins can typically be used, but for longer or more challenging fragments it can be helpful to use an alternative resin such as ChemMatrix® or TentaGel® that has high swelling in DCM and is optimized for use with challenging sequences.
If the resin is protected by an Fmoc group, deprotect by treatment 2 × 5 mL of 20% 4-methylpiperidine in DMF for 5 minutes each, followed by a 30 second DMF flow wash (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
-
Dissolve 0.5 mmol of Fmoc-MeDbz-OH in 1.25 mL of MeDbz coupling solution and allow to preactivate for 10 minutes.
The coupling solution used in this procedure employs HOAt in addition to that provided by HATU. Adding extra HOAt to coupling solution can serve to reduce the pH, which is especially necessary for syntheses using Fmoc-Dbz-OH rather than the second generation Fmoc-MeDbz-OH, so as to prevent acylation of the second aryl amine. The second generation MeDbz linker has significantly reduced tendency toward acylation, and so use of HOAt may not always be necessary but can still be used and should not significantly reduce the efficiency of most syntheses. 0.5 mmol is suggested due to the relative expense of protected MeDbz species relative to standard Fmoc-protected amino acids. If cost is not an issue then the standard 1 mmol (5eq) may be used, and liquid volumes for this coupling step should be doubled.
Add 138 μL (8 equivalents to resin) of neat DIEA to the dissolved Fmoc-MeDbz-OH to activate, vortex to mix and immediately add to resin.
-
Allow the MeDbz to couple for 1 hour, using a glass stir rod to mix the resin every 15 minutes. Follow with a 30 second DMF flow wash (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Coupling of MeDbz to the resin is usually quantitative after 1 hour and may be monitored with a standard ninhydrin test. If the test shows incomplete coupling, repeat steps 3–5 until the test shows no remaining free amine.
-
Synthesize the remainder of the peptide under standard Fmoc-SPPS conditions. A standard cycle consists of 2 × 5 mL of 20% 4-methylpiperidine in DMF for 5 minutes each, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total). During the Fmoc deprotection periods, preactivate 5 equivalents of N-Fmoc-protected amino acid in 5 equivalents of 0.4 M HATU in DMF for 5 minutes. Following the DMF flow wash after the Fmoc deprotection, add 8 equivalents of DIEA to the activated amino acid mixture and add the total mixture to the resin for coupling for 20 minutes. Follow with a standard DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
In general, standard Fmoc SPPS conditions are used, but it should be noted that the first coupling, onto the Dbz, cannot be monitored by the ninhydrin test. In addition, experimenters should not acetyl cap their peptides after every step. This will lead to acylation of the second Dbz amine, preventing the activation step from being performed. MeDbz is more resistant to this acylation, so a final N-terminal acetylation may be performed by 5-minute base-free treatment with 20% acetic anhydride in DMF. However, a small degree of acetylation of the Dbz may still occur in this case, so pre-acetylated amino acids may also be used.
Typically, a peptide of ~50 residues will be broken up into 3 full days of manual couplings, as the total time for one cycle will be approximately 30 minutes. The use of an automated peptide synthesizer at this step will reduce the chain assembly time to approximately 1 day, depending on the length of the peptide sequence.
When the peptide sequence is finished, perform the final Fmoc deprotection by treatment with 2 × 5 mL of 20% 4-methylpiperidine in DMF for 5 minutes each, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Perform a thorough flow wash with DCM for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total) to solvent-exchange the resin into DCM.
Add 10 mL of 100 mM 4-nitrophenyl chloroformate in DCM to the DCM swelled resin for 1 hour, using a glass stir rod to mix the resin every 15 minutes. Follow with a 30 second DCM flow wash (flow-through at the resin bed surface under vacuum, approx. 250 mL DCM total).
Perform a thorough flow wash with DMF for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total) to solvent-exchange the resin into DMF.
Add 10 mL of 0.5 M DIEA in DMF to the resin 30 minutes, using a glass stir rod to mix the resin every 15 minutes. Follow with a 30 second DMF flow wash (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Perform a thorough flow wash with DCM for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total) to solvent-exchange the resin into DCM. Dry the resin under vacuum for 1 hour.
-
Cleave the peptide from the resin with 10 mL of a standard TFA cleavage cocktail such as 95% TFA, 2.5% TIPS, and 2.5% H2O for 2 hours. Filter the cocktail from the resin using a disposable filter column, collecting the filtrate in a 50 mL Falcon tube. Evaporate the majority of the TFA from the cleavage cocktail by applying N2 gas for 30 minutes, or until <2 mL of TFA remains. Precipitate the peptide by adding 10 mL of ice-cold diethyl ether. Centrifuge the suspension for 5 minutes @ 4000 rpm. Pour off the diethyl ether and dissolve the peptide pellet in an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA.
The use of ethanedithiol (EDT) as a cleavage cocktail additive at this step is discouraged, as we have observed an elevated degree of hydrolysis of the activated Nbz species when EDT is employed during cleavage.
Lyophilize the crude peptide from 50% MeCN in H2O containing 0.05% TFA, or whichever HPLC buffer is readily employed by the experimenter. Proceed to analytical RP-HPLC, product identification by ESI-MS, and purification by preparatory RP-HPLC. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
BASIC PROTOCOL 3
NATIVE CHEMICAL LIGATION FROM POLYPEPTIDES CONTAINING HYDRAZIDE THIOESTER SURROGATES
This protocol describes the method for activating, thiol-exchanging and condensing a C-terminal peptide hydrazide for use in NCL through an acyl-pyrazole intermediate. Leveraging the Knorr Pyrazole Synthesis, C-terminal peptide hydrazides in acidic aqueous media can be efficiently and chemoselectively transformed into peptide acyl-pyrazoles upon the addition of of acetyl acetone (acac). Acyl-pyrazoles are known to be mild acylation agents and are readily exchanged for aryl-thiol catalysts in situ, generating peptide aryl-esters (Figure 6). These peptide thioesters can then be utilized in classic NCL conditions for the condensation of long peptide fragments containing native peptide bonds at their ligation site.
Figure 6:
Activation of a C-terminal peptide hydrazide through acid-catalyzed acyl pyrazole formation: the Knorr Pyrazole Synthesis. The C-terminal peptide pyrazole is rapidly exchanged to the arylthioester species for use in NCL by reaction with 4-mercaptophenylacetic acid (MPAA).
Materials
Ligation Buffer A: 6 M guanidine-HCl, pH 3 (see Reagents and Solutions)
Ligation Buffer B: 6 M guanidine-HCl, 200 mM Na2HPO4, pH 9 (see Reagents and Solutions)
Purified polypeptide with N-terminal Cys residue (lyophilized, >95% pure)
Purified polypeptide with C-terminal hydrazide functionality (lyophilized, >95% pure)
4-mercaptophenylacetic acid (MPAA)
Acetyl acetone (acac)
Tris(2-carboxyethyl)phosphine HCl (TCEP)
1 M NaOH solution
1 M HCl solution
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
1.5 mL Eppendorf tubes
pH meter with microprobe (Hamilton MiniTrobe pH electrode) or pH indicator strips (pH range 0–14, universal indicator)
0.22 μm syringe filters
Analytical RP-HPLC column (such as Phenomenex Jupiter Proteo®: 4 μm, 90 Å, 150 × 4.6 mm)
Preparative RP-HPLC column (such as Phenomenex Jupiter Proteo®: 10 μm, 90 Å, 250 × 21.20 mm)
Protocol Steps
-
Prepare the Ligation Buffers A and B as described in Reagents and Solutions.
The ligation buffers are shelf-stable for up to a year once prepared and pH balanced to either pH 3 or 9.
-
Weigh equimolar amounts of the fragments to be ligated (one containing an N-terminal Cys residue, one containing a C-terminal hydrazide) into SEPARATE appropriately sized vessels, such as a 1.5 mL Eppendorf tubes. For example, a test-sized ligation of two 36-mer fragments, both of molecular weight ~4000 Da, will utilize 1 mg of each fragment (0.25 μmol).
For a test-sized ligation of two polypeptides of equal length, ~1 mg of each fragment should be targeted, while a preparative-scale ligation will require ~10 mg or more of each fragment. The remainder of this protocol will be written at the scale of the test-sized reaction but can simply be scaled proportionately for preparatory scale. Additionally, if one peptide fragment is more expensive or challenging to prepare than the other, then the less expensive/challenging fragment can be used at 1.5 equivalents to ensure full consumption of the challenging fragment.
-
Dissolve the dry C-terminal hydrazide peptide in Ligation Buffer A to a concentration between 1 mM – 5 mM. Ideally, 2 mM or greater concentrations of peptide should be used if the fragment displays reasonable solubility. Add MPAA to a concentration of 200 mM (forming a saturated emulsion of MPAA). To this solution, add 1.5 equivalents of acetyl acetone (Acac) from a stock solution made in water (20 μL Acac in 200 μL water). Briefly vortex to mix.
Continuing from above, the test ligation of two equally sized 36-mer fragments would begin with 1 mg of the C-terminal hydrazide fragment (0.25 μmol) dissolved in 200 μL of Ligation Buffer A, which is a concentration of ~5 mg peptide per 1 mL of Ligation Buffer A. A preparative scale ligation of the same polypeptides would use 10 mg of each fragment dissolved together in 2 mL of Ligation Buffer A.
-
Allow the thioesterification to occur by leaving the tube at room temperature for 2–4 hours, gently agitating the emulsion periodically.
Thioesterification to a proline or a beta-branched residue such as valine or threonine will likely take at least 4 hours to reach completion.
-
Check the progress of the thioesterification by taking a 5 μL aliquot of the ligation (10–20 μg of peptide) and dilute the aliquot into 100 μL of an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA. Filter the sample through a 0.22 μm single-use syringe filter. Analyze the filtered sample by analytical LC-MS on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, to confirm the loss of the starting material hydrazide fragment and appearance of the desired thioesterified product.
Typically, the reaction reaches quantitative conversion to thioester product at 4 hours. If the reaction is not complete at 4 hours, check that the pH of the reaction mixture is acidic (pH=~3) and add an additional 1 equivalent of Acac. Leave the reaction another 4 hours and repeat step 5 to check its progress by LC-MS.
When thioesterification reaction is complete, dissolve the dry N-terminal cysteine peptide in Ligation Buffer B to a concentration between 1 mM – 5 mM. Ideally, 2 mM or greater concentrations of peptide should be used if the fragment displays reasonable solubility. Add TCEP to a final concentration of 50 mM and vortex to dissolve.
Combine the two dissolved peptides together and raise the pH to ~7.1 by using a pH meter equipped with a microprobe to verify the pH. Adjust the pH accordingly with 1 M NaOH by adding a 2 μL droplet on the interior of the Eppendorf cap and shaking vigorously before taking additional pH readings. Adding the droplet to the cap and shaking rather than into the reaction liquid itself will prevent immediate thioester hydrolysis where the droplet is added.
Allow the ligation to occur by leaving the tube at room temperature for between 4 and 48 hours. The tube can be flipped or raked across the Eppendorf tube rack to mix the components every few hours if desired.
For new ligation reactions, the first analytical timepoint should be checked at 4 hours. Check the progress of the ligation reaction by taking a 5 μL aliquot of the ligation (10–20 μg of peptide) and dilute the aliquot into 100 μL of an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA. After vortexing to mix, add one or two crystals of TCEP-HCl (<0.1 mg) from a spatula tip and shake until completely dissolved. Allow the aliquot to rest for 5 minutes, then filter the sample through a 0.22 μm single-use syringe filter.
-
Analyze the filtered sample by analytical LC-MS on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, to confirm the loss of starting material fragments and appearance of the desired protein product.
MPAA and MPAA disulfide will be observed as off-scale integrations but can be eluted by running 10–20% acetonitrile (isocratic) through the column for 10 minutes before starting the gradient elution program. The ligated product often appears with an elution time between the elution times of the two starting material fragments, but this is not always the case.
Repeat steps 9–10 until the ligation is complete (>90%). At this point, dilute the full reaction in 10 ligation reaction volumes of 50% MeCN in H2O containing 0.05% TFA, or whichever RP-HPLC buffer is readily employed by the experimenter. For example, a 500 μL ligation will be brought up to 5 mL with RP-HPLC buffer. Add 15 molar equivalents of TCEP (15 times the number of moles of the larger of the two polypeptides used in the ligation reaction), shake to dissolve, and allow to rest for 15 minutes. Filter the crude reaction through a 0.22 μm single-use syringe filter.
-
Purify the crude ligation mixture by preparative HPLC on a suitable RP-HPLC column, such as Phenomenex Jupiter Proteo®, guiding the choice of gradient by the known elution time from the analytical HPLC runs. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
Very high product yields (>90%) are typically obtained from the ligation step. In most cases, for peptides that exhibit complete solubility in the reaction medium, we observe complete conversion to the product (>95%) and achieve about 70% total yield after RP-HPLC purification and lyophilization.
If the target protein contains tertiary structure and/or disulfide bonds in the final product, follow Steps 11–13 of Basic Protocol 1.
SUPPORT PROTOCOL 3
CHEMICAL SYNTHESIS OF C-TERMINAL HYDRAZIDE PEPTIDES BY FMOC-SPPS
This protocol describes a method for the chain assembly of a peptide bearing a C-terminal hydrazide moiety by Fmoc-SPPS using 2-chlorotrityl chloride resin (Figure 7). 2-chlorotrityl chloride resin degrades over time by hydrolysis but may be regenerated to its original loading by a number of methods (García-Martín, Bayó-Puxan, Cruz, Bohling, & Albericio, 2007; Redwan & Grotli, 2012). Once liberated from resin by treatment with concentrated TFA, the C-terminus of the peptide contains a C-terminal hydrazide. The choice for the C-terminal amino acid is entirely unrestricted, but as with all other NCL methods, proline and beta branched amino acids such as threonine, valine and isoleucine should be avoided as ligation sites.
Figure 7:
Method for the chain assembly of a peptide bearing a C-terminal hydrazide moiety by Fmoc-SPPS using 2-chlorotrityl chloride resin. Following treatment with hydrazine hydrate, unreacted sites are capped with methanol. The resin then proceeds to routine Fmoc-SPPS protocols and TFA cleavage.
Materials
2-chlorotrityl chloride resin (such as TentaGel® R TRT Cl Resin Rapp Polymere; loading: 0.15–0.22 mmol/g)
Dimethyl formamide (DMF)
Dichloromethane (DCM)
Hydrazine monohydrate (NH2NH2 ⋅ H2O)
Methanol (MeOH)
Diisopropylethylamine (DIEA)
0.4 M solution of HATU in DMF
Trifluoroacetic acid (TFA)
Triisopropylsilane (TIPS)
Milli-Q purified H2O
N2 gas
Diethyl ether
20 mL Disposable filter columns (such as Bio-Rad® Econo-Pac® Chromatography columns)
50 mL Falcon® centrifuge tubes
Benchtop centrifuge (Sorvall XTR or equivalent)
RP-HPLC buffer: 50:50 H2O/MeCN, containing 0.05% TFA
Reaction vessel for Solid Phase Peptide Synthesis (fritted glass resin support, PTFE stopcock)
Glass stir rod
Diaphragm vacuum pump
Erlenmeyer filter flask (2 L capacity)
Black rubber vacuum tubing
Protocol Steps
Weigh a 0.2 mmol scale quantity of 2-chlorotrityl chloride resin into a reaction vessel. Swell the resin in 10 ml of DMF for 1 hour.
Freshly prepare 8 ml of 5% (vol/vol) NH2NH2 in DMF by adding 0.4 mL of hydrazine hydrate to 7.60 mL of DMF.
Add 4 mL of the 5% NH2NH2 solution to the resin for hydrazination. Gently agitate the mixture for 45 minutes at room temperature using a stir rod and then drain the solution under vacuum. Add 10 ml of DMF to the resin, gently agitate for 10 seconds, and then drain it under vacuum. Repeat this process with the remaining hydrazine solution to ensure complete hydrazination.
Flow wash the resin with DMF for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Freshly prepare 4 mL of 10% (vol/vol) MeOH/DMF by combining 0.4 mL of MeOH with 3.6 mL of DMF. Add the mixture to the resin to cap the unreacted sites. Gently agitate for 10 minutes, and then drain it under vacuum.
Flow wash the resin with DMF for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
-
Synthesize the peptide under standard Fmoc-SPPS conditions. A standard cycle consists of 2 × 5 mL of 20% 4-methylpiperidine in DMF for 5 minutes each, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total). During the Fmoc deprotection periods, preactivate 5 equivalents of N-Fmoc-protected amino acid in 5 equivalents of 0.4 M HATU in DMF for 5 minutes. Following the DMF flow wash after the Fmoc deprotection, add 8 equivalents of DIEA to the activated amino acid mixture and add the total mixture to the resin for coupling for 20 minutes. Follow with a standard DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Typically, a peptide of ~50 residues will be broken up into 3 full days of manual couplings, as the total time for one cycle will be approximately 30 minutes. The use of an automated peptide synthesizer at this step will reduce the chain assembly time to approximately 1 day, depending on the length of the peptide sequence.
When the peptide sequence is finished, perform the final Fmoc deprotection by treatment with 2 × 5 mL of 20% 4-methylpiperidine in DMF for 5 minutes each, followed by a thorough DMF flow wash for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total).
Perform a thorough flow wash with DCM for 30 seconds (flow-through at the resin bed surface under vacuum, approx. 250 mL DMF total) to solvent-exchange the resin into DCM. Dry the resin under vacuum for 1 hour.
Cleave the peptide from the resin with 10 mL of a standard TFA cleavage cocktail such as 95% TFA, 2.5% TIPS, and 2.5% H2O for 2 hours. Filter the cocktail from the resin using a disposable filter column, collecting the filtrate in a 50 mL Falcon tube. Evaporate the majority of the TFA from the cleavage cocktail by applying N2 gas for 30 minutes, or until <2 mL of TFA remains. Precipitate the peptide by adding 10 mL of ice-cold diethyl ether. Centrifuge the suspension for 5 minutes @ 4000 rpm. Pour off the diethyl ether and dissolve the peptide pellet in an RP-HPLC buffer of choice, such as 50% MeCN in H2O containing 0.05% TFA.
Lyophilize the crude peptide from 50% MeCN in H2O containing 0.05% TFA, or whichever HPLC buffer is readily employed by the experimenter. Proceed to analytical RP-HPLC, product identification by ESI-MS, and purification by preparatory RP-HPLC. Pool the fractions containing the target product mass at the highest purity (>95%) and proceed to lyophilization.
REAGENTS AND SOLUTIONS
Use Milli-Q purified water or equivalent for the preparation of all buffers.
Ligation Buffer (Basic Protocols 1 and 2, Alternate Protocol 1)
6 M Guanidine-HCl
200 mM Na2HPO4
Adjust pH to 8.5 with a 1 M NaOH.
Store up to 1 year at room temperature.
Thz Deprotection Buffer (Alternate Protocol 1)
6 M Guanidine-HCl
500 mM methoxylamine-HCl (CH3ONH2-HCl)
The pH will be approximately 1.9 upon dissolving the Guanidine-HCl. Adjust with 1 M HCl.
Freshly prepare this buffer on the day it is to be used.
Ligation Buffer A (Basic Protocol 3)
6 M Guanidine-HCl
The pH will be approximately 3 upon dissolving the Guanidine-HCl. Adjust with 1 M HCl.
Store up to 1 year at room temperature.
Ligation Buffer B (Basic Protocol 3)
6 M Guanidine-HCl
200 mM Na2HPO4
Adjust pH to 8.5 with 1 M NaOH.
Store up to 1 year at room temperature.
Folding Buffer (Basic Protocol 1)
6 M Guanidine-HCl
5 mM DTT
Adjust pH to 7 with 1 M NaOH.
Freshly prepare this buffer on the day it is to be used.
Dialysis Buffer (Basic Protocol 1)
6 M Guanidine-HCl
1 mM DTT
Adjust pH to 7 with 1 M NaOH.
Freshly prepare this buffer on the day it is to be used.
Disulfide Formation Buffer (Basic Protocol 1)
0.5 M Guanidine-HCl
100 mM Tris-HCl
Adjust pH to 8 with 1 M NaOH.
Freshly prepare this buffer on the day it is to be used.
COMMENTARY
Background Information
The introduction of solid-phase peptide synthesis (SPPS) in 1963 by Bruce Merrifield has enabled the facile synthesis of an enormous variety of small to medium length bioactive peptides and proteins. However, as the length of the peptide chain grows, incomplete deprotection and acylation reactions beyond ~50 residues result in diminishing yields and purity. Therefore, in order to access larger peptide and proteins, convergent synthetic strategies have been developed that rely on stitching together smaller peptide fragments to build large proteins, a class of reactions known as chemical ligations. Early efforts relied on the use of fully protected peptides to eliminate side reactions and yielded an unnatural linkage at the ligation site, such as an ester, with the reaction being performed in organic solvent. The advent of Native Chemical Ligation (NCL) in 1994 by Dawson and Kent revolutionized the field by enabling the use of fully deprotected peptides in the ligation reaction, relying on the bioorthogonal reactivity of a thioester motif and selectivity for an N-terminal Cysteine residue. The first step involves the chemoselective reaction of the free thiol of the N-terminal Cysteine with an unprotected C-terminal thioester group. Following transthioesterification and spontaneous S-to-N acyl transfer, a native amide bond is formed at the ligation site. The reaction is performed at near-neutral pH in aqueous buffer in the presence of thiol additives, such as thiophenol and 4-mercaptophenylacetic acid (MPAA), to keep cysteine side chains reduced and to generate a more reactive phenyl thioester from the starting material alkyl thioester through thiol exchange. For basic-attacking thiols such as the N-terminal Cysteine thiol, the formation of the intermediate occurs more rapidly when the departing thiol has a lower pKa, as studied by Jencks (Hupe & Jencks, 1977). The pKa of the departing alkythioester is above 9, while the pKa of the arylthioester is approximately 6.6 for both thiophenol and MPAA, explaining the rapid reactivity of the exchanged fragment (P. E. Dawson, Churchill, Ghadiri, & Kent, 1997; Johnson & Kent, 2006). The thiol additives also serve to maintain a reducing environment to prevent disulfide bond formation of the critical N-terminal Cys residue. With this reaction, functional proteins and protein domains can be reliably synthesized from their peptide fragment building blocks.
Critical Parameters and Troubleshooting
As mentioned in each Basic Protocol, the final pH of the ligation reaction is a critical parameter for success. At a pH of 8 or higher, thioester hydrolysis may compete with ligation, as well as promote the oxidation of twin N-terminal Cys residues towards disulfide-bonded fragment dimers. At a pH of 6 or lower, the reaction is exceedingly slow resulting from protonation of the nucleophilic cysteine thiolate. An incorrect final pH is the most common cause of a failed NCL reaction.
An additional important factor for a successful ligation reaction is the concentration and solubility of the two peptide fragments. In the case of peptide fragments of equal length, an ideal starting point is 10 mg of each fragment per 1 mL of ligation buffer. Practically, this will yield a concentration of 2–10 mM of each fragment in the ligation buffer, depending on the length of the fragments (1000 Da = 10 mM, 5000 Da = 2 mM, etc.). It will be a rare case to work with a peptide fragment that is not soluble to a minimum of 1 mM in 6 M Guanidine buffer. In the case of a difficult fragment, sonication will typically aid solubility by breaking up hydrophobic peptide aggregates. If sonication does not help, the concentration of peptide can be lowered to 0.5 mM, with more time afforded to the ligation reaction (typically overnight for dilute ligations). Co-solvents such as DMSO should be avoided, in favor of simply lowering the fragment concentrations. For such exceedingly slow ligations, heating to 40°C usually helps speed up the reaction, but is not required in the majority of cases, where the recommended temperature is simply room temperature (as described in each Basic Protocol unit).
As mentioned in the materials lists for each Basic Protocol, the use of high-purity fragments is paramount to successful ligations. >95% pure material by RP-HPLC peak area integration and confirmation by ESI-MS should be used, with the experimenter performing multiple purifications of starting material peptides if necessary.
A previously-purified and stored N-terminal Cys peptide, if checked by RP-HPLC for purity on the day of a ligation, may present as two clean peaks: one corresponding to the correct material, and one corresponding to the disulfide dimer. If this is the case, fully reduce the disulfide species by adding 2 equivalents of TCEP to the peptide brought up in a suitable volume of RP-HPLC buffer, allow the mixture to sit for 10 minutes, then filter and re-purify the peptide on preparatory RP-HPLC (as described in Basic Protocol 1, Step 10).
Understanding Results
Native Chemical Ligation has been successfully applied to the chemical synthesis of thousands of proteins and protein domains. Across these studies, very high yields of the protein product are typically obtained in the ligation step, with moderate losses coming from handling and subsequent purification steps. In most cases, the ligation of 2 fragments will result in near-total exhaustion of each starting material and the appearance of a single product peak by HPLC. In cases where multiple fragments are to be ligated in sequence, the intermediate thiazolidine opening and purification steps will lead to losses in overall yield, in addition to the final protein folding steps.
Time Considerations
Thioester resin (Support Protocol 1) can be prepared in one half day. The time to prepare the thioester-containing peptide will depend on the length of the fragment of interest. Typically, a peptide of ~50 residues can be prepared in 3 full days by manual synthesis or in 1 day by an automated peptide synthesizer.
The chemical ligation steps of Basic Protocol 1, 2 and 3 typically reach >95% completion overnight. The subsequent purification steps may be performed in a matter of hours. The lyophilization of the purified peptide or protein typically takes 1 day depending on the total volume of liquid being freeze dried.
Figure 4:
Synthesis of Fmoc-3-amino-4-(methylamino)benzoic acid (Fmoc-MeDbz) (4).
ACKNOWLEDGEMENT
This work was funded by the National Institutes of Health (NIH R01 AI113867 and NIH R01 NS087070).
LITERATURE CITED
- Behrendt R, White P, & Offer J (2016). Advances in Fmoc solid-phase peptide synthesis. J Pept Sci, 22(1), 4–27. doi: 10.1002/psc.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canosa JB, & Dawson PE (2008). An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed Engl, 47(36), 6851–6855. doi: 10.1002/anie.200705471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canosa JB, Nardone B, Albericio F, & Dawson PE (2015). Chemical Protein Synthesis Using a Second-Generation N-Acylurea Linker for the Preparation of Peptide-Thioester Precursors. J Am Chem Soc, 137(22), 7197–7209. doi: 10.1021/jacs.5b03504 [DOI] [PubMed] [Google Scholar]
- Camarero JA, & Muir TW (2001). Native chemical ligation of polypeptides. Curr Protoc Protein Sci, Chapter 18, Unit18 14. doi: 10.1002/0471140864.ps1804s15 [DOI] [PubMed] [Google Scholar]
- Dang B, Kubota T, Mandal K, Bezanilla F, & Kent SB (2013). Native chemical ligation at Asx-Cys, Glx-Cys: chemical synthesis and high-resolution X-ray structure of ShK toxin by racemic protein crystallography. J Am Chem Soc, 135(32), 11911–11919. doi: 10.1021/ja4046795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson P, Muir T, Clark-Lewis I, & Kent S (1994). Synthesis of proteins by native chemical ligation. Science, 266(5186), 776–779. doi: 10.1126/science.7973629 [DOI] [PubMed] [Google Scholar]
- Dawson PE, Churchill MJ, Ghadiri MR, & Kent SBH (1997). Modulation of reactivity in native chemical ligation through the use of thiol additives. Journal of the American Chemical Society, 119(19), 4325–4329. doi:DOI 10.1021/ja962656r [DOI] [Google Scholar]
- Erlanson DA, Chytil M, & Verdine GL (1996). The leucine zipper domain controls the orientation of AP-1 in the NFAT·AP-1·DNA complex. Chemistry & Biology, 3(12), 981–991. doi: 10.1016/s1074-5521(96)90165-9 [DOI] [PubMed] [Google Scholar]
- García-Martín F, Bayó-Puxan N, Cruz LJ, Bohling JC, & Albericio F (2007). Chlorotrityl Chloride (CTC) Resin as a Reusable Carboxyl Protecting Group. QSAR & Combinatorial Science, 26(10), 1027–1035. doi: 10.1002/qsar.200720015 [DOI] [Google Scholar]
- Gentle IE, De Souza DP, & Baca M (2004). Direct production of proteins with N-terminal cysteine for site-specific conjugation. Bioconjug Chem, 15(3), 658–663. doi: 10.1021/bc049965o [DOI] [PubMed] [Google Scholar]
- Hackeng TM, Griffin JH, & Dawson PE (1999). Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proceedings of the National Academy of Sciences, 96(18), 10068–10073. doi: 10.1073/pnas.96.18.10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo H, Kwon Y, Kakuta Y, Tsuda S, Tanaka I, Hikichi K, & Aimoto S (1993). Development of a Linker with an Enhanced Stability for the Preparation of Peptide Thioesters and Its Application to the Synthesis of a Stable-Isotope-Labelled HU-Type DNA-Binding Protein. Bulletin of the Chemical Society of Japan, 66(9), 2700–2706. doi: 10.1246/bcsj.66.2700 [DOI] [Google Scholar]
- Hupe DJ, & Jencks WP (1977). Nonlinear structure-reactivity correlations. Acyl transfer between sulfur and oxygen nucleophiles. Journal of the American Chemical Society, 99(2), 451–464. doi: 10.1021/ja00444a023 [DOI] [Google Scholar]
- Jaradat DMM (2018). Thirteen decades of peptide synthesis: key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids, 50(1), 39–68. doi: 10.1007/s00726-017-2516-0 [DOI] [PubMed] [Google Scholar]
- Johnson EC, & Kent SB (2006). Insights into the mechanism and catalysis of the native chemical ligation reaction. J Am Chem Soc, 128(20), 6640–6646. doi: 10.1021/ja058344i [DOI] [PubMed] [Google Scholar]
- Kuzniewski CN, Gertsch J, Wartmann M, & Altmann KH (2008). Total synthesis of hypermodified epothilone analogs with potent in vitro antitumor activity. Org Lett, 10(6), 1183–1186. doi: 10.1021/ol800089x [DOI] [PubMed] [Google Scholar]
- Li HX, & Dong SW (2017). Recent advances in the preparation of Fmoc-SPPS-based peptide thioester and its surrogates for NCL-type reactions. Science China-Chemistry, 60(2), 201–213. doi: 10.1007/s11426-016-0381-1 [DOI] [Google Scholar]
- Muir TW, Sondhi D, & Cole PA (1998). Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci U S A, 95(12), 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muttenthaler M, Albericio F, & Dawson PE (2015). Methods, setup and safe handling for anhydrous hydrogen fluoride cleavage in Boc solid-phase peptide synthesis. Nat Protoc, 10(7), 1067–1083. doi: 10.1038/nprot.2015.061 [DOI] [PubMed] [Google Scholar]
- Pollock SB, & Kent SB (2011). An investigation into the origin of the dramatically reduced reactivity of peptide-prolyl-thioesters in native chemical ligation. Chem Commun (Camb), 47(8), 2342–2344. doi: 10.1039/c0cc04120c [DOI] [PubMed] [Google Scholar]
- Redwan IN, & Grotli M (2012). Method for activation and recycling of trityl resins. J Org Chem, 77(16), 7071–7075. doi: 10.1021/jo300598d [DOI] [PubMed] [Google Scholar]
- Schnolzer M, Alewood P, Jones A, Alewood D, & Kent SB (1992). In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res, 40(3–4), 180–193. [DOI] [PubMed] [Google Scholar]
- Schnölzer M, Alewood P, Jones A, Alewood D, & Kent SBH (2007). In Situ Neutralization in Boc-chemistry Solid Phase Peptide Synthesis. International Journal of Peptide Research and Therapeutics, 13(1–2), 31–44. doi: 10.1007/s10989-006-9059-7 [DOI] [PubMed] [Google Scholar]
- Tolbert TJ, & Wong C-H (2002). New Methods for Proteomic Research: Preparation of Proteins with N-Terminal Cysteines for Labeling and Conjugation This research was supported by the NIH (R37 GM44154). Angewandte Chemie International Edition, 41(12). doi: 10.1002/1521-3773(20020617)41:12<2171::Aid-anie2171>3.0.Co;2-q [DOI] [PubMed] [Google Scholar]
- Vila-Perello M, Liu Z, Shah NH, Willis JA, Idoyaga J, & Muir TW (2013). Streamlined expressed protein ligation using split inteins. J Am Chem Soc, 135(1), 286–292. doi: 10.1021/ja309126m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain M, Gaertner H, & Botti P (2003). Native Chemical Ligation with Aspartic and Glutamic Acids asC-Terminal Residues: Scope and Limitations. European Journal of Organic Chemistry, 2003(17), 3267–3272. doi: 10.1002/ejoc.200300032 [DOI] [Google Scholar]
- Wang JX, Fang GM, He Y, Qu DL, Yu M, Hong ZY, & Liu L (2015). Peptide o-aminoanilides as crypto-thioesters for protein chemical synthesis. Angew Chem Int Ed Engl, 54(7), 2194–2198. doi: 10.1002/anie.201408078 [DOI] [PubMed] [Google Scholar]