Abstract.
Zonal segmentation of the prostate gland using magnetic resonance imaging (MRI) is clinically important for prostate cancer (PCa) diagnosis and image-guided treatments. A two-dimensional convolutional neural network (CNN) based on the U-net architecture was evaluated for segmentation of the central gland (CG) and peripheral zone (PZ) using a dataset of 40 patients (34 PCa positive and 6 PCa negative) scanned on two different MRI scanners (1.5T GE and 3T Siemens). Images were cropped around the prostate gland to exclude surrounding tissues, resampled to and -score normalized before being propagated through the CNN. Performance was evaluated using the Dice similarity coefficient (DSC) and mean absolute distance (MAD) in a fivefold cross-validation setup. Overall performance showed DSC of 0.794 and 0.692, and MADs of 3.349 and 2.993 for CG and PZ, respectively. Dividing the gland into apex, mid, and base showed higher DSC for the midgland compared to apex and base for both CG and PZ. We found no significant difference in DSC between the two scanners. A larger dataset, preferably with multivendor scanners, is necessary for validation of the proposed algorithm; however, our results are promising and have clinical potential.
Keywords: prostate cancer, magnetic resonance imaging, convolutional neural net, zonal segmentation
1. Introduction
Prostate cancer (PCa) is one of the leading causes of death from cancer worldwide.1 An early and accurate diagnosis improves the survival rate and reduces the costs of treatment.2 The use of magnetic resonance imaging (MRI) plays an essential role in the diagnosis of PCa patients to support biopsies, and for treatment decision and planning.3,4 A typical prostate MRI examination consists of an anatomical T2-weighted (T2W) imaging sequence combined with one or more functional imaging sequences.4 Usually, the examination is performed on a 1.5T or 3T MRI scanner with or without the use of an endorectal coil (ERC).5 The T2W sequence is used to determine the anatomical boundaries between the prostate zones and extend of tumor growth, e.g., extraprostatic extension or growth into the seminal vesicles.4 The functional sequences are especially advantageous in PCa detection, evaluation of treatment response, and staging.5
The prostate consists of four zones: the peripheral zone (PZ), transitional zone (TZ), central zone (CZ), and fibromuscular stroma.6 TZ and CZ are often considered as one central gland (CG) on MRI, as the zones exhibit similar imaging appearances.7
Segmentation of the prostate from surrounding tissues is useful, and sometimes even necessary, for a variety of clinical purposes, such as volume determination, MRI/ultrasound fusion systems for guided biopsies, lesion detection and scoring, accurate target delineation for radiotherapy treatment, or to obtain the region of interest for computer-aided detection of PCa.8–10 Manual delineation of the prostate on MRI is laborious and a time-consuming task with high risk of inter- and intraobserver variability.8 Automatic prostate segmentation algorithms have been an active research field for many years as they can greatly enhance the workflow in the clinic and reduce the subjectivity.8,11
Lately, there has been more focus on zonal segmentation of the prostate.12 The majority () of PCas occurs in the PZ; an accurate location of the PCa within the zones is extremely important and favors outcome.2,13,14 Furthermore, PCa located in the CG have significantly different biological behaviors compared to those in the PZ and may therefore be referred to different treatment options.7,15
Automatic zonal segmentation of the prostate is still relatively sparse in the literature. Initially, Makni et al. performed zonal prostate segmentation by classifying voxels as either PZ or CG using a -means clustering algorithm. Makni et al. yielded an average dice similarity coefficient (DSC) of 0.84 for CG and 0.73 for PZ.16 Litjens et al. used anatomical, textural, and intensity image features to segment the prostate into CG and PZ with promising results.17 However, the methods presented by Makni et al. and Litjens et al. required an initial manual segmentation of the whole gland (WG) as input, which is time-consuming and impractical. Toth et al. used an active appearance model (AAM) approach to segment the zones of the prostate using images from a 3T MR scanner using an ERC. The DSC achieved by this approach was 0.79 for the CG and 0.68 for the PZ.18 In a study by Chi et al., DSC of 0.83 and 0.52 were obtained for CZ and PZ, respectively, using a Gaussian mixture model in combination with atlas probability maps on 3T MRI examinations from eight patients.19 Chilali et al. investigated a voxel classification method combined with an atlas-based approach for zonal segmentation that resulted in DSC of 0.7 for TZ and 0.62 for PZ.20 The studies presented above all used anatomical MRI for the zonal segmentation. A semiautomatic atlas-based approach for WG and TZ segmentation using a functional imaging sequence, diffusion weighted (DWI), was presented by Zhang et al. with DSC of 0.85 (WG) and 0.77 (TZ).21 Another study used DWI for prostate zonal segmentation using a deep convolutional neural network (CNN) and obtained DSC of 0.93 and 0.88 for WG and TZ, respectively.22
CNNs have recently found great success achieving state-of-the-art performances for automatic medical image segmentation.23 These CNNs learn increasingly more complex features from the training images using multiple layers of adjustable filters taking into account the spatial information of the input. Usually, these networks require large amount of training data to learn and select features. Within medical imaging tasks, this is often beyond reach.24 The U-net, designed by Ronneberger et al., is a CNN architecture that has been developed for fast and precise segmentation of medical images and has shown great potential even with small datasets.25 Zhu et al., for example, did WG prostate segmentation using U-net on T2W images obtained by a 3T scanner from 80 patients and obtained a mean DSC of 0.865.26
CNNs have shown promising results within medical image segmentation but have not been investigated for zonal segmentation. In this study, we proposed a zonal segmentation algorithm using T2W MRI on prostate examinations from two different scanners [GE 1.5 (with ERC) and Siemens 3T] using a modified U-net architecture.
2. Methods
2.1. Data
The dataset used in this study is publicly available, published by Lemaître et al.27 The dataset contains multiparametric MRI examinations from a total of 40 patients with elevated PSA level, scanned on two different scanners—a General Electric (GE) 1.5T scanner (21 patients) with ERC, and a Siemens 3T scanner (19 patients). All MRI examinations include a T2W imaging sequence (and one or more functional sequences) along with ground truth (GT) images of WG, CG, and PZ made on the T2W image sequence performed by an experienced radiologist. The voxel spacing varied between patients, from 0.27 to 0.78 mm, with slice thickness of 3 mm for GE and 1.5 mm for Siemens. Of the 40 patients, six were PCa negative and 34 were PCa positive with varying lesion size (mean 4 cc, range 0 to 36 cc) and location (3 with CG lesion, 26 with PZ lesion, and 5 with CG and PZ lesion). Only the axial T2W image sequence was used for this study.
2.2. Preprocessing
In the dataset, the PZ GT delineation was obtained by subtracting the CG GT from the WG GT. Because the radiologist did not delineate the CG on all slices and relied on the interpolation performed by clinical system, the PZ GT equals the WG GT on some slices, see Fig. 1. Linear interpolation of the CG GT images was used to correct for this in order to have GT delineation on all slices for training.
Fig. 1.
Interpolation of GT images to account for the missing CG on some slices, (a) before and (b) after. White area is GT for CG and gray area is PZ.
Voxel dimensions and slice thickness differ between patients and were therefore resampled to using bicubic interpolation. Afterward, the images were cropped, to remove some of the surrounding tissues. As the size of the prostate varies between patients, the crop size and place were found based on the largest prostate in the dataset which resulted in an image size of (). Furthermore, image slices not containing GT delineation from either CG or PZ were removed. After resampling and cropping, the total number of image slices was 3015 (1842 for GE and 1173 for Siemens scanner). All images were -score normalized (zero mean and unit variance) to account for interpatient and interscanner variations. Figure 2 shows an example of an image slice after preprocessing with GT delineation of CG and PZ.
Fig. 2.
GT for patient 28 after preprocessing, yellow dashed line = GT for CG and white dashed line = GT for PZ.
2.3. Network Architecture
The original U-net architecture is shown in Fig. 3. The U-net is a fully convolutional network with skip connections which propagate the spatial location and combine it with coarse semantic information. The network has a contracting left side and an expansive right side. The contracting part resembles a typical CNN architecture, where convolutional layers ( kernels), with activation function units [rectified linear unit (ReLU)], are followed by a downsampling pooling layer ( pooling, strides = 2). After each downsampling layer, the number of feature channels doubles. The expansive part, which is almost symmetric to the contracting part, uses upconvolutions and combines features and spatial information from the contracting path. The input images are fed to the network and propagated through the network along all possible paths. The output is a segmentation map with the number of channels equal to the number of classes in the segmentation task.25
Fig. 3.
The original U-net architecture.25
To our implementation of the U-net some modifications were made; to keep the output prediction images of same size as the input images, zero padding was used in this study. Furthermore, we implemented batch normalization layers before max pooling layers in the contracting part of the network. Using batch normalization layers can significantly accelerate the training by normalizing the input distributions.28 The hyperparameters used for our implementation include: initial random weights (glorot_uniform), activations (elu), batch size (12), epochs (500), and optimization algorithm (Adam with default values and ) for the calculation of weight updates with initial learn rate ().
As loss function, a pixelwise cross entropy loss was used. The loss was weighted based on the class probability, to account for class imbalance. Class weights used = {CG:4.29, PZ:3.9, Background:1}. The weights were estimated from the dataset.
Data augmentation was applied on the fly on the training set, while validation and test sets were unchanged. The augmented images represent natural variations of the prostate and included horizontal flip, small rotations, width and height shift, zoom, and shearing.
2.4. Evaluation Metric
The model performance was evaluated using DSC and mean absolute distance (MAD) as measure. DSC and distance-based measures are widely used metrics within the field of medical image segmentation as evaluation measure. DSC is a spatial overlap index calculated as two times the intersection between the regions, divided by the sum of the two individual regions
| (1) |
MAD measures average minimum distance between two boundaries calculated as
| (2) |
where expresses the average minimum distance from all points on to .
In addition to overall DSC and MAD for CG and PZ, DSCs, and MADs are presented for prostate apex (most caudal aspect of the gland), base (most cranial aspect), and mid (sandwiched in between the apex and base). We defined the apex and base as the first and last 30% of the prostate in the -direction (superior-to-inferior direction), respectively, and prostate mid accounts for the remaining 40% as suggested in Ref. 29, see Fig. 4. Before calculation of DSC and MAD, postprocessing of the segmentations was done with the assumption that each zone only consists of one object. The postprocessing step removed small objects not connected to the largest three-dimensional (3-D) object in each patient for each zone.
Fig. 4.
Zonal and regional division of the prostate gland. The PZ extends from the base to the apex of the prostate. The CZ and transitional zone (TZ) together represent the CG. On the anterior surface, the anterior fibromuscular stroma is located. First 30% of the image sequence is apex, 40% mid-gland, and the last 30% for base. Modified from Ref. 30.
2.5. Validation
A stratified fivefold cross-validation setup was implemented for model evaluation, as depicted in Fig. 5. The dataset was randomly divided into five sets, stratified according to scanner (1.5 GE or 3T Siemens) which resulted in 32 patients in the training set, and eight patients in the test set for each fold. From the training set, two patients (one from each scanner) were used for validation during the training phase.
Fig. 5.
Fivefold stratified cross-validation setup used in this study. From the 40 patients, 32 patients are used for training and 8 for testing (marked in orange) for each fold. From the 32 training patients, 2 are used for validation (marked in red) during the training phase.
2.6. Implementation Details
All the experiments ran on a 64-bit Ubuntu 16.04 workstation with Intel Xeon E5620, 2.40 GHz CPU, and 12 GB of RAM. Preprocessing of the images was done in Matlab 2017b. After preprocessing, the images were imported into the open-source deep learning library Keras with Tensorflow as backend in Python 3.4. Training and testing of the network was done on a NVIDIA Geforce GTX1070 GPU with 8 GB of memory.
3. Results
The performance of the CNN multiclass segmentation was assessed quantitatively using DSC for overall performance, for the two scanners and for each region, and lastly by visual assessment.
The total number of trainable parameters in our implementation of U-net was 31,032,643. The computational time for training the network was for 500 epochs for each fold and the segmentation of one testing patient took less than 1 s, including preprocessing.
In Table 1, the overall mean DSC and MAD for all patients are presented, together with slice maximum DSC, maximum patient DSC, and minimum patient MAD, for CG and PZ. Overall mean DSC for CG was 0.794 and 0.692 for PZ. For CG the MAD was 3.349 mm and 2.992 mm for PZ. For most patients (34/40), the DSC was higher for the CG than for the PZ. The highest achieved DSC for a single slice was 0.992 and 0.991 for CG and PZ, respectively. Lowest patient MADs were 0.681 mm for CG and 0.695 mm for PZ.
Table 1.
Overall MAD and dice similarity coefficient (DSC) results for zonal segmentation of the prostate gland using T2W images. Overall DSC and MAD together with maximum slice and patient DSC, and minimum MAD are presented for the CG and PZ.
| Zone | MAD | DSC | |||
|---|---|---|---|---|---|
| Overall (mm) (std) | Minimum patient MAD (mm) | Mean DSC (std) | Maximum slice DSC | Maximum patient DSC | |
| CG | 3.349 () | 0.681 | 0.794 () | 0.992 | 0.9837 |
| PZ | 2.992 () | 0.695 | 0.692 () | 0.991 | 0.9669 |
One patient, number 15, obtained the highest patient DSC for both CG and PZ with close resemblance to the GT delineation. A coronal view of the GT and segmentation is shown in Fig. 6. The only visual difference between the GT and segmentation on the coronal view is a smoother contour from the segmentation compared to GT.
Fig. 6.
Patient 15, coronal view, and slice 60. Red solid line = predicted CG, yellow dashed line = GT CG, blue solid line = predicted PZ, and white dashed line = GT PZ.
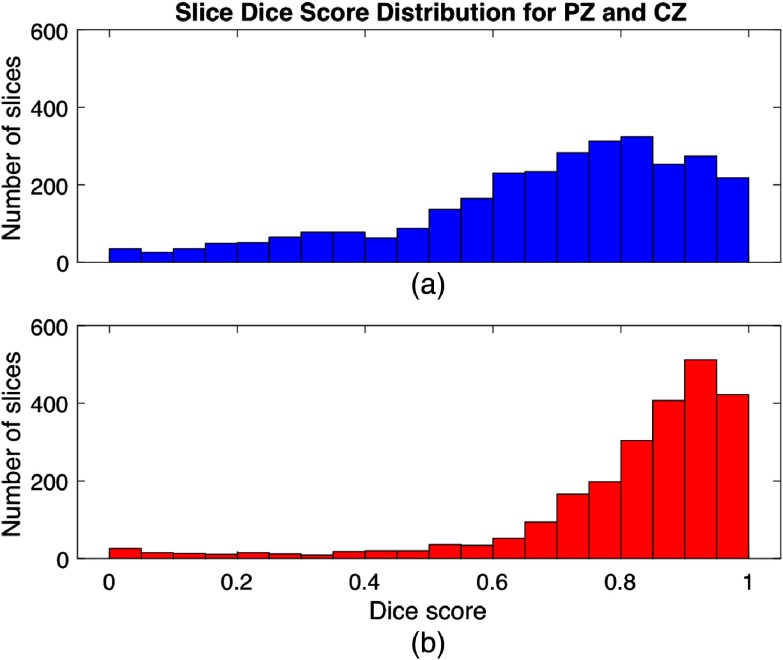
The distribution of slices with respect to DSC is shown in Fig. 7, with all DSCs represented but with most slices in the high end of the DSC scale for both CG and PZ. For the CG, the DSCs are generally higher than for the PZ. Examples of slices with low DSC will be presented and explained later in Sec. 3.3.
Fig. 7.
Histogram showing the number of slices versus dice score values for (a) PZ and (b) CG.
3.1. Comparison Between 1.5T GE and 3T Siemens Scanner
Variations in MRI protocol and scanner (field strength and vendor) can have large influence on algorithms for image processing. Therefore, we investigated differences in DSCs obtained from the data from the two different scanners, 1.5 T GE and 3 T Siemens, used for this study. In Table 2, the results for the two scanners are presented. The 1.5 T GE scanner showed slightly higher DSC for both zones; however, the difference was not statistically significant (-value ). No significance difference was found for MADs between the two scanners (-value ).
Table 2.
Results for prostate zonal segmentation for the two different scanners, GE and Siemens.
| Scanner | 1.5 T GE | 3 T Siemens | ||
|---|---|---|---|---|
| Zone | Central gland | Peripheral zone | Central gland | Peripheral zone |
| MAD (mm) (std) | 3.431 () | 2.985 () | 3.257 () | 3.000 () |
| Mean DSC (std) | 0.808 () | 0.694 () | 0.778 () | 0.690 () |
3.2. Regions Apex, Mid, and Base
At the apex and base of the prostate, the boundary of the gland is poorly defined with less discriminating gray level gradients and large shape variations of the regions.31,32 Results for the three regions are presented in Table 3. The mid-region shows the highest DSCs and lowest MADs for both zones and the prostate base the lowest DSC and highest MAD. There was a statistically significant difference between DSC for the mid-region and both apex and base in both zones (). No statistically significant difference was found for apex and base for neither CG nor PZ. The prostate base had significantly larger MAD compared to mid-gland in CG () and significantly larger MAD compared to apex and mid-gland in PZ ().
Table 3.
Results for each prostatic region in each zone.
| Region | Apex | Mid | Base | |||
|---|---|---|---|---|---|---|
| Zone | Central gland | Peripheral zone | Central gland | Peripheral zone | Central gland | Peripheral zone |
| MAD (mm) (std) | 3.224 () | 2.762 () | 2.671 () | 2.662 () | 3.663 () | 4.013 () |
| Mean DSC (std) | 0.718 () | 0.665 () | 0.859 () | 0.755 () | 0.736 () | 0.630 () |
3.3. Visual Assessment
Visual inspection of the output from the network showed some segmentation results close to the GT, like those shown in Fig. 8. Both zones show the correct size and shape as the GT, though with some differences, mostly for the PZ. For the boundary between CG and PZ in patient 5 [Fig. 8(a)], the network predicts the boundary to be located slightly lower (toward the rectum) than the GT, but still ensures that the boundaries of CG and PZ are closely aligned.
Fig. 8.
Examples of high dice scores from (a) patient 5 (GE scanner) and (b) patient 32 (Siemens scanner). Red solid line = predicted CG, yellow dashed line = GT CG, blue solid line = predicted PZ, and white dashed line = GT PZ.
Some slices showed zero or very low DSC, see Fig. 9. For example, patient 18 in Fig. 9(a) presents DSC of 0.000 for both zones. For this particular patient, the network predicts only a PZ, where there is actually CG and PZ. This is the case for several slices in -direction. Figure 9(b) shows slice 103 for the same patient where all of the prostate now belongs to PZ as the network also predicts, resulting in DSC of 0.890 for PZ.
Fig. 9.
Examples of low dice scores. (a, b) Patient 18 from GE scanner and (c, d) patient 28 from Siemens scanner. Red solid line = predicted CG, yellow dashed line = GT CG, blue solid line = predicted PZ, and white dashed line = GT PZ.
In patient 28 [Fig. 9(c)], the network predicts both CG and PZ where there is only a PZ for several slices. Nine slices away [Fig. 9(d)], the PZ divides into being partly PZ and CG, which is also predicted by the network. The cases shown in Fig. 9 are seen for several patients and slices, where the network struggles in the transition from one zone to two, or vice versa.
Examples where the network failed to do a satisfying segmentation was also seen during the visual inspection. Figures 10(a) and 10(b) show two examples where the segmentation result does not resemble the GT neither in term of size or shape of the two zones.
Fig. 10.
Examples of unsatisfactory segmentations. (a) Patient 23 from Siemens scanner and (b) patient 26 from Siemens scanner. Red solid line = predicted CG, yellow dashed line = ground truth CG, blue solid line = predicted PZ, and white dashed line = ground truth PZ.
4. Discussion and Conclusion
Accurate segmentation of the prostatic zones is important as the behavior of PCa lesions strongly correlates with its zonal location. Automatic segmentation of the zones can resolve the intra- and interobserver variability and reduce the time for manual delineation. Such automatic algorithms must be robust toward differences in the acquisition protocol, use of ERC, and magnetic field strength of the MRI scanner. In this study, we investigated a CNN for zonal segmentation using T2W MRI from 40 patients, scanned on two different scanners [1.5 T GE (with ERC) and 3 T Siemens] in a fivefold CV setup.
The overall performance of our network, DSC of 0.794 for CG and 0.692 for PZ, showed promising and comparable results to the current literature. Toth et al. obtained DSC of 0.79 for CG and 0.68 for PZ, which is very close to our results; however, they relied on a manual delineation of the WG as input for their multiple-levelset AAM approach and only included MR images acquired from one 3 T scanner. Toth et al. also investigated their approach without the use of a manual delineation of the WG and obtained DSC of 0.72 for CG and 0.6 for PZ with MADs of around 1.9 mm for CG and 1.3 for PZ which is lower MADs compared to our work (3.349 and 2.992 mm for CG and PZ, respectively).18 Qiu et al. investigated a global optimization-based approach for WG and zonal segmentation and achieved DSC of 0.823 and 0.693 for CG and PZ, respectively.33 They only used data from 15 patients scanned on a single 3T GE machine, but with promising results.
We found a higher DSC for the CG than the PZ for most patients, which is consistent with all the previously published studies found in the literature on zonal segmentation.18,19,34 This might be due to the PZ having more natural shape variations than CG and deformations caused by the ERC and differences in rectal filling.35 Even though the DSC obtained in this study is higher for CG than PZ, we like the study by Qui et al. achieved a larger MAD for CG than for PZ.33
We did not investigate the influence of PCa lesion size and location in this study. Since most PCa are located in the PZ, this could have affected the segmentation performance in this zone. PCa lesions can alter both prostate shape and appearance on MRI; however, an automatic segmentation method should be robust to these variations to be clinically useful.
Comparison of the results obtained for the two scanners used in this study showed no significant difference, suggesting that the network is robust against acquisition platform. Some of the earlier work on prostate WG/zonal segmentation have used data from only one scanner and might therefore not work on data from different scanners. Using a dataset with more than one vendor and field-strength scanner allow for more generic and robust algorithms.2,27
We found lower DSCs for apex and base compared to the mid-gland, which is consistent with all submitted algorithms in the PROMISE12 challenge on WG segmentation, and the zonal segmentation studies by Qiu et al.8,33,34 Due to unclear boundaries and partial volume effects, the segmentation of apex and base is more challenging, also for radiologists.26,33 Since the apex and base are close to sensitive organs during, for example, radiation therapy these regions are especially important to report for segmentation algorithms.36
We chose to implement a modified version of the U-net architecture for our zonal segmentation approach because it has proven useful for medical image segmentation even with a limited amount of data.25 Comparing our two-dimensional (2-D) input approach to the 3-D version of the U-net (3-D U-net) or the V-net could be interesting.37,38 The 3-D version could potentially be better at capturing the spatial context; however, processing 3-D images increases the complexity and computational expenses of the training process. One study investigated different 2-D and 3-D CNN architectures for cardiac MRI segmentation and found substantially higher DSCs for all evaluated 2-D architectures.39 Another approach to better capture spatial context is the use of recurrent neural networks, where the current input is depended on the previous one.26
Several hyperparameters can be optimized for CNNs, such as learning rate, optimization function, number of epochs, and batch size. Improvement in network performance might be seen if the parameters are optimized; however, these hyperparameters are believed to be of secondary importance, compared to network architecture and preprocessing.40 Potential future network experimentations could investigate the use of a dice loss function instead of the weighted pixelwise cross entropy we used for this study.
In our study, we did image augmentation in order to increase the amount of training data, minimize overfitting and to mimic shape and spatial location variations which could be present in the test set. Apart from horizontal flip, rotations, zoom, shear, and small height and width shifts as we used for the current study, other augmentations could be investigated such as histogram equalization or adding Gaussian noise. Another augmentation strategy could be oversampling the CG and PZ to account for the class imbalance.
We chose to use the T2W image sequence for our zonal segmentation, as it offers the best differentiation among the zones and visualization of prostate boundaries.2,14 One study, by Clark et al. did zonal segmentation on DWI with promising DSCs.22 Using multiple inputs for the CNN, e.g., T2W and DWI could potentially improve the results by adding additional information to the network. However, this would require an accurate coregistration to account for geometrical mismatch due to patient movement and/or bladder/rectum filling during image acquisition.
We acknowledge some limitations to this study; first, the delineation of the GT was only done by one radiologist. Since the task of prostate delineation is subjected to interobserver variability, a future study should include GT delineations from multiple radiologists to assess the variability, especially around the prostate apex and base.36 Second, the fixed crop size and location of the images were visually assessed from the dataset by determining the smallest possible box wherein all the prostates could fit in. To use a different dataset with our approach, one would have to validate that all prostates are actually within the cropping box. Since slices that do not include prostate gland were removed from the dataset as part of the preprocessing, we do not know the ability of the network to do segmentation in slices without any prostate tissue. For future studies, slices without prostate gland should be included in the dataset and the class probability weights adjusted accordingly.
As six of the patients were PCa negative and the CV setup was not stratified according to PCa positive/negative patients, there is a risk that not all folds will contain a patient without PCa, which can have a negative effect on the performance of the model. Optimally, the CV scheme had been performed, e.g., 5 to 10 times to account for this; however, this is very time demanding and not implemented in this study.
Lastly, a clear limitation of this study was the sample size. Forty patients from two different scanners do not include enough variation for the network to be used in the clinical workflow. A larger patient population, preferably with more than one radiologist to make the GT and more scanner vendors and image protocols, is warranted as validation to evaluate the true performance of the model.
This work presents preliminary results to support the use of CNN for zonal prostate segmentation. In conclusion, our results show that the U-net architecture can be used for prostate zonal segmentation with promising results comparable to the current literature. More work on the subject is necessary, ideally with data from multiple scanners and more than one experienced physician to do the validation GT.
Acknowledgments
The authors of this paper would like to acknowledge Lemaître et al. for making the dataset publicly available.
Biography
Biographies for the authors are not available.
Disclosures
No conflict of interest to report, neither financial or otherwise.
References
- 1.Miller K. D., et al. , “Cancer treatment and survivorship statistics, 2016,” CA Cancer J. Clin. 66(4), 271–289 (2016). 10.3322/caac.v66.4 [DOI] [PubMed] [Google Scholar]
- 2.Liu L., et al. , “Computer-aided detection of prostate cancer with MRI: technology and applications,” Acad. Radiol. 23(8), 1024–1046 (2016). 10.1016/j.acra.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhudd A., et al. , “The role of the multiparametric MRI in the diagnosis of prostate cancer in biopsy-naïve men,” Curr. Opin. Urol. 27(5), 488–494 (2017). 10.1097/MOU.0000000000000415 [DOI] [PubMed] [Google Scholar]
- 4.Weinreb J. C., et al. , “PI-RADS prostate imaging—reporting and data system: 2015, version 2,” Eur. Urol. 69(1), 16–40 (2016). 10.1016/j.eururo.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankineni S., Osman M., Choyke P. L., “Functional MRI in prostate cancer detection,” Biomed. Res. Int. 2014, 1–8 (2014). 10.1155/2014/590638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilanova J. C., et al. , Atlas of Multiparametric Prostate MRI: With PI-RADS Approach and Anatomic-MRI-Pathological Correlation, Springer International Publishing, Cham: (2017). 10.1007/978-3-319-61786-2 [DOI] [Google Scholar]
- 7.Viswanath S. E., et al. , “Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 tesla endorectal, in vivo T2-weighted magnetic resonance imagery,” J. Magn. Reson. Imaging 36(1), 213–224 (2012). 10.1002/jmri.23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litjens G., et al. , “Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge,” Med. Image Anal. 18(2), 359–373 (2014). 10.1016/j.media.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., et al. , “3D segmentation of the prostate via Poisson inverse gradient initialization,” in Int. Conf. Image Process., Vol. 5, pp. 25–28 (2007). 10.1109/ICIP.2007.4379756 [DOI] [Google Scholar]
- 10.Park S. H., et al. , “Interactive prostate segmentation using atlas-guided semi-supervised learning and adaptive feature selection,” Med. Phys. 41(11), 111715 (2014). 10.1118/1.4898200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschner M., Jung F., Wesarg S., “Automatic prostate segmentation in MR images with a probabilistic active shape model,” in MICCAI Grand Challenge: Prostate MR Image Segmentation (2012). [Google Scholar]
- 12.Garg G., Juneja M., “Cancer detection with prostate zonal segmentation: a review,” Lect. Notes Network Syst. 24, 829–835 (2018). 10.1007/978-981-10-6890-4 [DOI] [Google Scholar]
- 13.Lee J. J., et al. , “Biologic differences between peripheral and transition zone prostate cancer,” Prostate 75(2), 183–190 (2015). 10.1002/pros.v75.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmeri M. L., et al. , “B-mode and acoustic radiation force impulse (ARFI) imaging of prostate zonal anatomy: comparison with 3T T2-weighted MR imaging,” Ultrason. Imaging 37(1), 22–41 (2015). 10.1177/0161734614542177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vargas H. A., et al. , “Normal central zone of the prostate and central zone involvement by prostate cancer: clinical and MR imaging implications,” Radiology 262(3), 894–902 (2012). 10.1148/radiol.11110663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makni N., et al. , “Zonal segmentation of prostate using multispectral magnetic resonance images,” Med. Phys. 38(11), 6093–6105 (2011). 10.1118/1.3651610 [DOI] [PubMed] [Google Scholar]
- 17.Litjens G., et al. , “A pattern recognition approach to zonal segmentation of the prostate on MRI,” Lect. Notes Comput. Sci. 7511, 413–420 (2012). 10.1007/978-3-642-33418-4_51 [DOI] [PubMed] [Google Scholar]
- 18.Toth R., et al. , “Simultaneous segmentation of prostatic zones using active appearance models with multiple coupled levelsets,” Comput. Vision Image Understanding 117(9), 1051–1060 (2013). 10.1016/j.cviu.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi Y., et al. , “A compact method for prostate zonal segmentation on multiparametric MRIs,” Proc. SPIE 9036, 90360N (2014). 10.1117/12.2043334 [DOI] [Google Scholar]
- 20.Chilali O., et al. , “Gland and zonal segmentation of prostate on T2W MR images,” J. Digital Imaging 29(6), 730–736 (2016). 10.1007/s10278-016-9890-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., et al. , “A local ROI-specific atlas-based segmentation of prostate gland and transitional zone in diffusion MRI,” J. Comput. Vision Imaging Syst. 2(1), 2–4 (2016). 10.15353/vsnl.v2i1.113 [DOI] [Google Scholar]
- 22.Clark T., et al. , “Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks,” J. Med. Imaging 4(4), 041307 (2017). 10.1117/1.JMI.4.4.041307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G., et al. , “Interactive medical image segmentation using deep learning with image-specific fine-tuning,” IEEE Trans. Med. Imaging 37(7), 1562–1573 (2018). 10.1109/TMI.2018.2791721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen D., et al. , “Deep learning in medical image analysis,” Annu. Rev. Biomed. Eng. 19(1), 221–248 (2017). 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 26.Zhu Q., et al. , “Exploiting inter-slice correlation for MRI prostate image segmentation, from recursive neural networks aspect,” Complexity 2018, 4185279 (2018). 10.1155/2018/4185279 [DOI] [Google Scholar]
- 27.Lemaître G., et al. , “Computer-aided detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review,” Comput. Biol. Med. 60, 8–31 (2015). 10.1016/j.compbiomed.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 28.Ioffe S., Szegedy C., “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in Proc. 32nd Int. Conf. Mach. Learn. (2015). [Google Scholar]
- 29.Mahdavi S. S., et al. , “Semi-automatic segmentation for prostate interventions,” Med. Image Anal. 15(2), 226–237 (2011). 10.1016/j.media.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäderling F., “Preoperative local staging of prostate cancer aspects on predictive models, magnetic resonance imaging and interdisciplinary teamwork,” Karolinska Institutet; (2017). [Google Scholar]
- 31.Yan P., et al. , “Adaptively learning local shape statistics for prostate segmentation in ultrasound,” IEEE Trans. Biomed. Eng. 58(3), 633–641 (2011). 10.1109/TBME.2010.2094195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vikal S., et al. , “Prostate contouring in MRI guided biopsy,” Proc. SPIE 7259, 72594A (2009). 10.1117/12.812433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu W., et al. , “Efficient 3D multi-region prostate MRI segmentation using dual optimization,” Inf. Process. Med. Imaging 23, 304–315 (2013). 10.1007/978-3-642-38868-2 [DOI] [PubMed] [Google Scholar]
- 34.Qiu W., et al. , “Dual optimization based prostate zonal segmentation in 3D MR images,” Med. Image Anal. 18(4), 660–673 (2014). 10.1016/j.media.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 35.Lotfi M., et al. , “Evaluation of the changes in the shape and location of the prostate and pelvic organs due to bladder filling and rectal distension,” Iran. Red Crescent Med. J. 13(8), 564–573 (2011). [Google Scholar]
- 36.Shahedi M., et al. , “Accuracy validation of an automated method for prostate segmentation in magnetic resonance imaging,” J. Digit. Imaging 30(6), 782–795 (2017). 10.1007/s10278-017-9964-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Çiçek Ö., et al. , “3D U-net: learning dense volumetric segmentation from sparse annotation,” Lect. Notes Comput. Sci. 9901, 424–432 (2016). 10.1007/978-3-319-46723-8 [DOI] [Google Scholar]
- 38.Milletari F., Navab N., Ahmadi S.-A., “V-net: fully convolutional neural networks for volumetric medical image segmentation,” in Int. Conf. 3D Vision, pp. 1–11 (2016). 10.1109/3DV.2016.79 [DOI] [Google Scholar]
- 39.Baumgartner C. F., et al. , “An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation,” Lect. Notes Comput. Sci. 10663, 111–119 (2018). 10.1007/978-3-319-75541-0 [DOI] [Google Scholar]
- 40.Litjens G., et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal. 42, 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]