Abstract
Streptococcus mutans (S. mutans) is the primary etiological agent of dental caries. The S. mutans enzyme sortase A (SrtA) is responsible for anchoring bacterial cell wall surface proteins involved in host cell attachment and biofilm formation. Thus, SrtA is an attractive target for inhibiting dental caries caused by S. mutans-associated acid fermentation. In this study, we observed that astilbin, a flavanone compound extracted from Rhizoma Smilacis Glabrae, has potent inhibitory activity against the S. mutans SrtA, with an IC50 of 7.5 μg/mL. In addition, astilbin was proven to reduce the formation of biofilm while without affecting the growth of S. mutans. The results of a molecular dynamics simulation and a mutation analysis revealed that the Arg213, Leu111, and Leu116 of SrtA are important for the interaction between SrtA and astilbin. The results of this study demonstrate the potential of using astilbin as a nonbactericidal agent to modulate pathogenicity of S. mutans by inhibiting the activity of SrtA.
Keywords: Streptococcus mutans, astilbin, sortase A, biofilm, antivirulence
1. Introduction
Dental caries is a common chronic disease that causes considerable anxiety, pain, tooth loss, malnutrition, and even disability across all age groups [1]. The disease process mainly involves bacteria, including Streptococcus mutans, Streptococcus sanguis, Streptococcus sobrinus, Lactobacillus spp., and Streptococcus oralis [2,3]. Among these bacteria, the Gram-positive bacterium S. mutans is the primary etiological agent of human dental caries [4], while also infecting cardiac endothelial cells and causing infective endocarditis [5]. S. mutans uses two adhesion mechanisms—sucrose-independent adhesion and sucrose-dependent adhesion. In the presence of sucrose, cell-wall-associated glucosyltransferases convert extracellular sucrose into glucan, which, together with glucan-binding proteins (Gbps) on the bacterial surface, facilitate cell–cell aggregation and the subsequent formation of dental biofilms on the tooth surface [6]. In the absence of sucrose, the adhesion of S. mutans to the dental surface, or other bacteria in dental plaque, is mediated by several surface adhesins. One of the primary adhesins of S. mutans is streptococcal protein antigen P (SpaP, also known as antigen I/II or P1), which can bind to salivary agglutinin glycoprotein (SAG) [7]. S. mutans lacking SpaP exhibited diminished adhesion to SAG-coated surfaces or to salivary pellicles in vitro, and monkey or human subjects immunized with antigen I/II exhibit reduced colonization by S. mutans [8]. Subsequent studies showed that SpaP and another adhesin, wall-associated protein A (WapA), can mediate S. mutans binding to collagen [9], suggesting that they have a role in the bacterial attachment to oral and other tissues. In addition to SpaP and WapA, glucan-binding proteins A (GbpA) and C (GbpC) also play crucial roles in biofilm formation on the tooth surface [10,11].
The S. mutans surface adhesins are anchored to the bacterial cell surface by the highly conserved transpeptidase, sortase A (SrtA) [12]. SrtA recognizes the sorting signal of surface proteins containing a highly conserved LPXTG motif (where X represents any amino acid) at the carboxy-terminal end of the protein and cleaves peptide bonds after the threonine. The released carboxy-terminus of threonine is attached to the pentaglycine of lipid II-surface protein. Lastly, surface protein-lipid II complex is affixed to the cell wall peptidoglycan via transglycosylation and transpeptidation reactions [13]. Moreover, the SrtA-deficient S. mutans strain cannot anchor the protein to the bacterial cell surface, and exhibits lower adherence to oral mucosa or teeth and decreased biofilm biomass on the tooth surface, reducing the formation of caries [14]. Thus, SrtA has an important role in the formation of dental caries by regulating the sorting of the adhesion-related protein to the cell surface, and is a promising target for drug development to prevent or treat dental caries. Inhibition of bacterial adherence is an ideal strategy to combat biofilm-related infections, because it can prevent biofilm establishment without changing the ecological balance within the oral cavity.
To date, many SrtA inhibitors have been identified, including synthetic small molecules [15,16], rationally designed peptide-analogs [17,18], and natural products derived from plants [19,20,21,22]. Among them, many flavonoids extracted from medicinal plants display good inhibitory activity against SrtA, including quercetin, which inhibits the S. aureus SrtA [19], epigallocatechin gallate, which inhibits the S. pneumoniae SrtA [20], and formononetin, which was found to be a potent inhibitor of S. mutans SrtA [21]. Huang et al. reported that morin, a flavonoid constituent of numerous Chinese herbs, can restrain the SrtA of S. mutans, thereby inhibiting the adhesion of S. mutans and reducing the consequent formation of biofilm [22].
Astilbin is a naturally derived flavonoid compound isolated from Rhizoma Smilacis Glabrae (Figure 1A), which has been commonly used in traditional Chinese medical treatment. Astilbin has many properties, such as anti-S. sobrinus [23], anti-inflammatory [24], antioxidant [25], and immunosuppressive activities [26]. However, there are few reports on the inhibitory effects of astilbin on bacterial biofilms. In this study, we observed that astilbin can repress the activity of SrtA and the biofilm formation of S. mutans, indicating its potential for use as an oral biofilm inhibitor.
Figure 1.
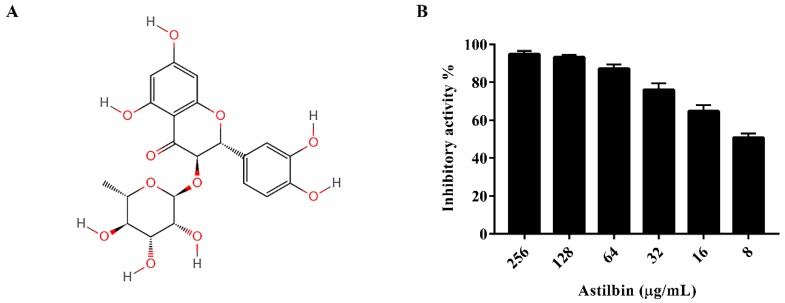
The structure of astilbin and the inhibition of S. mutans SrtA by astilbin in vitro. (A) The chemical structure of astilbin. (B) The inhibitory effect of astilbin against the SrtA of S. mutans. The purified SrtA was incubated with or without different concentrations of astilbin at 37 °C for 1 h, then the fluorescent substrate peptide was added, and the plate was incubated for 1 h. The fluorescence intensity of each well was measured with an excitation wavelength of 350 nm and an emission wavelength of 495 nm. The values indicate the mean values of three independent experiments. The error bars represent the standard deviations.
2. Results
2.1. Inhibition of S. mutans SrtA by Astilbin
The activity of SrtA was analyzed using a fluorescence resonance energy transfer (FRET) assay, as described in a previous study [27], with Abz-LPATG-Dap(Dnp)-NH2 used as the substrate peptide. The purified SrtA of S. mutans was incubated with the substrate peptide in the presence of various concentrations of astilbin in the reaction buffer. The results indicated that astilbin inhibited the activity of SrtA in a dose-dependent manner (Figure 1B), with an IC50 value of 7.5 μg/mL.
2.2. Antibacterial Activity of Astilbin
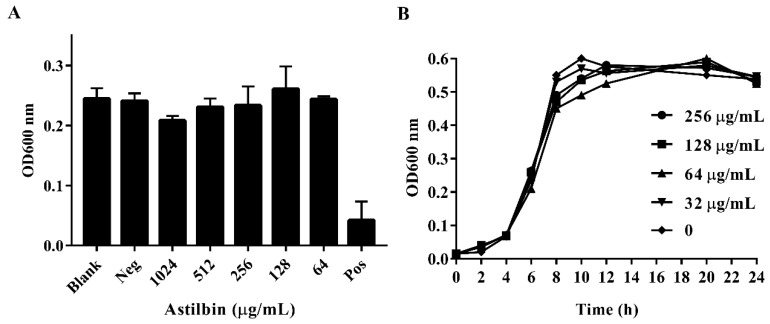
To determine if astilbin inhibits the growth of S. mutans, the minimum inhibitory concentration (MIC) of astilbin against S. mutans was determined, and S. mutans growth curves in the presence of astilbin were generated. As shown in Figure 2A, the MIC of astilbin against S. mutans was above 1024 μg/mL. Furthermore, the OD600 value of negative control (1% dimethyl sulfoxide (DMSO)) was similar to that of the blank control group, reflecting that there was no antimicrobial activity of the negative control. The growth curves showed that the growth of S. mutans treated with various concentrations of astilbin was similar to that of the untreated group (Figure 2B). These results suggest that astilbin does not affect the proliferation of S. mutans and will not lead to the development of bacterial drug resistance.
Figure 2.
The minimum inhibitory concentration (MIC) of astilbin against S. mutans and the growth curves of S. mutans treated with astilbin. (A) The growth state of S. mutans in the presence of different concentrations of astilbin. “Neg” represents the negative control group and “Pos” represents the positive control group. The blank group contained only the brain heart infusion (BHI) broth and the tested S. mutans, the negative control group was treated with 1% dimethyl sulfoxide (DMSO), and the positive control group was treated with 20 μg/mL of chlorhexidine. (B) The growth curves of S. mutans treated with different concentrations of astilbin. The initial inoculum was approximately 1 × 106 colony-forming units/mL. The growth rates were measured by determining the optical density (OD) every two hours for one day.
2.3. Inhibition of Biofilm Formation by Astilbin
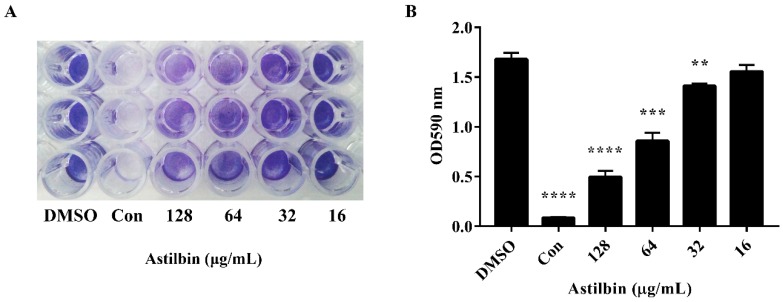
The attachment of S. mutans to tooth surfaces is the first step in biofilm formation [28], and SrtA inhibition should reduce the levels of cell surface proteins related to biofilm formation. Therefore, we further assessed the effect of astilbin on the S. mutans biofilm formation. The S. mutans biofilm biomass was measured after 18 h of growth on saliva-coated plates in a brain heart infusion (BHI) medium. As shown in Figure 3A, the experimental groups treated with 64 or 128 μg/mL of astilbin had a notable difference in biofilm integrity and thickness compared to the groups without astilbin. However, the biofilm treated with 32 and 16 μg/mL of astilbin did not show a difference from the controls. The quantitative results of biofilm formation were in line with the results of the crystal violet staining (Figure 3B). The groups treated with 64 μg/mL of astilbin showed a 49% reduction in biofilm formation, and the suppression effects of the 128 μg/mL groups (approximately 70%) were more obvious compared to the groups treated with 1% DMSO. The ability of S. mutans to form biofilms on plate surfaces was inhibited by astilbin in a dose-dependent manner.
Figure 3.
Inhibition of S. mutans biofilm formation by astilbin. (A) Photograph of S. mutans biofilms grown in the wells of a 96-well plate. Cells were cultured in BHI media containing different concentrations of astilbin for 16 h and stained with 0.1% crystal violet. The group treated with 128 μg/mL of morin served as a positive control (Con). The DMSO treatment group served as a negative control. (B) Quantification of the biomass of S. mutans treated with astilbin. The data were obtained from three independent experiments. Significant differences between groups were accepted at ** p < 0.01, *** p < 0.001, and ****p < 0.0001.
2.4. Determination of the Molecular Mechanism
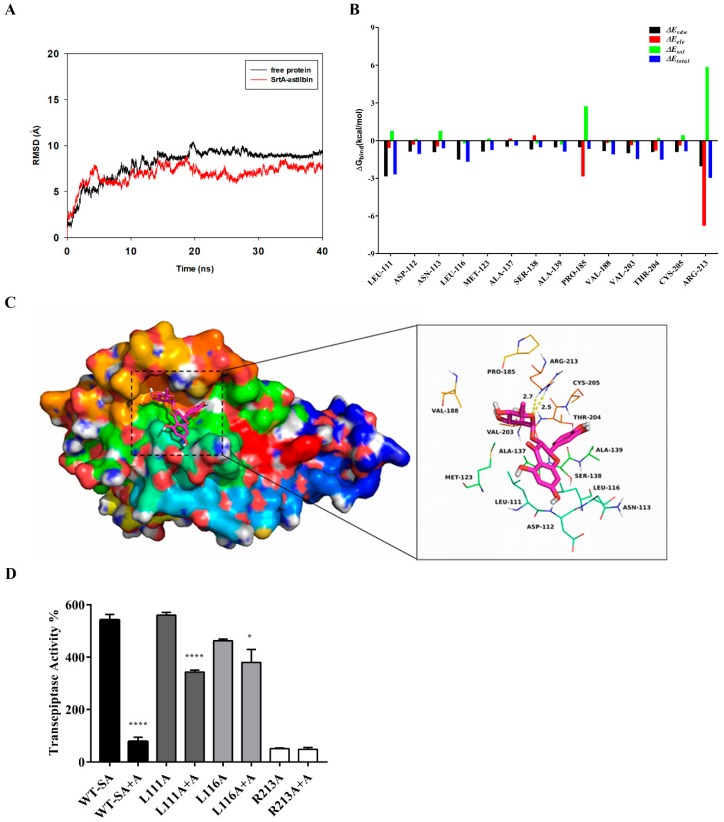
Based on the above results, we performed a molecular docking (MD) simulation of the SrtA-astilbin complex to investigate the underlying molecular mechanism that inhibits the SrtA activity by astilbin. The potential binding mode of astilbin to the active site of SrtA was assessed via a 40 ns MD simulation using Autodock vina 1.1.2 and Amber 14. As shown in Figure 4A, the results of the root-mean-square deviation (RMSD) assay showed that the system became an equilibrium state after 40 ns of the simulation. To obtain more information regarding the contributions of the residues surrounding the binding site to the system, the binding free energies (ΔGbind in kcal/moL) between these residues and astilbin were calculated. As shown in Figure 4B, in the SrtA-astilbin complex, the Arg213 residue has a strong electrostatic (∆Eele) interaction with astilbin, with an ∆Eele value of < −6.5 kcal/moL. Further analysis revealed that the Arg213 residue is oriented toward the rhamnose group of astilbin, leading to the formation of two strong hydrogen bonds (bond lengths of 2.5 and 2.7 Å) between SrtA and astilbin (Figure 4C). In addition, the residues Arg213, Leu111, and Leu116, with Van der Waals (∆Evdw) values of < −1.5 kcal/moL, have notable Van der Waals contributions due to their proximity to the astilbin molecule, indicating that these three residues are crucial for the binding of astilbin to SrtA. In addition, the total binding energy of the SrtA-astilbin complex was calculated, and a ∆Gbind value of –28.8 kcal/moL was determined for astilbin, suggesting that it can strongly bind to and interact with SrtA.
Figure 4.
The results of the molecular docking (MD) simulation of the SrtA-astilbin complex. (A) The root-mean-square deviations (RMSD) exposed by the backbone atoms of the protein during the MD simulation of the SrtA-astilbin complex. (B) Decomposition of the binding free energy on a per residue basis in the SrtA-astilbin complex. (C) The predicted interaction mode of astilbin with the amino acid residues of the catalytic center of SrtA. (D) The inhibition effect of astilbin (64 μg/mL) (A represents astilbin) on the activities of SrtA and its mutants L111A-SrtA, L116A-SrtA, and R213A-SrtA. Significant differences between groups were accepted at * p < 0.05 and **** p < 0.0001.
To further validate the results of the MD simulation, three mutants were constructed—L111A-SrtA, L116A-SrtA, and R213A-SrtA. The mutant proteins were subsequently expressed and purified, and the inhibitory activity of astilbin on these mutated proteins was assessed via a FRET assay. As shown in Figure 4D, the mutation of Arg213 resulted in a significant decrease in the transpeptidase activity, indicating that it is a crucial amino acid residue in SrtA. Moreover, the inhibitory activity of astilbin against the mutated proteins decreased sharply compared with the wild-type SrtA (WT-SA). These results were in agreement with those obtained by the MD simulation, validating the reliability of these results.
3. Discussion
Many studies have shown that isogenic SrtA knockout strains exhibit a notable reduction in the ability to anchor surface proteins containing the canonical LPXTG motif, and are less lethal than wild-type strains in various animal models of infection [29,30]. Additionally, the SrtA-mutants of S. mutans show a decreased ability to attach to human extracellular matrix proteins and to colonize the murine oral cavity and teeth [31]. As the primary aetiological agent of dental caries, S. mutans forms biofilm aggregates on the surfaces of teeth with the assistance of many cell surface-localized and secretory factors. Early studies demonstrated that a few compounds from Chinese traditional medicine, such as morin [22], curcumin [32], trans-chalcone [33], and metabolites from the flowers of Sophora japonica [34], can inhibit S. mutans biofilm formation by inhibiting SrtA activity. In this study, when SrtA of S. mutans was incubated with different concentrations of astilbin, the SrtA catalytic activity decreased in a concentration-dependent manner (Figure 1B). In addition, even increasing the concentration of astilbin to 1024 μg/mL, did not decrease the bacterial growth rate (Figure 2A). Thus, astilbin exerts little selective pressure on S. mutans, and is unlikely to induce the development of resistance. To further evaluate the inhibitory effects of astilbin on the biofilm formation of S. mutans, we used the crystal violet staining method to quantify biofilm biomass. The quantitative results (Figure 3) demonstrated that 64 μg/mL of astilbin could inhibit the biofilm formation of S. mutans in a concentration-dependent manner. There was a 50% and a 70% reduction of the biofilm biomass after 16 h in the presence of 64 and 128 μg/mL of astilbin, respectively. The in vitro IC50 value and biofilm inhibition concentrations are inconsistent, because SrtA activity is not completely inhibited at this concentration, so a small amount of surface protein is still present on the bacterial surface, indicating that a higher drug concentration may be required to completely inhibit biofilm formation.
The results of the MD simulation and the SrtA mutation analyses indicated that astilbin binds to SrtA via electrostatic, hydrogen bond, and Van der Waals interactions, and the amino acid residues Arg213, Leu111, and Leu116 play important roles in the interaction between SrtA and astilbin (Figure 4B–D). The relative positions of key active-site residues (His, Cys, Arg) are highly conserved in sortase enzymes from Gram-positive bacteria. Wallock-Richards et al. elucidated that, similar to other SrtA enzymes, the crystal structure of the SrtA of S. mutans showed that Cys205, Ala139, and Arg213 are located in three adjacent β-strands, which form a tunnel-like hydrophobic pocket [33]. The strong interaction of astilbin with Arg213 interferes with the catalytic activity of SrtA, and results in reduced anchoring of cell surface proteins.
The in vitro study here and other studies showed that the inhibition of SrtA can weaken the biofilm formation of S. mutans. However, as the pathogenesis of caries involves other bacterial species, the practical effects of in vivo use of SrtA inhibitors still needs further evaluation.
In summary, our results demonstrated that astilbin can inhibit the activity of SrtA by interacting with crucial amino acid residues and disrupt biofilm formation of S. mutans. Thus, astilbin is a promising lead compound to develop products that can be incorporated into oral care products, such as mouth rinses or toothpastes, to enhance their anticaries properties.
4. Materials and Methods
4.1. Bacteria, Chemicals, and Growth Conditions
The S. mutans strain (ATCC 25175) used throughout this study was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Bacteria were anaerobically cultured in a BHI broth (Sigma) at 37 °C with shaking. Astilbin was purchased from the Chengdu Herbpurify Corporation (Chengdu, China) with a purity > 98%. Astilbin was dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO, USA) to 102.4 mg/mL as a colorless, transparent solution, and stored at 4 °C until needed. The substrate peptide Abz-LPATG-Dap(Dnp)-NH2 (Abz: ortho-aminobenzoic acid; Dnp: 2,4-dinitrophenyl) was synthesized by GL Biochem (Shanghai, China).
4.2. Cloning and Expression of the SrtA and Its Mutants
A 741 bp DNA fragment encoding the SrtA gene was amplified by polymerase chain reaction (PCR) from the S. mutans chromosome using the primer pairs SmsrtA-F/SmsrtA-R, the amplified fragment was digested with BamHI and XhoI, and cloned into the vector pET28a, yielding pET28SmsrtA. Point mutations in the gene-encoding SrtA were generated using a QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA, USA) to yield L111A-SrtA, L116A-SrtA, and R213A-SrtA. The genes of point mutations were confirmed by sequencing by Sangon Biotech Co., Ltd (Shanghai, China). To obtain recombinant wild-type and mutant SrtA proteins, Escherichia coli strain BL21 (DE3) was transformed with pET28SmsrtA and the SrtA mutant constructs. Protein expression was induced with 0.5 mM isopropylthio-β-d-galactoside (IPTG) (Sigma) at the mid-log-phase for 12 h at 16 °C, respectively. The soluble His-tagged wild-type and mutant SrtA proteins were further purified using the Ni-NTA system, as described in a previous study [13]. All primers used in this study are listed in Table 1.
Table 1.
Primers used in this study.
| Primer Name | Sequences (5′–3′) |
|---|---|
| SmsrtA-F | CGCGGATCCATGAAAAAAGAACGTCAATCTAGGA |
| SmsrtA-R | CCGCTCGAGTTAAAATGATATTTGATTATAGGACTGCC |
| R213A-SrtA-F | TTGTTCATGGCACATATAAGGGGGAA |
| R213A-SrtA-R | TTGTTGCAGCAGTCGCCC |
| L111A-SrtA-F | AAGGAGCAGATAATGTTGGCTTAAC |
| L111A-SrtA-R | TGAAGATTGGTAAATTGATTTTTAAGTCTGG |
| L116A-SrtA-F | TTGGCGCAACATATGGTG |
| L116A-SrtA-R | ATTTATCTAATCCTTTGAAGATTGGTAAATTG |
The underlined basic groups represent restriction endonuclease recognition sites or mutated codons.
4.3. Determination of MIC
S. mutans was grown overnight on BHI agar plates under anaerobic conditions. The MIC values of astilbin against S. mutans were measured using the two-fold serial dilution method, following the Clinical Laboratory Standards Institute (CLSI) guidelines [35]. The astilbin stock solution was serially diluted with a BHI broth from 1024 to 64 μg/mL in a 96-well flat-bottomed plate under aseptic conditions. The BHI broth with 20 μg/mL of chlorhexidine was used as a positive control, and 1% DMSO served as a negative control. After adding 1 μL of the prepared bacterial suspension (1 × 107 colony-forming units (CFU)/mL) equally into each well, which contained 200 μL of the BHI broth and serial-diluted astilbin, the plate was placed in the anaerobic incubator at 37 °C for 16 h. Absorbance values at 600 nm were determined using a microplate reader (Infinite® F500, Tecan, Shanghai, China). The MIC was defined as the minimal concentration at which a visible growth of microorganisms was inhibited under defined growth conditions [36]. All experiments were repeated three times.
4.4. Growth Curves of S. mutans
S. mutans cultures were grown aerobically overnight at 37 °C in a BHI medium. After culturing, 100 μL of the bacterial cultures (1 × 107 CFU/mL) were transferred to aseptic test flasks containing 10 mL of a sterilized BHI medium and different concentrations of astilbin (0, 32, 64, 128, 256 μg/mL). All the flasks were aerobically cultured at 37 °C. The optical density of S. mutans was recorded at 600 nm every two hours for one day, with samples being vortexed before measuring.
4.5. SrtA Activity Assay
The SrtA activity inhibition assay was performed according to a previously published protocol, with slight modifications [13]. Briefly, the inhibition of SrtA activity of S. mutans by astilbin was determined by quantifying the fluorescence intensity, which changes with the cleavage of the fluorescent substrate peptide Abz-LPATG-Dap(Dnp)-NH2. The assay was conducted in a 200 μL reaction mixture containing buffer A (50 mM Tris-HCl, 300 mM NaCl, 5 mM CaCl2, pH 8.0), 7 μM of SrtA, and various concentrations of astilbin (256, 128, 64, 32, 16, 8 μg/mL). The reaction mixture was incubated for 1 h at 37 °C, after which 10 μM of Abz-LPATG-Dap(Dnp)-NH2 was added, and the reaction was incubated for another hour at the same temperature. To exclude the influence of the fluorescence of astilbin, the same reaction mixture without SrtA was used as a negative control and treated exactly in the same way. In addition, the blank control group (containing buffer A, 10 μM of substrate peptide, and 7 μM of SrtA) without astilbin was used to indicate the initial activity of SrtA. Each astilbin concentration, or well, had three parallel repetitions. The fluorescence values were measured and the percentage inhibition of SrtA by astilbin was calculated using the following equation: % inhibition = Fblank– (Fsample – Fnegative)/Fblank × 100%, where Fnegative is the fluorescence value of the negative control group, Fsample is the fluorescence value of the astilbin-treated group, and Fblank is the fluorescence value of the blank control [37]. The experiments were repeated at least three times independently.
4.6. Crystal Violet Biofilm Assay
Nonstimulated saliva from healthy rats was collected in sterile 50 mL centrifuge tubes, and clarified by centrifugation (16,000× g, 4 °C, 10 min). The supernatant after centrifugation was filtered with a 0.22 μm Millipore filter. The 96-well microtiter plates (Corning Costar Co., Cambridge, MA, USA) were coated with 200 μL of saliva-buffer mixture for 1 h at 37 °C, according to the procedure of Islam et al. [38] with minor modifications. Subsequently, the liquid was discarded aseptically, and 1 μL of an overnight culture of S. mutans (1 × 107 CFU/mL) was inoculated into 200 μL of sterile BHI broth supplemented with different concentrations of astilbin (128, 64, 32, 16 μg/mL). S. mutans cultured with the BHI broth with 128 μg/mL of morin served as a positive control. For each astilbin concentration, three parallel wells were assayed. After static incubation for 16 h at 37 °C anaerobically, the media and unattached cells were discarded gently, and the wells were washed three times with PBS. Then the biofilms were stained with 100 μL of 0.1% crystal violet for 15 min at 37 °C, after which the unbound component was rinsed twice with double distilled water. Subsequently, the crystal violet stain was dissolved in 200 μL of 33% acetic acid. The absorbance at 570 nm was determined using a microplate reader (Infinite® F500, Tecan, China) to quantify the biofilm biomass.
4.7. Molecular Docking and Molecular Dynamics Simulation
Autodock Vina 1.1.2 [39] was used to elucidate the binding mode between astilbin and S. mutans SrtA. The SrtA structure (PDB ID:4TQX) was obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank. The astilbin structure was generated using ChemBio3D Ultra 12.0 (ChemBioOffice, New Jersey, CA, USA). For Vina docking, the parameters were maintained at the default settings, unless otherwise mentioned. After docking, the most stable pose of the SrtA-astilbin complex that was obtained was used in an MD simulation analysis using Amber 14 [40,41,42]. The astilbin structure was first prepared using ACPYPE [43] to generate automatic topologies. The forcefield “leaprc.gaff” was selected for astilbin, while “leaprc.ff14SB” was selected to prepare SrtA. Before starting the simulation, equilibration of the solvated SrtA-astilbin complex was performed using a short minimization. Finally, the 40 ns MD simulation was performed on a Dell Precision T5500 workstation. Furthermore, the ligand-protein binding free energies (ΔGbinding in kcal/moL) were calculated using the analyze tool in AmberTools 15 [44]. For each complex, the binding free energy was calculated using the following equation: ΔGbinding = Gcomplex‒ Gprotein ‒ Gligand, where Gcomplex, Gprotein, and Gligand are the free energies of the SrtA-astilbin complex, the SrtA protein, and the ligand astilbin, respectively.
4.8. Statistical Analysis
All experiments were performed in triplicate. The statistical analysis of the data was performed using a one-way ANOVA and t-tests with GraphPad Prism7 software (GraphPad software Inc, La Jolla, CA, USA). All data of the three independent experiments were shown as mean ± SD. Statistical significance was determined when p < 0.05.
Author Contributions
D.W. and T.W. conceived and designed the experiments, J.W. and Y.S. performed and analyzed the experiments, H.D. and S.J. performed the molecular dynamics simulation, and J.W. and T.W. wrote the paper.
Funding
This work was supported by The National Key Technology R & D Program (No. 2016YFD05013) and Science Foundation of Jilin Province, China, (No.20180101276JC).
Conflicts of Interest
The authors declare no conflicts of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Van Houte J. Role of micro-organisms in caries etiology. J. Dent. Res. 1994;73:672. doi: 10.1177/00220345940730031301. [DOI] [PubMed] [Google Scholar]
- 2.Kreth J., Merritt J., Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi N., Nyvad B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 4.Loesche W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gendron R., Grenier D., Maheurobert L. The oral cavity as a reservoir of bacterial pathogens for focal infections. Microbes Infect. 2000;2:897–906. doi: 10.1016/S1286-4579(00)00391-9. [DOI] [PubMed] [Google Scholar]
- 6.Banas J.A., Vickerman M.M. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral. Biol. Med. 2003;14:89. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- 7.Jenkinson H.F., Demuth D.R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 2010;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 8.Ma J.K., Hunjan M., Smith R., Kelly C., Lehner T. An investigation into the mechanism of protection by local passive immunization with monoclonal antibodies against Streptococcus mutans. Infect. Immun. 1990;58:3407–3414. doi: 10.1128/iai.58.10.3407-3414.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AvilãS-Reyes A., Miller J.H., Lemos J.A., Abranches J. Collagen-binding proteins of Streptococcus mutans and related streptococci. Mol. Oral Microbiol. 2017;32:89–106. doi: 10.1111/omi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas W., Banas J.A. Ligand-binding properties of the carboxyl-terminal repeat domain of Streptococcus mutans glucan-binding protein A. J. Bacteriol. 2000;182:728. doi: 10.1128/JB.182.3.728-733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato Y., Yamamoto Y., Kizaki H. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 1997;65:668–675. doi: 10.1128/iai.65.2.668-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cossart P., Jonquières R. Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc. Natl. Acad. Sci. USA. 2000;97:5013–5015. doi: 10.1073/pnas.97.10.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonthat H., Liu G., Mazmanian S.K., Faull K.F., Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.F., Boran T.L. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 2003;71:676. doi: 10.1128/IAI.71.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maresso A.W., Ruiying W., Kern J.W., Rongguang Z., Dorota J., Missiakas D.M., Mark-Eugene D., Andrzej J., Olaf S. Activation of inhibitors by sortase triggers irreversible modification of the active site. J. Biol. Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel B.A., Matthew B., Kruger R.G., Mccafferty D.G. Vinyl sulfones: Inhibitors of SrtA, a transpeptidase required for cell wall protein anchoring and virulence in Staphylococcus aureus. J. Am. Chem. Soc. 2004;126:3404–3405. doi: 10.1021/ja0390294. [DOI] [PubMed] [Google Scholar]
- 17.Scott C.J., Andrew M.D., S Lorraine M., Lynas J.F., Koen V., Brian W. Irreversible inhibition of the bacterial cysteine protease-transpeptidase sortase (SrtA) by substrate-derived affinity labels. Biochem. J. 2002;366:953–958. doi: 10.1042/bj20020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Li H., Pan J., Dong J., Zhou X., Niu X., Deng X. Oligopeptide Targeting Sortase A as Potential Anti-infective Therapy forStaphylococcus aureus. Front. Microbiol. 2018;9:245. doi: 10.3389/fmicb.2018.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Chen F., Bi C., Wang L., Zhong X., Cai H., Deng X., Niu X., Wang D. Quercitrin, an Inhibitor of Sortase A, Interferes with the Adhesion of Staphylococcal aureus. Molecules. 2015;20:6533–6543. doi: 10.3390/molecules20046533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song M., Teng Z., Li M., Niu X., Wang J., Deng X. Epigallocatechin gallate inhibits Streptococcus pneumoniae virulence by simultaneously targeting pneumolysin and sortase A. J. Cell. Mol. Med. 2017;21:2586–2598. doi: 10.1111/jcmm.13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park W., Ahn C.H., Cho H., Kim C.K., Shin J., Oh K.B. Inhibitory effects of flavonoids from Spatholobus suberectus on sortase A and sortase A-mediated aggregation of Streptococcus mutans. J. Microbiol. Biotechnol. 2017;27:1457–1460. doi: 10.4014/jmb.1704.04001. [DOI] [PubMed] [Google Scholar]
- 22.Huang P., Hu P., Zhou S.Y., Li Q., Chen W.M. Morin inhibits sortase A and subsequent biofilm formation in Streptococcus mutans. Curr. Microbiol. 2014;68:47–52. doi: 10.1007/s00284-013-0439-x. [DOI] [PubMed] [Google Scholar]
- 23.Kuspradini H., Mitsunaga T., Ohashi H. Antimicrobial activity against Streptococcus sobrinus and glucosyltransferase inhibitory activity of taxifolin and some flavanonol rhamnosides from kempas (Koompassia malaccensis) extracts. J. Wood Sci. 2009;55:308–313. doi: 10.1007/s10086-009-1026-4. [DOI] [Google Scholar]
- 24.Huang H., Cheng Z., Shi H. Isolation and Characterization of Two Flavonoids, Engeletin and Astilbin, from the Leaves of Engelhardia roxburghiana and Their Potential Anti-inflammatory Properties. J. Agric. Food Chem. 2011;59:4562–4569. doi: 10.1021/jf2002969. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q.-F., Zhang Z.-R., Cheung H.-Y. Antioxidant activity of Rhizoma Smilacis Glabrae extracts and its key constituent-astilbin. Food Chem. 2009;115:297–303. doi: 10.1016/j.foodchem.2008.11.053. [DOI] [Google Scholar]
- 26.Jianming G., Feng Q., Jianxin L., Qiang X., Ting C. Identification of a new metabolite of astilbin, 3’-O-methylastilbin, and its immunosuppressive activity against contact dermatitis. Clin. Chem. 2007;53:465–471. doi: 10.1373/clinchem.2006.077297. [DOI] [PubMed] [Google Scholar]
- 27.Matayoshi E.D., Wang G.T., Krafft G.A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 28.Okahashi N., Sasakawa C., Yoshikawa M., Hamada S., Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol. Microbiol. 2010;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian S.K., Skaar E.P., Gaspar A.H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D.M., Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 30.Bolken T.C., Franke C.A., Jones K.F., Zeller G.O., Jones C.H., Dutton E.K., Hruby D.E. Inactivation of the srtA Gene in Streptococcus gordonii Inhibits Cell Wall Anchoring of Surface Proteins and Decreases In Vitro and In Vivo Adhesion. Infect. Immun. 2001;69:75–80. doi: 10.1128/IAI.69.1.75-80.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lévesque C.M., Voronejskaia E., Huang Y.C., Mair R.W., Ellen R.P., Cvitkovitch D.G. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 2005;73:3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu P., Huang P., Chen W.M. Curcumin inhibits the Sortase A activity of the Streptococcus mutans UA159. Appl. Biochem. Biotech. 2013;171:396–402. doi: 10.1007/s12010-013-0378-9. [DOI] [PubMed] [Google Scholar]
- 33.Wallock-Richards D.J., Jon M.W., Clarke D.J., Amarnath M., Michael D., Bryan H., Campopiano D.J. Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone. Chem. Commun. 2015;51:10483–10485. doi: 10.1039/c5cc01816a. [DOI] [PubMed] [Google Scholar]
- 34.Yang W.Y., Won T.H., Ahn C.H., Lee S.H., Yang H.C., Shin J., Oh K.B. Streptococcus mutans sortase A inhibitory metabolites from the flowers of Sophora japonica. Bioorg. Med. Chem. Lett. 2015;25:1394–1397. doi: 10.1016/j.bmcl.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 35.Goldstein E.J., Citron D.M., Warren Y.A., Tyrrell K.L., Merriam C.V., Fernandez H.T. In vitro activities of dalbavancin and 12 other agents against 329 aerobic and anaerobic gram-positive isolates recovered from diabetic foot infections. Antimicrob. Agents Chemother. 2006;50:2875. doi: 10.1128/AAC.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 37.Sebaugh J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011;10:128–134. doi: 10.1002/pst.426. [DOI] [PubMed] [Google Scholar]
- 38.Islam B., Khan S.N., Naeem A., Sharma V., Khan A.U. Novel effect of plant lectins on the inhibition of Streptococcus mutans biofilm formation on saliva-coated surface. J. App. Microbiol. 2010;106:1682–1689. doi: 10.1111/j.1365-2672.2008.04135.x. [DOI] [PubMed] [Google Scholar]
- 39.Oleg T., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Götz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce L.C., Salomon-ferrer R., Augusto F.d.O.C, Mccammon J.A., Walker R.C. Routine Access to Millisecond Time Scale Events with Accelerated Molecular Dynamics. J. Chem. Theory Comput. 2012;8:2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salomon-ferrer R., Götz A.W., Poole D., Le G.S., Walker R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 43.Sanner M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 44.Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3:198–210. doi: 10.1002/wcms.1121. [DOI] [Google Scholar]