Abstract
G-quadruplexes are four-stranded nucleic acids structures that can form in guanine-rich sequences. Following the observation that G-quadruplexes are particularly abundant in genomic regions related to cancer, such as telomeres and oncogenes promoters, several G-quadruplex-binding molecules have been developed for therapeutic purposes. Among them, naphthalene diimide derivatives have reported versatility, consistent selectivity and high affinity toward the G-quadruplex structures. In this review, we present the chemical features, synthesis and peculiar optoelectronic properties (absorption, emission, redox) that make naphtalene diimides so versatile for biomedical applications. We present the latest developments on naphthalene diimides as G-quadruplex ligands, focusing on their ability to bind G-quadruplexes at telomeres and oncogene promoters with consequent anticancer activity. Their different binding modes (reversible versus irreversible/covalent) towards G-quadruplexes and their additional use as antimicrobial agents are also presented and discussed.
Keywords: G-quadruplex, naphthalene diimide, telomeres, telomerase, oncogene promoters, ligands, anticancer activity, viruses, parasites, binding modes
1. Introduction
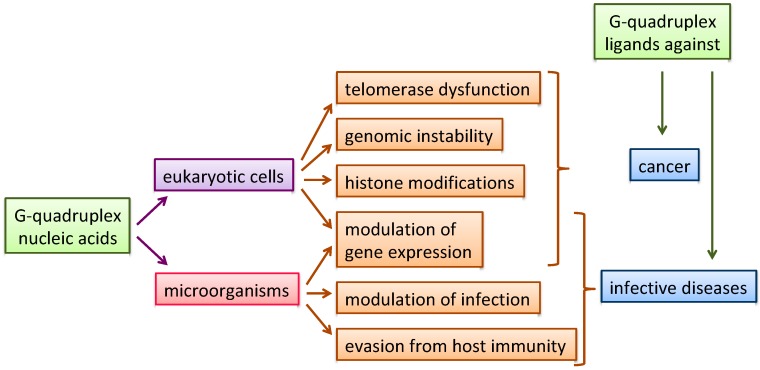
Nucleic acids (NAs) are key targets in medicinal chemistry; in particular, non-canonical NA structures, including G-quadruplex structures (G4s), have been proposed to be involved in genomic instability [1,2], telomerase dysfunction [3], histone modifications [4], modulation of gene expression [5,6,7,8] and viral transcription [9,10], providing possible targets in a number of different diseases. Moreover, G4 structures, from the earlier work of Hardin et al. [11] to the most recent evidence highlighted by Chowdhury [12], are arising as promising regulators of epigenetic modifications [13,14,15]. As a natural consequence, selective recognition and stabilization of G4s by small molecules is emerging as a new general strategy for innovative therapeutics (Figure 1) [16,17,18,19,20,21,22,23].
Figure 1.
Schematic representation of G4-modulated organisms, biological processes and fields of therapeutic applications.
In principle, any repetitive guanine-rich region may be able to fold into polymorphous four-NA-strand G4s. Four guanine bases can associate through Hoogsteen-type hydrogen bonds to form the G-quartet, a square planar structure. Two or more G-quartets can stack on top of each other, forming the G4, which is further stabilized by cations (Na+ or K+) located between the G-quartets [24].
Based on this peculiar common structure, G4-interacting compounds can be rationally designed according to the two following criteria: (1) Extended aromatic and electron poor surface (π-stacking interaction) and (2) cationic side chains (electrostatic attraction). Many small molecules that bind G4s have been developed, with several of them showing biological activity. Although there is still a long way to go in the development of drugs that selectively target one specific G4 structure, promising lead compounds that act in the cell have been presented. Among them, naphthalene diimides (NDIs) are a class of small molecules able to recognize, induce and stabilize G4 structures with high affinity, through π-stacking interactions (Figure 2).
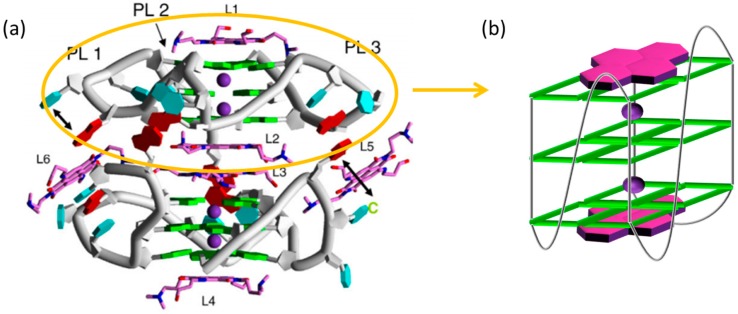
Figure 2.
(a) The crystal structure of the 23-mer intramolecular G4 d[TAGGG(TTAGGG)3] complexed with tetra-substituted naphthalene diimide ligands (Figure taken from [25]); (b) model of the NDI-G4 binding through π-stacking interactions.
Herein we report an overview on the NDI properties and their use as G4 ligands, with a particular focus on their anticancer achievements.
2. General Features, Synthesis and Applications of Naphthalene Diimides
1,4,5,8-Naphthalenediimides (NDIs) are perylene diimides (PDIs) smaller analogues (Figure 3) [26] composed of a single naphthalene core that bears two electron-withdrawing imide groups; the two nitrogen atoms can be substituted with a variety of alkyl or aryl chains. Unlike PDIs, which display outstanding absorption properties and are therefore of high significance as industrial pigments, naphthalene diimides without substituents at the aromatic core are colourless and therefore of no use as pigments. Nevertheless, NDIs are the most important PDI homologues. Addition of a single substituent at the aromatic core turns the NDI into a versatile large π-planar chromophore with interesting optoelectronic properties. Although the first reports on core-substituted naphthalene diimides (cNDIs) date back to 80 years ago, consistent research only began in the XXI century, when the synthetic route was improved and, more importantly, interesting chemical-physical properties were discovered. Due to their electron-deficient π-system, NDIs are now a class of compounds extensively studied in the field of supramolecular and materials chemistry. NDIs possess high electron affinity, good charge carrier mobility, and excellent thermal and oxidative stability. Thanks to their planar aromatic core, NDIs exhibit aromatic π-stacking and van der Waals interactions: the electron density on most aromatic rings creates a quadrupole moment with a partial negative charge above and below the plane and a partial positive charge around the periphery. This set of features makes NDIs promising candidates for a variety of applications in the field of material sciences; for example, they have been employed as n-semiconductors, photovoltaic devices, flexible displays and molecular tweezers [26,27,28,29,30,31]. Moreover, functionalization through core-substitution (2,6- or 2,3,6,7- to the core, Figure 3) produces cNDI analogues with modular absorption and fluorescence properties, positioning them among the most versatile and fascinating aromatic dyes [32].
Figure 3.
General structure of naphthalene dianhydride (NDA), naphthalene diimide (NDI) and perylene diimide (PDI).
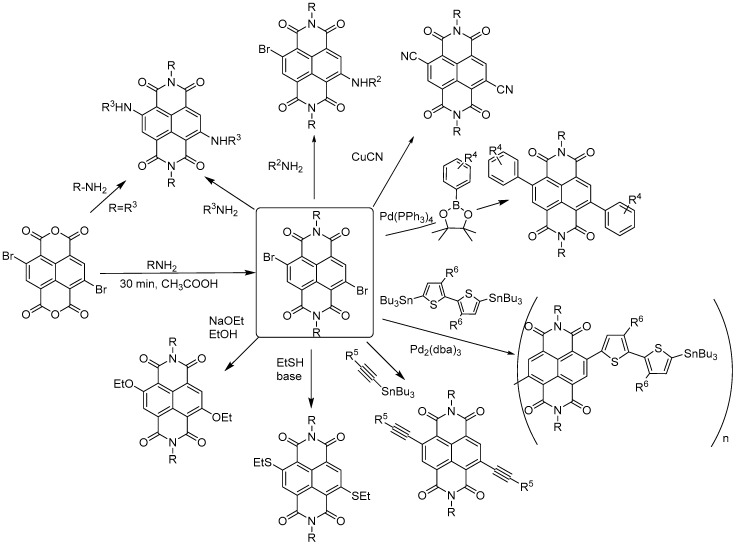
The synthesis of cNDIs starts from the halogenated naphthalene dianhydride (NDA) (Figure 4) prepared from pyrene. For the first half of the 20th century, the Vollmann’s method, the traditional synthetic route, employed a harsh oxidative halogenation to retrieve the di-chlorinated NDI in four steps [33]. Despite the fact that this method made halogenated NDI derivatives accessible for many years, the intense application prospects of cNDIs imposed the development of an alternative, more direct, and easier synthetic approach. Thalacker and Wurthner reported the first direct bromination of the commercially available NDA with one equivalent of dibromoisocyanuric acid (DBI) to afford the 2,6-dibromo dianhydride (Figure 4) [33]. The use of an excess of halogenating agent led to the 2,3,6,7-tetrabromo dianhydride. Starting from these two key intermediates, a multitude of chemical modifications were performed, as recently reviewed by Würthner and co-workers (Figure 5) [32].
Figure 4.
General synthetic route to access chloro and bromo cNDIs. (a) Cl2 (g), y = 36–38%; (b) KOH/EtOH, y = 96–97%; (c) HNO3 y = 32–45%; (d) HNO3/H2SO4, y = 45–50%; (e) oleum, 100 °C, y = 25%; (f) DBI, oleum, RT, y = 80%; (g) excess DBI, oleum, RT, y = 93%.
Figure 5.
Synthetic routes to core di-functionalization.
Four-site functionalization with the one substituent can be carried out in a single step in polar solvents at high temperature [34]. This set of conditions is particularly favourable for reductive dehalogenation [35]. Since the primary amine is an oxidizing agent, preservation of the four bromine atoms is difficult, unless bulky amines are used as reagents. By modulating the reaction conditions, it is possible to prevent reductive dehalogenation and introduce structural diversity to the naphthalene diimide core. The general protocol starts with the imidation reaction that leads to symmetric diimide substituents. It is then possible to obtain the orthogonal core-functionalization with stepwise nucleophilic substitutions [33]. The first step is carried out in acetic acid, ensuring optimal pH conditions for the catalytic protonation of the carbonyl and amine, in order to reduce nucleophilicity of the latter to a level that prevents aromatic substitutions [36]. Both chloro- and bromo-derivatives allow further nucleophilic aromatic substitutions that can be achieved through a non-catalysed process with classical nucleophiles, such as amines, alcohols, thiols, and cyanides. Alternatively, organometallic chemistry can be used, e.g., Suzuki–Miyaura coupling with boronate esters to attach phenyl groups or Sonogashira/Stille couplings to introduce alkynes and heterocycles (Figure 5) [27,32].
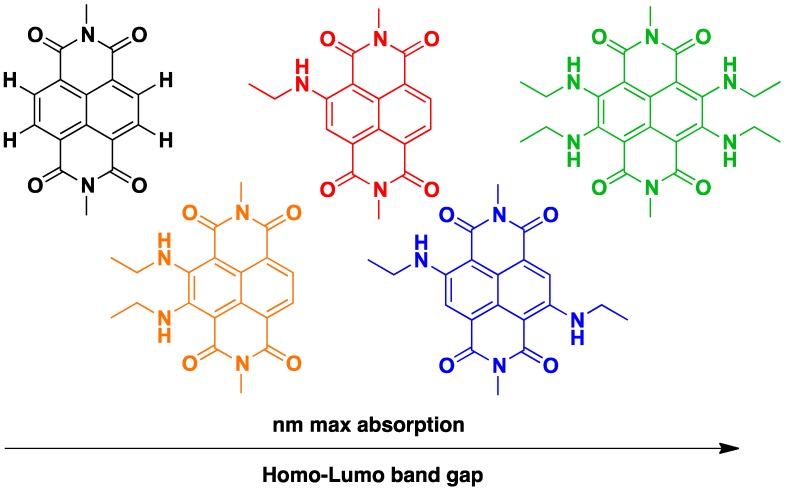
The easily tuneable synthesis of the cNDIs allows the conjugation of the NDI core to different heteroatoms. In particular, positions 2, 3, 6 and 7 of the cNDI aromatic core (Figure 3) can bear one or more substituents with different-extent electro-donor effect on the π system [32]. This in turn modulates the cNDI photophysical properties providing, for example, bright colouring and high fluorescence, both over a wide range of wavelengths [37]. The ability of cNDIs to change colour and redox properties without modifying their global features is one of their most attractive features. The typical colours of cNDIs originate from a charge-transfer band that, depending on the substituent, moves along the entire visible spectra. This property is related to the HOMO-LUMO “bandgap” (Figure 6) [38].
Figure 6.
A rainbow collection of cNDIs.
Besides their use as pigments and dyes, NDIs are also widely employed as main planar components of bio-polymers and supramolecular nanostructures. The existence of delocalized radical anions in the NDI π-stacks confers the ability to work as efficient electron transporters to these molecules. For this reason, cNDIs have been identified as a class of the few existing air-stable n-semiconductors used to generate high-performance organic thin-film transistors (OTFTs) [39,40]. The known spectroscopic properties matched to the ability to transport electrons make cNDIs the ideal systems for the photoinduced electron transport, supporting construction of photovoltaic cells and artificial photosynthesis. The photophysical properties of cNDIs are also well suited for sensing applications. In the literature, there are many examples of fluorescence sensor of different types, such as anion sensors, fluorescent chemosensors for zinc ions and pH sensors [41]. Their fluorescence associated with the flatness of the chromophore makes cNDIs useful for biomolecular recognition. Moreover, some tetrasubstituted NDIs can be used as photosensitizers for photodynamic therapy (PDT) [42,43]. These molecules exhibit large inter-system crossing quantum yields, whereby the generated triplet states are efficiently quenched by molecular oxygen to produce singlet molecular oxygen with high quantum yields.
In the field of biomedicine, cNDIs are used as scaffolds for G4-selective ligands. Their chemical accessibility, large planar electron-poor surface and the presence of protonable side chains are the most relevant features exploited for G4 binding. Thanks to the π acidity, a peculiar characteristic of the electron-deficient aromatic systems, the NDI core is prone to interact by π-π stacking with an electron-rich partner, like the G-quartet. In addition, the side chain substituents, which are often protected tertiary amines or hydroxyl groups, can form hydrogen bonds with the phosphate groups in the G4 grooves, thereby stabilizing the G4-NDI complex.
These properties have not only been investigated from the therapeutic point of view: Over the last decade, the NDI core has also become important in the field of diagnostics. By combining the cNDI outstanding fluorescent properties with its ability to interact and recognize the G4 structure, a wide range of selective chemosensors was generated [44,45,46,47,48,49]. Although most of the red-NIR G4 binders exhibit quenching upon binding, the cNDIs belonging to this class of sensors are able to selectively switch on their fluorescence properties upon G4 recognition. This selective response has made possible the use of one of these sensors in live cells, where excellent co-localization with an anti-G4-antibody (1H6) was obtained [50].
3. NDI Derivatives That Bind to Human Telomere G-Quadruplexes and Inhibit Telomerase Activity
3.1. Reversible NDI Ligands
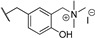
Around 10 years ago, Neidle’s group was the first to report stabilization of telomeric G4s by tri- and tetrasubstituted cNDIs, resulting in telomerase inhibition [34]. The presented library of NDIs, bearing different aliphatic tertiary amine or hydroxyl groups on the side chains (compounds 1–24 in Table 1), was investigated on two different DNA sequences: the human telomeric DNA (able to fold into G4) and a self-complementary duplex DNA hairpin. In general, an increase in the number of substituents corresponded to an increase in G4 stabilization: tetra-substituted cNDIs showed great selectivity for G4 vs duplex DNA in competition experiments. Moreover, enhanced G4 selectivity was obtained reducing the protonable sites in the side chains, e.g., with a morpholine cycle or hydroxyl group (compounds 13–16, 19, 20, 22, Table 1). These compounds reported melting values in telomeric G4 stabilization, measured by fluorescence resonance energy transfer (FRET) analysis, comparable to those obtained with well-known G4 ligands, such as BRACO-19. However, the cNDIs showed higher potency: the same degree of G4 stabilization was reached at 0.5 µM of cNDIs compared to 2.5 µM of BRACO-19. These cNDIs were able to inhibit telomerase with EC50 values in a µM range (compounds 1, 2, 5, 6, 9 and 11, Table 1), as measured by a modified telomeric repeat amplification protocol (TRAP) assay. The most potent cNDIs (compounds 2, 3, 5, 11 and 12, Table 1) arrested cell growth of MCF7 and A549 cancer cell lines with IC50 values of 5–20 nM, as assessed in the sulforhodamine B assay. As the corresponding IC50 values on a normal human fibroblast line (WI38) were typically greater than one order of magnitude, a potentially promising therapeutic index in the above cancer cell lines was indicated. The relation between G4 ligand activity at the telomere level and cellular senescence was supported by confocal microscopy experiments that showed exclusive cNDI localization in the nuclei, i.e., where telomerase acts.
Table 1.
Structures of NDIs used as reversible ligands to stabilize telomeric G4s.

| ||||
|---|---|---|---|---|
| NDI Core | ||||
| Name | R | X | Y | Ref. |
| 1 |
|
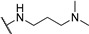
|
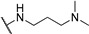
|
[34] |
| 2 |

|

|
-H | [34] |
| 3 |
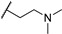
|
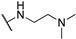
|

|
[34] |
| 4 |

|

|
-H | [34] |
| 5 |

|

|

|
[34] |
| 6 |

|

|
-H | [34] |
| 7 |

|
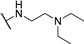
|

|
[34] |
| 8 |

|

|
-H | [34] |
| 9 |

|

|

|
[34] |
| 10 |

|

|
-H | [34] |
| 11 |
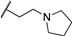
|
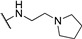
|

|
[34] |
| 12 |

|

|
-H | [34] |
| 13 |
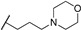
|

|

|
[34] |
| 14 |

|

|
-H | [34] |
| 15 |
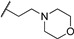
|

|

|
[34] |
| 16 |

|

|
-H | [34] |
| 17 |

|

|

|
[34] |
| 18 |

|

|
-H | [34] |
| 19 |

|
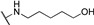
|

|
[34] |
| 20 |

|

|
-H | [34] |
| 21 |

|

|

|
[34] |
| 22 |

|
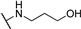
|

|
[34] |
| 23 |

|
-H | -H | [34] |
| 24 |

|
-H | -H | [34] |
| 25 |
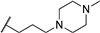
|
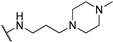
|

|
[53] |
| 26 |
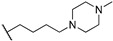
|
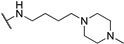
|

|
[53] |
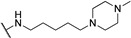
| 27 |
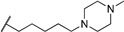
|
|

|
[53] |
| 28 |

|

|

|
[55] |
| 29 |

|
-H | -H | [56] |
| 30 |

|

|
-H | [57] |
| 31 |

|
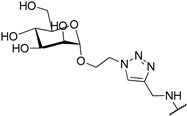
|
-H | [57] |
| 32 |

|
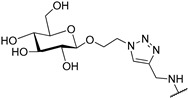
|
-H | [57] |
| 33 |

|
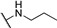
|
-H | [57] |
The quantitative interactions of cNDIs 1-9-22 (Table 1) were further analysed on human telomeric RNA (TERRA) quadruplex using electrospray mass spectrometry methods, comparing the obtained results with BRACO-19 [51]. While, all the three cNDIs bind to the telomeric DNA with high affinity, only 22 guarantees a strong formation of the NDI/RNA-G4 complex, showing equilibrium dissociation constants of Kd1 = 4.0 µM and Kd2 = 5.0 µM, respectively. These results were also justified by a molecular dynamics simulation [51,52].
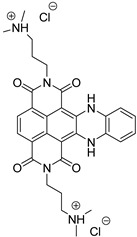
These molecules were particularly potent in G4 binding and telomerase inhibition; therefore, the NDI scaffold became the landmark for some of the most active small molecules able to efficiently target G4s. The NDI core was subsequently modified with manifold side chains, each with fundamental characteristics, in order to improve the selective interaction with the G4 target and the transition across the nuclear membrane.
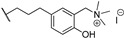
The NDI core functionalized with tetra N-methyl-piperazine end groups (compounds 25–27, Table 1) allowed to obtain crystallographic data of the cNDI-G4 complexes, highlighting a 1:1 stoichiometry, the interaction of positive charges with the groove, and the ability to induce the parallel topology of the human intramolecular telomeric G4 [53]. These small molecules presented high short-term cytotoxicity (IC50 around 0.1–0.2 µM) on a number of cancer cell lines (MCF7, A549, MIA-Pa-Ca-2, PANC-1, HPAC, BxPc-3) and resulted significantly less toxic on normal fibroblasts WI38 (IC50 increased of 2 order of magnitude) [54]. Based on the observation that the three-carbon atom linker (compound 25, Table 1) guaranteed the highest efficiency of cell entry, the mechanism of action of this compound was further studied. This molecule resulted in an effective stabilizer of the human telomeric G4s, able to displace the single-strand binding protein hPOT1 from telomeric ends. However, cell data suggested an additional contribution to its antitumor action: the possible simultaneous targeting of several G4s that induced a therapeutically beneficial HSP90 down-regulation.
Two N-methyl-piperazine units were subsequently replaced by morpholine groups (compound 28, Table 1) without loss of the intermolecular interaction of the cNDI side chains with the G4 grooves [55]. These cNDIs showed superior cell growth inhibition, in particular towards the pancreatic MIA PaCa-2 carcinoma cells (IC50 values around 10 nM), thus looking particularly promising for in vivo therapeutic applications. Unfortunately, no telomerase inhibition activity was found in this cell line at the nanomolar dosage level required to inhibit cellular proliferation.
Considering that the interaction with the G4 grooves is fundamental to improve G4-ligand binding, Czerwinska et al. proposed NDIs carrying a dipicolylamine unit as Zn2+ coordinating moiety (compound 29, Table 1) [56]. As hypothesized, the Zn2+ complex provided additional anchorages to the telomeric G4 through electrostatic interaction with the negative charges of the target. This ligand might be a good anticancer candidate (EC50 value around 5 µM), but further comparative studies will be necessary.
It has been shown that a large positive charge on one hand guarantees NDIs cellular uptake, on the other reduces the selectivity towards the target due to electrostatic interaction, which takes place also with the dsDNA. To solve this problem, Morales et al. engineered carbohydrate-cNDI conjugates (carb-cNDIs) that entered into cancer cells taking advantage of the overexpressed glucose transporter (GLUT) (compounds 30–32, Table 1) [57]. In particular, while aglycone cNDI 32 was quickly uptaken by passive diffusion or facilitated transport both in cancer cell lines HT-29 and MCF-7 and in the noncancerous cell line MRC-5, the uptake of compounds 30–32 in HT-29 and MCF-7 cells, where the GLUT is overexpressed, was more efficient. The compounds’ transport into cancer cell depended on the type of carbohydrate present on the NDI as well as on the compound incubation time and temperature.
3.2. Multimodal NDI Ligands
Taking into account the excellent binding properties of NDIs towards G4s, multimodal ligands that added the ability to selectively damage the targeted G4s were developed. The idea behind this approach is activation of a chemical reactive moiety on the NDI core only upon NDI recognition and stabilization of the therapeutic target G4s, in order to minimize typical off-target reactivity.
Freccero’s group developed hybrid structures that combine the G4-recognizing NDI core with quinone methide (QM) precursor units as activatable alkylating agents (compounds 34–48, Table 2) [58,59,60]. QMs are short-lived reactive Michael acceptors that can be generated in situ by mild thermal activation (40 °C), which is compatible with physiological conditions. The study of these alkylating ligands started with bis-substituted NDIs (34–36), where the activation of the quaternary ammonium salts was obtained by mild thermal incubation at 40 °C, pH 7, for less than 12 h [58]. Despite the presence of two symmetric QM precursors (QMP), only the mono alkylated telomeric G4 target was retrieved. The best compound of the series (35) formed a G4-NDI covalent adduct that was experimentally visible, starting from a compound concentration of 0.25 µM; at 2 µM, maximum alkylation (15%) over total DNA concentration was obtained. Direct comparison of compounds 34 and 36, indicated the key role of the alkyl-spacer in telomeric G4 alkylation. These compounds reported high selectivity towards G4-folded DNA vs dsDNA and a ss-scrambled-DNA unable to fold into G4.
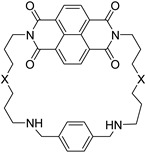
Table 2.
Structures of NDIs used as irreversible ligands to stabilize/modify telomeric G4s.
Subsequently, tri- and tetrasubstituted cNDIs containing only one QMP unit were synthetized to improve alkylation efficiency. In this case, the QMP was tethered to the aromatic core (compounds 37–48, Table 2) and two conformational flexible amines were introduced as solubilizing side chains. FRET-melting values obtained for the non-alkylating compounds (37–40) were in line with other tri- and tetrasubstituted cNDIs, with the best ligand of the series exhibiting ΔTm above 20 °C at 0.8 µM [59]. Interestingly, compound 37 inhibited human melanoma SKMeI-5 cell growth at 48 h with IC50 of 1.7 ± 0.5 µM. The marked reduction of telomeric 3′-overhang, the decrease in the amounts of both the telomere-associated proteins TRF2 and hPOT1 and the increase in γ-H2AX expression levels clearly indicated a noticeable perturbation at the telomeric level.
cNDI-QMPs analogues (41, 42) reported modest alkylation, with the highest percentage of alkylated adduct of 12% at 50 µM of compound 41. Formation of the alkylated adducts was detectable starting from 12 nM (compound 41) and 50 nM (compound 42) and only with the target telomeric G4. However, these compounds showed poor cellular permeability.
Further improvements were introduced in compounds cNDI-QMPs 43–48, in which a phenol moiety was introduced on the cNDI using alkyl-amido spacers [60]. The best alkylating agents of this series were 43 and 44, which presented alkylation starting from 30 nM compound concentration; alkylation maximum yields of 16.8% and 12.8%, respectively, were obtained at compound concentration of 10 µM. Alkylation was particularly selective for the G4 folded DNA structures and cytotoxicity towards two telomerase-positive human carcinoma cell lines (lung and colon) was 2–10 times higher in comparison with the effect observed on telomerase-negative human foreskin fibroblasts.
The above-described alkylating hybrid ligands constituted the first step towards the rational design and synthesis of new molecular devices capable of recognizing, stabilizing and selectively modifying the G4 targets with high efficiency. Freccero’s group next developed a cNDI functionalized with an intrinsically reactive oxirane unit (49, Table 2) [61]. Compound 49 formed adducts with G4 DNA that were stable up to 90 °C and selectively alkylated adenine versus guanine bases in the hTel G4 target. Adduct yield of 16% was obtained at the cNDI/G4 hTel ratio of 12:1, while non-significant alkylation was obtained on ss and ds DNA substrates.
In 2015, the same research group conducted further studies on a photoreactive G4 ligand (37, Table 2) formed by a trisubstituted cNDI dye conjugated to phenol moiety [62]. Upon green light (532 nm) laser excitation, compound 37 generated a phenoxyl radical by intramolecular photoinduced electron transfer (PeT). This carbon radical selectively reacted with thymine residues in the loops of the telomeric G4, inducing formation of 64% of covalent adduct over total DNA at compound concentration of 12.5 µM. The high adduct formation efficiency on the G4 was paralleled by hardly detectable adduct formation on a ss-DNA unable to fold into G4 and no adduct on ds-DNA (even using 200-fold compound excess). Moreover, compound 37 exhibited IC50 of 0.5 µM in the MCF7 breast cancer cell line under green light activation; the IC50 value was four times higher when the compound was maintained in the dark.
In order to verify singlet oxygen formation in PDT conditions, Freccero’s group synthetized a tetra-cationic quaternary ammonium cNDI. Compound 50 (Table 2) presented a singlet oxygen quantum yield of 0.30 upon excitation with red light [42] alongside good G4 binding abilities, similar to those of other tetrasubstituted cNDIs. Compound 50 accumulation into the nucleus of all analysed tumour cell lines (HeLa, BJEHLT and U2OS) was visualized by exploiting its intense emission band in the near-IR. Interestingly, the compound did not display cytotoxicity in the absence of red-light irradiation, a property that allows cNDI activation and hence DNA damage only when the compound is firmly linked to its therapeutic target. Reduction of cell viability up to 40% in BJEHLT and HeLa cell lines and a significant increase of cells expressing γH2AX foci (indicating DNA damage) were reported. However, the presence of four positive charges increased the general binding to nucleic acids due to electrostatic interactions. As a matter of fact, cNDI 50 was less selective towards the G4 structure, with dsDNA as an important off-target.
4. NDIs as G4-Mediated Downregulators of Gene Expression
G4s have been shown to be non-randomly distributed in the human genome: besides telomeres, G4s are mainly present in gene promoters and in regions that are close to transcription start sites (TSS) [63]. Recently, a Ch-IP sequencing-based genome-wide mapping of G4s showed enrichment of G4s in cancer-related genes, with MYC having the highest G4 density [64]. Altogether these data support a role of G4 structures in the progression of cancer [65].
These observations led to the hypothesis that G4 induction and stabilization by small molecules could be exploited to repress gene transcription. This hypothesis was first demonstrated with the porphyrin ligand TMPyP4, which was able to downregulate MYC expression [66], paving the way for the quest of specific G4-stabilizing molecules.
4.1. NDI Derivatives Targeting Oncogenes
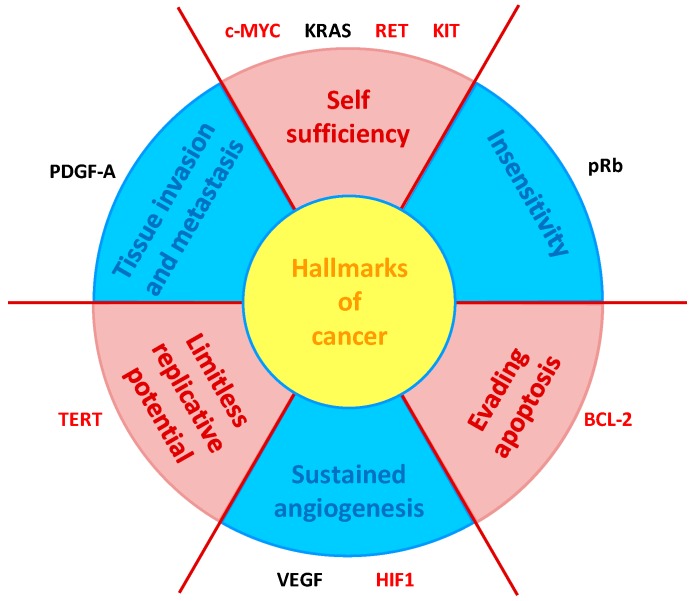
Based on the observation that all processes involved in oncogenic transformation and malignancy—the so-called hallmarks of cancer [67]—are represented by one or more genes containing G4s in their core or proximal promoter (Figure 7) [68], while tumor suppressor genes promoters notably lack G4s [69], a large part of G4 ligands has been designed to target oncogene promoters. In addition, given the multifactoriality of cancer and the involvement of G4s in all its hallmarks, the typical lack of specificity of the ligand towards one single G4 can become an advantage [70].
Figure 7.
Schematic representation of the six hallmarks of cancer [67], in relation with genes containing G4s in their promoters. Gene marked in red are targeted by one or more NDIs described in this session (adapted from [68]).
In this session, we will describe the NDIs developed against each oncogene promoter.
Table 1, Table 2 and Table 3 represent structures of the molecules considered in this section, while in Table 4, we have summarized and compared the in vitro and in cell activity of the presented NDI compounds. However, as a note of caution in interpreting this table, we wish to mention that the correlation between stabilization in vitro and cell activity is not always straightforward due to a series of reasons: the G4 target measured in vitro may not be the sole G4 targeted in cells (as a matter of fact not all G4 in cells have been characterized, therefore some may be targeted without even being recognized as targets). In addition, in cells additional factors come into play: these are related to the cells and their inherent variability. For example, a G4 ligand could be very active in vitro and be less active in cells due to poor entry or localization properties. Poorer or better cell entry may be found in different cell lines. Different cell lines may not be expressing the relevant target, or the same cell line at different cell life stages may be not be expressing the same G4 repertoire [64]. All these factors impact on the relationship (or absence of it) between the in vitro and in vivo results. In addition, activities measured in different labs can also be quite different based on the different experimental settings, and thus a direct comparison might not be straightforward.
Table 3.
Structures of NDIs used to target gene expression.
| Name | R | X | Y | Targeted Gene(s) | Ref. |
| 11 |
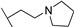
|
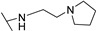
|

|
hTel, KIT, BCL-2 | [76,87] |
| 25 |
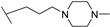
|
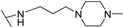
|

|
KIT, HIF1α | [77] |
| 55 |

|
-H | -H | KIT2 | [79] |
| 28 |

|
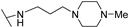
|

|
BCL-2, KRAS | [86] |
| 56 |
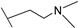
|

|
-H | TERT, BCL-2 | [89] |
| 37 |

|
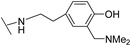
|
-H | TERT, BCL-2, RET, multi-targeting | [89,92] |
| 57 |

|

|

|
TERT, BCL-2 | [89] |
| 60 |

|
-H |

|
Multi-targeting | [95] |
| 58 |
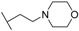
|

|

|
AR | [93] |
| 61 |

|
|

|
hTel, HSP90 | [98] |
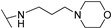
| Name | Structure | X | Targeted Gene(s) | Ref. | |
| 51 |
|
-NH- | hTel, KIT2 | [78] | |
| 52 | -NH(CH2)2NH- | ||||
| 53 | -NH(CH2)3NH- | ||||
| 54 | -NH(CH2)4NH- | ||||
| Name | Structure | Targeted Gene(s) | Ref. | ||
| 59 |
|
AR | [94] | ||
Table 4.
In vitro and cellular data of NDIs used to target gene expression.
| Name | Targeted Gene(s) | ΔTm (°C) a | IC50 (µM) b | mRNA Reduction c | Protein Reduction d | Ref. |
|---|---|---|---|---|---|---|
| 11 | hTel, KIT, BCL-2 | hTel 28.7/29.7, KIT-1 11.2, KIT-2 29.0, BCL-2 promoter 27.6, BCL-2 5’-UTR 10.7, dsDNA 5.7 | GIST882 * 1.6, GIST48 * 0.5, GIST62 * 0.4, HGC-27 * 0.04/0.1, HT-29 * 0.03/0.04 MCF7 * 0.02/0.01 | KIT ~90%, MYC 25–30% | KIT 17%, (in GIST48), KIT 90%, (in GIST882) | [76,87] |
| 25 | KIT, HIF1α | hTel 31.7, KIT-1 1.6, KIT-2 15.1, BCL-2 RNA 21.0, HIF1α 4.9, dsDNA 3.8 | [77] | |||
| 51 | hTel, KIT2 | hTel 12.6, KIT2 12.2, dsDNA 1.4 | A549 * 1.0, MCF7 * 0.8, MIA-PaCa * 0.4, PANC-1* 0.4, ALT * 1.0, WI38 * 1.3 | [78] | ||
| 54 | hTel, KIT2 | hTel 26.8, KIT2 33.1, dsDNA 8.6 | A549 * 10.4, MCF7 * > 25, MIA-PaCa * 19.3, PANC-1 * 13.6, ALT * 14.9, WI38 * > 25 | [78] | ||
| 55 | KIT2 | hTel 2.0, KIT-1 −1.5, KIT-2 14.6, MYC 0.1, dsDNA −2.5 | [79] | |||
| 28 | BCL-2, KRAS | KRAS1 22.5, KRAS2 19.8, BCL-2 26.4 | KRAS2 ~30%, BCL-2 ~40% | [86] | ||
| 56 | TERT, BCL-2 | U251 § 0.180, AG01522 § 1.020 | MYC slight, KIT slight, BCL-2 32.1% | [89] | ||
| 37 | TERT, BCL-2, RET, multi-targeting | U251 # 0.075, AG01522 # 0.430 | [89,92] | |||
| 57 | TERT, BCL-2 | U251 # 0.033, AG01522 # 0.820 | [89] | |||
| 60 | Multi-targeting | hTel 11.9, HSP90A 15.7, HSP90B 12.7, KRAS21 11.0, KRAS32 9.6, BCL-2 13.3, dsDNA 0.6 | A549 * 0.024, MCF7 * 0.159, MIA-PaCa2 * 0.007, PANC-1 * 0.018, WI38 * 1.190 | [95] | ||
| 58 | AR | hTel 25.8, AR3 14.9, dsDNA 0.2 | LNCaP $ 0.42, C4-2 $ 0.39, C4-2b $ 0.29, PC3 $ 0.68, DU145 $ 0.80, PNT1a $ 0.87 | AR 75% (in LNCaP) | AR 4-fold (in LNCaP) | [93] |
| 59 | AR | AR1 20.5, AR3 31.0, dsDNA 1.0 | LNCaP $ 0.14, PC3 $ 0.18, DU145 $ 0.08, PNT1a $ 0.28, 22Rv1 $ 0.01 | AR ~80% (in LNCaP) | AR ~55% (in LNCaP) | [93] |
| 61 | hTel, HSP90 | hTel 34, HSP90A 36, HSP90B 34, dsDNA 9 | RCC4 * 0.61, 786-O * 0.44, MCF7 * 0.014, MIA-PaCa2 * 0.050, A549 * 0.007, PANC-1 * 0.002, WI38 * 0.30 | [98] |
a assessed by FRET melting assay on the models of the indicated sequences; b short-term antiproliferative assay at 48 h (§), 72 h ($), 96 h (*) or 120 h (#); c assessed by RT-PCR; d assessed by Western blot.
4.1.1. KIT
The human proto-oncogene KIT encodes for a tyrosine kinase receptor that stimulates cell proliferation, differentiation and survival [71]. When mutated or overexpressed, KIT results in aberrant function and oncogenic cellular transformation leading to cellular self-sufficiency in growth signals. Activating mutations in KIT are responsible for most of the gastro-intestinal stromal tumors (GIST) and many of the current therapies that target KIT and PDGFRA proteins result in the onset of resistance. Thus, direct targeting of the KIT gene can be an alternative and fruitful approach.
The KIT promoter contains two G4-forming sequences upstream its TSS, namely KIT1 and KIT2 [72,73], whose NMR structures have been solved [74,75].
Several G4 stabilizers have been tested against KIT, including NDIs. The first report presented a tetrasubstituted cNDI (compound 11 in Table 1 and Table 3) [34] that stabilized both KIT-2 and the human telomeric sequence with similar ΔTm values (ΔTm 29.0 °C and 28.7 °C, respectively, Table 4): its activity was higher than that of other compounds such as BRACO-19 and TMPyP4, as assessed by FRET melting analysis [76]. Molecular modeling data of compound 11 on the human telomeric structure showed a higher affinity than the reference compound BRACO-19, paralleling the FRET melting data. Short-term antiproliferative activity showed values similar to imatinib, the reference drug for GIST treatment, in GIST cell lines (IC50 1.6 µM, Table 4); moreover, the NDI was able to reduce KIT transcription by 90% and to completely inhibit KIT expression at the same dosage level required to obtain antiproliferative effect in cells. Given the potent telomerase inhibition, assessed by TRAP-LIG assay, the authors hypothesized a dual mode of action, with inhibition of both telomerase activity and expression of the KIT gene.
A series of tetrasubstituted cNDIs, compounds 25–27 in Table 1, previously reported as stabilizers of the telomeric G4 and telomerase inhibitors [54], were able to stabilize also the G4s in the promoter region of the KIT and HIF-1α oncogenes, and the G4 in the mRNA of BCL-2 (Table 4) [77]. Compound 25 (Table 3) was able to initiate senescence in cancer cells, probably as a consequence of telomere uncapping through G4 stabilization, and subsequent DNA damage, as demonstrated by the detection of γ-H2AX foci. Furthermore, the compound led to telomeric chromosomal fusion, resulting in genomic instability and subsequent apoptosis. The closely related molecule 27 (Table 1) was able to enter cell cytoplasm and nucleoli, as assessed by confocal microscopy; it induced an increase of telomeric associations in cells, stimulating chromosomal instability.
Macrocyclic NDIs bearing spermidine- and spermine-like side chains (compounds 51–54, Table 3) [78] stabilized both the KIT2 and telomeric sequences (ΔTm 33.1 °C and 26.8 °C, respectively, Table 4) in biophysical assays, but they were not able to inhibit telomerase. Computational studies confirmed that the most stabilizing molecule was also the best G4 binder and proposed the electrostatic contribution of the side chains as the driving force for the binding process. However, in cells, the antiproliferative effect was not related to G4 stabilization (the higher the ΔTm, the lower the antiproliferative effect, and vice versa), and lacked cell-type selectivity: this was likely due to the physicochemical properties of the molecules, or to a different mechanism of action.
NDIs functionalized with disubstituted aminoacids were synthetized and investigated as G4 binders [79]: one of them (compound 55, Table 3) showed selective stabilization of the KIT2 sequence (ΔTm 14.6 °C, Table 4) coupled with a slight destabilization of the KIT 1 sequence in vitro.
4.1.2. BCL-2
BCL-2 is an antiapoptotic gene containing several G4-forming sequences in its promoter [80,81,82] and one in the 5′-UTR of its mRNA [83]. These sequences are able to fold into different intramolecular structures, having a critical role in BCL-2 gene expression regulation [84]. Over-expression of the BCL-2 protein prevents apopotosis and has been found to interfere with cancer therapeutics [85].
The tetrasubstituted cNDI derivative 28 (Table 1 and Table 3) belongs to a previously reported series of molecules that had been characterized as potent stabilizers of different G4s and antiproliferative agents in a panel of cancer cell lines, without being telomerase inhibitors [55]. Subsequently, 28 was reported to stabilize the KRAS1, KRAS2 and BCL-2 G4 sequences, with a slight selectivity for the latter (ΔTm 22.5 °C, 19.8 °C and 26.4 °C, respectively). In vivo studies showed nuclear localization in tumor cells, reduction of about 80% of the tumor growth in mice, and reduction of KRAS and BCL-2 protein levels in treated tumors (Table 4) [86].
In addition, the tetrasubstituted cNDI 11 (Table 1 and Table 3), active against KIT [76] as presented above, stabilized the BCL-2 G4 [87]. The compound showed high antiproliferative activity in a GIST-imatinib resistant cell line (GIST48) in short-term experiments (IC50 0.5 ± 0.05 µM, Table 4), did not affect telomerase activity in this cell line, and did not have a large effect on the KIT gene. Interestingly, this cNDI was able to strongly reduce (about 85%) the expression level of the BCL-2 protein both in cells and in a reporter system: hence, the authors inferred that the high potency of the molecule was mainly due to its activity on BLC-2 and that it acted both at the transcriptional (targeting DNA) and translational level (targeting RNA).
Recently, a series of three different G4-binding cNDI derivatives (compounds 56, 37, 57, Table 3) [59,62,88] were tested as inhibitors of cell proliferation in glioblastoma cells [89]. Interestingly, these cNDIs showed antiproliferative activity in the nanomolar range in U251MG cells, more than 10-times better than the reference compound RHPS4 (IC50 0.033 ± 0.006 and 0.460 ± 0.060 µM, respectively, Table 4) [90], while they had lower efficacy in targeting telomeres, as assessed by immunostaining and ChIP assay. This behavior, accompanied by the absence of radiosensitization in treated cells, supported the hypothesis of targets other than telomeres. Indeed, one of the molecules was able to induce BCL2 downregulation both at transcripts and protein levels and perturbation of cell cycle and apoptosis with a different mechanism of action with respect to RHPS4.
4.1.3. RET
RET (REarranged during Transfection) is a proto-oncogene associated with medullary thyroid cancer containing a G4 forming sequence in its promoter [91]. A previously published molecule (compound 37, Table 2 and Table 3) [59] was demonstrated to exert antiproliferative effect in cells (IC50 1.7 ± 0.2 µM, Table 4), coupled with a remarkable downregulation of RET expression. In particular, RET expression was markedly impaired, while the effect on MYC and BCL-2 proteins was negligible. The compound efficiently recognized and stabilized the RET G4 sequence in vitro, as assessed by FRET melting (ΔTm > 37.3 °C, measured by CD thermal unfolding at 10 mM K+), and Taq polymerase stop assay. Furthermore, in a reporter system, treatment with 37 showed a significant and dose-dependent inhibition of luciferase activity. Finally, in a mouse model of medullary thyroid cancer, treatment with this cNDI led to significant tumor growth delay and induced apoptosis [92].
4.1.4. AR
The androgen receptor (AR) has a role in the development of both benign and malignant prostate cancer, even after androgen deprivation. Because castration-resistant prostate cancer (CRPC) progresses in spite of androgen deprivation, transcriptional repression of AR signaling is a rational treatment approach.
A G4 forming region has been identified within the minimal promoter of the AR gene [93]: the three reported G4-forming sequences were characterized by means of biophysical techniques; one of them, showing the least heterogeneous NMR signal, was selected as target for molecule development. Among the seven cNDIs screened as ligands against this sequence, one of them (a tetrasubstituted cNDI, compound 58, Table 3) showed the ability to stabilize the AR G4, even if to a lower extent than the telomeric one (ΔTm 14.9 ± 0.4 °C and 25.8 ± 0.06 °C respectively, measured by FRET, Table 4). This compound also had an antiproliferative effect in the nanomolar range (IC50 0.29 ± 0.02 µM, Table 4) on prostate cancer cell lines and, very interestingly, a downregulation effect on AR transcription, coupled with perturbation of some AR’s upstream transcription factors.
Recently, a library of differently functionalized cNDIs (tri- and tetrasubstituted, core extended NDIs and NDI-dyads) was screened for the ability to stabilize the AR G4s [94]. A core extended NDI (compound 59 in Table 3) was identified as the best AR G4 stabilizer by means of FRET melting assay (ΔTm 31.0 ± 0.9 °C, Table 4): its affinity toward the AR G4 was in the low nanomolar range, as assessed by SPR analysis. Stabilization of the AR G4 was also confirmed by CD thermal folding analysis and Taq polymerase stop assay. The molecule had an antiproliferative effect which was greater in AR-positive (low nanomolar range, IC50 0.010 ± 0.001 µM, Table 4) than in AR-negative prostate cancer cell lines. Moreover, a marked decrease in the expression levels of AR but also of its downstream regulated gene KLK3 was observed, as well as marked downregulation of genes involved in tumor cell biology, such as cell cycle regulators, apoptosis inhibitors (BCL-2), KRAS, and genes involved in the PI3K/AKT/mTOR signaling pathway. Finally, the treatment of prostate cancer cell lines with this cNDI in combination with the AR antagonist Enzalutamide (used in second line therapies) resulted in a synergistic interaction in AR-positive but not in AR-negative cell lines.
4.1.5. Multi-Targeting NDIs
As already stated, tumor malignancies are multifactorial diseases, for which the lack of selectivity could be exploited as a positive feature. Indeed, a number of NDIs targeting multiple G4 structures and exerting antiproliferative activity has been reported.
A trisubstituted cNDI (compound 37, Table 1 and Table 3), previously indicated to stabilize the telomeric G4, cause telomere dysfunction and downregulate telomerase activity [59], was investigated to clarify its mechanism of action by analyzing its ability to interfere with oncogene expression [88]. The compound affected the expression levels of G4-containing genes, such as TERT, MYC, BCL-2 and KIT, and preferentially bound and greatly stabilized G4s in the promoter region of MYC and BCL-2 (Table 4). Gene expression profile analyses were performed in treated cells, showing a general and cell-type dependent perturbation of expression.
A recent paper presented a trisubstituted cNDI (compound 60, Table 3) able to affect human pancreatic ductal adenocarcinoma (PDAC) cell lines [95]. The molecule was designed by computer modeling starting from a co-crystal structure of a different cNDI in complex with an intramolecular telomeric G4 [55]: the rationally designed molecule stabilized G4s in vitro, even if less potently than the parental molecule, and was a potent antiproliferative agent, with IC50 in the low nanomolar range (IC50 0.007 ± 0.002 µM, Table 4) in PDAC cell lines. Moreover, in a mouse xenograft model of PDAC, the cNDI showed antitumor activity. Instead of checking the activity on a selected G4 target, the authors analyzed the global genome transcriptome by means of RNA-seq, in order to identify all genes affected by the molecule: the downregulated gene set was shown to be enriched in G4 forming sequences. Moreover, cell treatment with this molecule resulted in an increase in anti-G4 antibody (BG4) foci and DNA damage recorded as double strand breaks (DSB).
4.2. Naphthalene Diimides Targeting Other Genes
Targeting of G4s present within the promoter of other genes can also produce an anticancer effect: it is the case of the molecules reviewed in this section, targeting G4s in the promoter of the HSP90 gene.
HSP90
The molecular chaperone heat shock protein 90 (HSP90) stabilizes a number of oncogenic proteins involved in tumor proliferation and malignant progression and for this reason it is considered a rational target for anticancer therapy [96].
Two G4 forming sequences in the promoter of HSP90 gene have been reported and characterized [97]. A series of tetrasubstituted cNDIs, positional isomers of previously published molecules, has been screened for the ability to bind and stabilize the HSP90 G4s [98]. Many of these molecules exerted a marked stabilization of the two HSP90 G4s as well as the telomeric one (ΔTm 34 °C, 29 °C and 27 °C respectively, measured by FRET, Table 4); the most active of them (compound 61, Table 3) had a cytotoxic effect on pancreatic and lung cancer cell lines in the low nanomolar range (IC50 0.002 ± 0.001 µM, Table 4). Long-term telomere length studies showed no significant change in telomere length, indicating that the antiproliferative effect does not involve inhibition of telomerase. Finally, molecular dynamics studies helped the authors to understand if and how the positional isomers had any advantage in terms of G4 stabilization and inhibition of cell proliferation: in some cases, they found that more asymmetrically shaped molecules were more potent than their lead compounds, a fact that was likely due to subtle differences in the binding geometry to their targets.
5. Naphthalene Diimides as G4 Binders in Microorganisms
NDIs have also been employed as G4 binders in microorganisms: in particular, NDI derivatives were tested against the immunodeficiency virus 1 (HIV-1), the herpes simplex virus 1 (HSV-1), and a few parasites.
A core extended NDI derivative (compound 59, Table 3 and Table 4) exhibited a very promising antiviral activity in the low nanomolar range against different strains of HIV-1 (IC50 < 25 nM) [9], combined with a very low cytotoxicity and a very promising therapeutic window. Its high affinity for the viral G4s [20] and lower affinity for the telomeric one was accounted for in its antiviral activity. The same core extended NDI was also analyzed in the context of HSV-1 infection: the molecule showed a remarkable antiviral activity (IC50 = 0.018 ± 0.001 nM) and was more efficient than the reference drug acyclovir, the antiviral drug of choice for the treatment of HSV-1 infections [99]. In this case, the cNDI derivative did not exhibit a net preferentiality for the viral G4s over the cellular ones, but the higher content of HSV-1 G4s during the viral cycle was indicated to play a major role in the anti-herpetic effect.
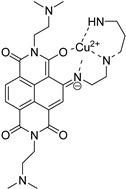
Recently, a diethylenetriamine (DETA) substituted cNDI (compound 62, Table 5) was shown to selectively chelate Cu2+ and to stabilize different G4 forming sequences [100]. In particular, taking advantage of the copper-catalyzed reduction of hydrogen peroxide in the presence of ascorbate, the molecule was able to generate reactive oxygen species (ROS) that did not diffuse in solution but reacted at specific sites of the target upon G4 binding: it was the case of the specific cleavage observed against the HIV-1 LTR-III [20] and HSV-1 un2 sequences [101]. The mechanism of action was characterized by mass spectrometry (MS and MS/MS): analysis of the cleavage products indicated H1′ and H4′ abstractions by the hydroxyl radical as the main mechanism of the G4 breaks. The complex of the molecule with LTR-III was also investigated by one-dimensional 1H-NMR: the ligand showed a binding preference for the duplex-quadruplex junction, a peculiar feature of this sequence, reported for the first time in a naturally occurring and biological relevant G4, and recently solved by NMR [102].
Table 5.
Structures of cNDIs used to target G4s in microorganisms.
Finally, a series of carbohydrate cNDI derivatives, previously evaluated as G4 binders and antitumor compounds [57], was tested against Trypanosoma brucei, Leishmania major and Plasmodium falciparum [103]. Two molecules (compounds 32 and 63, Table 5), which showed antiparasitic activity against the tested species, especially against T. brucei, greater than the G4-stabilizing reference compound BRACO-19, TmPyP4 and pyridostatin and accumulation in the parasite nucleus and kinetoplast, suggested that the G4 targets be mainly present in these locations. This series of carbohydrate cNDI derivatives was recently implemented, modifying both the spacer between the sugar unit and the NDI core, and the substituents [104]. Most of the new members of the family displayed IC50 values against T. brucei in the sub-µM range, coupled with a notable selectivity over control cells; moreover, all the carb-NDI conjugates showed a stabilization of the telomeric and the EBR1 sequences, greater than the previously reported molecules. Again, the selective localization in the T. brucei nucleus and kinetoplast, targets that harbor the putative G4 forming sequences, support the hypothesis of a novel G4-mediated antiparasitic approach.
6. Conclusions
NDI derivatives are molecules that, by virtue of their large aromatic core, selectively bind G4s. Their properties can be largely varied by the addition of specific substituents, making them amenable to developments as attractive anticancer and antimicrobial drugs and as G4 markers in cells.
In general, the parameters that describe the potency of G4 stabilization do not perfectly correlate with the IC50 anti-proliferative data. However, it is possible to envisage an overall rationalization since the best binders in general offer an outstanding biological activity. Starting from the first studied di-substituted cNDIs to the tetra-substituted one, an increase in the number of side chains corresponded to an increase in G4 stabilization. Considering that the interaction with the G4 grooves is fundamental to improve G4-ligand binding, many of the developed cNDIs bear a large positive charge on the side chains. This chemical property guarantees a greater interaction with the negative phosphate groups and good cellular permeability. However, due to the electrostatic interaction, an excess of positive charge reduces the selectivity of these NDIs towards the target, making them able to bind to other NA secondary structures as well. Enhanced G4 selectivity was thus obtained by reducing the protonable sites in the side chains and taking care not to lose the intermolecular interaction with the G4 grooves.
In order to optimize G4 stabilization and cell entry, another crucial parameter is the length of the functionalized side chains. Based on different observations, the three-carbon atom linker guaranteed the best compromise. Nevertheless, conjugation of active transport moieties improved cellular uptake.
Moreover, the extension of the cNDIs’ aromatic core is important: this modification greatly increases the affinity towards G4s, allowing the biological activity of these derivatives to reach the low nanomolar range.
In conclusion, the high potency and selectivity towards the NA G4 conformation make cNDI derivatives promising therapeutic agents, especially for cancer applications, where most of the G4s are involved in hallmarks of cancer. In this scenario, compounds not selective for a specific G4 could in some cases be advantageous. Conversely, for the treatment of diseases caused by infective agents, a discrete selectivity toward the target of choice would be more advisable. In this case, additional functional modifications will be needed. So far, compounds reported to have an increased selectivity for a specific G4 have added side chains that typically recognize flanking regions of the selected G4 [105]. Therefore, this may be a necessary route for the development of more selective compounds. With the compounds increasing in size, bioavailability may become an issue, which a prior accurate design of the side chains themselves could help overcome. Alternatively, a powerful screening or molecule construction towards and around the G4 target may yield small molecules with a reasonably small size that are selective for the G4 of choice [106,107]. In general, however, given that G4s demand that extensive planar moieties be optimally and selectively recognized, bioavailability of G4-ligands looks like the most impendent issue to be solved for the successful use of these compounds as therapeutic agents.
Acknowledgments
We thank E. Ruggiero for helpful discussion and advice, M. Zuffo for original idea in a figure edit and T. Agenda for information technology support.
Author Contributions
Literature review, Writing—Original Draft preparation, V.P., M.N. and F.D.; Writing—Review and Editing, S.N.R.
Funding
This research was funded by the European Research Council grant number (ERC Consolidator 615879).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansel-Hertsch R., Di Antonio M., Balasubramanian S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 3.Berardinelli F., Siteni S., Tanzarella C., Stevens M.F., Sgura A., Antoccia A. The G-quadruplex-stabilising agent RHPS4 induces telomeric dysfunction and enhances radiosensitivity in glioblastoma cells. DNA Repair. 2015;25:104–115. doi: 10.1016/j.dnarep.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Takahama K., Takada A., Tada S., Shimizu M., Sayama K., Kurokawa R., Oyoshi T. Regulation of Telomere Length by G-Quadruplex Telomere DNA- and TERRA-Binding Protein TLS/FUS. Chem. Biol. 2013;20:341–350. doi: 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Zeraati M., Moye A.L., Wong J.W.H., Perera D., Cowley M.J., Christ D.U., Bryan T.M., Dinger M.E. Cancer-associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression. Sci. Rep. 2017;7:708. doi: 10.1038/s41598-017-00739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin Y.J., Kumarasamy V., Camacho D., Sun D. Involvement of G-quadruplex structures in regulation of human RET gene expression by small molecules in human medullary thyroid carcinoma TT cells. Oncogene. 2015;34:1292–1299. doi: 10.1038/onc.2014.65. [DOI] [PubMed] [Google Scholar]
- 7.Gomez D., Guedin A., Mergny J.L., Salles B., Riou J.F., Teulade-Fichou M.P., Calsou P. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucl. Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwala P., Pandey S., Mapa K., Maiti S. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor beta2. Biochemistry. 2013;52:1528–1538. doi: 10.1021/bi301365g. [DOI] [PubMed] [Google Scholar]
- 9.Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M., et al. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015;58:9639–9652. doi: 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas B., Kandpal M., Vivekanandan P. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017;45:11268–11280. doi: 10.1093/nar/gkx823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardin C.C., Corregan M., Brown B.A., Frederick L.N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee A.K., Sharma S., Chowdhury S. Non-duplex G-Quadruplex Structures Emerge as Mediators of Epigenetic Modifications. Trends Genet. 2018 doi: 10.1016/j.tig.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J., Fleming A.M., Averill A.M., Burrows C.J., Wallace S.S. Nucleic Acids Res. Volume 43. Oxford University Press; Oxford, UK: 2015. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures; p. 7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X.S., Wu H., Krzisch M., Wu X., Graef J., Muffat J., Hnisz D., Li C.H., Yuan B., Xu C., et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell. 2018;172:979–992. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilbaud G., Murat P., Recolin B., Campbell B.C., Maiter A., Sale J.E., Balasubramanian S. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017;9:1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lammich S., Buell D., Zilow S., Ludwig A.K., Nuscher B., Lichtenthaler S.F., Prinzen C., Fahrenholz F., Haass C. Expression of the anti-amyloidogenic secretase ADAM10 is suppressed by its 5′-untranslated region. J. Biol. Chem. 2010;285:15753–15760. doi: 10.1074/jbc.M110.110742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lammich S., Kamp F., Wagner J., Nuscher B., Zilow S., Ludwig A.-K., Willem M., Haass C. Translational Repression of the Disintegrin and Metalloprotease ADAM10 by a Stable G-quadruplex Secondary Structure in Its 5′-Untranslated Region. J. Biol. Chem. 2011;286:45063–45072. doi: 10.1074/jbc.M111.296921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palù G., Richter S.N. A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 2013;56:6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016;5 doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimino-Reale G., Zaffaroni N., Folini M. Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy. Curr. Pharm. Des. 2016;22:6612–6624. doi: 10.2174/1381612822666160831101031. [DOI] [PubMed] [Google Scholar]
- 23.Kaulage M., Maji B., Bhat J., Iwasaki Y., Chatterjee S., Bhattacharya S., Muniyappa K. Discovery and Structural Characterization of G-quadruplex DNA in Human Acetyl-CoA Carboxylase Gene Promoters: Its Role in Transcriptional Regulation and as a Therapeutic Target for Human Disease. J. Med. Chem. 2016;59:5035–5050. doi: 10.1021/acs.jmedchem.6b00453. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S., Wu Y., Zhang W. G-quadruplex structures and their interaction diversity with ligands. ChemMedChem. 2014;9:899–911. doi: 10.1002/cmdc.201300566. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson G.N., Cuenca F., Neidle S. Topology Conservation and Loop Flexibility in Quadruplex–Drug Recognition: Crystal Structures of Inter- and Intramolecular Telomeric DNA Quadruplex–Drug Complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 26.Wurthner F. Perylene bisimide dyes as versatile building blocks for functional supramolecular architectures. Chem. Commun. Camb. 2004:1564–1579. doi: 10.1039/B401630K. [DOI] [PubMed] [Google Scholar]
- 27.Sakai N., Mareda J., Vauthey E., Matile S. Core-substituted naphthalenediimides. Chem. Commun. 2010;46:4225. doi: 10.1039/c0cc00078g. [DOI] [PubMed] [Google Scholar]
- 28.Wurthner F., Stolte M. Naphthalene and perylene diimides for organic transistors. Chem. Commun. Camb. 2011;47:5109–5115. doi: 10.1039/c1cc10321k. [DOI] [PubMed] [Google Scholar]
- 29.Zhan X., Facchetti A., Barlow S., Marks T.J., Ratner M.A., Wasielewski M.R., Marder S.R. Rylene and related diimides for organic electronics. Adv. Mater. 2011;23:268–284. doi: 10.1002/adma.201001402. [DOI] [PubMed] [Google Scholar]
- 30.Katz H.E., Lovinger A.J., Johnson J., Kloc C., Siegrist T., Li W., Lin Y.Y., Dodabalapur A. A soluble and air-stable organic semiconductor with high electron mobility. Nature. 2000;404:478–481. doi: 10.1038/35006603. [DOI] [PubMed] [Google Scholar]
- 31.Guha S., Goodson F.S., Corson L.J., Saha S. Boundaries of Anion/Naphthalenediimide Interactions: From Anion−π Interactions to Anion-Induced Charge-Transfer and Electron-Transfer Phenomena. J. Am. Chem. Soc. 2012;134:13679–13691. doi: 10.1021/ja303173n. [DOI] [PubMed] [Google Scholar]
- 32.Suraru S.L., Wurthner F. Strategies for the synthesis of functional naphthalene diimides. Angew. Chem. Int. Ed. Engl. 2014;53:7428–7448. doi: 10.1002/anie.201309746. [DOI] [PubMed] [Google Scholar]
- 33.Würthner F., Ahmed S., Thalacker C., Debaerdemaeker T. Core-Substituted Naphthalene Bisimides: New Fluorophors with Tunable Emission Wavelength for FRET Studies. Chem. Eur. J. 2002;8:4742–4750. doi: 10.1002/1521-3765(20021018)8:20<4742::AID-CHEM4742>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Cuenca F., Greciano O., Gunaratnam M., Haider S., Munnur D., Nanjunda R., Wilson W.D., Neidle S. Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands. Bioorg. Med. Chem. Lett. 2008;18:1668–1673. doi: 10.1016/j.bmcl.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 35.Doria F., di Antonio M., Benotti M., Verga D., Freccero M. Substituted heterocyclic naphthalene diimides with unexpected acidity. Synthesis, properties, and reactivity. J. Org. Chem. 2009;74:8616–8625. doi: 10.1021/jo9017342. [DOI] [PubMed] [Google Scholar]
- 36.Thalacker C., Roger C., Wurthner F. Synthesis and optical and redox properties of core-substituted naphthalene diimide dyes. J. Org. Chem. 2006;71:8098–8105. doi: 10.1021/jo0612269. [DOI] [PubMed] [Google Scholar]
- 37.Bhosale R., Misek J., Sakai N., Matile S. Supramolecular n/p-heterojunction photosystems with oriented multicolored antiparallel redox gradients (OMARG-SHJs) Chem. Soc. Rev. 2010;39:138–149. doi: 10.1039/B906115K. [DOI] [PubMed] [Google Scholar]
- 38.Kishore R.S., Kel O., Banerji N., Emery D., Bollot G., Mareda J., Gomez-Casado A., Jonkheijm P., Huskens J., Maroni P., et al. Ordered and oriented supramolecular n/p-heterojunction surface architectures: Completion of the primary color collection. J. Am. Chem. Soc. 2009;131:11106–11116. doi: 10.1021/ja9030648. [DOI] [PubMed] [Google Scholar]
- 39.Chen W., Zhang J., Long G., Liu Y., Zhang Q. From non-detectable to decent: Replacement of oxygen with sulfur in naphthalene diimide boosts electron transport in organic thin-film transistors (OTFT) J. Mater. Chem. C. 2015;3:8219–8224. doi: 10.1039/C5TC01519G. [DOI] [Google Scholar]
- 40.Shukla D., Nelson S.F., Freeman D.C., Rajeswaran M., Ahearn W.G., Meyer D.M., Carey J.T. Thin-Film Morphology Control in Naphthalene-Diimide-Based Semiconductors: High Mobility n-Type Semiconductor for Organic Thin-Film Transistors. Chem. Mater. 2008;20:7486–7491. doi: 10.1021/cm802071w. [DOI] [Google Scholar]
- 41.Kobaisi M.A., Bhosale S.V., Latham K., Raynor A.M. Functional Naphthalene Diimides: Synthesis, Properties, and Applications. Chem. Rev. 2016;116:11685–11796. doi: 10.1021/acs.chemrev.6b00160. [DOI] [PubMed] [Google Scholar]
- 42.Salvati E., Doria F., Manoli F., D’Angelo C., Biroccio A., Freccero M., Manet I. A bimodal fluorescent and photocytotoxic naphthalene diimide for theranostic applications. Org. Biomol. Chem. 2016;14:7238–7249. doi: 10.1039/C6OB00987E. [DOI] [PubMed] [Google Scholar]
- 43.Doria F., Manet I., Grande V., Monti S., Freccero M. Water-soluble naphthalene diimides as singlet oxygen sensitizers. J. Org. Chem. 2013;78:8065–8073. doi: 10.1021/jo401347z. [DOI] [PubMed] [Google Scholar]
- 44.Zuffo M., Guedin A., Leriche E.D., Doria F., Pirota V., Gabelica V., Mergny J.L., Freccero M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doria F., Oppi A., Manoli F., Botti S., Kandoth N., Grande V., Manet I., Freccero M. A naphthalene diimide dyad for fluorescence switch-on detection of G-quadruplexes. Chem. Commun. Camb. 2015;51:9105–9108. doi: 10.1039/C5CC01536G. [DOI] [PubMed] [Google Scholar]
- 46.Doria F., Nadai M., Sattin G., Pasotti L., Richter S.N., Freccero M. Water soluble extended naphthalene diimides as pH fluorescent sensors and G-quadruplex ligands. Org. Biomol. Chem. 2012;10:3830–3840. doi: 10.1039/c2ob07006e. [DOI] [PubMed] [Google Scholar]
- 47.Zuffo M., Ladame S., Doria F., Freccero M. Tuneable coumarin-NDI dyads as G-quadruplex specific light-up probes. Sensors Actuators B Chem. 2017;245:780–788. doi: 10.1016/j.snb.2017.01.144. [DOI] [Google Scholar]
- 48.Zuffo M., Doria F., Spalluto V., Ladame S., Freccero M. Red/NIR G-Quadruplex Sensing, Harvesting Blue Light by a Coumarin-Naphthalene Diimide Dyad. Chemistry. 2015;21:17596–17600. doi: 10.1002/chem.201503020. [DOI] [PubMed] [Google Scholar]
- 49.Zuffo M., Doria F., Botti S., Bergamaschi G., Freccero M. G-quadruplex fluorescence sensing by core-extended naphthalene diimides. Biochim Biophys Acta Gen. Subj. 2017;1861:1303–1311. doi: 10.1016/j.bbagen.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Doria F., Nadai M., Zuffo M., Perrone R., Freccero M., Richter S.N. A red-NIR fluorescent dye detecting nuclear DNA G-quadruplexes: In vitro analysis and cell imaging. Chem. Commun. Camb. 2017;53:2268–2271. doi: 10.1039/C6CC08492C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collie G., Reszka A.P., Haider S.M., Gabelica V., Parkinson G.N., Neidle S. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 52.Collie G.W., Haider S.M., Neidle S., Parkinson G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucl. Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collie G.W., Promontorio R., Hampel S.M., Micco M., Neidle S., Parkinson G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012;134:2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- 54.Hampel S.M., Sidibe A., Gunaratnam M., Riou J.F., Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg. Med. Chem. Lett. 2010;20:6459–6463. doi: 10.1016/j.bmcl.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 55.Micco M., Collie G.W., Dale A.G., Ohnmacht S.A., Pazitna I., Gunaratnam M., Reszka A.P., Neidle S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013;56:2959–2974. doi: 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- 56.Czerwinska I., Sato S., Takenaka S. Improving the affinity of naphthalene diimide ligand to telomeric DNA by incorporating Zn(2)(+) ions into its dipicolylamine groups. Bioorg. Med. Chem. 2012;20:6416–6422. doi: 10.1016/j.bmc.2012.08.055. [DOI] [PubMed] [Google Scholar]
- 57.Arevalo-Ruiz M., Doria F., Belmonte-Reche E., De Rache A., Campos-Salinas J., Lucas R., Falomir E., Carda M., Perez-Victoria J.M., Mergny J.L., et al. Synthesis, Binding Properties, and Differences in Cell Uptake of G-Quadruplex Ligands Based on Carbohydrate Naphthalene Diimide Conjugates. Chemistry. 2017;23:2157–2164. doi: 10.1002/chem.201604886. [DOI] [PubMed] [Google Scholar]
- 58.Di Antonio M., Doria F., Richter S.N., Bertipaglia C., Mella M., Sissi C., Palumbo M., Freccero M. Quinone methides tethered to naphthalene diimides as selective G-quadruplex alkylating agents. J. Am. Chem. Soc. 2009;131:13132–13141. doi: 10.1021/ja904876q. [DOI] [PubMed] [Google Scholar]
- 59.Doria F., Nadai M., Folini M., Di Antonio M., Germani L., Percivalle C., Sissi C., Zaffaroni N., Alcaro S., Artese A., et al. Hybrid ligand-alkylating agents targeting telomeric G-quadruplex structures. Org. Biomol. Chem. 2012;10:2798–2806. doi: 10.1039/c2ob06816h. [DOI] [PubMed] [Google Scholar]
- 60.Nadai M., Doria F., Di Antonio M., Sattin G., Germani L., Percivalle C., Palumbo M., Richter S.N., Freccero M. Naphthalene diimide scaffolds with dual reversible and covalent interaction properties towards G-quadruplex. Biochimie. 2011;93:1328–1340. doi: 10.1016/j.biochi.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 61.Doria F., Nadai M., Folini M., Scalabrin M., Germani L., Sattin G., Mella M., Palumbo M., Zaffaroni N., Fabris D., et al. Targeting loop adenines in G-quadruplex by a selective oxirane. Chemistry. 2013;19:78–81. doi: 10.1002/chem.201203097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nadai M., Doria F., Germani L., Richter S.N., Freccero M. A photoreactive G-quadruplex ligand triggered by green light. Chemistry. 2015;21:2330–2334. doi: 10.1002/chem.201405215. [DOI] [PubMed] [Google Scholar]
- 63.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M., et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 65.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 68.Brooks T.A., Kendrick S., Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maizels N. G4-associated human diseases. Embo Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017;1 doi: 10.1038/s41570-017-0041. [DOI] [Google Scholar]
- 71.Yarden Y., Kuang W.J., Yangfeng T., Coussens L., Munemitsu S., Dull T.J., Chen E., Schlessinger J., Francke U., Ullrich A. Human Protooncogene C-Kit-a New Cell-Surface Receptor Tyrosine Kinase for an Unidentified Ligand. Embo J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernando H., Reszka A.P., Huppert J., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phan A.T., Kuryavyi V., Burge S., Neidle S., Patel D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Todd A.K., Haider S.M., Parkinson G.N., Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gunaratnam M., Swank S., Haider S.M., Galesa K., Reszka A.P., Beltran M., Cuenca F., Fletcher J.A., Neidle S. Targeting Human Gastrointestinal Stromal Tumor Cells with a Quadruplex-Binding Small Molecule. J. Med. Chem. 2009;52:3774–3783. doi: 10.1021/jm900424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hampel S.M., Pepe A., Greulich-Bode K.M., Malhotra S.V., Reszka A.P., Veith S., Boukamp P., Neidle S. Mechanism of the Antiproliferative Activity of Some Naphthalene Diimide G-Quadruplex Ligands. Mol. Pharmacol. 2013;83:470–480. doi: 10.1124/mol.112.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchetti C., Minarini A., Tumiatti V., Moraca F., Parrotta L., Alcaro S., Rigo R., Sissi C., Gunaratnam M., Ohnmacht S.A., et al. Macrocyclic naphthalene diimides as G-quadruplex binders. Bioorg. Med. Chem. 2015;23:3819–3830. doi: 10.1016/j.bmc.2015.03.076. [DOI] [PubMed] [Google Scholar]
- 79.Rasadean D.M., Sheng B., Dash J., Pantos G.D. Amino-Acid-Derived Naphthalenediimides as Versatile G-Quadruplex Binders. Chem.-Eur. J. 2017;23:8491–8499. doi: 10.1002/chem.201700957. [DOI] [PubMed] [Google Scholar]
- 80.Dai J.X., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D.Z. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai J.X., Chen D., Jones R.A., Hurley L.H., Yang D.Z. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agrawal P., Lin C., Mathad R.I., Carver M., Yang D.Z. The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-nt Loop in K+ Solution. J. Am. Chem. Soc. 2014;136:1750–1753. doi: 10.1021/ja4118945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bugaut A., Balasubramanian S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Onel B., Carver M., Wu G.H., Timonina D., Kalarn S., Larriva M., Yang D.Z. A New G-Quadruplex with Hairpin Loop Immediately Upstream of the Human BCL2 P1 Promoter Modulates Transcription. J. Am. Chem. Soc. 2016;138:2563–2570. doi: 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reed J.C., Kitada S., Takayama S., Miyashita T. Regulation of Chemoresistance by the Bcl-2 Oncoprotein in Non-Hodgkins-Lymphoma and Lymphocytic-Leukemia Cell-Lines. Ann. Oncol. 1994;5:S61–S65. doi: 10.1093/annonc/5.suppl_1.S61. [DOI] [PubMed] [Google Scholar]
- 86.Ohnmacht S.A., Marchetti C., Gunaratnam M., Besser R.J., Haider S.M., Di Vita G., Lowe H.L., Mellinas-Gomez M., Diocou S., Robson M., et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015;5 doi: 10.1038/srep11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gunaratnam M., Collie G.W., Reszka A.P., Todd A.K., Parkinson G.N., Neidle S. A naphthalene diimide G-quadruplex ligand inhibits cell growth and down-regulates BCL-2 expression in an imatinib-resistant gastrointestinal cancer cell line. Bioorg. Med. Chem. 2018;26:2958–2964. doi: 10.1016/j.bmc.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 88.Nadai M., Cimino-Reale G., Sattin G., Doria F., Butovskaya E., Zaffaroni N., Freccero M., Palumbo M., Richter S.N., Folini M. Assessment of gene promoter G-quadruplex binding and modulation by a naphthalene diimide derivative in tumor cells. Int. J. Oncol. 2015;46:369–380. doi: 10.3892/ijo.2014.2723. [DOI] [PubMed] [Google Scholar]
- 89.Muoio D., Berardinelli F., Leone S., Coluzzi E., di Masi A., Doria F., Freccero M., Sgura A., Folini M., Antoccia A. Naphthalene diimide-derivatives G-quadruplex ligands induce cell proliferation inhibition, mild telomeric dysfunction and cell cycle perturbation in U251MG glioma cells. FEBS J. 2018;285:3769–3785. doi: 10.1111/febs.14628. [DOI] [PubMed] [Google Scholar]
- 90.Gowan S.M., Heald R., Stevens M.F.G., Kelland L.R. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 91.Guo K., Pourpak A., Beetz-Rogers K., Gokhale V., Sun D., Hurley L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lopergolo A., Perrone R., Tortoreto M., Doria F., Beretta G.L., Zuco V., Freccero M., Borrello M.G., Lanzi C., Richter S.N., et al. Targeting of RET oncogene by naphthalene diimide-mediated gene promoter G-quadruplex stabilization exerts anti-tumor activity in oncogene-addicted human medullary thyroid cancer. Oncotarget. 2016;7:49649–49663. doi: 10.18632/oncotarget.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mitchell T., Ramos-Montoya A., Di Antonio M., Murat P., Ohnmacht S., Micco M., Jurmeister S., Fryer L., Balasubramanian S., Neidle S., et al. Downregulation of Androgen Receptor Transcription by Promoter G-Quadruplex Stabilization as a Potential Alternative Treatment for Castrate-Resistant Prostate Cancer. Biochemistry. 2013;52:1429–1436. doi: 10.1021/bi301349c. [DOI] [PubMed] [Google Scholar]
- 94.Tassinari M., Cimino-Reale G., Nadai M., Doria F., Butovskaya E., Recagni M., Freccero M., Zaffaroni N., Richter S.N., Folini M. Down-Regulation of the Androgen Receptor by G-Quadruplex Ligands Sensitizes Castration-Resistant Prostate Cancer Cells to Enzalutamide. J. Med. Chem. 2018;61:8625–8638. doi: 10.1021/acs.jmedchem.8b00502. [DOI] [PubMed] [Google Scholar]
- 95.Marchetti C., Zyner K.G., Ohnmacht S.A., Robson M., Haider S.M., Morton J.P., Marsico G., Vo T., Laughlin-Toth S., Ahmed A.A., et al. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J. Med. Chem. 2018;61:2500–2517. doi: 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maloney A., Workman P. HSP90 as a new therapeutic target for cancer therapy: The story unfolds. Expert Opin. Biol. Ther. 2002;2:3–24. doi: 10.1517/14712598.2.1.3. [DOI] [PubMed] [Google Scholar]
- 97.Ohnmacht S.A., Micco M., Petrucci V., Todd A.K., Reszka A.P., Gunaratnam M., Carvalho M.A., Zloh M., Neidle S. Sequences in the HSP90 promoter form G-quadruplex structures with selectivity for disubstituted phenyl bis-oxazole derivatives. Bioorg. Med. Chem. Lett. 2012;22:5930–5935. doi: 10.1016/j.bmcl.2012.07.065. [DOI] [PubMed] [Google Scholar]
- 98.Mpima S., Ohnmacht S.A., Barletta M., Husby J., Pett L.C., Gunaratnam M., Hilton S.T., Neidle S. The influence of positional isomerism on G-quadruplex binding and anti-proliferative activity of tetra-substituted naphthalene diimide compounds. Bioorg. Med. Chem. 2013;21:6162–6170. doi: 10.1016/j.bmc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 99.Callegaro S., Perrone R., Scalabrin M., Doria F., Palu G., Richter S.N. A core extended naphtalene diimide G-quadruplex ligand potently inhibits herpes simplex virus 1 replication. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-02667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nadai M., Doria F., Scalabrin M., Pirota V., Grande V., Bergamaschi G., Amendola V., Winnerdy F.R., Phan A.T., Richter S.N., et al. A Catalytic and Selective Scissoring Molecular Tool for Quadruplex Nucleic Acids. J. Am. Chem. Soc. 2018;140:14528–14532. doi: 10.1021/jacs.8b05337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Artusi S., Nadai M., Perrone R., Biasolo M.A., Palu G., Flamand L., Calistri A., Richter S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antivir. Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Butovskaya E., Heddi B., Bakalar B., Richter S.N., Phan A.T. Major G-Quadruplex Form of HIV-1 LTR Reveals a (3+1) Folding Topology Containing a Stem-Loop. J. Am. Chem. Soc. 2018;140:13654–13662. doi: 10.1021/jacs.8b05332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Belmonte-Reche E., Martinez-Garcia M., Guedin A., Zuffo M., Arevalo-Ruiz M., Doria F., Campos-Salinas J., Maynadier M., Lopez-Rubio J.J., Freccero M., et al. G-Quadruplex Identification in the Genome of Protozoan Parasites Points to Naphthalene Diimide Ligands as New Antiparasitic Agents. J. Med. Chem. 2018;61:1231–1240. doi: 10.1021/acs.jmedchem.7b01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zuffo M., Stucchi A., Campos-Salinas J., Cabello-Donayre M., Martínez-García M., Belmonte-Reche E., Pérez-Victoria J.M., Mergny J.L., Freccero M., Morales J.C., et al. Carbohydrate-naphthalene diimide conjugates as potential antiparasitic drugs: Synthesis, evaluation and structure-activity studies. Eur. J. Med. Chem. 2019;163:54–66. doi: 10.1016/j.ejmech.2018.11.043. [DOI] [PubMed] [Google Scholar]
- 105.Asamitsu S., Obata S., Phan A.T., Hashiya K., Bando T., Sugiyama H. Simultaneous Binding of Hybrid Molecules Constructed with Dual DNA-Binding Components to a G-Quadruplex and Its Proximal Duplex. Chemistry. 2018;24:4428–4435. doi: 10.1002/chem.201705945. [DOI] [PubMed] [Google Scholar]
- 106.Calabrese D.R., Chen X., Leon E.C., Gaikwad S.M., Phyo Z., Hewitt W.M., Alden S., Hilimire T.A., He F., Michalowski A.M., et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018;9:4229. doi: 10.1038/s41467-018-06315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Di Antonio M., Biffi G., Mariani A., Raiber E.A., Rodriguez R., Balasubramanian S. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2012;51:11073–11078. doi: 10.1002/anie.201206281. [DOI] [PMC free article] [PubMed] [Google Scholar]