Abstract
Chloride and sodium constitute as the major ions in most saline soils, contributing to salt-induced damage in plants. Research on salt tolerance has mostly concentrated on the sodium toxicity; however, chloride toxicity also needs to be considered to understand the physiological, biochemical, and metabolite changes under individual and additive salts. In this study, we investigated the effect of individual Na+ and/or Cl− ions (equimolar 100 mM NaCl, Na+ and Cl− salts) using in vitro cultures of four soybean genotypes with contrasting salt tolerance. In general, all the treatments significantly induced antioxidant enzymes activities such as catalase, ascorbate peroxidase, glutathione reductase, guaiacol peroxidase, and superoxide dismutase and osmolytes including proline, glycine betaine, and total soluble sugar (TSS). Both individual (Na+, Cl−) and additive (NaCl) stresses induced more pronounced activation of antioxidant enzyme machinery and osmolytes accumulation in the tolerant genotypes (MAUS-47 and Bragg). The sensitive genotypes (Gujosoya-2 and SL-295) showed higher accumulation of Na+ and Cl−, while the tolerant genotypes were found to maintain a low Na+/K+ and high Ca2+ level in combination with enhanced antioxidant defense and osmotic adjustment. Gas chromatography–mass spectrometry (GC–MS)-based metabolomic profiling depicted the association of certain metabolites under individualistic and additive salt effects. The genotype-specific metabolic changes indicated probable involvement of azetidine, 2-furanmethanol, 1,4-dioxin, 3-fluorothiophene, decanoic acid and 2-propenoic acid methyl ester in salt-tolerance mechanism of soybean.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1599-6) contains supplementary material, which is available to authorized users.
Keywords: Soybean, Na-dominant, Cl-dominant, NaCl stress, GC–MS
Introduction
Soil salinity is a major threat to crop productivity and overall crop yield. It causes osmotic, ionic, and oxidative stress, which leads to decline in growth and plant development (Hossain and Dietz 2016). Globally, salinity affects almost 0.8 billion hectares of land, which is almost 6% of total land area (Muchate et al. 2016a; Nikalje et al. 2017). Accumulation of toxic Na+ and Cl− ions leads to decrease in activity of stomata and electron transport system (ETS), which generate excess reactive oxygen species (ROS) (Negrao et al. 2017). The ROS causes oxidation of proteins, carbohydrates, lipids, chlorophyll and nucleic acids, and this results in cell death (Hossain and Dietz 2016). To combat salinity, plants have developed adaptive defense mechanism which includes synthesis of compatible solutes, compartmentalization of toxic ions, and induction of ROS-scavenging system (Abogadallah 2010).
Saline soil is rich in cations like calcium (Ca2+), magnesium (Mg2+), and sodium (Na+), and anions like carbonates (including bicarbonates), chloride (Cl−), and sulfate (SO42−) (Parihar et al. 2015). Sodium chloride is the most toxic salt comprising of sodium and chloride ions, which constitute more than 50% of total soluble salts (Li et al. 2017). The Na+ and Cl− ions dominate in the saline soils (Ismail et al. 2014). It is well established that the toxic Na+ triggers stress to most plant species, but, in some plants, Cl− ion is more toxic than Na+ (Li et al. 2017). Apart from toxicity, Cl− ions have regulatory role in the generation of turgor, stability of enzymes, alteration in membrane potential, pH, balance of charge, volume control, osmoregulation, and stomatal conductance which leads to prevention of water loss, high water use and photosynthetic efficiency (Franco-Navarro et al. 2016; Li et al. 2017). Therefore, it is essential to consider toxicity of both the ions, and study the effect of Cl− and Na+ ions alone and combination with NaCl form. In Medicago, Lotus, Faba bean, Barley, Chrysanthemum, Cucumber, and rice, it has been shown that controlled chloride transport and exclusion from shoot are well correlated (Sibole et al. 2003; Teakle et al. 2007; Tavakkoli et al. 2010, 2011; Guan et al. 2012; Huang et al. 2015; Khare et al. 2015). Most of the studies at cellular level have been carried out in the presence of NaCl salt (Rahnama et al. 2003; Rahnama and Ebrahimzadeh 2004; Kusvuran et al. 2016; Bezirganoglu 2017), but very meager information is available using individual Na+ or Cl− salts. In vitro culture techniques offer a significant tool to evaluate genotypes for salt tolerance in less time and space at cellular level under uniform and controlled growth and treatment conditions (Nikam et al. 2014; Piwowarczyk et al. 2016; Bezirganoglu 2017; Boamponsem et al. 2018). Physiological and biochemical mechanisms of salinity tolerance could be different at both cellular and whole plant levels (Nikam et al. 2014; Zhao et al. 2018). In addition, salinity tolerance related traits expressed at cellular level could be used to select tolerant cell lines which provide an important tool to understand mechanisms of salt tolerance (Rania et al. 2015). The differential responses to salinity in plants are depicted in terms of altered growth, compatible solutes and antioxidants (Kumar et al. 2017; Muchate et al. 2016a).
Soybean (Glycine max (L.) Merr.), one of the important leading economic crops, contains almost 20% oil and 40% protein in seeds (Amirjani 2010). It is also an attractive crop for biodiesel production and protein source for humans and other animals worldwide. Soybean enriches soil through symbiotic nitrogen fixation, which improves the growth and development of next crop (Piwowarczyk et al. 2016). Despite its wide adaptability to different agro-climatic zones, the productivity of Soybean is affected drastically due to salinity. The crop is categorized as moderately salt sensitive (Munns and Tester 2008; Cao et al. 2017). However, regarding salt tolerance and salt sensitivity, there is large variation among the soybean genotypes necessitating screening of available germplasm (Shelke et al. 2017). In vitro screening for salt tolerance can provide the most methodical, rapid, and resourceful route to isolate stress tolerant genotypes. Information on salt tolerance under individual and additive salt in soybean crop is limited. The present investigation was aimed to study toxicity caused by individual Na+ or Cl− and NaCl salt in callus cultures of different soybean genotypes with contrasting salt tolerance. Different physiological and biochemical parameters were considered for salt-sensitive or salt-tolerant response. Metabolic analysis using GC–MS was done to reveal the involvement of major metabolites responsible for salt-tolerance mechanism of soybean at cellular level.
Materials and methods
Plant material, seed germination, and callus formation
Soybean seeds (MAUS-47 and Bragg- salt tolerant and Gujosoya-2 and SL-295- salt sensitive) were collected from National Institute of Soybean Research, Indore, MP, India. Seeds were surface sterilized using HgCl2 (0.1%, w/v) followed by rinsing in distilled water and inoculated on Murashige and Skoog medium consisting of 0.1% sucrose and 0.8% agar as a solidifying agent in test tubes. After 3 days of dark incubation, test tube-containing seeds were transferred to light for next 7 days for seed germination at 25 ± 2 °C and 14 h light (36 µmol m−2 s−1) in culture room. After seed germination, the cotyledons were excised and inoculated on callus induction medium (MS + 10.74 µM NAA). After 45 days of growth callus was subjected to Na-dominant, Cl-dominant, and NaCl salt treatments.
Differential salt treatment
About 0.5 g callus was subjected to 100 mM Na-dominant (Na+) (EC 10.4 ds m−1), Cl-dominant (Cl−) (EC 11 ds m−1), and NaCl (EC 11.4 ds m−1) salt-enriched callus induction medium. The callus grown on only callus induction medium was used as control. The 100 mM sodium dominant salt solution was prepared using Na2SO4 (15 mM), Na2HPO4 (15 mM), and NaNO3 (40 mM). The chloride dominant salt solution was prepared using MgCl2 (15 mM), CaCl2 (15 mM), and KCl (40 mM) (Kumar and Khare 2016). After 15 days of salt treatment, callus was harvested for analysis.
Determination of growth attributes
The changes in growth attributes (fresh weight, dry weight, and %TWC) were measured to study callus growth. After harvesting, the fresh weight of the callus was immediately recorded and subjected to oven drying (at 60 °C) until constant dry weight. The %TWC (% tissue water content) was measured as per Muchate et al. (2016b).
Extraction and estimation of antioxidant enzymes
Fresh callus (0.5 g) was homogenized using chilled sodium phosphate buffer (50 mM, pH 7.0), EDTA (0.1 mM) and PVP (1%). The homogenate was centrifuged (15,000 rpm, 20 min, 4 °C) and supernatant was used as enzyme source. The Lowry’s method (1951) was used to estimate soluble protein content.
Estimation of superoxide dismutase (SOD)
The activity of SOD was recorded as per Beauchamp and Fridovich (1971). The assay mixture consisted of phosphate buffer (50 mM, pH 7.8), EDTA (0.1 mM), methionine (14.3 mM), NBT (82.5 µM), riboflavin (2.2 µM), and enzyme source (25 µl). After addition of enzyme source, the assay mixture was exposed to light (2000 lux light intensity) along with light blank (assay mixture without enzyme source) for 30 min. The assay mixture consisted of enzyme source served as dark blank and incubated in dark. The reduction of NBT was observed spectrophotometrically at 560 nm (Shimadzu UV-1800) and enzyme activity was expressed as Units SOD activity mg−1 protein.
Estimation of catalase (CAT)
The activity was assayed as per Cakmak and Marschner (1992) method. The assay mixture contained phosphate buffer (50 mM, pH 7.0) and H2O2 (15 mM). After addition of enzyme source (50 µl), the activity was measured as decrease in O.D. at 240 nm and expressed as µkat mg−1 protein (extinction coefficient = 36 mM−1 cm−1).
Estimation of ascorbate peroxidase (APX)
The method described by Nakano and Asada (1981) was used for APX assay. The assay mixture (3 ml) consisted of phosphate buffer (50 mM, pH 7.0), ascorbate (0.5 mM), EDTA (0.1 mM), and enzyme source (100 µl). The decline in O.D. was observed and enzyme activity was expressed as µkat mg−1 protein (extinction coefficient = 2.8 mM−1 cm−1).
Estimation of guaiacol peroxidase (GPOX)
The Hemeda and Klein (1990) method was used for the measurement of GPOX activity. The assay mixture (3 ml) consisted of phosphate buffer (50 mM, pH 6.6), guaiacol (1%, (w/v)), H2O2 (0.3%), and enzyme source (25 µl). Increase in O.D. (at 470 nm) was observed and enzyme activity was expressed as units mg−1 protein (1 unit = 1 µmol of guaiacol oxidized min−1).
Estimation of glutathione reductase (GR)
The method described by Smith et al. (1988) was used for the measurement of GR activity. The assay mixture (3 ml) consisted of phosphate buffer (50 mM, pH 7.5), DTNB (3 mM), EDTA (1 mM), H2O2 (0.1 mM), NADPH (2 mM), enzyme source (25 µl), and GSSG (20 mM). Increase in O.D. (at 421 nm) was observed and enzyme activity was expressed as units mg−1 protein (1 unit = 1 µmol of GSSG reduced min−1).
Extraction and estimation of osmolytes
Estimation of glycine betaine (GB)
The glycine betaine content was measured as per the method of Grieve and Grattan (1983). The fresh callus (0.5 g) was homogenized in 20 ml distilled water and placed on rotary shaker for 16 h. The homogenate was centrifuged at 1000 rpm for 10 min. The supernatant was mixed with 2N sulphuric acid (1:1) and mixture was incubated on ice water bath for 1 h followed by the addition of freshly prepared I2-KI reagent. This mixture was incubated at 4 °C for 16 h and then centrifuged (10,000 rpm, 15 min at 0 °C). The supernatant was discarded, and the residue was recovered carefully and dissolved in 1,2-dichloroethane. After 2 h of incubation at room temperature, the O.D. was measured at 365 nm.
Estimation of proline
The homogenate was prepared from fresh callus (0.5 g) in aqueous 5-sulphosalicylic acid (3%) and was centrifuged at 10,000 rpm (for 10 min at 4 °C). The supernatant was mixed with glacial acetic acid and acid ninhydrin reagent (2 ml of each). The reaction mixture was incubated in boiling water bath for 1 h and reaction was terminated on ice bath. Then, 4 ml of toluene was added in cold reaction mixture and vortexed robustly for 15 s. The reaction blend lacking extract was used as a blank and the O.D. was measured at 520 nm. The content of proline was calculated using the standard curve and expressed as mg of proline g−1FW (Bates et al. 1973).
Estimation of total soluble sugars (TSS)
The fresh callus (0.2 g) was homogenized in ice-chilled 80% ethanol. The homogenate was centrifuged (5000 rpm for 10 min at 4 °C) and supernatant and anthrone reagent (1:3) incubated in water bath (100 °C) for 10 min. The reaction was terminated on ice bath and O.D. was measured at 620 nm. TSS content was determined using the standard D-glucose curve and expressed as mg glucose g−1FW (Watanabe et al. 2000).
Estimation of ion content
The dry calli (50 mg) was digested in concentrated nitric acid for 24 h. Then, acid was evaporated at 100 °C for 2 h. The digested samples were dissolved in 10 ml deionized distilled water and used for the estimation of cation (Na+, K+, Ca2+, Mn2+, Fe2+, and Mg2+) content. The ion content was measured using atomic absorption flame photometer (Agilant, India). The Cl− content was estimated as per Chapman and Pratt (1961) by potentiometric method.
GC–MS analysis
The fresh callus (5 g) was crushed in liquid nitrogen. The resultant powder was soaked in 10 ml methanol (95%) and placed on rotary shaker for 12 h, followed by sonication for 25 min (CD-4820 Ultrasonic cleaner). To this, sodium sulfate (2 g) was mixed thoroughly and the mixture was filtered using pre-wetted Whatman filter paper no. 1 with 95% ethanol. The resultant aliquot was passed through 0.2 µm syringe micro-filters. The aliquots (2 µl) were analyzed using the GC–MS system (Agilent technologies 7890B GC and 5977A MSD system) equipped with HP5 column. The helium was used as a carrier gas with 1 ml min−1 flow rate. The oven temperature of GC was raised successively from 110 °C to 280 °C at 5 °C min−1 except at former temperature held for 2 min and later for 9 min. The electron ionization and detector was used to obtain mass spectrum of compounds with mass scan range 45–450 amu. Total 34 min running time 45–450 Da fragments were analyzed in each 0.5-s scan period. Identification of compounds was done based on the calculated fragments, molecular structure, and molecular mass. The test material compound name, molecular formula, RT, similarity index, and peak area of the components were assessed (Supplementary Table 1). The Turbomas 5.2 software was used to compare the spectrum of unknown compounds with spectrum of the components deposited in the updated version of NIST library (Keskes et al. 2017).
Statistical analysis
The experiments were setup in completely randomized design with 20 replicates to study differential impact of Cl− and Na+ ions on callus growth of 4 soybean genotypes. The one-way analysis of variance was evaluated using statistical software SPSS 16.0. The data represented as mean ± standard error of biological triplicate, and means were compared using DMRT (Duncan’s 1955) at P ≤ 0.05.
Results
Influence of Na+, Cl−, and NaCl on growth and water status
The effect of individual ionic (Na+ and Cl−) and additive NaCl stress on callus cultures of four contrasting soybean genotypes under 100 mM of salt concentration was studied. A significant reduction was observed in callus FW and DW under Na-dominant, Cl-dominant, and NaCl treatments as compared to control, in the four soybean genotypes (Table 1). In all the genotypes, significantly higher reduction of %TWC was observed in Na-dominant treatment rather than NaCl and Cl-dominant treatments as compared to their respective controls. In the genotypes MAUS-47 and Bragg, %TWC was found to be better maintained than Gujosoya-2 and SL-295 under Na-dominant, Cl-dominant, and NaCl salt treatments.
Table 1.
Physiological parameters under Na+, Cl−, and NaCl stress of four soybean genotypes in callus culture system
| Genotypes | Treatment | FW | DW | %TWC |
|---|---|---|---|---|
| MAUS-47 | Control | 1.6 ± 0.2aB | 0.1 ± 0aB | 94 ± 0.2aA |
| Na-dominant | 0.64 ± 0bA | 0.06 ± 0bAB | 90 ± 0.7cA | |
| Cl-dominant | 0.75 ± 0bA | 0.06 ± 0bBC | 92 ± 0.2abA | |
| NaCl | 0.71 ± 0bA | 0.06 ± 0bA | 91 ± 0.2bcA | |
| Bragg | Control | 1.54 ± 0.05aB | 0.087 ± 0aB | 94 ± 0.1aA |
| Na-dominant | 0.52 ± 0.02bB | 0.057 ± 0bB | 89 ± 0.6cA | |
| Cl-dominant | 0.67 ± 0.03bB | 0.059 ± 0bC | 91 ± 0.1bA | |
| NaCl | 0.6 ± 0.03bA | 0.057 ± 0bA | 90 ± 0.7bAB | |
| Gujosoya-2 | Control | 1.99 ± 0.1aA | 0.11 ± 0aA | 94 ± 0.2aA |
| Na-dominant | 0.47 ± 0bBC | 0.07 ± 0bA | 84 ± 0.5cB | |
| Cl-dominant | 0.56 ± 0bB | 0.06 ± 0cAB | 90 ± 0.7bB | |
| NaCl | 0.57 ± 0bB | 0.07 ± 0bcA | 88 ± 0.4bB | |
| SL-295 | Control | 1.45 ± 0.1aB | 0.1 ± 0aB | 93 ± 0.2aB |
| Na-dominant | 0.4 ± 0bC | 0.08 ± 0bA | 81 ± 1.7cC | |
| Cl-dominant | 0.56 ± 0bB | 0.06 ± 0bA | 90 ± 0.6abC | |
| NaCl | 0.52 ± 0bB | 0.07 ± 0bA | 86 ± 0.4bcB |
Each value is the mean (± SE) of 20 replicates (Duncan’s test, P ≤ 0.05) and different letters for each factor in each column indicate significant difference. Small letter denotes significant difference between treatments, and capital letter denotes significant difference between genotypes in each treatment at 0.05% significance level
Influence of Na+, Cl−, and NaCl on antioxidant enzyme activity
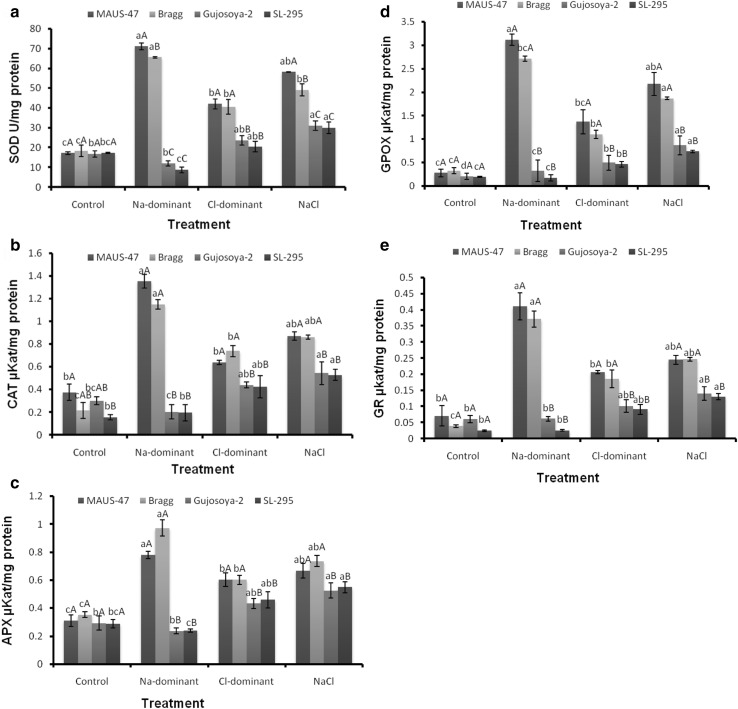
The analysis of variance (ANOVA) showed differential response of soybean genotypes to Na-dominant, Cl-dominant, and NaCl salt treatments for the studied antioxidant enzymes activity. This indicated a significant interaction between the genotypes and applied stresses. The Na-dominant salt treatment significantly increased SOD activity in MAUS-47 and Bragg (4.1 and 3.6-fold, respectively), while it decreased in Gujosoya-2 and SL-295 (1.4- and 2-fold, respectively) genotypes compared to their control. The Cl-dominant and NaCl salt also significantly increased the enzyme activity in MAUS-47 (2.5- and 3.4-fold, respectively), Bragg (2.2- and 2.7-fold, respectively), Gujosoya-2 (1.4- and 1.9-fold, respectively), and SL-295 (1.2- and 1.7-fold, respectively) genotypes (Fig. 1a).
Fig. 1.
Impact of individual (Na+ and Cl−) and additive (NaCl) salt on induction of a SOD, b CAT, c APX, d GPOX, and e GR activity of four soybean genotypes callus. Small letters denotes significant difference between treatment and capital letter denotes significant difference between genotypes in each treatment at 0.05% significance level. SOD superoxide dismutase, CAT catalase, APX ascorbic peroxidase, GPOX guaiacol peroxidase, GR glutathione reductase
The CAT activity varied significantly among the control and treatments in the tested genotypes. The Na-dominant treatment significantly induced CAT activity in MAUS-47 (3.6-fold) and Bragg (5.3-fold), while, in Gujosoya-2 and SL-295, the enzyme activity was decreased by 1.5- and 1.3-fold, respectively. The Cl-dominant and NaCl treatment also significantly induced the CAT activity in all the soybean genotypes (1.5- and 2.3-fold in MAUS-47, 3.4- and 4-fold in Bragg, 1.5- and 1.8-fold in Gujosoya-2, and 1.6 and 2-fold in SL-295, respectively) (Fig. 1b).
A similar trend was observed for APX activity, which was also varied significantly in all the tested genotypes under Na-dominant, Cl-dominant, and NaCl salt treatments. The Na-dominant salt treatment significantly induced higher APX activity in MAUS-47 (2.5-fold) and Bragg (2.8-fold), while it decreased in Gujosoya-2 (1.2-fold) and SL-295 (1.2-fold) genotypes compared to its control. The Cl-dominant and NaCl salt also significantly increased this enzyme activity in MAUS-47 (1.9- and 2.2-fold, respectively), Bragg (1.7- and 2.1-fold, respectively), Gujosoya-2 (1.5- and 1.8-fold, respectively), and SL-295 (1.6- and 1.9-fold, respectively) genotypes (Fig. 1c).
The GPOX activity under Na-dominant salt was significantly increased in MAUS-47 (11.2-fold) and Bragg (8.2-fold) as compared to Gujosoya-2 (1.6), while, in SL-295, the enzyme activity was decreased by 1.1-fold as compared to their respective controls. In Cl-dominant salt treatment, all the genotypes showed increase in GPOX activity as compared to their respective controls. Within genotypes, MAUS-47 (4.9-fold) showed highest induction followed by Bragg (4.9-fold), Gujosoya-2, and SL-295 (2.4-fold). Significantly increased GPOX activity was recorded under NaCl treatment in all the genotypes; genotypes MAUS-47 (7.8-fold) and Bragg (5.7-fold) showed the highest GPOX activity than Gujosoya-2 (4.1-fold) and SL-295 (3.8-fold) (Fig. 1d).
The GR activity was the highest in MAUS-47 (5.8-fold) and Bragg (9.5-fold), while, in Gujosoya-2, no significant change was observed. However, decrease in GR activity (1.8-fold) under Na-dominant salt treatment was observed in SL-295. In the Cl-dominant salt treatment, the GR activity was significantly increased in MAUS-47 (2.9-fold) and Bragg (4.8) as compared to Gujosoya-2 (1.7-fold) and SL-295 (twofold). Similar to Cl-dominant, under NaCl treatment, MAUS-47 (3.4-fold) and Bragg (5.5-fold) genotypes showed increased GR activity as compared to Gujosoya-2 (2.3-fold) and SL-295 (2.9-fold) (Fig. 1e). Among the genotypes, MAUS-47 and Bragg showed significant induction of antioxidant enzyme activity than Gujosoya-2 and SL-295 under individual or additive salt treatments (Fig. 1).
Influence of Na+, Cl−, and NaCl on osmolytes accumulation
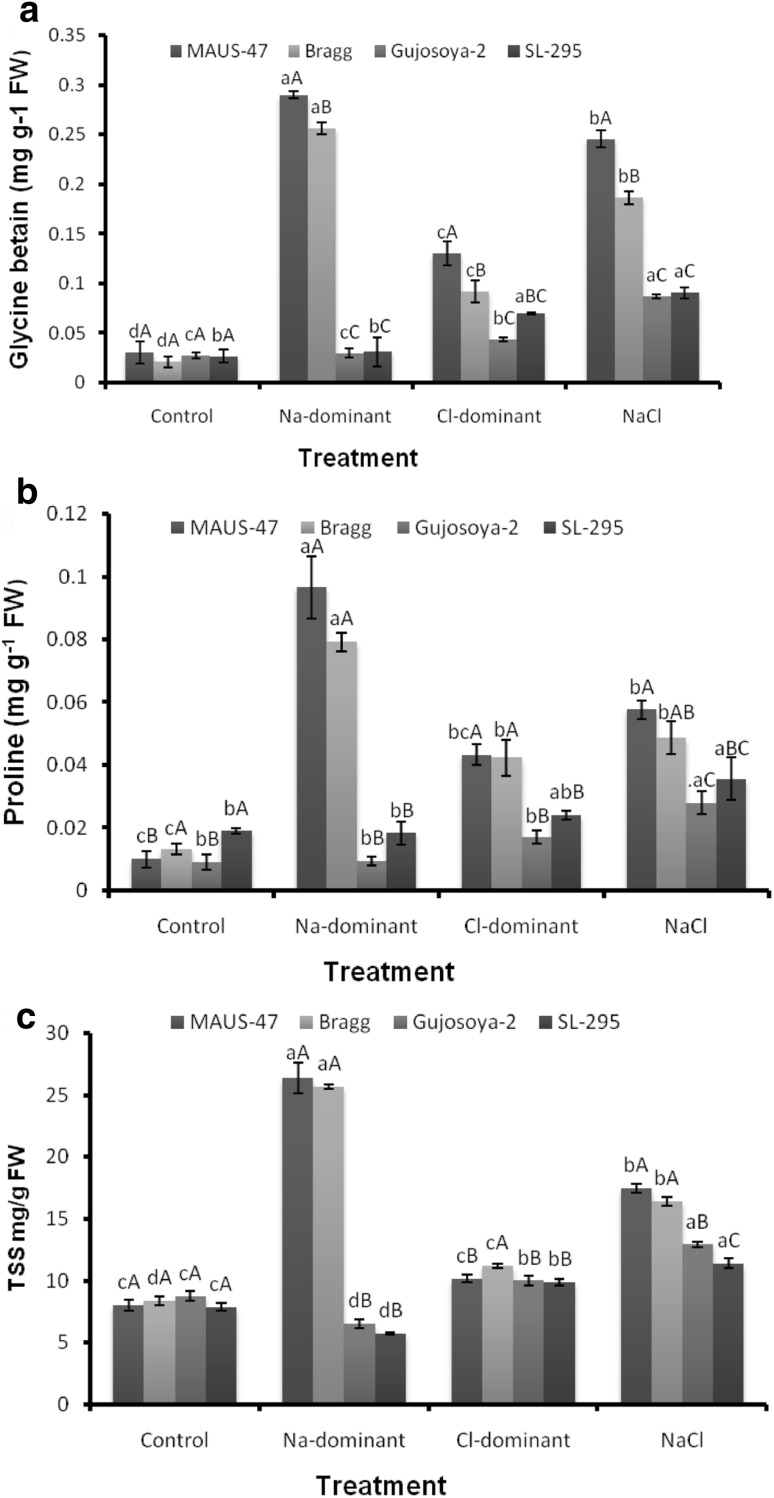
Comparative study of the accumulation pattern of glycine betaine, proline, and total soluble sugars in soybean genotypes revealed significant differences between the Na-dominant, Cl-dominant, and NaCl treatments. The increase in glycine betaine content was about 9.4- and 12.1-fold in MAUS-47 and Bragg genotypes respectively in Na-dominant salt treatments (Fig. 2a). The Cl-dominant and NaCl salts also significantly increased glycine betaine content in MAUS-47 (4.2- and 8-fold, respectively), Bragg (4.3- and 8.8-fold, respectively), Gujosoya-2 (1.6- and 3.2-fold respectively), and SL-295 (2.6- and 3.4-fold, respectively) genotypes (Fig. 2a).
Fig. 2.
Impact of individual (Na+, Cl−) and combined (NaCl) salt on accumulation of a glycine betaine, b proline, and c TSS of four Soybean genotypes callus. Small letter denotes a significant difference between treatments, and capital letter denotes significant difference between genotypes in each treatment at 0.05% significance level
The proline content in callus cultures of control and treatments differed significantly among the four tested genotypes. The Na-dominant salt treatment significantly induced higher proline content in MAUS-47 (9.7-fold), Bragg (sixfold), and Gujosoya-2 (onefold), while it was decreased in SL-295 (onefold) genotype compared to their control. The Cl-dominant and NaCl salt treatment also significantly increased the proline content in all the soybean genotypes. Proline content was 4.3- and 5.8-fold in MAUS-47; 3.2- and 3.7-fold in Bragg, 1.9- and 3.1-fold in Gujosoya-2; and 1.3 and 1.9-fold in SL-295, respectively (Fig. 2b). In general, in all the treatments, Gujosoya-2 and SL-295 showed significantly lower accumulation of proline.
A similar trend was observed for TSS accumulation, which also increased significantly in all the tested genotypes under Na-dominant, Cl-dominant, and NaCl salt treatments (Fig. 2c). The Na-dominant salt treatment significantly increased TSS content in MAUS-47 (3.3-fold) and Bragg (3.1-fold), while it was decreased in Gujosoya-2 (1.3-fold) and SL-295 (1.4-fold) genotypes compared to its control. The Cl-dominant and NaCl salt also significantly increased TSS in MAUS-47 (1.3- and 2.2-fold, respectively), Bragg (1.3- and 2-fold, respectively), Gujosoya-2 (1.1- and 1.5-fold, respectively), and SL-295 (1.3- and 1.5-fold, respectively) genotypes (Fig. 2c). To summarize, the data showed that, in comparison with all the genotypes and treatments, Gujosoya-2 and SL-295 showed significantly lower glycine betaine, proline, and TSS accumulation than MAUS-47 and Bragg.
Influence of Na+, Cl−, and NaCl on ion content
The effects of Na-dominant, Cl-dominant, and NaCl salt treatments on ions homeostasis in callus cultures of four soybean genotypes are presented in Table 2. There was a significant Na+ and Cl− ion accumulation (up to 10.4- and 32.6-fold, respectively) in the four tested genotypes. The Na-dominant salts treatment caused 7.2- and 5.6-fold higher Na+ ion accumulation in callus cells of MAUS-47 and Bragg, while it was 10.4- and 7.8-fold in Gujosoya-2 and SL-295 genotypes. A similar trend was observed with Cl-dominant salt treatment. The accumulation was 6.3- and 7.7-fold high Cl− ion in MAUS-47 and Bragg, while 28.8- and 32.6-fold in Gujosoya-2 and SL-295 genotypes. On the other hand, the additive (NaCl) salt treatment induced 6.4- and 5.1-fold higher Na+ and 3.3 and 5.2-fold Cl− in MAUS-47 and Bragg genotypes, while 10.8 and 7.6-fold high Na+ and 18.2 and 21.6-fold Cl− was recorded in Gujosoya-2 and SL-295, respectively. The MAUS-47 and Bragg genotypes displayed significantly lower Na+ and Cl− accumulation than Gujosoya-2 and SL-295 genotypes. The MAUS-47 and Bragg genotypes showed efficient regulation of Na+ influx under Na-dominant and NaCl treatment which, in turn, could maintain lower Na+/K+ ratio, while uncontrolled influx in Gujosoya-2 (3.2 and 2.1 ratio, respectively) and SL-295 (2.1 and 1.8 ratio, respectively) could results in failure in maintaining the influx. In Na+ treatment, not only K+ but also Ca2+ content decreased significantly under Na-dominant and NaCl stress. The genotypes, Gujosoya-2, and SL-295 reported significantly lower accumulation of K+ and Ca2+ in all the salt treatments. Therefore, the results suggested that MAUS-47 and Bragg genotypes showed a better regulation of these ions than Gujosoya-2 and SL-295 genotypes. The analysis of Mg2+, Mn2+, and Fe2+ showed insignificant difference among the treatments except Cl-dominant treatment for Mg2+ in all tested genotypes (Table 2).
Table 2.
Ion accumulation in callus (mg g−1 DW) of four soybean genotypes under Na+, Cl−, and NaCl stress
| Genotypes | Treatment | Na+ | K+ | Na+/K+ | Ca2+ | Mg2+ | Mn2+ | Fe2+ | Cl− |
|---|---|---|---|---|---|---|---|---|---|
| MAUS-47 | Control | 0.28 ± 0.02bAB | 2.55 ± 0.06bA | 0.1 ± 0.01bA | 0.93 ± 0.08aA | 0.125 ± 0.04bA | 0.051 ± 0aA | 0.31 ± 0.01aA | 12.9 ± 2.1cA |
| Na-dominant | 1.98 ± 0.28aA | 1.98 ± 0.17cA | 1 ± 0.1aB | 0.42 ± 0.02bA | 0.127 ± 0.02bA | 0.075 ± 0aA | 0.45 ± 0.1aA | 10.8 ± 2.1cA | |
| Cl-dominant | 0.38 ± 0.11bA | 4.88 ± 0.19aA | 0.1 ± 0.03bA | 0.81 ± 0.06aAB | 0.258 ± 0.01aA | 0.058 ± 0.0aA | 0.49 ± 0.09aA | 81.5 ± 7.5aC | |
| NaCl | 1.76 ± 0.33aA | 2.18 ± 0.07bcA | 0.8 ± 0.14aB | 0.48 ± 0.11bA | 0.134 ± 0.03bA | 0.068 ± 0.01aA | 0.33 ± 0.0aA | 42 ± 5.5bD | |
| Bragg | Control | 0.34 ± 0.01bA | 2.94 ± 0.29bA | 0.1 ± 0.02bA | 0.94 ± 0.1aA | 0.116 ± 0.02bA | 0.057 ± 0.01aA | 0.36 ± 0.04bA | 15 ± 3.6cA |
| Na-dominant | 1.91 ± 0.18aA | 2.08 ± 0.13bA | 0.9 ± 0.14aB | 0.44 ± 0.08bA | 0.131 ± 0.02bA | 0.076 ± 0.01aA | 0.5 ± 0.07aA | 18.3 ± 1.7cA | |
| Cl-dominant | 0.3 ± 0.02bA | 5.35 ± 0.47aA | 0.1 ± 0bA | 0.85 ± 0.04aA | 0.274 ± 0.02aA | 0.057 ± 0.01aA | 0.43 ± 0.02abA | 114.8 ± 6.2aB | |
| NaCl | 1.84 ± 0.01aA | 2.1 ± 0.11bA | 0.8 ± 0.12aB | 0.5 ± 0.1bA | 0.134 ± 0.03bA | 0.068 ± 0.01aA | 0.33 ± 0.0bA | 77.8 ± 6.2bC | |
| Gujosoya-2 | Control | 0.21 ± 0.01bB | 2.81 ± 0.63aA | 0.1 ± 0.02bA | 0.74 ± 0.09aA | 0.077 ± 0.03bA | 0.052 ± 0.0aA | 0.37 ± 0.01aA | 6.7 ± 2.08cA |
| Na-dominant | 2.17 ± 0.14aA | 0.69 ± 0.31cB | 3.2 ± 0.55aA | 0.25 ± 0.01cB | 0.105 ± 0bA | 0.067 ± 0.0aA | 0.36 ± 0.05aA | 15 ± 3.6cA | |
| Cl-dominant | 0.35 ± 0.1bA | 3.06 ± 0.33aB | 0.1 ± 0.03bA | 0.5 ± 0.1abC | 0.255 ± 0.02aA | 0.053 ± 0.01aA | 0.46 ± 0.01aA | 191.8 ± 5.5aA | |
| NaCl | 2.24 ± 0.12aA | 1.1 ± 0.11bB | 2.1 ± 0.18abA | 0.35 ± 0.07cA | 0.08 ± 0.02bA | 0.063 ± 0.0aA | 0.37 ± 0.05aA | 121 ± 9.5bB | |
| SL-295 | Control | 0.3 ± 0.07bAB | 2.74 ± 0.18aA | 0.1 ± 0.02bA | 0.85 ± 0.03aA | 0.091 ± 0.01bA | 0.045 ± 0.0aA | 0.37 ± 0.02aA | 6.7 ± 4.2cA |
| Na-dominant | 2.34 ± 0.03aA | 1.09 ± 0.19bB | 2.3 ± 0.51aA | 0.26 ± 0.02cB | 0.085 ± 0.02bA | 0.067 ± 0.01aA | 0.32 ± 0.01aA | 17.1 ± 2.1cA | |
| Cl-dominant | 0.2 ± 0.01bA | 3.16 ± 0.36aB | 0.1 ± 0.01bA | 0.58 ± 0.07bBC | 0.242 ± 0.04aA | 0.046 ± 0.0aA | 0.46 ± 0.02aA | 216.7 ± 11.5aA | |
| NaCl | 2.27 ± 0.28aA | 1.24 ± 0.24bB | 1.9 ± 0.24aA | 0.36 ± 0.04cA | 0.098 ± 0.01bA | 0.068 ± 0.01aA | 0.38 ± 0.03aA | 143.9 ± 4.2bA |
Each value is the mean (± SE) of three replicates (Duncan’s test, P ≤ 0.05) and different letters for each factor in each column indicate significant difference. Small letter denotes significant difference between treatments, and capital letter denotes significant difference between genotypes in each treatment at 0.05% significance level
GC–MS-based metabolite profiling
To analyze the salt induced metabolic perturbations in the four contrasting soybean genotypes, we compared the GC–MS-based metabolic profile of MAUS-47, Bragg, Gujosoya-2, and SL-295 genotypes (Table 3). The identified peaks contained metabolites such as alcohols, fatty acids, carboxylic acids, sugars, and ketones. The 2-furan-carboxaldehyde was observed in MAUS-47 under control, NaCl, Na+, and Cl− stress, while 2-Furancarboxaldehyde, 5-methyl- were observed both in control and NaCl conditions. The 2-Furanmethanol was observed only in NaCl and Na+ salt. The phenylacetaldehyde was observed only under control condition, while (2H)-furan-3-one, 2-propenoic acid, methyl ester, 3-oxabicyclo[3.1.0]hexan-2-one, and pyrazole-4-carbaldehyde were detected in NaCl treatment. The dimethyl 2-(N-formyl-methylamino)butane-1,4-dioate, azetidine, dimethyl 2-hydroxy-4-[((tetrahydropyran-2-yl)oxy)methyl]benzene-1,3-dicarboxylate, and hexadecanoic acid were observed only in Na+ salt and (E)-5-Benzyloxypent-3-en-1-yne, silanediol, dimethyl- in Cl− salt (Table 3). In Bragg, 2-Furanmethanol was observed in all the salt treatments, while N-Methylcyclopentane-1,1-dicarboximide and phenylacetaldehyde was found in the control and NaCl salt. 2-Propenoic acid, methyl ester was observed in NaCl, Na+, and Cl−. The 2-furancarboxaldehyde, 5-methyl- was recorded in NaCl and Cl− and decanoic acid in NaCl and Na+ salt. The isoflavonoid azetidine was found in Na+ and Cl− salt treatments in Bragg genotypes, while in Na+ salt in MAUS-47. The 1,3-dioxane-5-d, 2-(1,1-dimethylethyl)-4-methyl-, [2S-(2.alpha.,4.alpha.,5.alpha.)]-, 1,4-ethanonaphthalene-5,8-dione, 1,4,4a,8a-tetrahydro-1-methyl-4-(1-methylethyl)- and 3-pyridinamine were observed only under control condition of Bragg genotype, while 2-furan-carboxaldehyde, cyclohexan-2,2,3,3,4-d5-ol, 4-methyl-, (1S-trans)-, N-Benzylindole, and 2-Octanone were found under NaCl treatment. The 1,4-dioxin, 2,4(1H,3H)-Pyrimidinedione, 5-methyl-, 3-Fluorothiophene, and syn-2-(2-Hydroxypropyl)-6-methyldihydropyran-3(6H)-ol were observed only under Na+ salt. The (4S)-3,3-Dideuterio-4-methylcyclohexanone, (E)-6-Biphenyl-4-yl-4-oxohexanoic acid ethanolamide, (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide, 7-Methoxy-2,3-dihydro-2-phenyl-4-quinolone, Ethanol, 2,2,2-trichloro-, propanoate, Methyl (2R)-2-[(tert-Butoxy)carbonylamino]-5,5-dimethyl-5-silahexanoate, and 2(5H)-furanone were observed under Cl− salt (Table 3).
Table 3.
GC–MS-based metabolite profiling of MAUS-47, Bragg, Gujosoya-2, and SL-295 soybean genotypes callus under controlled and exposed to Na+, Cl− and NaCl salt
| Genotypes | Control | NaCl | Na-dominant (Na+) | Cl-dominant (Cl−) |
|---|---|---|---|---|
| MAUS-47 | 2-Furan-carboxaldehyde | 2-Furan-carboxaldehyde | 2-Furan-carboxaldehyde | 2-Furan-carboxaldehyde |
| 2-Furancarboxaldehyde, 5-methyl- | 2-Furancarboxaldehyde, 5-methyl- | Dimethyl 2-(N-formyl-methylamino)butane-1,4-dioate | (E)-5-Benzyloxypent-3-en-1-yne | |
| Phenylacetaldehyde | (2H)-Furan-3-one | Azetidine | Silanediol, dimethyl- | |
| 2-Furanmethanol | 2-Furanmethanol | |||
| 2-Propenoic acid, methyl ester | Dimethyl 2-hydroxy-4-[((tetrahydropyran-2-yl)oxy)methyl]benzene-1,3-dicarboxylate | |||
| 3-Oxabicyclo[3.1.0]hexan-2-one | Hexadecanoic acid | |||
| Pyrazole-4-carbaldehyde | ||||
| Bragg | 1,3-Dioxane-5-d, 2-(1,1-dimethylethyl)-4-methyl-, [2S-(2.alpha.,4.alpha.,5.alpha.)]- | (+/−)Bis(2-aminobut-3-enyl)disulphide | 1,4-Dioxin | (+/−)Bis(2-aminobut-3-enyl)disulphide |
| 1,4-Ethanonaphthalene-5,8-dione, 1,4,4a,8a-tetrahydro-1-methyl-4-(1-methylethyl)- | 2-Furan-carboxaldehyde | 2,4(1H,3H)-Pyrimidinedione, 5-methyl- | (4S)-3,3-Dideuterio-4-methylcyclohexanone | |
| 2-Furanmethanol | 2-Furanmethanol | 2-Furanmethanol | 2-Furanmethanol | |
| 3-Pyridinamine | 2-Propenoic acid, methyl ester | 2-Propenoic acid, methyl ester | 2-Propenoic acid, methyl ester | |
| N-Methylcyclopentane-1,1-dicarboximide | N-Methylcyclopentane-1,1-dicarboximide | 3-Fluorothiophene | N-Methylcyclopentane-1,1-dicarboximide | |
| Phenylacetaldehyde | Phenyl acetaldehyde | Azetidine | Azetidine | |
| Decanoic acid | Decanoic acid | (E)-6-Biphenyl-4-yl-4-oxohexanoic acid ethanolamide | ||
| Cyclohexan-2,2,3,3,4-d5-ol, 4-methyl-, (1S-trans)- | Syn-2-(2-Hydroxypropyl)-6-methyldihydropyran-3(6H)-ol | (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide | ||
| N-Benzylindole | 7-Methoxy-2,3-dihydro-2-phenyl-4-quinolone | |||
| 2-Furancarboxaldehyde, 5-methyl- | 2-Furancarboxaldehyde, 5-methyl- | |||
| 2-Octanone | Ethanol, 2,2,2-trichloro-, propanoate | |||
| Methyl (2R)-2-[(tert-Butoxy)carbonylamino]-5,5-dimethyl-5-silahexanoate | ||||
| 2(5H)-Furanone | ||||
| Gujosoya-2 | (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide | N-Methylcyclopentane-1,1-dicarboximide | (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide | (R)-16-Hydroxy-3-methylhexadecanoic acid |
| Phenylacetaldehyde | Phenylacetaldehyde | Phenylacetaldehyde | Butyl 2,4-dimethyl-2-nitro-4-pentenoate | |
| 2-Furanmethanol | 2-Furanmethanol | Pentane, 3-bromo- | ||
| 2-Propenoic acid, methyl ester | 2-Propenoic acid, methyl ester | |||
| 4-Amino-3,5-bis(dimethoxymethyl)-4H-1,2,4-triazole | 4-Amino-3,5-bis (dimethoxymethyl)-4H-1,2,4-triazole | |||
| 6-Methylhexahydrocycloprop[a]pentalen-3a,6-diol | N-Methylcyclopentane-1,1-dicarboximide | |||
| Pent-4-enal | Pentane, 3-bromo- | |||
| 2-Furancarboxaldehyde, 5-methyl- | 1-(2,3,4,5,6-pentafluorophenyl)sulfanylhex-5-en-2-one | |||
| SL-295 | 2-Propenoic acid, methyl ester | 2-Propenoic acid, methyl ester | (+/−)Bis(2-aminobut-3-enyl) disulphide | (+/−)Bis(2-aminobut-3-enyl) disulphide |
| 3-Oxabicyclo[3.1.0]hexan-2-one | 3-Oxabicyclo[3.1.0]hexan-2-one | (E)-6-Biphenyl-4-yl-4-oxohexanoic acid ethanolamide | (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide | |
| Phenylacetaldehyde | Phenyl acetaldehyde | Phenyl acetaldehyde | Benzeneacetaldehyde | |
| 1,1,4,4,7,7,10,10,13,13,16,16-Dodecachloro-hexasila-18-crow-6 | 4-endo-Amino-2-oxabicyclo[3.3.0]oct-7-en-3-one | 2(5H)-Furanone | 2-Furanmethanol | |
| 4-Amino-3,5-bis(dimethoxymethyl)-4H-1,2,4-triazole | N-Methylcyclopentane-1,1-dicarboximide | 2-Furancarboxaldehyde, 5-methyl- | 2-Furancarboxaldehyde, 5-methyl- | |
| N-Methylcyclopentane-1,1-dicarboximide | Pentane, 3-bromo- | 3,5-Dideutero-aniline | Cyclooctylhydroperoxide | |
| Pent-4-enal | 2-Furanmethanol | 2-Furanmethanol | Methyl 3-hydroxy-2-(p-toluenesulfonyloxy)methyl-10-tetrahydropyranyloxydecanoate | |
| 1,4-Ethanonaphthalene-5,8-dione, 1,4,4a,8a-tetrahydro-1-methyl-4-(1-methylethyl)- | 3-Fluorothiophene | |||
| syn-2-(2-Hydroxypropyl)-6-methyldihydropyran-3(6H)-ol | (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide |
In Gujosoya-2, phenylacetaldehyde was a common metabolite in control, NaCl, and Na+ treatment. The (E)-6-Naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide, 2-Furanmethanol, 2-Propenoic acid, methyl ester, and 4-Amino-3,5-bis (dimethoxymethyl)-4H-1,2,4-triazole were observed in control and Na+ treatments. N-Methylcyclopentane-1,1-dicarboximide was found under NaCl and Na+ salt. Pentane, 3-bromo- only observed in Na+ and Cl− salt. The 6-Methylhexahydrocycloprop[a]pentalen-3a,6-diol, Pent-4-enal and 2-Furancarboxaldehyde, 5-methyl- were observed only under controlled condition. The compound 1-(2,3,4,5,6-pentafluorophenyl)sulfanylhex-5-en-2-one was found only in Na+ salt and (R)-16-Hydroxy-3-methylhexadecanoic acid, and butyl 2,4-dimethyl-2-nitro-4-pentenoate which were observed under Cl− treatment (Table 3). In SL-295, phenylacetaldehyde was observed in control, NaCl, and Na+, while 2-Furanmethanol in NaCl, Na+, and Cl− salt. The 2-Propenoic acid, methyl ester, and 3-Oxabicyclo[3.1.0]hexan-2-one were observed in control and NaCl treatment. The (+/−)bis(2-aminobut-3-enyl) disulphide, (E)- 6-naphthalen-2-yl-4-oxohex-5-enoic acid diethanolamide, and 2-Furancarboxaldehyde 5-methyl- were synthesized in Na+ and Cl− salt. The metabolites 1,1,4,4,7,7,10,10,13,13,16,16-Dodecachloro-hexasila-18-crow-6, 4-Amino-3,5-bis(dimethoxymethyl)-4H-1,2,4-triazole, N-Methylcyclopentane-1,1-dicarboximide, Pent-4-enal, 1,4-Ethanonaphthalene-5,8-dione, 1,4,4a,8a-tetrahydro-1-methyl-4-(1-methylethyl), and syn-2-(2-Hydroxypropyl)-6-methyldihydropyran-3(6H)-ol were observed only under control condition, while 4-endo-Amino-2-oxabicyclo[3.3.0]oct-7-en-3-one, N-Methylcyclopentane-1,1-dicarboximide, Pentane, 3-bromo were observed only under the NaCl treatment. The metabolites (E)-6-Biphenyl-4-yl-4-oxohexanoic acid ethanolamide, 2(5H)-furanone, 3,5-Dideutero-Aniline- and 3-Fluorothiophene observed only under Na+ salt, while benzeneacetaldehyde, cyclooctylhydroperoxide, and methyl 3-hydroxy-2-(p-toluenesulfonyloxy)methyl-10-tetrahydropyranyloxydecanoate were found under Cl− salt (Table 3). The metabolic variation at certain level was observed in between genotypes and within treatment of each genotype.
Discussion
Studies on plant salt tolerance have contributed a great deal of information on plant responses and management of salt (NaCl) stress, and in most cases, effects of sodium toxicity have been the major focus. However, simultaneous comparison of the individual Na+, Cl− ions, and NaCl salt effects are scarcely reported in field crops (Kumar and Khare 2016; Li et al. 2017). It is, therefore, important to study the individual effects of Cl− and Na+ ions on physiological and biochemical aspects of growth and development. In the present study, the relative impact of individual Cl− and Na+ and additive NaCl salt was analyzed using in vitro callus cultures of four contrasting soybean genotypes that were exposed to equimolar Na+, Cl−, and NaCl salt. Previous studies have shown that in vitro culture systems provide controlled environmental conditions to analyze the salt-stress effects at cellular level (Nikam et al. 2014; Rania et al. 2015). Both Cl− and Na+ exerted negative impact on dry matter (Table 1) and all the salt treatments caused reduction in FW and DW in all the genotypes as compared to their respective controls. NaCl stress-induced reduction in fresh and dry weight and TWC has been shown previously (Forooghian and Esfarayeni 2013; Balen et al. 2013). In the present study, both the ionic stresses were found to be genotype and treatment specific, and significantly affected FW, DW, and %TWC (Table 1). Among salt treatments, Na-dominant salt treatment showed significantly higher reduction in %TWC than NaCl and Cl-dominant. Gujosoya-2 and SL-295 showed more reduction in %TWC than MAUS-47 and Bragg genotypes in all the salt treatments. Such NaCl-induced reduction in %TWC in callus cultures was also reported in Guizotia abyssinica (Ghane et al. 2014) and Saccharum officinarum (Patade et al. 2008; Nikam et al. 2014).
Increasing salinity stress causes reactive oxygen species (ROS) burst in plant cells. To combat the ROS induced damage, plants deploy both non-enzymatic and enzymatic antioxidant systems. Several studies revealed that efficient and timely induction of these antioxidant systems plays a vital role in protection of plants from various abiotic stresses by scavenging and detoxifying ROS molecules (Hossain et al. 2007; Niknam et al. 2011; Kiani-Pouya 2015). The salt-tolerance ability of tolerant germplasm is often associated with effective antioxidant machinery as compared to their susceptible genotypes relatives (Yasar et al. 2013; Kiani-Pouya 2015). The present study revealed that MAUS-47 and Bragg genotypes showed significantly higher SOD, CAT, APX, GPOX, and GR activities than Gujosoya-2 and SL-295 (Fig. 1). Using in vitro callus cultures, several researchers analyzed salt-stress-induced differential antioxidant activities in callus of different plant species such as Sugarcane (Patade et al. 2012), Melon (Kusvuran et al. 2016), and Triticale (Bezirganoglu 2017). The Gujosoya-2 and SL-295 genotypes were unable to maintain higher enzymatic antioxidant activities to cope with Na+ stress. The results suggested that Na+ caused severe damage in Gujosoya-2 and SL-295 callus which could not be prevented by the induction of antioxidant activities. Similarly, under salt stress, in salt-sensitive sunflower callus, the increase in activities of antioxidant enzymes were observed (Davenport et al. 2003). The NaCl and Cl− also induced significantly higher enzyme activities in MAUS-47 and Bragg genotypes than Gujosoya-2 and SL-295. The enhanced enzyme activities are well associated with salt-tolerance ability of genotypes (Gandonou et al. 2006; Sharma and Ramawat 2013). Based on our results, the stress-induced responses are suggested to be more pronounced under the Na+ treatment followed by NaCl and Cl−.
Increased accumulation of osmolytes is an essential adaptive response to salt stress (Ghane et al. 2014; Suprasanna et al. 2016). Osmotically dynamic compounds like sucrose, glycine betaine, proline, etc. are involved in salt-stress amelioration via maintenance of osmotic adjustment, stabilization of different proteins and their complexes, and scavenging the ROS under salt stress (Slama et al. 2015). In the present investigation, osmolyte accumulation (glycine betaine, proline, and TSS) was studied in the four genotypes for their ability to tolerate individual and combined salts. The results showed noticeably higher contents of glycine betaine, proline, and TSS in MAUS-47 and Bragg under Na+ salt followed by NaCl and Cl−. In Gujosoya-2 and SL-295 proline, glycine betaine and TSS contents were found low and equal to control and thus callus failed to combat against Na+ and other stress treatments. The increased osmolyte content under salinity stress is a significant trait of the salt-tolerant genotypes (Rai et al. 2011; Patade et al. 2012; Nikam et al. 2014). Our results are indicative that the tolerant genotypes showed higher accumulation of osmolytes than sensitive genotypes, and the differential osmolyte accumulation was more under Na+ salt than NaCl and Cl− salt treatment in soybean.
Plant adaptation to salt stress, essentially, is dependent on efficient cellular ion homeostasis of net intracellular Na+ and Cl− uptake and its compartmentalization (Munns and Tester 2008). In the present study, increased Na+ and Cl− contents were observed under NaCl, Na+, and Cl− salt, but there was a significant variation among the soybean genotypes (Table 2). The increased Na+ content under Na+ and NaCl salt resulted in a significant drop in K+ and Ca2+ levels (Table 2). The toxic impact of Na+ in many plant species because of its role in nutrient imbalance has been demonstrated during salt stress (Ahmad et al. 2007; Kumar et al. 2008). Ability of Na+ to compete with K+ ions for absorption and binding sites of enzymes and subsequent inactivation of essential enzymes is the root cause of toxicity (Balen et al. 2013). Under saline condition, callus experienced dual cellular damage with low K+ concentrations and high Na+ toxicity (Errabii et al. 2007; Balen et al. 2013). In our study, both the sensitive genotypes, Gujosoya-2 and SL-295 exhibited an elevated Na+/K+ ratio compared with tolerant genotypes MAUS-47 and Bragg. This indicated inefficiency of the former to restrict the influx of toxic ions into cell during saline conditions. In contrast, tolerant genotypes (MAUS-47 and Bragg) successfully employ ion exclusion, maintain better cellular K+ level under NaCl and Na+ stress, and achieve higher salt tolerance. Interaction between Na+ and Ca2+ ions under salt stress disturbs ion transport and cell membrane properties, and leads to change in Ca2+ activity in cytoplasm. This resulted in altered physiological and biochemical attributes related to callus growth, nutrition, osmolyte, ion accumulation, and water transport (Summart et al. 2010; Ghane et al. 2014). In the present investigation, Ca2+ markedly decreased in Na+, Cl−, and NaCl stress, and it exhibited genotype-specific variation. Under these treatments, Gujosoya-2 and SL-295 showed lower accumulation of K+ and Ca2+, compared to treatment under Cl− stress (because of KCl and CaCl2 application) which showed significantly high accumulation of K+ and Ca2+ which may render lower cellular toxicity. Similar results were revealed in sugarcane calli where the gathering of Na+ and Cl− content decreased K+ and Ca2+ during salt stress (Patade et al. 2008). A higher concentration of Ca2+ in MAUS-47 and Bragg genotypes could help the replacement of displaced Ca2+, which results in restoration of plasma membrane integrity and cell wall stability, facilitating higher Na+/K+ selectivity, increased Na+ exclusion, and ultimately improvement in salt tolerance (Errabii et al. 2007; Summart et al. 2010; Nikam et al. 2014). The salt stress affects the allocation of nutrient elements and its uptake in callus tissues (Ahmad et al. 2009; Kumar et al. 2008).
Extensive research has been conducted on plant metabolomic responses under high salinity (Widodo et al. 2009; Zhang et al. 2016; Guo et al. 2017), but these studies were mostly limited to NaCl-induced stress. In the present investigation, involvement of certain metabolites under individual and additive salt effects of NaCl was studied. The capability of a plant to withstand against salt stress is primarily based on the induction of plant hormones, synthesis of osmolytes, induction of ROS scavengers, and modifications of cell membrane, and ensuing metabolite changes that could be useful in metabolic analysis to discriminate soybean genotypes. In the present study, among the metabolites, aldehydes and furans such as benzeneacetaldehyde, 2-furan-carboxaldehyde, 2-furancarboxaldehyde 5-methyl-, phenyl acetaldehyde, and 2-furanmethanol were found in salt-stressed calli. Some of these metabolites were previously observed in soybean due to lipid oxidation, degradation, and sugar dehydration (Pokorny et al. 2000; Lee and Ahn 2009). The present results showed that, under different salt treatments, soybean callus synthesized aldehydes and alcohol. The aldehydes and alcohol were previously shown to be induced in daffodil flowers under salinity stress (Koksal et al. 2015). Aldehydes are important intermediate of carbohydrates, vitamins, steroids, amino acids, and lipids anabolic and catabolic pathways (Kirch et al. 2004). The various stresses induced the excess accumulation of aldehydes (Feder and Hofmann 1999) which negatively affect plant growth (Kotchoni et al. 2010). Plants regulate the cellular aldehyde level by limiting membrane peroxidation events by exploring enzymatic (SOD, CAT, and POD) antioxidant properties and directly through aldehyde dehydrogenases enzyme activity (Waheed et al. 2018). Some aldehydes and alcohol derivatives act like reactive molecules in plants have damaging effect on cell wall and cell membranes (Huang et al. 2011). The higher molecular weight methyl esters and acids were observed in differentially stressed calli samples which may be because of esterification of alcohol and free fatty acids (Lee and Ahn 2009). The fatty acids synthesized in N. tangutorum Bobr. suspension cells showed improved salinity stress tolerance (Ni et al. 2015). Ketones were also observed in differentially stressed calli samples possibly due to the degradation of lipids and/or amino acids (Su 1986). The MAUS-47 and Bragg under Na+ and Cl− salt was found to have azetidine isoflavonoid. Previously, it was reported that the proline analog azetidine-2-carboxylic acid accelerates the assembly of class I small heat shock proteins (sHSPs), which provides thermo-tolerance in soybean seedlings (Jinn et al. 2004), but it will be worthwhile to further explore any such role of this proline analog under individual and additive salt treatments. In this study, we have observed different ketones, aldehydes, and alcohols which were also detected. It was observed that lipoxygenases (LOXs) are an important class of non-heme iron enzymes yield flavor precursors hydroperoxides and further hydroperoxide lyase convert it in to flavor compounds such as ketones, aldehydes, and alcohols (Fauconnier and Marlier 1997). In our study, 1,4-dioxin and 2-octanone were detected in Bragg genotype under Na+ and NaCl treatments, respectively. Vega et al. (2005) reported that activity of LOXs was reliant on the cosolvent concentration, which was augmented with 1,4-dioxane up to 5% (v/v), while its higher concentration decreased its activity and 2-octanone also showed its inhibitory effect on activity of LOXs. The data also showed the presence of 3-fluorothiophene compound under Na+ salt stress in callus of Bragg genotypes. It was reported that it plays an inhibitory role in xanthine oxidase enzymes (patents/EP1783124A1). Xanthine oxidase (XOD) contributes to H2O2 production in plants during saline stress (Sharma et al. 2012). Sauter et al. (2002) reported improved stress tolerance under high oxidative stress through overexpression of ROS quenchers like chlorothiophene as a thiophene derivative. The 3-Fluorothiophene detected in tolerant genotypes may prompt further studies on its role in salinity tolerance. In view of the present results, we hypothesized that these metabolic changes could be used to unravel the salt tolerence mechanism in soybean and other crop plants.
Conclusion
In conclusion, our results on the effects of individual and additive sodium and chloride ions revealed that soybean genotypes differ in their physiological and biochemical responses. The tolerance mechanism in soybean genotypes MAUS-47 and Bragg was due to increased activity of antioxidant system, accumulation of osmolytes, and maintenance of Na+/K+ ratio and Ca2+ level. On the contrary, the response to salt stress was declined in sensitive genotypes SL-295 and Gujosoya-2. Among the salts, Na+ was more detrimental followed by NaCl and Cl−. Since the toxicity of Cl− was enough to reduce plant growth and development, further investigation is needed to study the molecular mechanism. The genotype-specific occurrence of azetidine, 2-furanmethanol, 1,4-dioxin, 3-fluorothiophene, decanoic acid and 2-propenoic acid, and methyl ester metabolites indicate their probable role in salt tolerance. Detailed studies are warranted on the role of specific salt responsive metabolites in soybean which could be used in the improvement of salt-tolerant genotypes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the help of National Institute of Soybean Research, Indore, MP, India, for supply of soybean genotypes seeds, and the research facilities available at Department of Botany, Savitribai Phule Pune University, Pune, under the research grants from DST-PURSE, DST-FIST, and UGC DSA-I program of Government of India.
Author contributions
The authors TDN, SP, and BNZ have developed and supervised the research work. The authors DBS, GCN, and MRC have carried out lab experiment, data collection, analysis, and drafting of manuscript. SP and TDN revised the MS.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Abogadallah GM. Antioxidative defense under salt stress. Plant Signal Behav. 2010;5(4):369–374. doi: 10.4161/psb.5.4.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad MSA, Javed F, Ashraf M. Iso-osmotic effect of NaCl and PEG on growth, cations and free proline accumulation in callus tissue of two indica rice (Oryza sativa L.) genotypes. Plant Growth Regul. 2007;53:53–63. [Google Scholar]
- Ahmad MSA, Javed F, Javed S, Alvi AK. Relationship between callus growth and mineral nutrients uptake in salt-stressed indica rice callus. J Plant Nutr. 2009;32:382–394. [Google Scholar]
- Amirjani MR. Effect of salinity stress on growth, mineral composition, proline content, antioxidant enzymes of soybean. Am Plant Physiol. 2010;5:350–360. [Google Scholar]
- Balen B, Tkalec M, Rogic T, Simac M, Stefanic PP, Roncevic S, Svedruzic LP, Krsnik-Rasol M. Effects of iso-osmotic NaCl and mannitol on growth, proline content, and antioxidant defense in Mammillaria gracilis Pfeiff. in vitro-grown cultures. In Vitro Cell Dev Biol Plant. 2013;49:421–432. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–208. [Google Scholar]
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44(1):276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bezirganoglu I. Response of five triticale genotypes to salt stress in in vitro culture. Turk J Agric For. 2017;41:372–380. [Google Scholar]
- Boamponsem GA, Leung DWM, Lister C. Relationships among iron deficit-induced potato callus growth inhibition, Fe distribution, chlorosis, and oxidative stress amplified by reduced antioxidative enzyme activities. Plant Cell Tissue Organ Cult. 2018;132:393–412. [Google Scholar]
- Cakmak I, Marschner H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase and glutathione reductase in bean leaves. Plant Physiol. 1992;98:1222–1227. doi: 10.1104/pp.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Li Y, Liu B, Kong F, Tran LP. Adaptive mechanisms of soybean grown on salt-affected soils. Land Degrad Dev. 2017;29:1054–1064. [Google Scholar]
- Chapman HD, Pratt PF. Method for analysis of soil, plants and waters. Berkeley: University of California; 1961. [Google Scholar]
- Davenport SB, Gallego SM, Benavides MP, Tomaro ML. Behaviour of antioxidant defense system in the adaptive response to salt stress in Helianthus annuus L. cells. Plant Growth Regul. 2003;40:81–88. [Google Scholar]
- Duncan DB. Multiple range and multiple F tests. Biometrics. 1955;11:1. [Google Scholar]
- Errabii T, Gandonou CB, Essalmani H, Abrini J, Idaomar M, Skali-Senhaji N. Effects of NaCl and mannitol induced stress on sugarcane (Saccharum sp.) callus cultures. Acta Physiol Plant. 2007;29:95–102. [Google Scholar]
- Fauconnier ML, Marlier M. Fatty acid hydroperoxides pathways in plants. A review. Grasas Aceites. 1997;48:30–37. [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Forooghian S, Esfarayeni S. An evaluation of effect of salt stress on callus induction in different potato cultivars. Am-Euras. J Agric Environ Sci. 2013;13(8):1135–1140. [Google Scholar]
- Franco-Navarro JD, Brumos J, Rosales MA, Cubero-Font P, Talón M, Colmenero-Flores JM. Chloride regulates leaf cell size and water relations in tobacco plants. J Exp Bot. 2016;67:873–891. doi: 10.1093/jxb/erv502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandonou CB, Errabii T, Abrini J, Idaomar M, Senhaji SN. Selection of callus cultures of sugarcane (Saccharum sp.) tolerant to NaCl and their response to salt stress. Plant Cell Tissue Organ Cult. 2006;87:9–16. [Google Scholar]
- Ghane SG, Lokhande VH, Nikam TD. Growth, physiological, and biochemical responses in relation to salinity tolerance for in vitro selection in oil seed crop Guizotia abyssinica Cass. J Crop Sci Biotechnol. 2014;17(1):11–20. [Google Scholar]
- Grieve CM, Grattan SR. Rapid assay for determination of water soluble quaternary ammonium compounds. Plant Soil. 1983;70:303–307. [Google Scholar]
- Guan Z, Chen S, Chen F, Liu Z, Fang W, Tang J. Comparision of stress effect of NaCl, Na+ and Cl− on two Chrysanthemum species. Acta Hortic. 2012;937:369–375. [Google Scholar]
- Guo R, Shi LX, Yan C, Zhong X, Gu F, Liu Q, Xia X, Li H. Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol. 2017;17:41. doi: 10.1186/s12870-017-0994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemeda HM, Klein BP. Effects of naturally occurring antioxidants on peroxidase activity of vegetable extracts. J Food Sci. 1990;55:184–186. [Google Scholar]
- Hossain MS, Dietz K. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front Plant Sci. 2016;7:548. doi: 10.3389/fpls.2016.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z, Mandal AKA, Datta SK, Biswas AK. Development of NaCl-tolerant line in Chrysanthemum morifolium Ramat., through shoot organogenesis of selected callus line. J Biotechnol. 2007;129:658–667. doi: 10.1016/j.jbiotec.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 2011;7:e1001335. doi: 10.1371/journal.ppat.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chen L, Zhen A, Liu ZX, Lei B, Kong QS, Bie ZL. Effects of iso-osmotic Na+, Cl and NaCl stress on the plant growth and physiological parameters of grafted cucumber. Acta Hortic. 2015;1086:153–160. [Google Scholar]
- Ismail A, Takeda S, Nick P. Life and death under salt stress: same players, different timing. J Exp Bot. 2014;65(12):2963–2979. doi: 10.1093/jxb/eru159. [DOI] [PubMed] [Google Scholar]
- Jinn TL, Chiu CC, Song WW, Chen YM, Lin CY. Azetidine-induced accumulation of class I small heat shock proteins in the soluble fraction provides thermotolerance in soybean seedlings. Plant Cell Physiol. 2004;45:1759–1767. doi: 10.1093/pcp/pch193. [DOI] [PubMed] [Google Scholar]
- Keskes H, Belhadj S, Jlail L, Feki AE, Damak M, Sayadi S, Allouche N. LC-MS–MS and GC–MS analyses of biologically active extracts and fractions from Tunisian Juniperus phoenice leaves. Pharm Biol. 2017;55(1):88–95. doi: 10.1080/13880209.2016.1230139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Kumar V, Kavi Kishor PB. Na+ and Cl ions show additive effects under NaCl stress on induction of oxidative stress and the responsive antioxidative defense in rice. Protoplasma. 2015;252:1149–1165. doi: 10.1007/s00709-014-0749-2. [DOI] [PubMed] [Google Scholar]
- Kiani-Pouya A. Changes in activities of antioxidant enzymes and photosynthetic attributes in triticale (× Triticosecale Wittmack) genotypes in response to long-term salt stress at two distinct growth stages. Acta Physiol Plant. 2015;37:72. [Google Scholar]
- Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004;9:371–377. doi: 10.1016/j.tplants.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Koksal N, Kafkas E, Sadighazadi S, Kulahlioglu I. Floral fragrances of daffodil under salinity stress. Romanian Biotechnol Lett. 2015;20:4. [Google Scholar]
- Kotchoni SO, Jimenez-Lopez JC, Gao D, Edwards V, Gachomo EW, Margam VM, Seufferheld MJ. Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS One. 2010;5:e11516. doi: 10.1371/journal.pone.0011516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Khare T. Differential growth and yield responses of salt-tolerant and susceptible rice cultivars to individual (Na+ and Cl) and additive stress effects of NaCl. Acta Physiol Plant. 2016 [Google Scholar]
- Kumar N, Pamidimarri S, Kaur M, Boricha G, Reddy MP. Effects of NaCl on growth, ion accumulation, protein, proline contents and antioxidant enzymes activity in callus cultures of Jatropha curcas. Biologia. 2008;63(3):378–382. [Google Scholar]
- Kumar S, Beena AS, Awana M, Singh A. Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front Plant Sci. 2017;8:1151. doi: 10.3389/fpls.2017.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusvuran S, Ellialtıoglu SS, Talhouni M, Sonmez K, Kıran S. Effects of salt and drought stresses on physiological and biochemical changes in callus tissues of melon cultivars. Acta Hortic. 2016;1142:239–246. [Google Scholar]
- Lee SJ, Ahn B. Comparison of volatile components in fermented soybean pastes using simultaneous distillation and extraction (SDE) with sensory characterization. Food Chem. 2009;114:600–609. [Google Scholar]
- Li B, Tester M, Gilliham M. Chloride on the move. Trends Plant Sci. 2017;22(3):236–248. doi: 10.1016/j.tplants.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosenbrough NJ, Far AL, Randall RJ. Protein measurement with the Folin-phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Muchate NS, Nikalje GC, Rajurkar NS, Suprasanna P, Nikam TD. Plant salt stress: adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot Rev. 2016 [Google Scholar]
- Muchate NS, Nikalje GC, Rajurkar NS, Suprasanna P, Nikam TD. Physiological responses of the halophyte Sesuvium portulacastrum to salt stress and their relevance for saline soil bio-reclamation. Flora. 2016;224:96–105. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxide in spinach chloroplasts. Plant Cell Physiol. 1981;22:867–880. [Google Scholar]
- Negrao S, Schmockel SM, Tester M. Evaluating physiological responses of plants to salinity stress. Ann Bot. 2017;119:1–11. doi: 10.1093/aob/mcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Yang X, Zhu J, Liu Z, Ni Y, Wu H, Zhang H, Liu T. Salinity-induced metabolic profile changes in Nitraria tangutorum Bobr. suspension cells. Plant Cell Tissue Organ Cult. 2015;122:239–248. [Google Scholar]
- Nikalje GC, Srivastava AK, Pandey GK, Suprasanna P. Halophytes in biosaline agriculture: mechanism, utilization and value addition. Land Degrad Dev. 2017 [Google Scholar]
- Nikam AA, Devarumath RM, Shitole MG, Ghole VS, Tawar PN, Suprasanna P. Gamma radiation, in vitro selection for salt (NaCl) tolerance, and characterization of mutants in sugarcane (Saccharum officinarum L.) In Vitro Cell Dev Biol Plant. 2014;50:766–776. [Google Scholar]
- Niknam V, Meratan AA, Ghaffari SM. The effect of salt stress on lipid peroxidation and antioxidative enzymes in callus of two Acanthophyllum species. In Vitro Cell Dev Biol Plant. 2011;47:297–308. [Google Scholar]
- Parihar P, Singh S, Singh R, Singh VP, Prasad SM. Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res. 2015;22:4056–4075. doi: 10.1007/s11356-014-3739-1. [DOI] [PubMed] [Google Scholar]
- Patade VY, Suprasanna P, Bapat VA. Effects of salt stress in relation to osmotic adjustment on sugarcane (Saccharum officinarum L.) callus cultures. Plant Growth Regul. 2008;55:169–173. [Google Scholar]
- Patade VY, Bhargava S, Suprasanna P. Effects of NaCl and iso-osmotic PEG stress on growth, osmolytes accumulation and antioxidant defense in cultured sugarcane cells. Plant Cell Tissue Organ Cult. 2012;108:279–286. [Google Scholar]
- Patent: 2-Phenylthiophene derivative EP 1783124 A1, PCT/JP2005/015550, patents/EP1783124A1?cl=en
- Piwowarczyk B, Tokarz K, Kaminska I. Responses of grass pea seedlings to salinity stress in in vitro culture conditions. Plant Cell Tissue Organ Cult. 2016;124:227–240. [Google Scholar]
- Pokorny J, Mansour AF, Pudil F, Vaclav J. Effect of defatted soybean flour on the flavour of extruded mixtures with wheat flour. Czech J Food Sci. 2000;20:229–236. [Google Scholar]
- Rahnama H, Ebrahimzadeh H. The effect of NaCl on proline accumulation in potato seedlings and calli. Acta Physiol Plant. 2004;26:263–270. [Google Scholar]
- Rahnama H, Ebrahimzadeh H, Ghareyazie B. Antioxidant enzyme responses to NaCl stress in calli of four potato cultivar. Pak J Bot. 2003;35:579–586. [Google Scholar]
- Rai MK, Kalia RK, Singh R, Gangola MP, Dhawan AK. Developing stress tolerant plants through in vitro selection—an overview of the recent progress. Environ Exp Bot. 2011;71:89–98. [Google Scholar]
- Rania JK, Safa C, Wiem E, Mohamed NS, Noureddine D, Radhia GB, Oumema NE. Investigation of the response to salinity and to oxidative stress of interspecific potato somatic hybrids grown in a greenhouse. Plant Cell Tissue Organ Cult. 2015;120:933–947. [Google Scholar]
- Sauter A, Dietz KJ, Hartung W. A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ. 2002;25:223–228. doi: 10.1046/j.1365-3040.2002.00747.x. [DOI] [PubMed] [Google Scholar]
- Sharma V, Ramawat KG. Salinity-induced modulation of growth and antioxidant activity in the callus cultures of miswak (Salvadora persica) 3 Biotech. 2013;3:11–17. doi: 10.1007/s13205-012-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot. 2012;217037:26. [Google Scholar]
- Shelke DB, Pandey M, Nikalje GC, Zaware BN, Suprasanna P, Nikam TD. Salt responsive physiological, photosynthetic and biochemical attributes at early seedling stage for screening soybean genotypes. Plant Physiol Biochem. 2017;118:519–528. doi: 10.1016/j.plaphy.2017.07.013. [DOI] [PubMed] [Google Scholar]
- Sibole JV, Cabot C, Poschenrieder C, Barcelo J. Efficient leaf ion partitioning, an overriding condition for abscisic acid-controlled stomatal and leaf growth responses to NaCl salinization in two legumes. J Exp Bot. 2003;390:2111–2119. doi: 10.1093/jxb/erg231. [DOI] [PubMed] [Google Scholar]
- Slama I, Abdelly C, Bouchereau A, Flowers T, Savoure A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann Bot. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IK, Vierheller TL, Thurne CA. Assay of glutathione reductase in crude tissue homogenates using 5,5-dithiobis (2-nitrobenzoic acid) Anal Biochem. 1988;175:408–413. doi: 10.1016/0003-2697(88)90564-7. [DOI] [PubMed] [Google Scholar]
- Su YC. Sufu. In: Reddy NR, Pierson MD, editors. Legume-based fermented foods. Boca Raton: CRC Press; 1986. pp. 69–83. [Google Scholar]
- Summart J, Thanonkeo P, Panichajakul S, Prathepha P, McManus MT. Effect of salt stress on growth, inorganic ion and proline accumulation in Thai aromatic rice, Khao Dawk Mali 105, callus culture. Afr J Biotechnol. 2010;9(2):145–152. [Google Scholar]
- Suprasanna P, Nikalje GC, Rai AN. Osmolyte accumulation and implications in plant abiotic stress tolerance. In: Iqbal N, Nazar R, Khan NA, editors. Osmolytes and plants acclimation to changing environment: emerging omics technologies. New Delhi: Springer; 2016. pp. 1–12. [Google Scholar]
- Tavakkoli E, Rengasamy P, Mcdonald GK. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of Faba bean under salinity stress. J Exp Bot. 2010;61:4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E, Fatehi F, Coventry S, Rengasamy P, Mcdonald GK. Additive effects of Na+ and Cl– ions on barley growth under salinity stress. J Exp Bot. 2011;62:2189–2203. doi: 10.1093/jxb/erq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teakle NL, Flowers TJ, Real D, Colmer TD. Lotus tenuis tolerates the interactive effects of salinity and water logging by ‘excluding’ Na+ and Cl from the xylem. J Exp Bot. 2007;58:2169–2180. doi: 10.1093/jxb/erm102. [DOI] [PubMed] [Google Scholar]
- Vega M, Karboune S, Kermasha S. Stability of immobilized soybean lipoxygenase in selected organic solvent media. Appl Biochem Biotechnol. 2005;127:0029. doi: 10.1385/abab:127:1:029. [DOI] [PubMed] [Google Scholar]
- Waheed A, Sami RA, Lili L, Malota S, Xiaomin C, Frankline JO, Tao Y, Yahong L, Justice N, Zonghua W. Family-four aldehyde dehydrogenases play an indispensable role in the pathogenesis of Magnaporthe oryzae. Front Plant Sci. 2018;9:980. doi: 10.3389/fpls.2018.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Kojima K, Ide Y, Sasaki S. Effect of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell Tissue Organ Cult. 2000;63:199–206. [Google Scholar]
- Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J Exp Bot. 2009;60:4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasar F, Uzal O, Yasar O. Determination of antioxidative enzyme activities in callus culture of the salt-tolerant and salt-sensitive watermelon (Citrullus lanatus (Thunb.) Mansf.) genotypes under salt stress. J Food Agric Environ. 2013;11:1437–1440. [Google Scholar]
- Zhang J, Yang D, Li M, Shi L. Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS ONE. 2016;11(7):e0159622. doi: 10.1371/journal.pone.0159622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhao X, Li M, Jiang Y, Xu J, Jin J, Li K. Ectopic expression of Limonium bicolor (Bag.) Kuntze DREB (LbDREB) results in enhanced salt stress tolerance of transgenic Populus ussuriensis Kom. Plant Cell Tissue Organ Cult. 2018;132:123–136. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.