Abstract

A recently discovered photodecarboxylase from Chlorella variabilis NC64A (CvFAP) bears the promise for the efficient and selective synthesis of hydrocarbons from carboxylic acids. CvFAP, however, exhibits a clear preference for long-chain fatty acids thereby limiting its broad applicability. In this contribution, we demonstrate that the decoy molecule approach enables conversion of a broad range of carboxylic acids by filling up the vacant substrate access channel of the photodecarboxylase. These results not only demonstrate a practical application of a unique, photoactivated enzyme but also pave the way to selective production of short-chain alkanes from waste carboxylic acids under mild reaction conditions.
Introduction
Valorization of biobased feedstock into chemicals1−4 and fuels5−7 represents one of the pillars of a more sustainable society. The last few decades have seen an explosion of synthetic methodologies to transform natural products into building blocks for the chemical industry.1−4
Catalytic methodologies for the conversion of biobased carboxylic acids, however, are largely limited to esterification or amidation reactions.8 More recently, chemical9−12 and biocatalytic13−17 reduction reactions of carboxylic acids have been developed as well as some approaches for the oxidative decarboxylation into terminal alkenes.18−21
In addition, redox-neutral decarboxylation of carboxylic acids into the corresponding alkanes represents an attractive method for the synthesis of alkanes for fuel and synthetic applications. Conventional methods, however, require harsh reaction conditions and high loadings of precious metal catalysts. This implies a significant environmental impact of these reactions and questions their economic feasibility. In addition, the selectivity of the reported methods mostly is poor due to the formation of Kolbe- and Hofer–Moest-byproducts.22,23
To overcome some of these drawbacks, enzymes have been gaining an increasing interest for the direct formation of hydrocarbons.24−27 Recently, a new fatty acid photodecarboxylase from the unicellular photosynthetic green microalga Chlorella variabilis NC64A (CvFAP) was reported by Beisson and co-workers.28CvFAP mediates the blue-light driven decarboxylation of various fatty acids to the corresponding (C1-shortened) alkanes (Figure 1a). CvFAP, however, shows a marked preference for long-chain fatty acids (C16–C17) with its catalytic activity dropping dramatically with shorter chain carboxylic acids. Presumably, shorter substrates are less efficiently stabilized in the long, narrow substrate access channel of the decarboxylase.28
Figure 1.

CvFAP-catalyzed photobiocatalytic decarboxylation reaction. (a) general reaction scheme and reaction conditions; (b) schematic representation of acetic acid coordinating to the catalytic flavin group together with tetradecane as decoy molecule filling up the substrate access channel. CvFAP (PDB: 5NCC) is presented as a green cartoon showing the substrate channel as transparent gray surface, while FAD (yellow), acetic acid (blue) and tetradecane (magenta) are represented as sticks. The distances in angstroms from the carboxylate oxygen atom of acetic acid to FAD and from the C1-atom of tetradecane to both carbon atoms of acetic acid are indicated. First, acetic acid was docked into 5NCC, then tetradecane was docked into the minimized result of the first docking using VINA in Yasara (www.yasara.org). The figure was prepared using PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
A similar phenomenon has been observed with the P450 monooxygenase from Bacillus megaterium, which exhibits (almost) no oxyfunctionalisation activity toward small substrate molecules. The groups of Reetz29 and Watanabe30 independently proposed to fill up the vacant space left by small substrate molecules with unreactive, perfluorinated fatty acids. Inspired by their work we reasoned that a similar strategy might be applicable to enlarge the substrate scope of CvFAP (Figure 1b).
We propose using simple alkanes as decoy molecules, as cocatalysts, to accelerate the CvFAP-catalyzed decarboxylation of short-chain carboxylic acids.
Results and Discussion
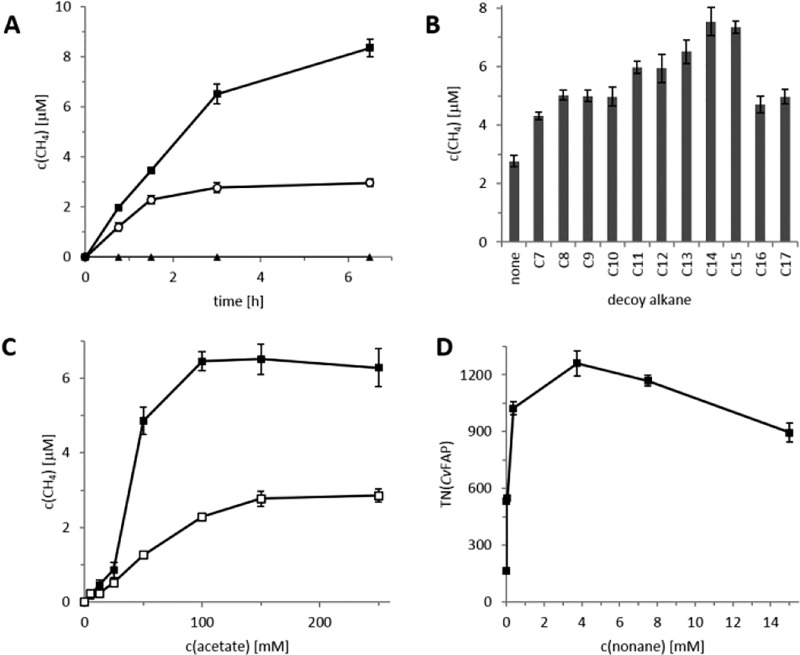
We first evaluated CvFAP as a photobiocatalyst for the decarboxylation of acetic acid to methane in the presence of various decoy alkanes (Figure 2). For this the enzyme was overproduced in Escherichia coli BL21 (DE3). A typical time course of the reaction is shown in Figure 2a. No methane could be detected in the absence of CvFAP and only minor amounts were formed in the absence of active illumination (possibly due to the incomplete light-insulation of the reaction vessel). Moreover, no background activity was found in crude extracts prepared from E. coli cells containing an empty vector. 13CH3–CO2H was converted into 13CH4 (Figure S6). Most interestingly, the rate of methane accumulation was almost doubled in the presence of tridecane as decoy molecule, while also the robustness of the reaction increased significantly leading to more than a 3-fold increase of methane yield. The highest acceleration of methane production was achieved in the presence of tetradecane (Figure 2b). Both in the absence and presence of a decoy molecule, the enzyme reaction rate depended on the concentration of acetic acid (Figure 2c). In both cases half-maximal rates were obtained at comparable acetic acid concentrations (around 50 mM), suggesting that the decoy molecule did not facilitate the binding of the carboxylic acid but rather increased the enzyme reaction rate. When varying the concentration of the decoy molecule (Figure 2d), a saturation-type dependency of the reaction rate was observed. Furthermore, at elevated concentrations of the decoy molecule an inhibitory effect was observed, which may be indicative for an ordered mode of binding of substrate and decoy molecule.
Figure 2.
Photoenzymatic decarboxylation of acetic acid to methane (A-C). (A) Time course of methane formation performed with (■) and without decoy molecule tridecane (○), and in the absence of CvFAP (▲). Reaction conditions: [decoy molecule] = 15 mM, [CvFAP] = 6 μM, [acetic acid] = 150 mM, Tris–HCl buffer pH 8.5 (100 mM), 20% DMSO, 30 °C, blue light (450 nm). Methane concentration refers to product in the headspace of the reaction vial (4.0 mL of headspace plus 1.0 mL of liquid). (B) Effect of carbon length of decoy alkane on the methane formation. Reaction time: 3h. (C) The rate-dependency of the methane production rate on the acetic acid concentration with (■) and without tridecane (□). Reaction time: 3 h. (D) Turnover number (TON) of CvFAP in the production of butane using nonane as decoy molecule. TON = molbutane × molCvFAP–1. The contribution of reaction without using nonane was subtracted. Condition: [CvFAP] = 6 μM, [pentanoic acid] = 150 mM, Tris–HCl buffer pH 8.5 (100 mM), 20% DMSO, 30 °C, 3 h. Error bars indicate the standard deviation of duplicate experiments (n = 2).
The rate of the photobiocatalytic decarboxylation reaction was linearly dependent on both the enzyme concentration (Figure S7) and the light intensity (Figure S8). The rate of the reaction in D2O was approximately 1.5 times faster than the same reaction in H2O (Figure S9). This inverse kinetic isotope effect most probably is due to an increased lifetime of the photoexcited flavin cofactor of CvFAP in D2O, as well documented previously for other flavoproteins.31−33 We interpret these findings with the lifetime of the photoexcited flavin of CvFAP being overall rate-limiting.
Encouraged by these promising results, we further evaluated the decarboxylation of a broader range of short-chain carboxylic acids (Table 1). The substrate scope of CvFAP was not limited to straight-chain carboxylic acids as also some branched and unsaturated carboxylic acids were converted at significant rates. Decarboxylation of acrylic acid and propionic acid was only possible with the assistance of decoy molecules. Notably, hydrogen gas was formed from the decarboxylation of formic acid. It also becomes clear from Table 1 (and Figures S11–19) that the most suited decoy molecule depends on the carboxylic acid used. In general the number of carbon atoms of substrate and decoy molecule should sum up to 16, which is in line with the previously reported optimal fatty acid chain length.28 It is also interesting to note that the accelerating effect of the decoy molecule decreased with the chain length of the substrate carboxylic acids.
Table 1. Substrate Scope of the Photobiocatalytic Decarboxylation of Short-Chain Carboxylic Acidsa.
Reaction conditions: [decoy molecule] = 7.5 mM, [CvFAP] = 6 μM, [substrate] = 150 mM, Tris–HCl pH 8.5 (100 mM), 20% DMSO, 30 °C, 3 h, blue light (450 nm), photon flux 13.8 μE/Ls. The standard deviation is based on duplicate experiments (n = 2). The concentration refers to product in the headspace of the reaction vial (4.0 mL of headspace plus 1.0 mL of liquid).
Selectivity % = [target product]/([target product]+[other product]) × 100, calculation based on GC.
0.4% propane, 0.038% ethane, 0.01% ethane, 0.2% isopentane and 0.05% propane were observed from Entries 3–7, accordingly.
Reaction was performed at 40 °C.
The quantum efficiency of the photobiocatalytic decarboxylation reactions was determined. Using acetic acid, butyric acid, pentanoic acid and hexanoic acid, the quantum efficiency (at photon flux 13.8 μE L–1s–1) was 0.008%, 1.3%, 3.2% and 2.7%, respectively (Figure S10). It should be noted that a direct comparison with the numbers reported in the initial report is difficult due to the lack of all necessary data.
A further insight into the decarboxylation mechanism was obtained in the framework of the density functional theory (DFT) and time-dependent (TD-DFT) density functional theory (PBE1PBE+D3(BJ)/6-311+G(d,p)+PCM(water)) analysis. These calculations were carried out on a representative model system mimicking the reactive complex of the flavin active site of the FDA and acetate anion as the representative substrate. Our calculations reveal the crucial role of the prearrangement of acetate in the vicinity of the N(5) site of the FAD (CH3COO–···FAD, Figure S32). This van der Waals complex closely resembles the configuration found from the crystal structure analysis shown in Figure 1. Importantly, in the absence of the confinement caused by the protein residues of CvFAP, an alternative coordination mode dominated by H-bonding between the acetate anion and the N–H moiety of FAD is favored by ca. 20 kJ/mol. However, we were not able to identify a favorable decarboxylation path initiated from this structure, which is in agreement with the lack of decarboxylation activity of the free flavin (Figure S33).
When acetate substrate prearrangement is effective, the decarboxylation path starts from the CH3COO–···FAD van der Waals complex exhibiting an absorption maximum at 467 nm.33 Analysis of the respective frontier orbitals (HOMO, LUMO, Figure S32a) shows that photoexcitation of this molecular complex yields a CH3COO•···FAD• radical pair, from which the decarboxylation of the CH3COO· proceeds rapidly via a highly favorable and low-barrier noncatalytic process. Computational studies propose that the generated CH3 radical further binds the anionic flavin semiquinone resulting in the relaxation of the excited state to the original S0 and the formation of a covalent dearomatized methylquinone flavin adduct. The overall decarboxylation reaction is slightly thermodynamically favorable (ΔGo = −5 kJ·mol–1) allowing thus for a smooth continuation of the catalytic cycle. The flavin adduct intermediate (CH3–FAD−) is then protonated to release the alkane product and regenerate the oxidized FAD closing the catalytic cycle (Figure S32b). Such a reaction pathway is also in agreement with the observed excellent selectivity of the photobiocatalytic decarboxylation reactions with all substrates used. This high selectivity makes the occurrence of free, C-centered radicals (at least for a prolonged period) highly unlikely.
Conclusion
Overall, in the present study we have demonstrated the photobiocatalytic decarboxylation of short-chain carboxylic acids using the fatty acid photodecarboxylase from Chlorella variabilis NC64A (CvFAP). While the wild-type enzyme to some extent can convert also shorter substrates, the use of so-called decoy molecules significantly accelerates the reaction rate. Admittedly, the present reactions showed modest turnover numbers. However, we expect that engineering of both the enzyme and the decoy molecules will further increase the reaction rate and eventually render the CvFAP-catalyzed decarboxylation of a large scope of (waste) carboxylic acids into a viable approach for the generation of light hydrocarbons. Given the recent success in the photodecarboxylation of long fatty acids,28,34 this work also demonstrates the broad potential of this photodecarboxylase for the application in biofuel synthesis and thus valorization of organic waste streams.
Acknowledgments
The Netherlands Organisation for Scientific Research is gratefully acknowledged for financial support through a VICI grant (no. 724.014.003). We thank Dr. Linda G. Otten and Dr. Fabio Tonin for support with the modelling.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b12282.
Experimental details including the preparation of the cell free extracts, photoenzymatic reactions, length of decoy molecule on decarboxylation activity, GC chromatogram and DFC calculations (PDF)
Author Contributions
∇ W.Z., M.M. and M.M.E.H. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Bozell J. J.; Petersen G. R. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539–554. 10.1039/b922014c. [DOI] [Google Scholar]
- Sheldon R. A. Green and sustainable manufacture of chemicals from biomass: state of the art. Green Chem. 2014, 16, 950–963. 10.1039/C3GC41935E. [DOI] [Google Scholar]
- Corma A.; Iborra S.; Velty A. Chemical routes for the transformation of biomass into chemicals. Chem. Rev. 2007, 107, 2411–2502. 10.1021/cr050989d. [DOI] [PubMed] [Google Scholar]
- Gallezot P. Conversion of biomass to selected chemical products. Chem. Soc. Rev. 2012, 41, 1538–1558. 10.1039/C1CS15147A. [DOI] [PubMed] [Google Scholar]
- Chu S.; Majumdar A. Opportunities and challenges for a sustainable energy future. Nature 2012, 488, 294–303. 10.1038/nature11475. [DOI] [PubMed] [Google Scholar]
- Potocnik J. Renewable energy sources and the realities of setting an energy agenda. Science 2007, 315, 810–811. 10.1126/science.1139086. [DOI] [PubMed] [Google Scholar]
- Obama B. The irreversible momentum of clean energy. Science 2017, 355, 126–129. 10.1126/science.aam6284. [DOI] [PubMed] [Google Scholar]
- Aransiola E. F.; Ojumu T. V.; Oyekola O. O.; Madzimbamuto T. F.; Ikhu-Omoregbe D. I. O. A review of current technology for biodiesel production: State of the art. Biomass Bioenergy 2014, 61, 276–297. 10.1016/j.biombioe.2013.11.014. [DOI] [Google Scholar]
- Das S.; Join B.; Junge K.; Beller M. A general and selective copper-catalyzed reduction of secondary amides. Chem. Commun. 2012, 48, 2683–2685. 10.1039/c2cc17209g. [DOI] [PubMed] [Google Scholar]
- Das S.; Möller K.; Junge K.; Beller M. Zinc-catalyzed chemoselective reduction of esters to alcohols. Chem. - Eur. J. 2011, 17, 7414–7417. 10.1002/chem.201100800. [DOI] [PubMed] [Google Scholar]
- Addis D.; Das S.; Junge K.; Beller M. Selective Reduction of Carboxylic Acid Derivatives by Catalytic Hydrosilylation. Angew. Chem., Int. Ed. 2011, 50, 6004–6011. 10.1002/anie.201100145. [DOI] [PubMed] [Google Scholar]
- Das S.; Addis D.; Zhou S.; Junge K.; Beller M. Zinc-Catalyzed Reduction of Amides: Unprecedented Selectivity and Functional Group Tolerance. J. Am. Chem. Soc. 2010, 132, 1770–1771. 10.1021/ja910083q. [DOI] [PubMed] [Google Scholar]
- Winkler M. Carboxylic acid reductase enzymes (CARs). Curr. Opin. Chem. Biol. 2018, 43, 23–29. 10.1016/j.cbpa.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Finnigan W.; Thomas A.; Cromar H.; Gough B.; Snajdrova R.; Adams J. P.; Littlechild J. A.; Harmer N. J. Characterization of Carboxylic Acid Reductases as Enzymes in the Toolbox for Synthetic Chemistry. ChemCatChem 2017, 9, 1005–1017. 10.1002/cctc.201601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. K.; Turner N. J.; Jones P. R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 87–92. 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitasubramanian P.; Daniels L.; Rosazza J. P. N. Reduction of Carboxylic Acids by Nocardia Aldehyde Oxidoreductase Requires a Phosphopantetheinylated Enzyme. J. Biol. Chem. 2007, 282, 478–485. 10.1074/jbc.M607980200. [DOI] [PubMed] [Google Scholar]
- Ni Y.; Hagedoorn P. L.; Xu J. H.; Arends I.; Hollmann F. A biocatalytic hydrogenation of carboxylic acids. Chem. Commun. 2012, 48, 12056–12058. 10.1039/c2cc36479d. [DOI] [PubMed] [Google Scholar]
- Lu C.; Shen F.; Wang S.; Wang Y.; Liu J.; Bai W.-J.; Wang X. An Engineered Self-Sufficient Biocatalyst Enables Scalable Production of Linear α-Olefins from Carboxylic Acids. ACS Catal. 2018, 8, 5794–5798. 10.1021/acscatal.8b01313. [DOI] [Google Scholar]
- Dennig A.; Kurakin S.; Kuhn M.; Dordic A.; Hall M.; Faber K. Enzymatic Oxidative Tandem Decarboxylation of Dioic Acids to Terminal Dienes. Eur. J. Org. Chem. 2016, 2016, 3473–3477. 10.1002/ejoc.201600358. [DOI] [Google Scholar]
- Zachos L.; Gassmeyer S. K.; Bauer D.; Sieber V.; Hollmann F.; Kourist R. Photobiocatalytic decarboxylation for olefin synthesis. Chem. Commun. 2015, 51, 1918–1921. 10.1039/C4CC07276F. [DOI] [PubMed] [Google Scholar]
- Dennig A.; Kuhn M.; Tassoti S.; Thiessenhusen A.; Gilch S.; Bülter T.; Haas T.; Hall M.; Faber K. Oxidative Decarboxylation of Short-Chain Fatty Acids to 1-Alkenes. Angew. Chem., Int. Ed. 2015, 54, 8819–8822. 10.1002/anie.201502925. [DOI] [PubMed] [Google Scholar]
- Hamid S.; Ivanova I.; Jeon T. H.; Dillert R.; Choi W.; Bahnemann D. W. Photocatalytic conversion of acetate into molecular hydrogen and hydrocarbons over Pt/TiO2: pH dependent formation of Kolbe and Hofer-Moest products. J. Catal. 2017, 349, 128–135. 10.1016/j.jcat.2017.02.033. [DOI] [Google Scholar]
- Esteves L. M.; Brijaldo M. H.; Passos F. B. Decomposition of acetic acid for hydrogen production over Pd/Al2O3 and Pd/TiO2: Influence of metal precursor. J. Mol. Catal. A: Chem. 2016, 422, 275–288. 10.1016/j.molcata.2016.02.001. [DOI] [Google Scholar]
- Rebelein J. G.; Lee C. C.; Hu Y.; Ribbe M. W. The in vivo hydrocarbon formation by vanadium nitrogenase follows a secondary metabolic pathway. Nat. Commun. 2016, 7, 13641. 10.1038/ncomms13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller H. R.; Rodrigues A. V.; Zargar K.; Wu Y.-W.; Saini A. K.; Saville R. M.; Pereira J. H.; Adams P. D.; Tringe S. G.; Petzold C. J.; Keasling J. D. Discovery of enzymes for toluene synthesis from anoxic microbial communities. Nat. Chem. Biol. 2018, 14, 451–457. 10.1038/s41589-018-0017-4. [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya P. P.; Zhang F.; del Cardayre S. B.; Keasling J. D. Microbial engineering for the production of advanced biofuels. Nature 2012, 488, 320. 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U. T.; Huisman G. W.; Kazlauskas R. J.; Lutz S.; Moore J. C.; Robins K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- Sorigué D.; Légeret B.; Cuiné S.; Blangy S.; Moulin S.; Billon E.; Richaud P.; Brugière S.; Couté Y.; Nurizzo D.; Müller P.; Brettel K.; Pignol D.; Arnoux P.; Li-Beisson Y.; Peltier G.; Beisson F. An algal photoenzyme converts fatty acids to hydrocarbons. Science 2017, 357, 903–907. 10.1126/science.aan6349. [DOI] [PubMed] [Google Scholar]
- Zilly F. E.; Acevedo J. P.; Augustyniak W.; Deege A.; Häusig U. W.; Reetz M. T. Tuning a P450 Enzyme for Methane Oxidation. Angew. Chem., Int. Ed. 2011, 50, 2720–2724. 10.1002/anie.201006587. [DOI] [PubMed] [Google Scholar]
- Kawakami N.; Shoji O.; Watanabe Y. Use of Perfluorocarboxylic Acids To Trick Cytochrome P450BM3 into Initiating the Hydroxylation of Gaseous Alkanes. Angew. Chem., Int. Ed. 2011, 50, 5315–5318. 10.1002/anie.201007975. [DOI] [PubMed] [Google Scholar]
- Staudt H.; Oesterhelt D.; Grininger M.; Wachtveitl J. Ultrafast Excited-state Deactivation of Flavins Bound to Dodecin. J. Biol. Chem. 2012, 287, 17637–17644. 10.1074/jbc.M111.331652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire R.; Feldman I. The quenching of tyrosine and tryptophan fluorescence by H2O and D2O. Photochem. Photobiol. 1973, 18, 119–124. 10.1111/j.1751-1097.1973.tb06401.x. [DOI] [PubMed] [Google Scholar]
- Bonetti C.; Mathes T.; van Stokkum I. H. M.; Mullen K. M.; Groot M.-L.; van Grondelle R.; Hegemann P.; Kennis J. T. M. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys. J. 2008, 95, 4790–4802. 10.1529/biophysj.108.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers M. M. E.; Zhang W.; Tonin F.; Hollmann F. Light-Driven Enzymatic Decarboxylation of Fatty Acids. Angew. Chem., Int. Ed. 2018, 57, 13648–13651. 10.1002/anie.201807119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.