Abstract
Glioblastoma (GBM) is an aggressive tumor of the central nervous system that has poor prognosis despite extensive therapy. Therefore, it is essential to identify a gene expression-based signature for predicting GBM prognosis. The RNA sequencing data of GBM patients from the Chinese Glioma Genome Atlas (CGGA) and The Cancer Genome Atlas (TCGA) databases were employed in our study. The univariate and multivariate regression models were utilized to assess the relative contribution of each gene to survival prediction in both cohorts, and the common genes in two cohorts were identified as a final prognostic model. A prognostic risk score was calculated based on the prognostic gene signature. This prognostic signature stratified the patients into the low- and high-risk groups. Multivariate regression and stratification analyses were implemented to determine whether the gene signature was an independent prognostic factor. We identified a 6-gene signature through univariate and multivariate regression models. This prognostic signature stratified the patients into the low- and high-risk groups, implying improved and poor outcomes respectively. Multivariate regression and stratification analyses demonstrated that the predictive value of the 6-gene signature was independent of other clinical factors. This study highlights the significant implications of having a gene signature as a prognostic predictor in GBM, and its potential application in personalized therapy.
Introduction
Glioblastoma (GBM) is the most common and aggressive tumor of the central nervous system in adults. Currently, standard therapy for newly diagnosed GBM is surgical resection to the viable extent, followed by radiotherapy and adjuvant chemotherapy1,2. Although the survival of GBM patients has improved due to advances in modern combination therapies, GBM still has the worst 5-year overall survival rates among all human cancers3, with a dismal median duration of 14 months4,5. Accumulating evidence in recent years shows that tumors consist of multiple cancer cell populations, each harboring specific genetic abnormalities6. Large-scale transcriptome studies on GBM have indicated the possible mechanisms underlying its aggressive behavior7,8, and substantial effort has focused on identifying those genetic alterations that might predict patient prognosis and therapeutic response9.
GBM is a highly heterogeneous disease consisting of multiple molecular alterations7. The differential molecular characteristics of histologically similar tumors make it challenging to predict the clinical outcomes and select the optimum treatment strategies. Currently, prognosis and risk stratification of GBM patients are largely based on the clinico-pathological features including age, tumor size, performance status and grade10. Given the heterogeneity of GBM and the multitude of factors influencing disease progression, conventional clinical characteristics are insufficient to accurately predict individual outcomes and survival. Therefore, it is necessary to identify different GBM specific genomic signatures to improve prognostic and therapeutic success.
The advances in sequencing and bioinformatics technologies have permitted genome-wide sequence analyses in many cancers including GBM. Sequencing data of a multitude of GBM samples have been archived in the open access databases, such as the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) and The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/). The aim of this study was to identify a prognostic signature of GBM using the RNAseq data of GBM patients from the CGGA and TCGA databases. A reliable genomic prognostic signature can complement the conventional clinical prognostic factors, and further enable personalized therapy.
Results
Identification of prognostic gene signature
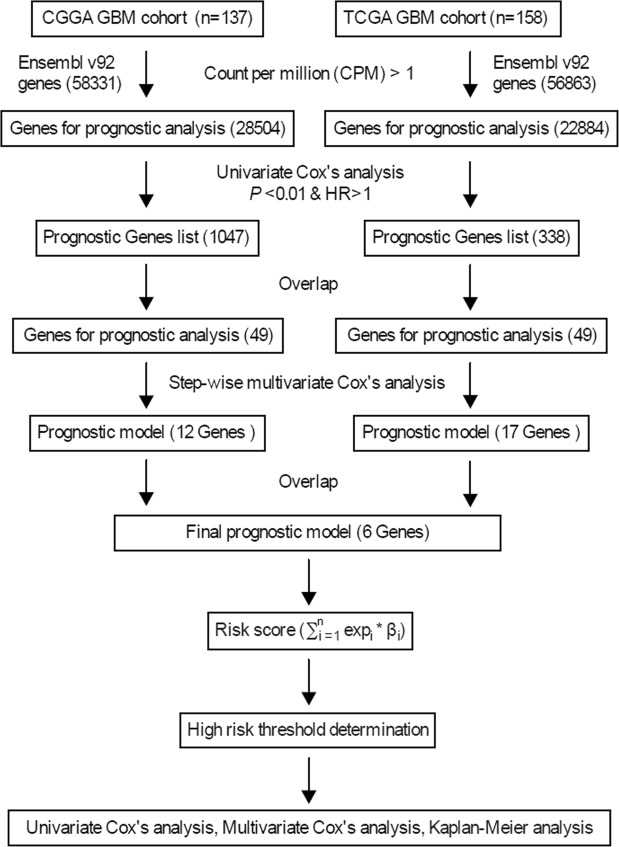
There were 58,331 and 56,863 genes in the CGGA and TCGA cohorts respectively, and after filtering, 28,504 and 22,884 genes were used for Cox’s regression analyses to screen for candidate genes associated with overall survival. Using the cut-off values of P < 0.01 and HR > 1, we identified 1047 and 338 candidate genes in the CGGA and TCGA cohorts respectively, of which 49 were common in both. Step-wise multivariate Cox’s regression analysis identified 12 and 17 genes respectively in the CGGA and TCGA cohorts that were independent survival predictors, of which CD79B, MAP2K3, IMPDH1, SLC16A3, MPZL3 and APOBR were common to both and thus subsequently were analyzed (Fig. 1). The 6 genes are summarized in Table 1.
Figure 1.
Study outline.
Table 1.
General information of 6 genes for constructing the prognostic signature.
| Gene stable ID | Gene name | Gene type | Chromosome | Gene start (bp) | Gene end (bp) |
|---|---|---|---|---|---|
| ENSG00000007312 | CD79B | protein_coding | 17 | 63928740 | 63932354 |
| ENSG00000034152 | MAP2K3 | protein_coding | 17 | 21284672 | 21315240 |
| ENSG00000106348 | IMPDH1 | protein_coding | 7 | 128392277 | 128410252 |
| ENSG00000141526 | SLC16A3 | protein_coding | 17 | 82228397 | 82261129 |
| ENSG00000160588 | MPZL3 | protein_coding | 11 | 118226690 | 118252350 |
| ENSG00000184730 | APOBR | protein_coding | 16 | 28494649 | 28498970 |
Validation of the 6-gene signature in the two independent GBM cohorts
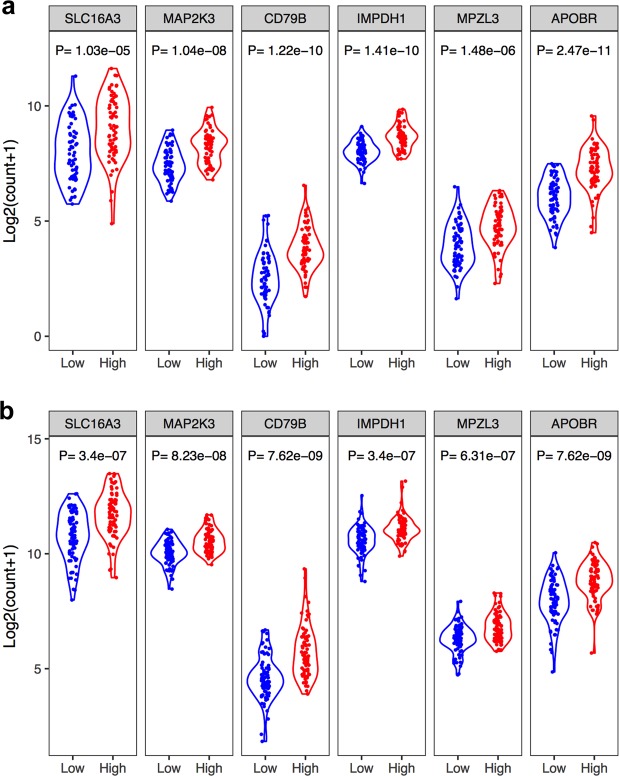
Based on the expression levels of the 6 survival-relevant genes and their relative contributions as per the multivariate regression analysis, we developed a prognostic model by the risk score method for survival prediction. This 6-gene signature-based prognostic model endowed a risk score for each GBM patient, and was validated in the CGGA and TCGA cohorts. In both the CGGA and TCGA cohorts, patients were divided into the low- and high-risk groups using the median risk score as cutoff threshold. The distribution of risk scores, the expression values of the six genes and the survival status of patients ranked according to the risk scores are shown in Fig. 2a–c. Kaplan-Meier survival curves with log-rank test showed that patients in the high-risk group had a significantly shorter survival duration compared to the low-risk group (Fig. 2d, log-rank P = 9.35e-06 for CGGA cohort; log-rank P = 4.36e-04 for TCGA cohort). High risk score was therefore an adverse prognostic factor for GBM patients (HR = 2.6, 95% CI = 1.68–4.03 for CGGA cohort; HR = 1.98, 95% CI = 1.34–2.91 for TCGA cohort). The 1-year and 2-year survival as predicted by the risk scores are shown in Fig. 2e,f, with AUC values of 0.699 and 0.779 for CGGA cohort, 0.718 and 0.704 for TCGA cohort, respectively, implying that the 6-gene signature had high specificity and sensitivity in predicting survival, and was competent for predicting the survival of GBM patients. Finally, the expression levels of all the six genes were significantly higher in the high-risk compared to the low-risk groups in both cohorts (Fig. 3).
Figure 2.
Correlation between the 6-gene signature and overall survival of patients. (a) The distribution of risk scores. (b) The expression heatmap of the 6 prognostic genes. (c) Scatterplot of patient survival status. (d) Kaplan-Meier curves of overall survival of the low- and high-risk groups. (e) ROC curve for 1-year survival prediction by the 6-gene signature. (f) ROC curve for 2-year survival prediction by the 6-gene signature. The black dotted line in a and c represents the median risk score cutoff dividing patients into the low- and high-risk groups.
Figure 3.
The expression levels of the individual genes of the 6-gene signature. Violin plot showing the expression levels of the genes between low- and high-risk groups in the CGGA cohort (a) and TCGA cohort (b).
The 6-gene signature is an independent prognostic factor of survival
Cox’s regression models were used to determine whether the prognostic value of the 6-gene signature was independent of other clinical factors in each cohort. The results are shown in Table 2. Univariate regression analysis of the CGGA cohort indicated that the 6-gene signature-based risk score (HR = 2.60, 95% CI = 1.68–4.03; P = 1.82e-05), radiation therapy (HR = 0.41, 95% CI = 0.26–0.66; P = 1.85e-04), MGMT expression (HR = 1.37, 95% CI = 1.12–1.68; P = 2.19e-03) and chemotherapy (HR = 0.34, 95% CI = 0.21–0.53; P = 2.30e-06) were significantly associated with patient prognosis, while age, gender and other gene mutations showed no significant association with overall survival (P > 0.05). Multivariate regression analysis showed a significant correlation of the 6-gene signature with survival after adjusting for other clinical factors. The 6-gene risk score (HR = 2.40, 95% CI = 1.42–4.06; P = 1.11e-03) was an independent adverse prognostic factor, while radiation therapy (HR = 0.42, 95% CI = 0.25–0.69; P = 7.13e-04) and chemotherapy (HR = 0.35, 95% CI = 0.20–0.59; P = 8.50e-05) were independent favorable factors.
Table 2.
Univariate and multivariate Cox’s regression analyses of the gene signature in two independent cohorts.
| Variables | Patients (N) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| CGGA | ||||||
| Age | <= 49/ > 49 | 73/64 | 1.00 (0.99–1.02) | 5.76e-01 | 0.99 (0.96–1.02) | 4.06e-01 |
| Gender | Female/Male | 47/90 | 1.22 (0.79–1.88) | 3.75e-01 | 1.59 (0.88–2.86) | 1.23e-01 |
| Radiation therapy | No/Yes | 44/79 | 0.41 (0.26–0.66) | 1.85e-04 | 0.42 (0.25–0.69) | 7.13e-04 |
| Chemotherapy | No/Yes | 41/82 | 0.34 (0.21–0.53) | 2.30e-06 | 0.35 (0.20–0.59) | 8.50e-05 |
| IDH1 mutation | No/Yes | 104/33 | 0.64 (0.38–1.07) | 8.57e-02 | 1.24 (0.54–2.83) | 6.16e-01 |
| TP53 mutation | No/Yes | 62/75 | 0.70 (0.46–1.05) | 8.64e-02 | 0.87 (0.54–1.41) | 5.72e-01 |
| EGFR mutation | No/Yes | 101/36 | 0.74 (0.46–1.21) | 2.34e-01 | 0.57 (0.31–1.03) | 6.64e-02 |
| ATRX mutation | No/Yes | 125/12 | 0.79 (0.38–1.63) | 5.17e-01 | 0.76 (0.26–2.28) | 6.28e-01 |
| EZH2 mutation | No/Yes | 115/22 | 1.13 (0.65–1.98) | 6.68e-01 | 1.57 (0.81–3.06) | 1.84e-01 |
| MGMT expression | Low/High | 69/68 | 1.37 (1.12–1.68) | 2.19E-03 | 1.23 (0.94–1.62) | 1.30E-01 |
| Risk score | Low/High | 69/68 | 2.60 (1.68–4.03) | 1.82e-05 | 2.40 (1.42–4.06) | 1.11e-03 |
| TCGA | ||||||
| Age | <= 60/ > 60 | 80/78 | 1.03 (1.01–1.04) | 1.26e-03 | 1.01 (0.99–1.03) | 2.33e-01 |
| Gender | Female/Male | 56/102 | 0.95 (0.65–1.41) | 8.08e-01 | 1.22 (0.77–1.95) | 4.03e-01 |
| Radiation therapy | No/Yes | 21/130 | 0.16 (0.09–0.26) | 1.34e-12 | 0.12 (0.06–0.23) | 7.76e-11 |
| MGMT expression | Low/High | 79/79 | 1.20 (1.01–1.43) | 3.81E-02 | 1.26 (1.05–1.51) | 1.10E-02 |
| Risk score | Low/High | 81/77 | 1.98 (1.34–2.91) | 5.56e-04 | 1.70 (1.10–2.63) | 1.73e-02 |
In the TCGA cohort, univariate analysis showed that age (HR = 1.03, 95% CI = 1.01–1.04; P = 1.26e-03), 6-gene risk score (HR = 1.98, 95% CI = 1.34–2.91; P = 5.56e-04), MGMT expression (HR = 1.20, 95% CI = 1.01–1.43; P = 3.81e-02), and radiation therapy (HR = 0.16, 95% CI = 0.09–0.26; P = 1.34e-12) were significantly associated with overall survival, while gender had no significant correlation (P > 0.05). Multivariate Cox’s regression analysis showed that age was not an independent factor. Furthermore, the 6-gene signature had a significant prognostic value after adjusting for the other clinical factors. Taken together, the 6-gene risk score (HR = 1.70, 95% CI = 1.10–2.63; P = 1.73e-02), and MGMT expression (HR = 1.26, 95% CI = 1.05–1.51; P = 1.10e-02) were independent adverse prognostic factors, while radiation therapy (HR = 0.12, 95% CI = 0.06–0.23; P = 7.76e-11) was an independent favorable factor.
Stratification analysis: prognostic value of 6-gene signature stratified by clinical factors
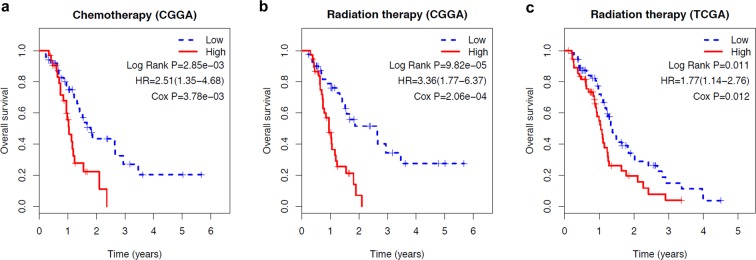
In this study, radiation therapy, chemotherapy, and MGMT expression were identified as survival-associated factors. Therefore, patients were further stratified on the basis of whether they received radio-/chemotherapy and the MGMT expression level, in order to assess the prognostic value of the 6-gene signature. The patients in each cohort were first stratified into subgroups (such as chemotherapy (no/yes), radiation therapy (no/yes), and MGMT expression (low/high)), and each subgroup was further divided into the low- and high-risk groups using the 6-gene signature. In the radio-/chemotherapy subgroups of the CGGA cohort, patients in the high-risk group had a significantly shorter survival duration compared to those in the low-risk group (Fig. 4a,b, P < 0.05), indicating that the 6-gene risk score was an adverse prognostic factor and could predict the survival of patients receiving chemotherapy (HR = 2.51, 95% CI = 1.35–4.68) or radiotherapy (HR = 3.36, 95% CI = 1.77–6.37). In the radiation therapy and MGMT expression subgroups of the TCGA cohort, patients in the high-risk group had significantly poorer prognosis than those in the low-risk group (Fig. 4c, P < 0.05 for radiation therapy and Fig. 5c, P < 0.05 for MGMT expression), further validating that the 6-gene risk score can predict survival in patients receiving radiation therapy (HR = 1.77, 95% CI = 1.14–2.76) or with high/low MGMT expression.
Figure 4.
Kaplan-Meier analysis of overall survival of patients stratified by treatment. (a) Chemotherapy stratification analysis in CGGA cohort. (b) Radiation therapy stratification analysis in CGGA cohort. c: Radiation therapy stratification analysis in TCGA cohort.
Figure 5.
Kaplan-Meier analysis of overall survival of patients stratified by gene mutations or different GBM types. (a) Stratification analysis in CGGA cohort based on gene mutations (IDH1, TP53, EGFR, ATRX, EZH2, and MGMT). (b) Stratification analysis for primary, secondary, and recurrent GBMs in CGGA cohort. (c) Stratification analysis in TCGA cohort based on MGMT mutations.
Then, we analyzed the relationship of risk score with gene mutations including IDH1, TP53, EGFR, ATRX, EZH2, and MGMT. To begin with, the patients in the CGGA cohort were firstly divided into two subgroups based on whether these gene were mutated or not, and next each subgroup was further separated into high- and low-risk group relying on the 6-gene signature. In the subgroups (except ATRX subgroup), patients in the high-risk group had a significantly worse prognosis, relative to the low-risk group (Fig. 5a, P < 0.05), implicating that the 6-gene risk score can predict the survival status in GBM patients.
Taken together, our findings suggested that the prognostic value of the 6-gene signature was independent of other clinical features for predicting survival in GBM patients.
Comparison of selected genes expression between primary and secondary GBM
Only primary GBM data are included in the TCGA database, and there are primary GBM data, recurrent GBM data, and secondary data in the CGGA database. In our analysis, we used all GBM cases to establish a prognosis signature. However, primary and secondary GBMs are different diseases from the molecular point of view. Thus, we performed the grouping analysis for primary, secondary, and recurrent GBM data. The analysis results (Fig. 5b) show that this 6-gene signature can be used as a prognostic indicator for primary and secondary GBM. However, for recurrent GBM patients, there is no difference in survival in different risk score groups, suggesting that this 6-gene prognosis signature might not be applicable for recurrent GBM.
Discussion
Studies show that gene alterations play significant roles in tumorigenesis and patient prognosis, indicating a potential application of characteristic gene signatures in cancer diagnosis and prognosis. Several reports are available on the correlation between altered genes and GBM prognosis. For example, Nicolasjilwan et al.11 analyzed the genetic biomarkers to predict the survival of GBM patients in the TCGA database based on clinical factors and imaging characteristics, but the sensitivity and specificity of the gene signature in survival prediction were not measured. Another study also using TCGA GBM data to detect an inverse correlation between IL-13Rα1 and α2 expression and unsatisfied prognosis12. Moreover, Jia et al.13 have identified prognosis-related genes for GBM based on TCGA and CGGA databases. However, these studies have focused on single GBM-related genes, which have limited prognostic and predicative power. Therefore, we tested the predictive power of a panel of GBM-associated genes using regression analyses. We developed a risk score model based on 6 genes to predict clinical outcomes of GBM patients by analyzing the RNAseq data from two open access databases. The 6-gene signature competitively predicted patient survival, and according to the univariate and multivariate analyses, was an independent prognostic factor in addition to chemotherapy therapy, radiation therapy, and MGMT expression.
Due to extensive heterogeneity of GBM, even patients with similar histo-pathological characteristics differ in their genetic landscape, making it challenging to effectively target different patients using the same protocol. Therefore, individualized risk stratification and treatment are urgently needed. Omics-based patient-specific therapy is based on the genomics, transcriptomics and proteomics data of individual patients. An understanding of patient-specific mutations would help design the most effective therapeutic strategy for that particular patient. Omics-based techniques can be used at both pre- and post-treatment stages to track specific mutations as well as development of resistance, and help modify the treatment accordingly.
Univariate analysis showed that age was not significantly associated with overall survival in the CGGA cohort, while a significant association was observed in the TCGA cohort. Age at diagnosis is a major predictor of patient survival, and younger patients tend to survive longer than the older patients8,14,15. However, age alone is not a predictor for survival in GBM because older patients are less likely to receive adjuvant treatment16. Multivariate analysis confirmed that age was not an independent predictor for survival in our cohorts as well. Currently, surgical resection, radiotherapy and adjuvant chemotherapy are the standard treatment options in GBM patients. For elderly patients, radiotherapy can improve survival without reducing life quality or cognition17. However, the survival is significantly reduced if radiotherapy is not initiated within 6 weeks after complete resection18. Furthermore, radiotherapy plus temozolomide provides a significant survival benefit compared to radiotherapy alone in treating GBM19,20. We also found that radiation therapy and chemotherapy were independent favorable prognostic factors. The 6-gene signature stratified the treated and untreated patients into the low- and high-risk groups that had significantly different prognosis, indicating that the 6-gene signature can improve survival prediction and can also identify high-risk patients for adjuvant therapy in addition to the standard regimen.
Several gene mutations have been identified which correlate with the pathogenesis of GBM21. EGFR mutations confer enhanced tumorigenicity by increasing proliferation and reducing apoptosis of GBM cells, especially in secondary GBM22. Mutations in IDH1 predict a more favorable prognosis23–25, and are often associated with TP53 and ATRX mutations26,27. The CGGA database includes gene mutations and gene expression profiles from a number of GBM patients. However, univariate and multivariate analyses showed that these mutations had no significant prognostic value, possibly because most of them are more common in patients with secondary GBM while most subjects in our study had primary GBM.
The results about the relationship of risk score with IDH1 mutations showed patients in the high-risk group had a significantly worse prognosis, relative to the low-risk group, which implicated that the 6-gene risk score is an independent prognostic factor independent of IDH1 mutation.
As we all know, MGMT promoter methylation ultimately determines the expression of MGMT, thus, we analyzed the relationship of prognosis and MGMT expression level in the CGGA and TCGA databases. Multivariate analysis exhibited that in the CGGA database, the expression of MGMT was not an independent prognostic factor, while MGMT was an independent prognostic factor in the TCGA database. The subgroup analysis based on MGMT expression level to reveal the relation between our risk score and MGMT expression showed that patients in the high-risk group had a significantly worse prognosis, relative to the low-risk group, which demonstrated that the 6-gene risk score is an independent prognostic factor independent of MGMT expression.
Our 6 prognostic panel genes have established roles in oncogenesis. For example, CD79B has been associated with reduced survival, and is considered a suitable immunotherapeutic target for leukemia and lymphoma28,29. Several studies have highlighted the critical role for MAP2K3 in tumor progression and targeted therapies30,31. MAP2K3 inhibition sensitizes tumor cells to chemotherapy31, and has been identified as a putative target for molecular therapy against GBM32. SLC16A3 is typically over-expressed in GBM and patients with higher SLC16A3 levels have significantly poorer survival33,34, making it a potential prognostic biomarker and metabolic target in GBM35. Based on the genomic locations for three genes (CD79B, MAP2K3, SLC16A3), we find that CD79B locates at chromosome 17q23.3, MAP2K3 locates at chromosome 17q11.2, and SLC16A3 locates at chromosome 17q25.3. Although these all locate on chromosome 17, these 3 genes are not close to one another. Chromosome 17, as one of 23 pairs of human chromosomes, whose abnormities and roles that have been investigated about the expression of genes in this chromosome influence the nervous system, especially cell differentiation and maturation. Deletions in the region of chromosome 17 are the most common abnormal event in primary solid tumors36. Petitjean et al.37 have suggested that the deletion of the region of chromosome 17 is greatly important in carcinogenesis, as found in other examples including breast cancer, colon cancer, liver cancer, medulloblastoma and in head tumors36,38. Further, Sunpaweravong et al.39 have implicated that gains of cytoband 17q25.3 are found in 61% lung cancer patients, underscoring a potential biological role for the genes within this region in the progression of lung cancer, which further substantiates the fact that there are many oncogenes located at chromosome 17. Significantly, loss of heterozygosity for the short arm of chromosome 17 has been demonstrated in a 4-year-old boy with GBM40. According to the importance of chromosome 17 described above, losses or changes in genes in chromosome 17 might potentially involve in GBM. In our study, it is more convinced that CD79B, MAP2K3, and SLC16A3 on chromosome 17 play important roles in GBM.
IMPDH1, one isotype of IMPDH, is ubiquitously expressed in mammals41. IMPDH1 up-regulation has been found in many types of cancer, such as bladder cancer, brain cancer, lung cancer, ovarian cancer, and GBM42. Increased MPZL3 is found to exert important function in ovarian cancer via regulating metablolism43. Moreover, RNA expression of MPZL3 has been reported to be highly expressed in radio-resistant rectal cancer cell lines44. APOBR mutation is observed in hepatocellular carcinoma45. In our study, this 6-gene signature would independently predict overall survival in GBM patients. The advantage of this work was to combine the clinical characteristics and CGGC/TCGA data to assess the GBM patients survival through establishing a genes-associated risk score. Therefore, the combination of these 6 genes (CD79B, MAP2K3, IMPDH1, SLC16A3, MPZL3 and APOBR) could be regarded as a novel risk factor that might function as a prognosis indicator for GBM patients.
To summarize, we combined 6 GBM specific genes into a single diagnostic panel using regression analyses, and established its predictive value in overall survival of the patients. We selected only those survival-related genes that were common to both the CGGA and TCGA cohorts, which is more reliable and stable relative to single cohort analysis.
In conclusion, we identified a 6-gene signature for predicting survival in GBM by analyzing RNAseq-based gene expression profiles in the CGGA and TCGA patients. Multivariate and stratified analysis demonstrated that the gene panel was independent of other clinical and pathological features, and therefore is a potential prognostic biomarker of GBM.
Methods
Patients
The transcriptomic data of GBM patients were obtained from two independent data portals (CGGA and TCGA). As part of the CGGA project, Zhao et al.46 provided the RNA-seq transcriptomic profiles of 325 gliomas samples. The raw RNA-seq data were downloaded from the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) using the accession numbers SRP027383 and SRP091303. The raw fastq files were converted from SRA using Fastq-dump app (version: 2.8.1), and the sam and read count data were obtained by the hisat2 (version 2.1.0) and htseq software (version 0.10.0), respectively. A total of 137 GBM samples with clinical data were selected from the CGGA cohort. Data of an independent cohort of 158 patients was obtained from the TCGA database. The transcriptomic profiles (level 3 data) and corresponding clinical data of these patients were downloaded from the Data Coordinating Center. All RNA-seq libraries were sequenced using the Illumina HiSeq2000 Systems. The raw count data were normalized by the trimmed mean of M-values (TMM) method47, and genes with extremely low total abundance (count per million <1) were filtered out. The data preprocessing was performed using edgeR package48 (version 3.20.9) in Bioconductor.
Screening for the prognostic gene signature
The schematics of our study are illustrated in Fig. 1. Univariate Cox’s proportional hazards regression analysis was used to evaluate the correlation between the expression level of each gene and overall survival in both cohorts. Only genes with P values < 0.01 and hazard ratio (HR) > 1 were considered as candidate genes for the correlation analysis, and those common genes to both CGGA and TCGA cohorts were used to construct the predictive model. The common candidate genes were then fitted in step-wise multivariate Cox’s regression model to assess the relative contribution of each gene to survival prediction in both cohorts. The genes that correlated with survival and were common to both cohorts were included in the prognostic signature. According to the estimated regression coefficients in multivariate Cox’s regression model, a prognostic risk score for predicting overall survival was then calculated as follows,
where n is the number of prognostic genes, expi is the expression level of prognostic gene i, and βi is the regression coefficient of gene i. Using the median risk score in CGGA cohort as the cutoff value, GBM patients in each cohort were divided into low- and high-risk groups.
Statistical analysis
Kaplan-Meier analysis was used to determine the survival in the low- and high-risk groups in each cohort, and log-rank test was used to assess the statistical significance. Multivariate Cox’s regression model and stratification analysis were implemented to determine whether the gene signature was independent of other clinical features. The time-dependent receiver operating characteristic (ROC) curve was used to compare the sensitivity and specificity of the gene signature in survival prediction. The area under the curve (AUC) was calculated from the ROC curve. For both log-rank test and Cox regression analysis, the significance was set at P value < 0.05.
Acknowledgements
This work was funded by Natural Science Foundation of Henan Province (No. 162300410040), National Natural Science Foundation of China (No. 81301963) and Outstanding Youth Science Foundation of Henan university (No. yqpy20140036).
Author Contributions
Study concept and design: Shuguang Zuo; Interpretation of data and statistical analysis: Xinhong Zhang and Liping Wang; Manuscript preparation and editing: Xinhong Zhang and Liping Wang; Figure and Table preparation: Xinhong Zhang and Liping Wang; Critical revision of the manuscript for important intellectual content: Shuguang Zuo. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuguang Zuo, Xinhong Zhang and Liping Wang contributed equally.
References
- 1.Lee JH, et al. Performance status during and after radiotherapy plus concomitant and adjuvant temozolomide in elderly patients with glioblastoma multiforme. J Clin Neurosci. 2013;20:503–508. doi: 10.1016/j.jocn.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 2.Komotar RJ, Otten ML, Moise G, Connolly ES., Jr. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma-a critical review. Clin Med Oncol. 2008;2:421–422. doi: 10.4137/cmo.s390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tykocki, T. & Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J Clin Neurosci, 10.1016/j.jocn.2018.05.002 (2018). [DOI] [PubMed]
- 4.Buckner, J. C. Factors influencing survival in high-grade gliomas. Semin Oncol30, 10–14, S0093775403005992 (2003). [DOI] [PubMed]
- 5.Van Meir EG, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.2006960/3/166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 10.Delgado-Lopez PD, Corrales-Garcia EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 11.Nicolasjilwan M, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42:212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing H, Puri RK. Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor α1 and α2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme. Journal of Neuro-Oncology. 2018;136:463–474. doi: 10.1007/s11060-017-2680-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia, D. et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging10 (2018). [DOI] [PMC free article] [PubMed]
- 14.Morgan ER, Norman A, Laing K, Seal MD. Treatment and outcomes for glioblastoma in elderly compared with non-elderly patients: a population-based study. Curr Oncol. 2017;24:e92–e98. doi: 10.3747/co.24.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris G, et al. Survival Outcomes of Elderly Patients With Glioblastoma Multiforme in Their 75th Year or Older Treated With Adjuvant Therapy. Int J Radiat Oncol Biol Phys. 2017;98:802–810. doi: 10.1016/j.ijrobp.2017.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Gately L, Collins A, Murphy M, Dowling A. Age alone is not a predictor for survival in glioblastoma. J Neurooncol. 2016;129:479–485. doi: 10.1007/s11060-016-2194-x. [DOI] [PubMed] [Google Scholar]
- 17.Keime-Guibert F, et al. Radiotherapy for glioblastoma in the elderly. N Engl J Med. 2007;356:1527–1535. doi: 10.1056/NEJMoa065901. [DOI] [PubMed] [Google Scholar]
- 18.Valduvieco I, et al. Impact of radiotherapy delay on survival in glioblastoma. Clin Transl Oncol. 2013;15:278–282. doi: 10.1007/s12094-012-0916-x. [DOI] [PubMed] [Google Scholar]
- 19.Yang LJ, Zhou CF, Lin ZX. Temozolomide and radiotherapy for newly diagnosed glioblastoma multiforme: a systematic review. Cancer Invest. 2014;32:31–36. doi: 10.3109/07357907.2013.861474. [DOI] [PubMed] [Google Scholar]
- 20.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 21.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro Oncol. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai A, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bleeker FE, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 25.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 26.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.11643821164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Y, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahn L, et al. Therapeutic targeting of the BCR-associated protein CD79b in a TCR-based approach is hampered by aberrant expression of CD79b. Blood. 2015;125:949–958. doi: 10.1182/blood-2014-07-587840. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, et al. CD79B and MYD88 mutations in diffuse large B-cell lymphoma. Hum Pathol. 2014;45:556–564. doi: 10.1016/j.humpath.2013.10.023S0046-8177(13)00454-1. [DOI] [PubMed] [Google Scholar]
- 30.Bossi G. MKK3 as oncotarget. Aging (Albany NY) 2016;8:1–2. doi: 10.18632/aging.100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldari S, Ubertini V, Garufi A, D’Orazi G, Bossi G. Targeting MKK3 as a novel anticancer strategy: molecular mechanisms and therapeutical implications. Cell Death Dis. 2015;6:e1621. doi: 10.1038/cddis.2014.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu T, et al. ss-Elemene inhibits proliferation of human glioblastoma cells and causes cell-cycle G0/G1 arrest via mutually compensatory activation of MKK3 and MKK6. Int J Oncol. 2011;38:419–426. doi: 10.3892/ijo.2010.855. [DOI] [PubMed] [Google Scholar]
- 33.Beckner ME, Pollack IF, Nordberg ML, Hamilton RL. Glioblastomas with copy number gains in EGFR and RNF139 show increased expressions of carbonic anhydrase genes transformed by ENO1. BBA Clin. 2016;5:1–15. doi: 10.1016/j.bbacli.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaumeil MM, et al. Hyperpolarized (13)C MR imaging detects no lactate production in mutant IDH1 gliomas: Implications for diagnosis and response monitoring. Neuroimage Clin. 2016;12:180–189. doi: 10.1016/j.nicl.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim KS, et al. Inhibition of monocarboxylate transporter-4 depletes stem-like glioblastoma cells and inhibits HIF transcriptional response in a lactate-independent manner. Oncogene. 2014;33:4433–4441. doi: 10.1038/onc.2013.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olivier M, et al. Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Therapy. 2009;16:1–12. doi: 10.1038/cgt.2008.69. [DOI] [PubMed] [Google Scholar]
- 37.Petitjean A, Achatz MIW, Borresendale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 38.López-Ochoa S, Ramírez-García M, Castro-Sierra E, Arenas-Huertero F. Analysis of Chromosome 17 miRNAs and Their Importance in Medulloblastomas. Biomed Research International. 2015;2015:1–12. doi: 10.1155/2015/717509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunpaweravong P, Thu KL, Wan LL, Mai S. Assessment of the clinical relevance of 17q25.3 copy number and three-dimensional telomere organization in non-small lung cancer patients. Journal of Cancer Research & Clinical Oncology. 2016;142:749–756. doi: 10.1007/s00432-015-2080-5. [DOI] [PubMed] [Google Scholar]
- 40.Cotter, J. A. et al. Transmission of a TP53 germline mutation from unaffected male carrier associated with pediatric glioblastoma in his child and gestational choriocarcinoma in his female partner. Cold Spring Harb Mol Case Stud (2018). [DOI] [PMC free article] [PubMed]
- 41.Carr SF, Papp E, Wu JC, Natsumeda Y. Characterization of human type I and type II IMP dehydrogenases. Journal of Biological Chemistry. 1993;268:27286–27290. [PubMed] [Google Scholar]
- 42.However. A Review of the Potential Utility of Mycophenolate Mofetil as a Cancer Therapeutic. Journal of Cancer Research,2014,(2014-7-8)2014, 1–12 (2014).
- 43.Worley B, et al. The Role of MPZL3 as a Metabolic Regulator in Ovarian Cancer. Free Radical Biology and Medicine. 2017;112:178–179. doi: 10.1016/j.freeradbiomed.2017.10.280. [DOI] [Google Scholar]
- 44.Kim SC, Shin YK, Kim YA, Jang SG, Ku JL. Identification of genes inducing resistance to ionizing radiation in human rectal cancer cell lines: re-sensitization of radio-resistant rectal cancer cells through down regulating NDRG1. BMC Cancer. 2018;18:594. doi: 10.1186/s12885-018-4514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ally A, et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Z, et al. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci Data. 2017;4:170024. doi: 10.1038/sdata.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]