Abstract
The most common type of head and neck cancer, head and neck squamous cell carcinoma (HNSCC), can develop therapeutic resistance that complicates its treatment. The 5-y survival rate for HNSCC remains at ~50%, and improving these outcomes requires a better understanding of the pathogenesis of HNSCC. Studies of HNSCC using in vitro, ex vivo, and in vivo approaches provide a novel conceptual framework based on epigenetic mechanisms for developing future clinical applications. Normal oral tissues are influenced by environmental factors that induce pathological changes affecting the network of epigenetic enzymes and signaling pathways to induce HNSCC growth and metastasis. Although various epigenetic regulator families, such as DNA methyltransferases, ten-eleven translocation proteins, histone acetyltransferases, histone deacetylases, BET bromodomain proteins, protein arginine methyltransferases, histone lysine methyltransferases, and histone lysine demethylases, have a role in diverse cancers, specific members have a function in HNSCC. Recently, lysine-specific demethylases have been identified as a potential, attractive, and novel target of HNSCC. Lysine-specific demethylase 1 (LSD1) expression is inappropriately upregulated in HNSCC and an orthotopic HNSCC mouse model. LSD1 can demethylate lysine at specific histone positions to repress gene expression or stimulate transcription, indicating a dual and context-dependent role in transcriptional regulation. Our study showed that LSD1 promotes HNSCC growth and metastasis. Pharmacological attenuation of LSD1 inhibits orthotopic and patient-derived HNSCC xenograft growth-specific target genes and signaling pathways. This review provides recent evidence demonstrating the function of epigenetic regulator enzymes in HNSCC progression, including potential therapeutic applications for such enzymes in combination and immunotherapy.
Keywords: epigenetics, mouth neoplasms, head and neck neoplasms, histone demethylases, LSD1, translational medical research
Introduction
Head and neck squamous cell carcinoma (HNSCC) is an aggressive cancer that predominantly arises from a preneoplastic dysplasia that undergoes genetic and epigenetic modifications to progress to malignancy. Epigenetic modifications can also result in acquired therapeutic resistance, leading to poor outcomes among HNSCC patients—the 5-y survival rate for HNSCC is just ~50%. Experimental data indicate that oncogenic factors, such as human papillomavirus (HPV), alcohol, tobacco, environmental carcinogens, and lifestyle changes, could directly induce epigenetic changes and modifications in epigenetic enzymes, which themselves could promote HNSCC growth and metastasis (Boscolo-Rizzo et al. 2017) (Figs. 1–3). However, the mechanisms underlying these modifications are not well understood, hindering further progress in therapeutic development.
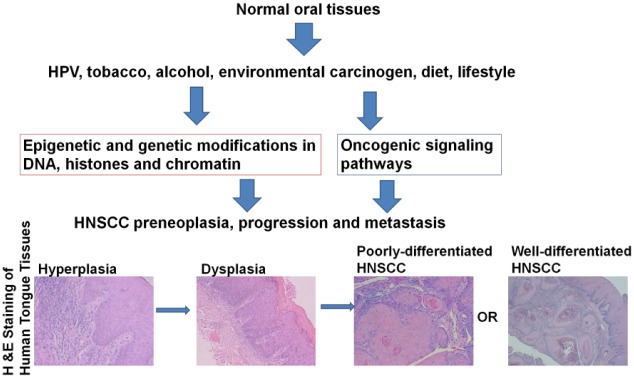
Figure 1.
Impact of epigenetic regulation on head and neck squamous cell carcinoma (HNSCC) growth and metastasis. Normal oral tissues are influenced by oncogenic factors, such as human papillomavirus (HPV), tobacco, alcohol, environmental carcinogens, diet, and lifestyle-related changes. Hematoxylin and eosin–stained sections of human tongue tissue demonstrate that oncogenic factors promote pathological changes in normal oral mucosa, leading to hyperplasia, dysplasia, poorly differentiated HNSCC, and well-differentiated HNSCC.
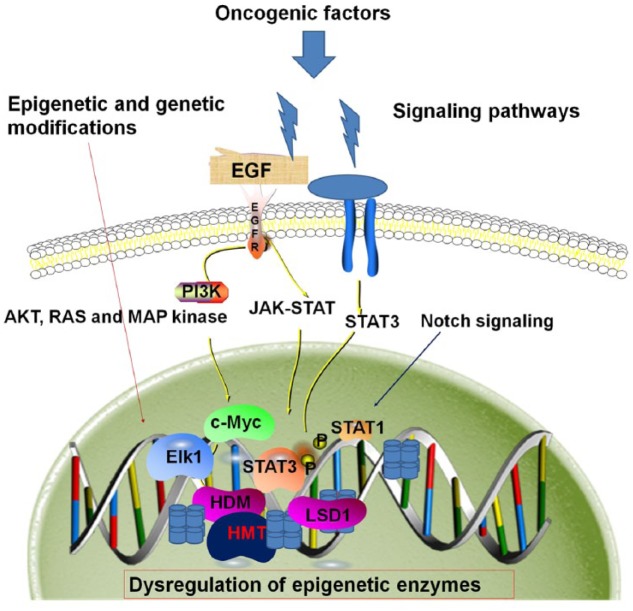
Figure 2.
Epigenetic and genetic modifications and signaling pathways leading to head and neck squamous cell carcinoma (HNSCC). Oncogenic factors, mutations, cancer stem cells, or immune invasion can lead to genetic and epigenetic changes or alterations in signaling pathways, such as EGFR, NOTCH, and JAK/STAT, that lead to dysregulated epigenetic enzymes, shifting the balance toward oncogenic transformation in HNSCC growth and metastasis.
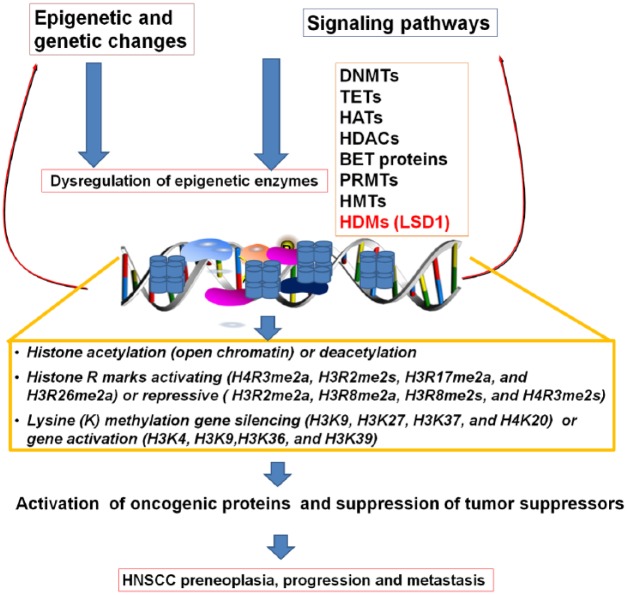
Figure 3.
Dysregulation of epigenetic enzymes and its downstream effectors leading to head and neck squamous cell carcinoma (HNSCC). Epigenetic and genetic changes and activated signaling pathways dysregulate epigenetic enzymes, which affects acetylation, methylation of lysine and arginine, and other epigenetic modifications, promoting oncogenic factors and inhibition of tumor suppressors potentially leading to HNSCC.
Epigenetic modifications occur within nuclear chromatin. Chromatin falls into 2 categories: heterochromatin contains inactive genes, is tightly condensed, and is late to replicate; euchromatin contains most of the active genes and is relatively open. The basic functional unit in which nuclear chromatin is packaged is the nucleosome. The nucleosome contains a 147–base pair stretch of DNA wrapped around a histone octamer, with 2 each of histones H2A, H2B, H3, and H4. Epigenetic modifications—mainly acetylation, methylation, and demethylation—are governed by specific modifier enzymes at the DNA, histone, or chromatin level, and these enzymes are categorized as writers, erasers, or readers. There are 4 types of methylated DNA: 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. Histone modifications include acetylation/deacetylation, methylation/demethylation (lysine, arginine), phosphorylation (serine/threonine, tyrosine), ubiquitylation/deubiquitylation, sumoylation, adenosine diphosphate (ADP) ribosylation, deimination, proline isomerization, crotonylation, propionylation, butyrylation, formylation, hydroxylation, O-GlcNAcylation, and other serine and threonine modifications (Dawson and Kouzarides 2012).
We recently showed that the epigenetic regulator lysine-specific demethylase 1 (LSD1) has increased expression in dysplasias, advanced tumor grades and stages, and mouse models of HNSCC (Bais et al. 2015; Alsaqer et al. 2017). Furthermore, our studies reveal that attenuation of LSD1 inhibits EGFR signaling and downregulates key oncogenic factors in HNSCC (Alsaqer et al. 2017). Here, we summarize advances in understanding epigenetic enzyme functions and roles in HNSCC therapy resistance (Figs. 1–3), as well as review selected drugs in clinical applications for HNSCC (Table).
Table.
The Selected Epidrugs Alone or in Combination for HNSCC Clinical Trials.
| NCT No. | Interventions | Phase | Statusa | Study Completion | Description/Goal |
|---|---|---|---|---|---|
| NCT03019003 | Azacitidine + durvalumab + tremelimumab | I, II | R | 07/01/24 | Nonrandomized, open-label, phase Ib/II study of treatment of patients with recurrent and/or metastatic HNSCC who progressed after treatment with anti–PD-1, anti–PD-L1, or anti–CTLA-4 monotherapy. (https://clinicaltrials.gov/ct2/show/NCT03019003) |
| NCT00004062 | Azacitidine + liothyronine sodium + iodine I-131 | I | C | 08/01/06 | Azacitidine may help thyroid cancer cells regain the ability to take up iodine, which would allow cancer to be detected and treated with radioactive iodine. (https://clinicaltrials.gov/ct2/show/NCT00004062) |
| NCT00901537 | Azacitidine + cisplatin | I | T | 10/01/11 | Azacitidine has been shown to be a cisplatin “helper”—that is, it makes cisplatin more effective in inhibiting growth of head and neck or lung cancer. (Trial terminated due to no enrollments within last 4 mo.) (https://clinicaltrials.gov/ct2/show/NCT00901537) |
| NCT00443261 | Azacitidine + cisplatin | I | T | 06/01/08 | To evaluate the safety and toxicity of azacitidine (5-azacitidine, Vidaza) and cisplatin combination in patients with HNSCC. (Study terminated due to poor accrual; only enrolled patient died.) (https://clinicaltrials.gov/ct2/show/NCT00443261) |
| NCT02269943 | Azacitidine | II | C | 04/20/17 | Multicenter, international, single-arm study to assess the safety and efficacy of single-agent oral azacitidine (cc-486) in previously treated subjects with locally advanced or metastatic nasopharyngeal carcinoma. (https://clinicaltrials.gov/ct2/show/NCT02269943) |
| NCT02178072 | Azacitidine + CUDC-101 + cisplatin + radiation | II | R | 12/01/18 | Assess the activity of azacitidine in patients with HPV-positive and HPV-negative HNSCC. The study will evaluate TP53 and interferon levels in tumors. (https://clinicaltrials.gov/ct2/show/NCT02178072) |
| NCT01384799 | CUDC-101 + cisplatin + radiation | I | C | 10/01/13 | Dose escalation study of CUDC-101 in combination with concurrent cisplatin and radiation therapy in patients with locally advanced head and neck cancer. CUDC-101 is a multitargeted agent designed to inhibit EGFR, HER2, and HDAC. (https://clinicaltrials.gov/ct2/show/NCT01384799) |
| NCT00738751 | Panobinostat + erlotinib | I | C | 02/01/15 | Phase I study of panobinostat in combination with erlotinib for advanced aerodigestive tract cancers. (https://clinicaltrials.gov/ct2/show/NCT00738751) |
| NCT01171924 | CUDC-101 | I | C | 10/01/11 | Phase Ib open-label expansion study of CUDC-101 in patients with advanced head and neck, gastric, breast, liver, and non–small cell lung cancer tumors. (https://clinicaltrials.gov/ct2/show/NCT01171924) |
| NCT01695122 | Valproic acid | II | C | 11/01/14 | Evaluate if adding valproic acid to standard platinum-based chemoradiation for locally advanced HNSCC can improve treatment outcomes, such as response rate. (https://clinicaltrials.gov/ct2/show/NCT01695122) |
| NCT00670553 | Panobinostat | I | C | 01/01/10 | Study of panobinostat in combination with external beam radiotherapy to treat prostate, esophageal, and head and neck cancer. (https://clinicaltrials.gov/ct2/show/NCT00670553) |
| NCT02538510 | Pembrolizumab + vorinostat | I, II | A, NR | 06/30/19 | Study the side effects of pembrolizumab (Biologics) and vorinostat in treating patients with squamous cell head and neck cancer or salivary gland cancer that has returned, spread to other places in the body, and/or cannot be removed by surgery. mAbs, such as pembrolizumab, may interfere with the ability of tumor cells to grow and spread. Vorinostat may stop growth of tumor cells by blocking some of the enzymes needed for cell growth. The combination may be a better treatment for head and neck or salivary gland cancer. (https://clinicaltrials.gov/ct2/show/NCT02538510) |
| NCT01267240 | Capecitabine + vorinostat | II | T | 10/01/16 | Partially randomized trial for capecitabine and vorinostat to treat patients with head and neck cancer that has come back after previous treatment or that has spread to other areas in the body. Drugs used in chemotherapy, such as capecitabine, work in different ways to stop growth of tumor cells, either by killing the cells or by stopping them from dividing. Vorinostat may stop growth of tumor cells by blocking some of the enzymes needed for cell growth. It is not yet known whether giving capecitabine together with vorinostat is more effective than capecitabine alone to treat patients with head and neck cancer. (The trial was terminated because study treatment did not show clinical activity.) (https://clinicaltrials.gov/ct2/show/NCT01267240) |
| NCT02608736 | Valproic acid placebo | I | C | 07/01/17 | This study evaluates the addition of valproic acid (randomized valproic acid compared to placebo) as a chemopreventive drug in HNSCC patients who do not have signs of recurrence or residual disease. The primary outcome is to document histone acetylation and DNMT in saliva. (https://clinicaltrials.gov/ct2/show/NCT02608736) |
| NCT00413322 | Belinostat + 5-fluorouracil (5-FU) | I | C | 06/01/08 | Assess the combination of belinostat (PXD101) and 5-FU in patients with advanced solid tumors. The primary goal is to understand safety, antitumor activity, and drug behavior when administered with 5-FU. (https://clinicaltrials.gov/ct2/show/NCT00413322) |
| NCT00978250 | 5-Fluoro-2-deoxycytidine (FdCyd) | II | A, NR | 07/22/19 | Trials with 2 experimental drugs, FdCyd (also called 5-fluoro-2′-deoxcytidine), and THU (also called tetrahydrouridine), are undergoing trials to test their effectiveness in treating cancer that has not responded to standard therapies. FdCyd is thought to work by changing how genes function in cancer cells. THU does not have any anticancer effects on its own, but it helps keep the other drug, FdCyd, from being broken down by the body. (https://clinicaltrials.gov/ct2/show/NCT00978250) |
A, active; C, completed; DNMT, DNA methyltransferase; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; mAbs, monoclonal antibodies; NR, not recruiting; R, recruiting; T, terminated.
Epigenetic Regulators and Their Role in HNSCC
DNA Methyltransferase
DNA methyltransferases (DNMTs) are a conserved family of cytosine methylases. DNMTs are involved in alternative splicing, gene duplication, gene loss, gene silencing, transcriptional activation, and posttranscriptional regulation. The regulatory mechanisms of DNMT protein also include molecular interactions with other proteins (Lyko 2018). In addition, DNMTs are associated with cancers, including HNSCC, and aberrant DNMT promoter methylation is considered a major mechanism underlying inactivation of tumor-related genes. DNMT3A and DNMT3B levels are increased in various cancer tissues and cell lines, which may partly account for the hypermethylation of CpG-rich regions in tumor suppressor gene promoters in HNSCC (Subramaniam et al. 2014).
Ten-Eleven Translocation Protein
Ten-eleven translocation protein 1 (TET1), TET2, and TET3 are potential DNA demethylases—the enzymes oxidize 5-methylcytosines and promote locus-specific reversal of DNA methylation (Rasmussen and Helin 2016). Mutations and functional inactivation of TET genes contribute to HNSCC (Misawa et al. 2018; Song et al. 2018). DNA methylation levels and specific patterns are regulated by the balance between DNA methyltransferases, such as DNMT1, DNMT3A, and DNMT3B, and demethylating proteins, such as TET1, TET2, and TET3. Therefore, functional inactivation of TET proteins could allow DNMTs to induce neoplastic transformation (Rasmussen and Helin 2016).
Histone Acetyltransferase and Histone Deacetylase
The opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) control histone acetylation. Histone acetylation is associated with a more “open” chromatin conformation—acetylation neutralizes lysine’s positive charge and may consequently weaken the electrostatic interaction between histones and negatively charged DNA. Eighteen human HDACs are grouped into 4 classes on the basis of their homology: class I (HDAC1, 2, 3, and 8), class IIa (HDAC4, 5, 7, and 9) and IIb (HDAC6 and 10), class III (7 sirtuins [SIRTs]), and class IV (HDAC11) (Li and Seto 2016). HDAC1 and HDAC2 are overexpressed in tongue squamous cell carcinomas (Theocharis et al. 2011). The HAT CBP/p300 has an oncogenic role in HNSCC, as pharmacological attenuation inhibits xenograft growth (Albrengues et al. 2015; Selvi et al. 2015). Furthermore, increased HDAC9 messenger RNA (mRNA) and protein expression in clinical HNSCC is associated with significantly reduced overall survival, and HDAC9 knockdown suppresses cell proliferation, increases apoptosis, and induces G0/G1 cell cycle arrest in HNSCC cells (Rastogi et al. 2016). SIRT3 and SIRT5 have demonstrated tumor suppressor as well as tumor promoter properties under different cellular conditions, tumor stages, and tissues of origin. In addition, mitochondrial SIRTs have an emerging role in HNSCC and other cancers (George and Ahmad 2016).
Bromodomain and Extra-Terminal Domain Proteins
Bromodomain and extra-terminal domain (BET) proteins are epigenetic readers characterized by the presence of 2 tandem bromodomains (BD1 and BD2), an extra-terminal domain (ET), and a C-terminal domain (CTD). They comprise the ubiquitously expressed BRD2, BRD3, and BRD4 and testis-restricted BRDT, and they mainly recognize acetylated lysine of histone 4. A recent study showed that genetic and pharmacologic inhibition of BRD4 reduces cell viability in models of acquired and intrinsic cetuximab resistance. Furthermore, a combination of cetuximab and bromodomain inhibitor JQ1 delays acquired resistance in patient-derived xenograft mouse models of HNSCC, indicating the potential for cetuximab and epidrugs for HNSCC (Leonard et al. 2018).
Protein Arginine Methyltransferase
Arginine methylation plays a major role in gene regulation because of the ability of protein arginine methyltransferases (PRMTs) to deposit key activating (histone H4R3me2a, H3R2me2s, H3R17me2a, and H3R26me2a) or repressive (histone H3R2me2a, H3R8me2a, H3R8me2s, and H4R3me2s) histone marks. However, there is limited evidence to demonstrate the role of PRMTs in HNSCC. A member of the PRMT family, PRMT5, has weak and progressive expression in the cytoplasm and nucleus of dysplastic and cancer cells. Furthermore, PRMT5 expression correlates with loss of E-cadherin and cytokeratin 17 and upregulation of vimentin, features that are indicative of an epithelial-to-mesenchymal transition (EMT) (Amano et al. 2018).
Histone Lysine Methyltransferase (KMT/HMT)
Histones can be mono-, di-, or trimethylated at lysines and arginines by histone lysine methyltransferases (HMTs), which transfer the methyl group from S-adenosyl methionine to the histone substrate, generating S-adenosyl homocysteine. HMTs are the “writers” of methylation marks on histones (detailed biochemistry reviewed in Mentch and Locasale 2016). Lysine (K) methylation at specific histone (H) positions (i.e., H3K9, H3K27, H3K37, and H4K20) is linked to the formation of tightly packed chromatin and gene silencing. In contrast, methylation of H3K4, H3K36, and H3K39 is associated with actively transcribed regions and gene activation. HMTs, such as NSD1, EZH2, KMT5A, SMYD2, and MLL, promote HNSCC growth, activate codependent oncogenic signaling pathways, and are involved in chemoresistance (Wend et al. 2013; Huang et al. 2016; Luo et al. 2016; Ohtomo-Oda et al. 2016; Sun et al. 2016; Brennan et al. 2017; Bui et al. 2018; Liao et al. 2018). Therefore, HMTs are extensively studied as therapeutic targets.
Histone Lysine Demethylase (KDM/HDM)
Two classes of histone lysine demethylase (KDMs) are the amine oxidases like histone demethylases (lysine-specific demethylases LSD1/2 or KDM1A/KDM1B) and the other the iron- and α-ketoglutarate dependent JmJC-domain containing proteins (KDM 2–6). LSD1 and LSD2, non-JmJC-domain containing proteins require the presence of an intermediate iminium cation in their respective catalytic cycle. However, the demethylases containing a JmjC domain has a different reaction mechanism, which also allows for removal of a trimethyl mark, which is not possible for LSD1-type demethylases. LSD1 can demethylate lysine at specific histone positions to repress gene expression (e.g., H3K4) or stimulate transcription (e.g., H3K9), indicating a dual and context-dependent role in transcriptional regulation (Cai et al. 2014). LSD1 promotes cancer initiation, progression, and relapse through several mechanisms (Alsaqer et al. 2017): 1) increasing expression of pluripotency-related genes, including SOX2, OCT4, and NANOG, and thus supporting cancer-initiating cells; 2) regulating expression of tumor suppressors, such as E-cadherin and TP53; and 3) demethylating lysine residues in several nonhistone substrates, such as TP53, DNMT1, and E2F1. Pumilio (PUM) translational repressor complex posttranscriptionally regulates LSD1 family protein levels as a feedback loop and suggested its functionally important role during development and in human malignancies (Miles et al. 2015). LSD1 expression among esophageal carcinoma subtypes is highly variable; statistical analysis reveals low confidence for differences noted in some subtypes (Yu et al. 2013). The role of LSD2 (KDM1B) in HNSCC has not been characterized. Recently, LSD2 has been shown to promote small cell lung cancer by regulating TFPI2 expression through the mediation of DNMT3B expression or through the regulation of the demethylation of H3K4me1 in the promoter region of the TFPI2 gene (Cao et al. 2018).
Our group recently explored the therapeutic potential of LSD1 by evaluating its ability to target HNSCC tumors undergoing EMT (Alsaqer et al. 2017). Microarray analyses indicate that treatment of epithelial and mesenchymal patient-derived head and neck cancer cells (tonsillar, myoepithelial, and oral osteosarcoma cells) with GSK-LSD1 helps reduce key downstream signaling molecules, including JAK1, CD274 (PD-L1), AKT1, MYC, CTGF, c-Kit, and PIK3CA. There are differences in the gene signatures downregulated. However, some specific questions remain unanswered in our and other published studies. Does LSD1 expression differentiate dysplasias, which develop into malignant lesions, from ones that do not progress to malignancy? Can LSD1 expression be regarded as a biomarker for malignancy, and if so, what are the cutoff points for LSD1 since it is expressed in normal tissues? Answering these questions is important for dissecting the contributions of LSD1 in HNSCC.
Jumonji C (JmjC) domain-containing histone demethylases require unique cofactors, for example, Fe(II) and α-ketoglutarate. Although JmjC domain histone demethylase could be an attractive target for HNSCC, only 2 studies have identified the role in HNSCC. KDM5B overexpression is predicted as a biomarker and indicator of poor prognosis in HNSCC (Huang et al. 2018). KDM5B (JARID1B) has also been shown to maintain stem-like cell populations (Facompre et al. 2016). The other JmjC domain demethylases, including KDM2, KDM3, KDM4, and KDM6, are characterized in various cancers, but their functional role, molecular targets, and mechanism are not known in HNSCC. KDM2A has been shown to promote lung tumorigenesis by enhancing ERK1/2 signaling. KDM2B is involved in leukemic stem cells. KDM2B induces the EZH2 and PRC2 complex to promote bladder cancer angiogenesis. KDM3A has been shown to regulate breast cancer cell invasion and apoptosis. KDM3B is thought to play a role in transcriptional activation of the LMO2 oncogene in leukemia as well as in tumor suppression. KDM4A and KDM4B interact with ERα to promote transcription of MYC. KDM5A is overexpressed in multiple cancers. KDM5B is overexpressed in prostate cancer, with potential coactivation of the androgen receptor. KDM5B has a role in luminal breast cancer and testis cancer. KDM6A is mutated in cancers. Thus, it could potentially have a role in dysregulation of the cell cycle as its target gene RB-binding proteins. KDM6B is proposed to have a context-dependent role in cancers; in breast cancer cells, it is involved in activating transcription of antiapoptotic protein Bcl2 in an ERα-dependent manner. KDM6B expression was found to stabilize nuclear p53 and induced expression of p21 to aid in partial cell cycle arrest and differentiation in germline stem cells, preventing tumorigenesis (D’Oto et al. 2016).
Epigenetic Regulators in Malignant Transformation
The different classes of epigenetic regulator enzymes could impart different functions in dysplasia leading to malignancy and in dysplasia that does not progress to malignancy, as well as HNSCC grades and stages. The histological analysis showed that LSD1 expression is higher in dysplasia compared to adjacent normal tissues and progresses with HNSCC grade. However, retrospective clinical studies by histological and molecular analysis are not reported or performed to determine if these dysplasias progress to malignancy. In addition, The Cancer Genome Atlas (TCGA) data analysis showed that LSD1 expression increases with progressive stage and grade (Alsaqer et al. 2017). Although 4% to 25% of dysplasias progress to malignancy, clinical findings of histological dysplasia are alone inadequate predictors. The series of studies proposed that the combination of histological findings along with molecular signatures is needed. The extensive list of predictive molecular markers for premaligancy is reviewed (Nikitakis et al. 2018), which includes DNA ploidy status, loss of heterogeneity (3 p14 (FHIT), 9 p21 (p16 INK4 a/CDKN2 A), 17 p (TP53), 4 q), cell cycle regulation (Ki-67, PCNA, MCM2, cyclin D1, p16 INK4 A, TP53, MDM2, pRb, surviving), immortalization status (telomerase, hTERT, hTERC), angiogenesis (VEGF-A, VEGFR2), cell adhesion (E-cadherin, catenins, CD44 v6), matrix metalloproteinases (MMPs; MMPs 1, 2, 9, 20), inflammation (COX-2), molecular signaling (EGFR/TGF-α, PI3 K/Akt, mTOR/pS6), DNA methylation (p16 INK4 A, p15, p14 ARK, FOXM1, MGMT, RARB2, CDH1, COX-2, TSPYL5, CLDN11, NKX2-3, RBP4, CMTM3, TRPC4, MAP6, KIF1 A, EDNRB, HOXA9, DCC, ZNF582 m PAX1 m, DAPK, GSTP1, LATS1, DBCR1), histone modification (H3K4, 9, 18, 27, 36), miRNAs ( upregulation of miR-21, miR-31; downregulation of miR-125b, miR-145; and overexpressed miR-208b-3p), stem cell–related proteins (PDPN, ABCG2, ALDH1, CD133), DNA damage (γ-H2 AX, phospho-CHK2 Thr68), and S100 proteins (Nikitakis et al. 2018). Furthermore, the studies have proposed that plastic chromatin may allow premalignant or malignant cells to sample alternative transcriptional states, gene pathways, or developmental programs, a subset of which may be pro-oncogenic or clonally adaptive (Flavahan et al. 2017). KDMs are responsive to signals from the tumor microenvironment and have a role in erasing epigenetic memory. It is predicted that deregulation of these epigenetic enzymes may confer plasticity and facilitate reprogramming of premalignant or cancer cells (Flavahan et al. 2017). Thus, these genetic and epigenetic markers in combination with clinical, pathological features and grade of oral dysplasia are expected to be more helpful in predicting malignant transformation risk. These markers could potentially provide knowledge about patient stratification and application of specific therapeutic intervention to dysplasia progressing to malignancy and HNSCC.
Epigenetic Regulators in Combination and Immunotherapy
Epigenetic regulatory enzymes are involved in the development of resistance to therapy by tumor cells. Dysregulation of epigenetic enzymes could lead to acquired drug resistance, which could be overcome by targeting these enzymes (e.g., bromodomain in combination with cetuximab in HNSCC xenografts) (Leonard et al. 2018). The studies have shown that several candidate epigenetic enzymes, including MGMT, KDM5A, and others, are involved in acquired drug resistance and epigenetically poised states (Brown et al. 2014). Cancer stem cells have self-renewal and tumor-initiating properties and are involved in chemotherapeutic drug resistance (Brown et al. 2014). Epigenetic drugs could reprogram these cancer stem cells and sensitize to chemotherapy.
Nonetheless, epigenetic regulation is a key driver of most malignancies. Therefore, epigenetic drugs, or epidrugs, are extensively used in combination with chemotherapy or immunotherapy. For instance, EGF mutation or stimulation activates immune checkpoint regulator expression in HNSCC (Concha-Benavente et al. 2016; Zhang et al. 2017). Thus, a combination of epidrugs along with EGF signaling pathway mediators, standard chemotherapy, or a specific immune checkpoint regulator inhibitor could provide a synergistic effect for HNSCC therapy. The strategies under investigation are blocking programmed cell death protein 1 (PD1), PD1 ligand 1 (PD-L1), and cytotoxic T lymphocyte antigen 4 (CTLA4), which enables patients to produce an effective antitumor response by blocking immunoinhibitory signals (Mahoney et al. 2015). Epidrugs could be used in combination with these immune checkpoint blockades to reset the epigenome of exhausted T cells and achieve durable immune responses. PD-L1 expression localizes in the membrane and/or cytoplasm and in the nucleus, where expression is significantly associated with reduced patient survival (Satelli et al. 2016), and PD-L1 expression directly protects tumors from cytolytic T-cell killing (Lau et al. 2017). Therefore, the expression of PD-L1 within a tumor, either by tumor cells or tumor-infiltrating cells, explains the therapeutic effect of PD1/PD-L1 blocking antibodies (Juneja et al. 2017). Recently, epigenetic inhibitors were shown to downregulate expression of PD1 or PD-L1, alleviating T-cell exhaustion (Barrero 2017). The bromodomain inhibitor JQ1 also suppresses PD-L1 expression in ovarian cancer cell lines and a mouse model, and it reverts PD-L1 induction that is mediated by infiltrating T-cell–secreted IFNγ (Zhu et al. 2016). In addition, both tumor- and host-derived PD-L1 can play critical roles in immunosuppression (Juneja et al. 2017). Therefore, appropriate immune regulation could help avoid side effects of and resistance to immunotherapy applications (Moskovitz and Ferris 2018). A basic understanding of how epigenetic DNA and histone regulators mediate expression of immune checkpoints will provide novel potential applications for combination immunotherapy for oral squamous cell carcinoma.
Epidrugs in HNSCC Clinical Trials
Although several drugs are in preclinical development, few drugs are in phase I and II trials alone or in combination (Table). The most extensively studied DNMT inhibitor/DNA methylation agents are cytidine analogues 5-azacitidine (5-aza-CR; azacitidine) and 5-aza-2′-deoxycytidine (5-aza-CdR; decitabine) (Murgo 2005). Azacitidine has been used in various preclinical and clinical studies (Viet et al. 2014; Biktasova et al. 2017).
Several antibody-based therapeutics also are being explored for cancers. Durvalumab is an anti–PD-L1 antibody in anticancer therapy pipelines (Vanella et al. 2017). Tremelimumab is an anti-CTLA4 humanized monoclonal antibody used for various cancers (Rini et al. 2011) and has been tested in a clinical trial for advanced stage IV non–small cell lung cancer (NSCLC) (Peters et al. 2016). Furthermore, a combination of azacitidine, durvalumab, and tremelimumab is in clinical trial for HNSCC. CC-486 is an oral azacitidine used alone or in combination with cytotoxic agents carboplatin or nab-paclitaxel in patients with advanced unresectable solid tumors (Von Hoff et al. 2018). CUDC-101, a multitarget inhibitor of HDACs, EGFR, and HER2, combined with chemoradiation, has been shown to be clinically beneficial in 9 of 12 HNSCC patients (Galloway et al. 2015). When combined with erlotinib, the HDAC inhibitor panobinostat is tolerable in patients with advanced NSCLC and HNSCC (Gray et al. 2014). In addition, a phase II clinical study showed that valproic acid plus cisplatin and cetuximab in recurrent and/or metastatic HNSCC is a less toxic and more effective first-line chemotherapy regimen (Caponigro et al. 2016). HDAC inhibitor vorinostat demonstrated efficacy in a phase II study of locally advanced, recurrent, or metastatic adenoid cystic carcinoma, supporting the potential of HDAC inhibitors in future therapeutic studies (Goncalves et al. 2017). Furthermore, HDAC inhibitor PXD101 (belinostat) synergizes with 5-fluorouracil to inhibit colon cancer cell growth in vitro, in vivo (Tumber et al. 2007), and in a clinical trial for HNSCC. In addition, DNMT inhibitor 5-fluoro-2′-deoxycytidine is being investigated in preclinical studies for anticancer activity (Beumer et al. 2008; Morfouace et al. 2016) and is in a clinical trial for HNSCC. Interestingly, some of these trials (Table) represent a broad umbrella approach, for example, NCT00004062 for thyroid and head and neck (HN) cancer, NCT02269943 for nasopharyngeal neoplasms and HN cancer, and NCT00738751 for lung and HN cancer.
Conclusions and Perspectives
A balance of epigenetic regulators is critical to maintaining healthy cells and tissues—for example, the balance between HATs and HDACs and between HMTs and HDMs maintains appropriate tissue-specific regulation. Dysregulation, mutation, overexpression, or attenuation of key epigenetic regulators can shift the balance toward oncogenic transformation, leading to HNSCC growth and metastasis. Furthermore, the level of these epigenetic modulators varies with the type of HNSCC. Since these epigenetic enzymes impart regulation in normal tissues, there would be a possibility of off-target effects from systemically administered therapies, as is the case in other therapies. However, the current studies have shown beneficial effects. Valproic acid has been prescribed for epilepsy patients for several decades now, without reporting serious side effects (Amitai et al. 2015); this drug also has anticancer effects. Azacitidine was tested on myeloid dysplastic syndrome patients and did not induce serious side effects, so there may be more concern about acquired resistance than intrinsic toxicity (Altucci and Rots 2016). Therefore, basic understanding of this epigenetic balance is critical for rational drug design and precision therapy.
We have extensively used orthotopic and patient-derived oral cancer mouse models to test epigenetic regulators and other candidates in HNSCC (Bais et al. 2015; Hiemer et al. 2015; Ozdener et al. 2016; Stanford et al. 2016; Alsaqer et al. 2017; Kartha et al. 2018). Recent advances in genetically modified and humanized NSG mouse models could be useful for additional epigenetic preclinical studies (Morton et al. 2016). Furthermore, recently developed epigenetic tools—such as reporter mice (Stelzer et al. 2015), fusion of Tet1 or Dnmt3a with a catalytically inactive Cas9 that can edit DNA methylation in mice (Liu et al. 2016), and single-cell epigenomics (Stelzer et al. 2015; Clark et al. 2016)—can be applied to future HNSCC studies. Although we have achieved some molecular understanding, further delineation of the key differences in the epigenetic regulation of HNSCC compared to other cancer types and subtypes will improve future opportunities to treat this pernicious disease.
Author Contributions
M.V. Bais, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. The author gave final approval and agrees to be accountable for all aspects of the work.
Acknowledgments
The author acknowledges National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grants R21DE026892 and R03DE025274, Oral Cancer Affinity Research Collaborative ARC (EPOC ARC account 9950000118), and CTSI-Boston University pilot funding (1UL1TR001430) to Manish V. Bais.
Footnotes
The author declares no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: M.V. Bais  https://orcid.org/0000-0003-4198-7377
https://orcid.org/0000-0003-4198-7377
References
- Albrengues J, Bertero T, Grasset E, Bonan S, Maiel M, Bourget I, Philippe C, Herraiz Serrano C, Benamar S, Croce O, et al. 2015. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat Commun. 6:10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaqer SF, Tashkandi MM, Kartha VK, Yang YT, Alkheriji Y, Salama A, Varelas X, Kukuruzinska M, Monti S, Bais MV. 2017. Inhibition of LSD1 epigenetically attenuates oral cancer growth and metastasis. Oncotarget. 8(43):73372–73386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altucci L, Rots MG. 2016. Epigenetic drugs: from chemistry via biology to medicine and back. Clin Epigenetics. 8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Matsubara D, Yoshimoto T, Tamura T, Nishino H, Mori Y, Niki T. 2018. Expression of protein arginine methyltransferase-5 in oral squamous cell carcinoma and its significance in epithelial-to-mesenchymal transition. Pathol Int. 68(6):359–366. [DOI] [PubMed] [Google Scholar]
- Amitai M, Sachs E, Zivony A, Remez R, Ben Baruch R, Amit BH, Kronenberg S, Apter A, Shoval G, Weizman A, et al. 2015. Effects of long-term valproic acid treatment on hematological and biochemical parameters in adolescent psychiatric inpatients: A retrospective naturalistic study. Int Clin Psychopharmacol. 30(5):241–248. [DOI] [PubMed] [Google Scholar]
- Bais MV, Kukuruzinska M, Trackman PC. 2015. Orthotopic non-metastatic and metastatic oral cancer mouse models. Oral Oncol. 51(5):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero MJ. 2017. Epigenetic strategies to boost cancer immunotherapies. Int J Mol Sci. 18(6). pii: E1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, Egorin MJ. 2008. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemother Pharmacol. 62(2):363–368. [DOI] [PubMed] [Google Scholar]
- Biktasova A, Hajek M, Sewell A, Gary C, Bellinger G, Deshpande HA, Bhatia A, Burtness B, Judson B, Mehra S, et al. 2017. Demethylation therapy as a targeted treatment for human papillomavirus-associated head and neck cancer. Clin Cancer Res. 23(23):7276–7287. [DOI] [PubMed] [Google Scholar]
- Boscolo-Rizzo P, Furlan C, Lupato V, Polesel J, Fratta E. 2017. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: role of HPV and lifestyle factors. Clin Epigenetics. 9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan K, Shin JH, Tay JK, Prunello M, Gentles AJ, Sunwoo JB, Gevaert O. 2017. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep. 7(1):17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. 2014. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 14(11):747–753. [DOI] [PubMed] [Google Scholar]
- Bui N, Huang JK, Bojorquez-Gomez A, Licon K, Sanchez KS, Tang SN, Beckett AN, Wang T, Zhang W, Shen JP, et al. 2018. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther. 17(7):1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, Chen S, Chen MW, Zhang J, Ahmed M, et al. 2014. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 9(5):1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Guo C, Yin Y, Li X, Zhou L. 2018. Lysine-specific demethylase 2 contributes to the proliferation of small cell lung cancer by regulating the expression of TFPI-2. Mol Med Rep. 18(1):733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro F, Di Gennaro E, Ionna F, Longo F, Aversa C, Pavone E, Maglione MG, Di Marzo M, Muto P, Cavalcanti E, et al. 2016. Phase II clinical study of valproic acid plus cisplatin and cetuximab in recurrent and/or metastatic squamous cell carcinoma of Head and Neck-V-CHANCE trial. BMC Cancer. 16(1):918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. 2016. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, Freeman GJ, Ferris RL. 2016. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNgamma that induce PD-L1 expression in head and neck cancer. Cancer Res. 76(5):1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Oto A, Tian QW, Davidoff AM, Yang J. 2016. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 1(2):34–40. [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: from mechanism to therapy. Cell. 150(1):12–27. [DOI] [PubMed] [Google Scholar]
- Facompre ND, Harmeyer KM, Sole X, Kabraji S, Belden Z, Sahu V, Whelan K, Tanaka K, Weinstein GS, Montone KT, et al. 2016. Jarid1b enables transit between distinct states of the stem-like cell population in oral cancers. Cancer Res. 76(18):5538–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science. 357(6348):eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway TJ, Wirth LJ, Colevas AD, Gilbert J, Bauman JE, Saba NF, Raben D, Mehra R, Ma AW, Atoyan R, et al. 2015. A phase I study of CUDC-101, a multitarget inhibitor of HDACs, EGFR, and HER2, in combination with chemoradiation in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 21(7):1566–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Ahmad N. 2016. Mitochondrial sirtuins in cancer: emerging roles and therapeutic potential. Cancer Res. 76(9):2500–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves PH, Heilbrun LK, Barrett MT, Kummar S, Hansen AR, Siu LL, Piekarz RL, Sukari AW, Chao J, Pilat MJ, et al. 2017. A phase 2 study of vorinostat in locally advanced, recurrent, or metastatic adenoid cystic carcinoma. Oncotarget. 8(20):32918–32929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JE, Haura E, Chiappori A, Tanvetyanon T, Williams CC, Pinder-Schenck M, Kish JA, Kreahling J, Lush R, Neuger A, et al. 2014. A phase I, pharmacokinetic, and pharmacodynamic study of panobinostat, an HDAC inhibitor, combined with erlotinib in patients with advanced aerodigestive tract tumors. Clin Cancer Res. 20(6):1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemer SE, Zhang L, Kartha VK, Packer TS, Almershed M, Noonan V, Kukuruzinska M, Bais MV, Monti S, Varelas X. 2015. A YAP/TAZ-regulated molecular signature is associated with oral squamous cell carcinoma. Mol Cancer Res. 13(6):957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Qiu Y, Li G, Liu C, She L, Zhang D, Chen X, Zhu G, Zhang X, Tian Y, et al. 2018. Kdm5b overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J Cancer. 9(1):198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou L, Chen H, Wu C, Duo Z, Zhang Y. 2016. EZH2 is overexpressed in laryngeal squamous cell carcinoma and enhances the stem-like properties of AMC-HN-8 cells. Oncol Lett. 12(2):837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH. 2017. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 214(4):895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha VK, Alamoud KA, Sadykov K, Nguyen BC, Laroche F, Feng H, Lee J, Pai SI, Varelas X, Egloff AM, et al. 2018. Functional and genomic analyses reveal therapeutic potential of targeting beta-catenin/CBP activity in head and neck cancer. Genome Med. 10(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J, Cheung J, Navarro A, Lianoglou S, Haley B, Totpal K, Sanders L, Koeppen H, Caplazi P, McBride J, et al. 2017. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat Commun. 8:14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard B, Brand TM, O’Keefe RA, Lee ED, Zeng Y, Kemmer JD, Li H, Grandis JR, Bhola NE. 2018. Bet inhibition overcomes receptor tyrosine kinase-mediated cetuximab resistance in HNSCC. Cancer Res. 78(15):4331–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Seto E. 2016. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med. 6(10):a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao T, Wang YJ, Hu JQ, Wang Y, Han LT, Ma B, Shi RL, Qu N, Wei WJ, Guan Q, et al. 2018. Histone methyltransferase KMT5a gene modulates oncogenesis and lipid metabolism of papillary thyroid cancer in vitro. Oncol Rep. 39(5):2185–2192. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. 2016. Editing DNA methylation in the mammalian genome. Cell. 167(1):233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Jiang Y, Ma S, Chang H, Yi C, Cao H, Gao Y, Guo H, Hou J, Yan J, et al. 2016. EZH2 promotes invasion and metastasis of laryngeal squamous cells carcinoma via epithelial-mesenchymal transition through H3K27me3. Biochem Biophys Res Commun. 479(2):253–259. [DOI] [PubMed] [Google Scholar]
- Lyko F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 19(2):81–92. [DOI] [PubMed] [Google Scholar]
- Mahoney KM, Rennert PD, Freeman GJ. 2015. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 14(8):561–584. [DOI] [PubMed] [Google Scholar]
- Mentch SJ, Locasale JW. 2016. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci. 1363:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles WO, Lepesant JM, Bourdeaux J, Texier M, Kerenyi MA, Nakakido M, Hamamoto R, Orkin SH, Dyson NJ, Di Stefano L. 2015. The lsd1 family of histone demethylases and the pumilio posttranscriptional repressor function in a complex regulatory feedback loop. Mol Cell Biol. 35(24):4199–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa K, Imai A, Mochizuki D, Mima M, Endo S, Misawa Y, Kanazawa T, Mineta H. 2018. Association of TET3 epigenetic inactivation with head and neck cancer. Oncotarget. 9(36):24480–24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfouace M, Nimmervoll B, Boulos N, Patel YT, Shelat A, Freeman BB, III, Robinson GW, Wright K, Gajjar A, Stewart CF, et al. 2016. Preclinical studies of 5-fluoro-2′-deoxycytidine and tetrahydrouridine in pediatric brain tumors. J Neurooncol. 126(2):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JJ, Bird G, Keysar SB, Astling DP, Lyons TR, Anderson RT, Glogowska MJ, Estes P, Eagles JR, Le PN, et al. 2016. Xactmice: humanizing mouse bone marrow enables microenvironment reconstitution in a patient-derived xenograft model of head and neck cancer. Oncogene. 35(3):290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz JM, Ferris RL. 2018. Tumor immunology and immunotherapy for head and neck squamous cell carcinoma. J Dent Res. 97(6):622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgo AJ. 2005. Innovative approaches to the clinical development of DNA methylation inhibitors as epigenetic remodeling drugs. Semin Oncol. 32(5):458–464. [DOI] [PubMed] [Google Scholar]
- Nikitakis NG, Pentenero M, Georgaki M, Poh CF, Peterson DE, Edwards P, Lingen M, Sauk JJ. 2018. Molecular markers associated with development and progression of potentially premalignant oral epithelial lesions: current knowledge and future implications. Oral Surg Oral Med Oral Pathol Oral Radiol. 125(6):650–669. [DOI] [PubMed] [Google Scholar]
- Ohtomo-Oda R, Komatsu S, Mori T, Sekine S, Hirajima S, Yoshimoto S, Kanai Y, Otsuji E, Ikeda E, Tsuda H. 2016. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Hum Pathol. 49:145–155. [DOI] [PubMed] [Google Scholar]
- Ozdener GB, Bais MV, Trackman PC. 2016. Determination of cell uptake pathways for tumor inhibitor lysyl oxidase propeptide. Mol Oncol. 10(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S, Antonia S, Goldberg SB, Heymach JV, Kim ES, Nakagawa K, Papadimitrakopoulou V, Mukhopadhyay P, McIntosh S, Rizvi NA. 2016. 191TiP: MYSTIC: A global, phase 3 study of durvalumab (MEDI4736) plus tremelimumab combination therapy or durvalumab monotherapy versus platinum-based chemotherapy (CT) in the first-line treatment of patients (pts) with advanced stage IV NSCLC. J Thorac Oncol. 11(4 Suppl):S139–S140. [Google Scholar]
- Rasmussen KD, Helin K. 2016. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30(7):733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi B, Raut SK, Panda NK, Rattan V, Radotra BD, Khullar M. 2016. Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol Cell Biochem. 415(1–2):183–196. [DOI] [PubMed] [Google Scholar]
- Rini BI, Stein M, Shannon P, Eddy S, Tyler A, Stephenson JJ, Jr., Catlett L, Huang B, Healey D, Gordon M. 2011. Phase 1 dose-escalation trial of tremelimumab plus sunitinib in patients with metastatic renal cell carcinoma. Cancer. 117(4):758–767. [DOI] [PubMed] [Google Scholar]
- Satelli A, Batth IS, Brownlee Z, Rojas C, Meng QH, Kopetz S, Li S. 2016. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep. 6:28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi RB, Swaminathan A, Chatterjee S, Shanmugam MK, Li F, Ramakrishnan GB, Siveen KS, Chinnathambi A, Zayed ME, Alharbi SA, et al. 2015. Inhibition of p300 lysine acetyltransferase activity by luteolin reduces tumor growth in head and neck squamous cell carcinoma (HNSCC) xenograft mouse model. Oncotarget. 6(41):43806–43818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Wang L, Wu X, Wang K, Xie D, Xiao Q, Li S, Jiang K, Liao L, Yates JR, III, et al. 2018. PML recruits TET2 to regulate DNA modification and cell proliferation in response to chemotherapeutic agent. Cancer Res. 78(10):2475–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford EA, Ramirez-Cardenas A, Wang Z, Novikov O, Alamoud K, Koutrakis P, Mizgerd JP, Genco CA, Kukuruzinska M, Monti S, et al. 2016. Role for the aryl hydrocarbon receptor and diverse ligands in oral squamous cell carcinoma migration and tumorigenesis. Mol Cancer Res. 14(8):696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y, Shivalila CS, Soldner F, Markoulaki S, Jaenisch R. 2015. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 163(1):218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam D, Thombre R, Dhar A, Anant S. 2014. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yu F, Zhang L, Zhou X. 2016. EZH2, an on-off valve in signal network of tumor cells. Cell Signal. 28(5):481–487. [DOI] [PubMed] [Google Scholar]
- Theocharis S, Klijanienko J, Giaginis C, Rodriguez J, Jouffroy T, Girod A, Alexandrou P, Sastre-Garau X. 2011. Histone deacetylase-1 and -2 expression in mobile tongue squamous cell carcinoma: associations with clinicopathological parameters and patients’ survival. J Oral Pathol Med. 40(9):706–714. [DOI] [PubMed] [Google Scholar]
- Tumber A, Collins LS, Petersen KD, Thougaard A, Christiansen SJ, Dejligbjerg M, Jensen PB, Sehested M, Ritchie JW. 2007. The histone deacetylase inhibitor PXD101 synergises with 5-fluorouracil to inhibit colon cancer cell growth in vitro and in vivo. Cancer Chemother Pharmacol. 60(2):275–283. [DOI] [PubMed] [Google Scholar]
- Vanella V, Festino L, Strudel M, Simeone E, Grimaldi AM, Ascierto PA. 2017. PD-L1 inhibitors in the pipeline: promise and progress. Oncoimmunology. 7(1):e1365209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet CT, Dang D, Achdjian S, Ye Y, Katz SG, Schmidt BL. 2014. Decitabine rescues cisplatin resistance in head and neck squamous cell carcinoma. PLoS One. 9(11):e112880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Rasco DW, Heath EI, Munster PN, Schellens JHM, Isambert N, Le Tourneau C, O’Neil BH, Mathijssen RH, Lopez-Martin JA, et al. 2018. Phase I study of CC-486 alone and in combination with carboplatin or nabpaclitaxel in patients with relapsed or refractory solid tumors. Clin Cancer Res. 24(17):4072–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend P, Fang L, Zhu Q, Schipper JH, Loddenkemper C, Kosel F, Brinkmann V, Eckert K, Hindersin S, Holland JD, et al. 2013. Wnt/beta-catenin signalling induces MLL to create epigenetic changes in salivary gland tumours. EMBO J. 32(14):1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang B, Zhang K, Lei Z, Guo Y, Xiao H, Wang J, Fan L, Lan C, Wei Y, et al. 2013. High expression of lysine-specific demethylase 1 correlates with poor prognosis of patients with esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 437(2):192–198. [DOI] [PubMed] [Google Scholar]
- Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, Liu X, Yuan Z, Wang P, Xiao Z. 2017. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 10:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, et al. 2016. BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 16(11):2829–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]