 By designing and engineering photo-conditional viral inhibitors, spatiotemporal control of the transporter associated with antigen processing TAP was sustained, allowing the on-demand antigen translocation in human immune cell lines and primary cells by light.
By designing and engineering photo-conditional viral inhibitors, spatiotemporal control of the transporter associated with antigen processing TAP was sustained, allowing the on-demand antigen translocation in human immune cell lines and primary cells by light.
Abstract
The immune system makes use of major histocompatibility complex class I (MHC I) molecules to present peptides to other immune cells, which can evoke an immune response. Within this process of antigen presentation, the MHC I peptide loading complex, consisting of a transporter associated with antigen processing TAP, MHC I, and chaperones, is key to the initiation of immune response by shuttling peptides from the cytosol into the ER lumen. However, it is still enigmatic how the flux of antigens is precisely coordinated in time and space, limiting our understanding of antigen presentation pathways. Here, we report on the development of a synthetic viral TAP inhibitor that can be cleaved by light. This photo-conditional inhibitor shows temporal blockade of TAP-mediated antigen translocation, which is unleashed upon illumination. The recovery of TAP activity was monitored at single-cell resolution both in human immune cell lines and primary cells. The development of a photo-conditional TAP inhibitor thus expands the repertoire of chemical intervention tools for immunological processes.
Introduction
To avoid threats from pathogens and malignantly transformed cells, the immune system elaborated sophisticated ways to process antigens for their presentation to different immune cell subsets via MHC I molecules.1,2 The transporter associated with antigen processing (TAP) is a central assembly unit of the peptide-loading complex (PLC). TAP translocates cytosolic antigenic peptides into the endoplasmic reticulum (ER) for subsequent loading onto MHC I molecules.3,4 Peptide–MHC complexes are then released from the PLC and traffic to the cell surface. An important step in the understanding of MHC I antigen presentation pathways was recently achieved by the structure determination of the human PLC.5 As bait, the TAP inhibitor ICP47 was employed to arrest and isolate endogenous PLCs.
Antigen processing comprises a number of crucial steps that shape the immune response. Within the PLC, TAP is a major target for tumor and viral immune evasion.6,7 The viral inhibitor ICP47 encoded by herpes simplex virus represents one of the most potent immune suppressors.8–10 Its active domain is located in the N-terminal region (residue 2–34), which inhibits MHC I antigen presentation by blocking TAP and thus the supply of antigenic peptides.11,12 Upon binding to TAP, the active domain of ICP47 adopts two α-helices connected by a flexible loop.13–15 The consecutive region (residues 35–55) is required for stabilization of TAP by a conformational arrest.16 Although many steps in antigen processing have been elucidated,1 it is less clear how the events are linked and how the flux of peptides is spatiotemporally coordinated. Hence, conditional tools are required to control distinctive check-points in the antigen processing pathway with high accuracy in time and space.
Optogenetics and optochemical biology have emerged to modulate biological processes. Light enables a spatiotemporal regulation of bioactivity. So far, the repertoire of photo-responsive biomolecules ranges from caged ATP, second messengers, and neurotransmitters to peptides and proteins.17–19 Protein caging is typically achieved by masking amino acids, which are essential for activity or ligand interaction. Genetic code expansion promoted the incorporation of caged unnatural amino acids by amber stop codon, but is restricted to side-chain functionalities.20–22 In contrast, chemical protein synthesis allows for the flexible integration of photo-sensitive moieties and generation of caged proteins equipped with light-sensitive units.
With the PLC being one of the key checkpoints in MHC I antigen presentation, we targeted TAP as its central translocation unit to control its functional activity by light. We designed a photo-sensitive ICP47 to selectively arrest TAP in situ, which results in the conditional blockage of the substrate-binding pocket and thus inhibits peptide transport from the cytosol into the ER lumen. Photo-conditional ICP47 (pc-ICP47) variants were designed to specifically block antigen supply by TAP inhibition and instantaneously initiate TAP activity upon pc-ICP47 photo-cleavage (Fig. 1). Inhibition of peptide translocation and its activation by light was analyzed by a series of peptide binding and translocation assays. In-cell applications demonstrate that pc-ICP47 blocks TAP-dependent peptide translocation, which is reversed after illumination. We report on the development of pc-ICP47, which can be used as a molecular plug for the PLC in order to enable control of peptide translocation in time and space by light.
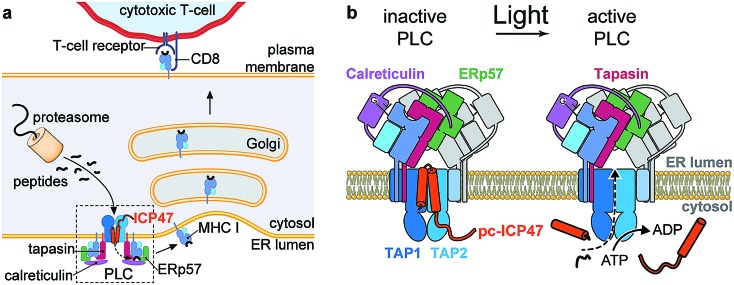
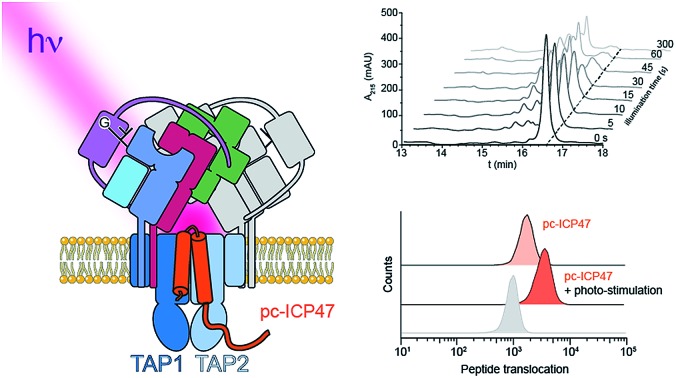
Fig. 1. Photo-cleavable TAP inhibitor ICP47 in MHC I antigen presentation pathway. (a) The transporter associated with antigen processing (TAP1/2) translocates proteasomal degradation products from the cytosol into the ER lumen. Peptide loading of MHC I molecules is orchestrated by the peptide-loading complex (PLC), composed of TAP, the chaperone tapasin, the disulfide-reductase ERp57, the lectin calreticulin, the MHC I heavy chain, and β2-microglobulin. Upon successful loading, editing, and proof-reading, stable peptide/MHC I complexes are released and transported to the cell surface to present their antigenic cargo to cytotoxic T-lymphocytes. Antigen processing can be blocked by the TAP inhibitor ICP47 derived from the herpes simplex virus. (b) Photo-conditional ICP47 (pc-ICP47) blocks peptide translocation into the ER lumen. Photolysis induces fragmentation of the pc-ICP47 and subsequently reactivates TAP.
Results and discussion
Design and photo-scission of photo-conditional TAP inhibitors
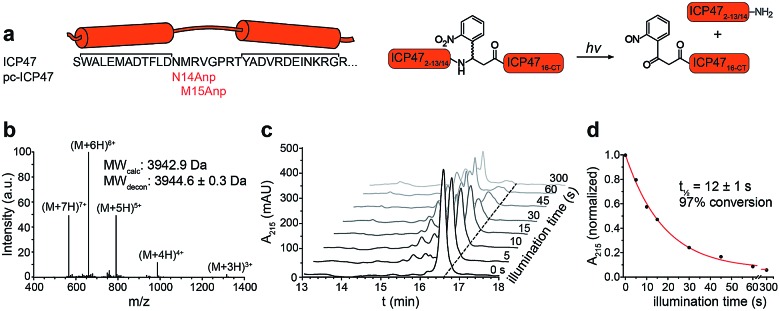
To establish a synthetic route providing access to backbone-modified ICP47, we employed 9-fluorenyl-methoxycarbonyl (Fmoc) solid-phase peptide synthesis. Since the site-specific incorporation of photo-labile amino acids in backbone-modified proteins cannot be achieved by genetic or enzymatic approaches, the synthetic strategy was adjusted to engineer pc-ICP47 variants in good yield. We rationalized that positions in the flexible loop of the active region of ICP47 are likely to tolerate substitutions in contrast to the two α-helices.11,12 We therefore replaced residue Asn14 or Met15 by the photo-sensitive 3-amino-3-(2-nitrophenyl)propionic acid (Anp) in either S or R stereo configuration (Fig. 2a). It is worth mentioning that the additional rotatable bond of the photo-labile β3-amino acid Anp increases the backbone flexibility and enables an adaptation in the binding cavity of TAP. In total, we synthesized a set of photo-conditional variants based on the active inhibitory region (ICP472–34, pc-ICP4714R-Anp2–34, pc-ICP4714S-Anp2–34, pc-ICP4715R-Anp2–34, and pc-ICP4715S-Anp2–34) or the full-interacting inhibitor (ICP472–55 and pc-ICP4715R-Anp2–55). After purification, the identity of all variants was confirmed by high-resolution mass spectrometry (Fig. S1–S4 and Table S1†). The photo-conditional ICP47 variants displayed an α-helical conformation similar to unmodified ICP47 (Fig. S5 and Table S2†).13,14,23
Fig. 2. Design and characterization of pc-ICP47. (a) pc-ICP47 was designed by substituting either Asn14 or Met15 with the photo-cleavable amino acid Anp. Upon illumination at 365 nm, the high-affinity viral inhibitor is split into two fragments, which do not display a detectable affinity for TAP. (b) Identity of purified pc-ICP4715R-Anp2–34 was confirmed by ESI-MS, revealing a molecular weight M + H of 3944.6 ± 0.3 Da (calc. 3942.9 Da). (c) Time-dependent photolysis of pc-ICP4715R-Anp2–34 monitored by RP-C18 HPLC. (d) Exponential photo-decay of pc-ICP4715R-Anp2–34 was determined with t1/2 = 12 ± 1 s and 97% conversion.
We next examined the photo-cleavage kinetics of the pc-ICP47 variants. Illumination at 365 nm (185 mW cm–2) for various intervals led to clean fragmentation of all photo-labile variants (Fig. 2b, S6 and S7†). All pc-ICP47 variants displayed a similar photo-scission rate of t1/2 ≈ 12 s, and the photolytic fragmentation was virtually completed within 60 s, leading to more than 90% fragmentation (Fig. 2b and Table S3†). The position and stereo-configuration of the photo-cleavable amino acid had no significant influence on the photolytic kinetics. The fast photo-activation along with a clean fragmentation is essential for a precise control of cellular processes.
Peptide binding to TAP by light
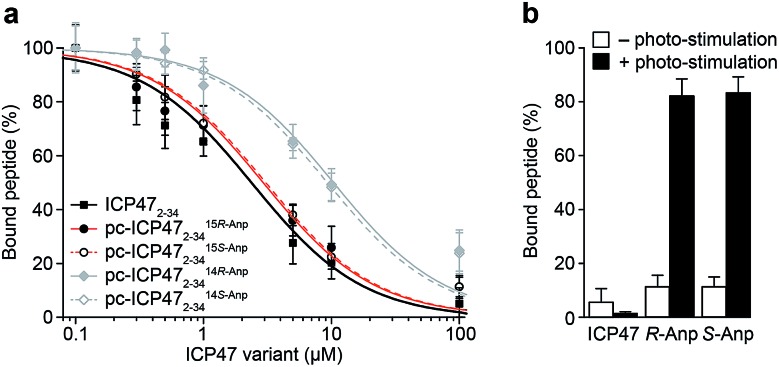
We first examined the inhibitory activity of all pc-ICP47 variants prior to photo-cleavage (Fig. 3). A defined concentration of the fluorescein (FL) labeled reporter peptide (RRYCFLKSTEL, named C4F) was assayed for TAP binding with rising concentrations of the respective pc-ICP47. Incorporation of Anp at position 15 in either R or S configuration (pc-ICP4715Anp2–34) displayed similar IC50 values (15R-Anp, Ki = 121 ± 9 nM; 15S-Anp, Ki = 125 ± 8 nM) to unmodified ICP472–34 (Ki = 100 ± 11 nM). In contrast, substitution at position 14 led to a significant lower affinity (14R-Anp, Ki = 550 ± 35 nM; 14S-Anp, Ki = 498 ± 22 nM), indicating that the position of the photo-labile amino acid has a critical impact on the inhibitory activity (Fig. 3a and Table S4†). We next exploited peptide binding after photo-illumination. It is important to mention that short fragments of ICP47 generated upon photo-cleavage display no detectable affinity for TAP.12 Indeed, after illumination at 365 nm (185 mW cm–2) for 300 s, peptide binding to TAP was restored (>80%) for all pc-ICP47 variants (Fig. 3b). In contrast, the activity of TAP alone or in complex with unmodified ICP472–34 was not affected by illumination.
Fig. 3. Photo-controlled TAP inhibition by pc-ICP47 variants. (a) Binding of the reporter peptide C4F to TAP was competed with increasing concentrations of ICP472–34 or pc-ICP472–34 variants. Unmodified ICP47 (Ki = 100 ± 11 nM) and pc-ICP47 with the photo-cleavable amino acid at position 15 have very similar IC50 values (15R-Anp, Ki = 121 ± 9 nM; 15S-Anp, Ki = 125 ± 8 nM), whereas lower inhibition constants were observed for pc-ICP4714Anp variants (14R-Anp, Ki = 550 ± 35 nM; 14S-Anp, Ki = 489 ± 22 nM). (b) Peptide binding was blocked with 100 μM of pc-ICP4715Anp2–34 and unmodified ICP472–34. After photo-cleavage at 365 nm, peptide binding to TAP was restored (>80%) for both pc-ICP4715Anp2–34, while illumination of unmodified ICP472–34 did not recover the TAP activity.
Peptide translocation triggered by light
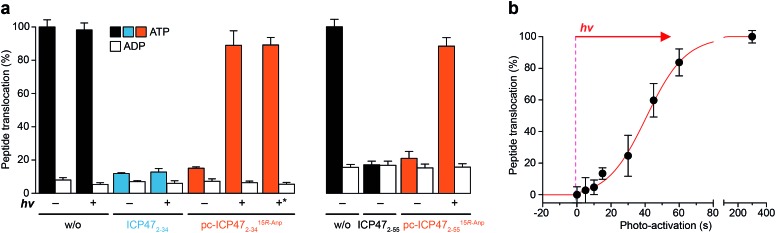
We next analyzed whether the antigen translocation complex could be reactivated upon illumination. TAP-dependent transport of a fluorescent peptide (RRYQNSTCFLL, named NST-F) was monitored using microsomes isolated from human Burkitt's lymphoma (Raji) cells (Fig. 4). After translocation, the NST-F peptide is specifically N-core glycosylated in the ER lumen based on the glycosylation targeting sequence (NST).24,25 Before illumination, TAP-dependent peptide translocation was blocked by pc-ICP472–34 to a level similar to unmodified ICP472–34 (>85% inhibition; Fig. 4a). The inhibition was dependent on the position of the photo-cleavable amino acid as demonstrated by a higher IC50 value of ICP472–34 modified with the photo-cleavable amino acid at position 14 compared to position 15. Consistent with the peptide binding data, the blockage was not affected by the stereochemistry of the photo-labile amino acid, due to the conformational freedom of the β3-amino acid Anp (Fig. S8†). Upon photo-stimulation, peptide translocation of pc-ICP47 blocked TAP was restored (>90%). A slightly lower efficiency was achieved for pc-ICP47 modified at position 14 (∼85%). Photo-activation of TAP resulted in a fast restoration of peptide transport into the ER lumen (Fig. 4b). Importantly, the light induced recovery of peptide translocation occurred at a rate comparable to the one obtained for photo-cleavage of free pc-ICP472–34, demonstrating that the photolysis kinetics of pc-ICP472–34 in the unbound and bound state are similar.
Fig. 4. Photo-modulated peptide transport of TAP. (a) Peptide transport was carried out in the presence of 3 mM of Mg-ATP, 1 μM of NST-F, 5 μM of either ICP472–34, ICP472–55 or their photo-conditional variants in Raji microsomes for 10 min at 37 °C. The amount of N-core glycosylated reporter peptides was quantified. Translocation was normalized to peptide transport in the absence of ICP472–34. In the presence of unmodified ICP472–34 and its photo-conditional variants, peptide translocation was effectively blocked. Upon photolysis of pc-ICP4715R-Anp2–34 as well as pc-ICP4715R-Anp2–55, peptide transport was recovered to >90%. *: photo-cleavage of pc-ICP4715R-Anp2–34 prior to its use. (b) Fast and dose-controlled photo-stimulation of pc-ICP4715R-Anp2–34 was analyzed by peptide translocation. All data represent mean ± SD (n = 3).
Based on the optimal position and stereochemistry of the photo-labile amino acid, we also synthesized a full-interacting photo-conditional inhibitor, which is required to stabilize the TAP complex and to isolate the native PLC.5,16 For pc-ICP4715R-Anp2–55, similar inhibition constants, photo-fragmentation kinetics, and photo-activation of TAP were obtained (Fig. 4a, S9–S11 and Table S4†). Once arrested either by photo-conditional or unmodified viral inhibitors, the TAP complex is stabilized. However, after photo-exposure, the pc-ICP472–55 arrested TAP complex disassembles, whereas the complex containing the unmodified ICP472–55 remains stable (Fig. S12†).
Light-controlled antigen translocation at single-cell resolution
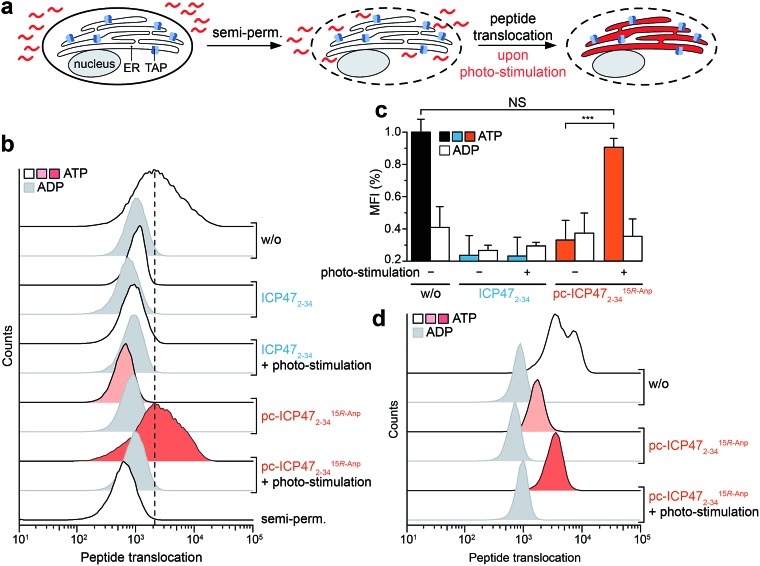
Finally, we examined pc-ICP472–34 on TAP activity in B cells (Raji), a frequently employed lymphoma cell line as a model for antigen presentation. The TAP-dependent translocation of fluorescent peptides in the ER was monitored by flow cytometry (Fig. 5a). The plasma membrane was semi-permeabilized by streptolysin O (SLO) while ER membranes stayed intact, which has been previously evaluated in cells expressing cytosolic GFP and ER-resident mCherry.25 Defined concentrations of SLO led to a loss of cytosolic GFP but retained an intact mCherry signal, demonstrating that only the plasma membrane but not the ER membrane is semi-permeabilized. Cells pre-incubated either with unmodified ICP472–34 or pc-ICP472–34 showed no peptide transport activity above the background signal of ATP depleted cells.
Fig. 5. Light-triggered peptide translocation in human immune cells. (a) Raji cells or primary human monocytes were semi-permeabilized with streptolysin O to provide a fluorescent peptide (30 nM of NST-AF647). Translocation was carried out for 15 min at 37 °C in the presence of ATP or ADP (10 mM each) and analyzed by flow cytometry. (b) Peptide translocation in Raji cells controlled by light. In the presence of ICP472–34 and pc-ICP4715R-Anp2–34, peptide translocation into the ER was blocked to background level. After illumination at 365 nm, photo-fragmentation of pc-ICP4715R-Anp2–34 restored TAP activity and peptide translocation into the ER lumen. In contrast, photo-stimulation of unmodified ICP47 did not reset peptide translocation. (c) Bar diagram summarizing the light-triggered peptide translocation in Raji cells. The data represent two individual experiments, each in biological duplicates with mean ± SD, utilizing a one-way analysis of variance (ANOVA) followed by Tukey's test (P < 0.001). (d) Photo-stimulated peptide translocation in human donor-derived monocytes. In the presence of pc-ICP4715R-Anp2–34, peptide translocation was reduced to background level. Photo-cleavage of pc-ICP4715R-Anp2–34 restored peptide translocation in primary human immune cells.
After illumination, peptide translocation was recovered for TAP blocked by pc-ICP472–34, whereas unmodified ICP472–34 keeps TAP in its arrested state (Fig. 5b and c). After photo-cleavage of pc-ICP472–34, peptide translocation was restored to the level of active TAP. Statistical analysis of independent experiments further corroborated the results, accentuating that TAP and peptide translocation into the ER can be controlled by light using pc-ICP472–34 (>90%; Fig. 5c). Importantly, the cell viability is not affected by the photo-activation (Fig. S13†).
To generalize the versatile use of photo-conditional inhibitors, we assigned the single-cell based antigen translocation assay to primary cells of human donors. Hence, we isolated human blood-derived monocytes that show high TAP activity among other immune cell subsets.25 In the presence of pc-ICP472–34, peptide translocation was blocked to a similar extent as with the unmodified TAP inhibitor. Upon illumination, TAP and thus peptide translocation was restored in monocytes pre-incubated with pc-ICP472–34, but not in cells with unmodified ICP472–34 (Fig. 5d and S14†). Thus, we have demonstrated that pc-ICP472–34 blocked TAP in different cell types and antigen translocation can be unleashed by light.
Conclusions
We developed a methodology to control antigen processing via a photo-conditional checkpoint blockade of the antigen translocation complex TAP. We report on the design and characterization of the photo-conditional TAP inhibitor ICP47 to activate the PLC and subsequent TAP-dependent peptide translocation. The engineered ICP47 possessed a high light-sensitivity and fast photo-cleavage, which proved to be crucial for in situ activation of the PLC. Peptide translocation, mediated by TAP within the PLC, was markedly inhibited by synthetic pc-ICP47 and exclusively re-activated upon illumination in human cell lines and primary immune cells.
As the synthetic TAP inhibitor ICP47 provides new angles to study immune surveillance mechanisms, we foresee that the orthogonal modality will be a valuable approach to probe and manipulate the PLC. For instance, the role of the PLC in different antigen presentation pathways is not yet fully understood, and more comprehensive studies will require precise spatiotemporal control of its cellular activity. The photo-conditional viral inhibitors will help to elucidate the influence of the PLC function in primary immune cells and to probe antigen processing pathways in situ. The photo-cleavable inhibitors may open new avenues for time-resolved cryo-EM and super-resolution microscopic analyses to reveal the transient states of the assembly and disassembly of the PLC. To date, no light-controllable synthetic inhibitors are available to influence TAP-dependent antigen processing with high temporal resolution. The development of an intervention toolbox to modulate checkpoint in antigen processing continues to be an important aspect of our research.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The German Research Foundation (GRK 1986 to R. W. and R. T., SFB 807 – Membrane Transport and Communication to R. T., EXC 114 – Macromolecular Complexes to R. W. and R. T., and SPP 1623 to R. T.) and the European Research Council (ERC Advanced Grant, Life Sciences to R. T.) supported the work. We thank Dr Elisa Lehnert for providing TAP-containing membranes from P. pastoris, Michael Urban for nanoparticle tracking, and Dr Stefan H. Hüttenhain (Darmstadt University of Applied Sciences) for the help with the LC-MS analyses.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc04863k
References
- Rock K. L., Reits E., Neefjes J. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts C., Robinson B. W., Knolle P. A. Nat. Rev. Immunol. 2010;10:403–414. doi: 10.1038/nri2780. [DOI] [PubMed] [Google Scholar]
- Blum J. S., Wearsch P. A., Cresswell P. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Tampé R. Front. Immunol. 2017;8:65. doi: 10.3389/fimmu.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blees A., Januliene D., Hofmann T., Koller N., Schmidt C., Trowitzsch S., Moeller A., Tampé R. Nature. 2017;551:525–528. doi: 10.1038/nature24627. [DOI] [PubMed] [Google Scholar]
- Parcej D., Tampé R. Nat. Chem. Biol. 2010;6:572–580. doi: 10.1038/nchembio.410. [DOI] [PubMed] [Google Scholar]
- Verweij M. C., Horst D., Griffin B. D., Luteijn R. D., Davison A. J., Ressing M. E., Wiertz E. J. PLoS Pathog. 2015;11:e1004743. doi: 10.1371/journal.ppat.1004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh K., Ahn K., Djaballah H., Sempé P., van Endert P. M., Tampé R., Peterson P. A., Yang Y. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- Hill A., Jugovic P., York I., Russ G., Bennink J., Yewdell J., Ploegh H., Johnson D. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- Ahn K., Meyer T. H., Uebel S., Sempe P., Djaballah H., Yang Y., Peterson P. A., Früh K., Tampé R. EMBO J. 1996;15:3247–3255. [PMC free article] [PubMed] [Google Scholar]
- Galocha B., Hill A., Barnett B. C., Dolan A., Raimondi A., Cook R. F., Brunner J., McGeoch D. J., Ploegh H. L. J. Exp. Med. 1997;185:1565–1572. doi: 10.1084/jem.185.9.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L., Kraas W., Uebel S., Jung G., Tampé R. J. Mol. Biol. 1997;272:484–492. doi: 10.1006/jmbi.1997.1282. [DOI] [PubMed] [Google Scholar]
- Pfänder R., Neumann L., Zweckstetter M., Seger C., Holak T. A., Tampé R. Biochemistry. 1999;38:13692–13698. doi: 10.1021/bi9909647. [DOI] [PubMed] [Google Scholar]
- Aisenbrey C., Sizun C., Koch J., Herget M., Abele R., Bechinger B., Tampé R. J. Biol. Chem. 2006;281:30365–30372. doi: 10.1074/jbc.M603000200. [DOI] [PubMed] [Google Scholar]
- Oldham M. L., Grigorieff N., Chen J. eLife. 2016;5:e21829. doi: 10.7554/eLife.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbring V., Bäucker A., Trowitzsch S., Tampé R. Sci. Rep. 2016;6:36907. doi: 10.1038/srep36907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieke C., Rohrbach F., Gottschalk A., Mayer G., Heckel A. Angew. Chem., Int. Ed. 2012;51:8446–8476. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
- Baker A. S., Deiters A. ACS Chem. Biol. 2014;9:1398–1407. doi: 10.1021/cb500176x. [DOI] [PubMed] [Google Scholar]
- Ankenbruck N., Courtney T., Naro Y., Deiters A. Angew. Chem., Int. Ed. Engl. 2018;57:2768–2798. doi: 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T., Suga H. Trends Biochem. Sci. 2014;39:400–408. doi: 10.1016/j.tibs.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Lang K., Chin J. W. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- Courtney T., Deiters A. Curr. Opin. Chem. Biol. 2018;46:99–107. doi: 10.1016/j.cbpa.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert D., Neumann L., Uebel S., Tampé R. Biochemistry. 1997;36:4694–4700. doi: 10.1021/bi962940v. [DOI] [PubMed] [Google Scholar]
- Neefjes J. J., Momburg F., Hämmerling G. J. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- Fischbach H., Doring M., Nikles D., Lehnert E., Baldauf C., Kalinke U., Tampé R. Nat. Commun. 2015;6:6199. doi: 10.1038/ncomms7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.