Abstract
Synthetic oligonucleotides (ODNs) containing CpG motifs stimulate human plasmacytoid dendritic cells (pDCs) to produce type-1 interferons (IFNs) and proinflammatory cytokines. Previous studies demonstrated that interferon regulatory factors (IRFs) play a central role in mediating CpG-induced pDC activation. This work explores the inverse effects of IRF5 and IRF8 (also known as IFN consensus sequence-binding protein) on CpG-dependent gene expression in the human CAL-1 pDC cell line. This cell line shares many of the phenotypic and functional properties of freshly isolated human pDCs. Results from RNA interference and microarray studies indicate that IRF5 upregulates TLR9-driven gene expression whereas IRF8 downregulates the same genes. Several findings support the conclusion that IRF8 inhibits TLR9-dependent gene expression by directly blocking the activity of IRF5. First, the inhibitory activity of IRF8 is only observed when IRF5 is present. Second, proximity ligation analysis shows that IRF8 and IRF5 colocalize within the cytoplasm of resting human pDCs and cotranslocate to the nucleus after CpG stimulation. Taken together, these findings suggest that IRF5 and IRF8, two transcription factors with opposing functions, control TLR9 signaling in human pDCs.
Keywords: CpG oligonucleotide, Dendritic cell, IRF8, IRF5, TLR9
Introduction
Plasmacytoid dendritic cells (pDCs) function at the interface between the innate and adaptive immune systems and play a critical role in the host’s response to infectious pathogens [1, 2]. Considerable effort has been invested in clarifying the molecular mechanisms through which pDCs perform these vital functions. In humans, pDCs constitutively express TLR 9 enabling them to sense unmethylated CpG motifs expressed by microbial pathogens [3–5].
Synthetic oligodeoxynucleotides expressing such CpG motifs mimic the ability of bacterial DNA to stimulate pDCs [6]. In particular, “K” class phosphorothioate oligonucleotide expressing immunostimulatory CpG motif (CpG ODN) have been extensively studied in clinical trials and generate a response characterized by the production of type-1 IFNs, proinflammatory cytokines, and chemokines [7, 8]. The release of type-1 IFNs is mediated in a cell type and species-specific manner by “IFN regulating factors” (IRFs)[9]. Recent studies examined which members of the human IRF family regulated the IFN response of CpG-stimulated human pDCs. IRF5 was found to play a vital role in the upregulation of type-1 IFNs and proinflammatory cytokines, represented by IFN-β, and IL-6, respectively [10]. Unexpectedly, IRF8 was found to inhibit the stimulatory activity of IRF5, a result inconsistent with reports showing that IRF8 boosted rather than inhibited CpG-induced DC activation in mice [11].
Since IRF8 influences the development of DCs in both humans and mice [11, 12], this work further examines the effect of IRF5 and IRF8 on human pDCs. Targeted RNAi technology was used to silence specific IRF genes and microarrays then employed to monitor the effect of CpG stimulation on the silenced cells. Studies were performed on the human CAL-1 pDC cell line since efforts to isolate resting primary human pDCs were stymied by the rarity of such cells (<0.5% of PBMC pool) and their propensity to activate during the purification process [13, 14]. Previous work established that CAL-1 cells shared many of the phenotypic and functional properties of freshly isolated human pDCs and mirrored the response of such cells to CpG stimulation [10, 15, 16].
Silencing either IRF8 of IRF5 had widespread effects on the expression of genes activated via TLR9. Silencing IRF8 resulted in a 60% increase in the number of genes activated by CpG stimulation. These genes were members of the same canonical pathways activated when CAL-1 cells were stimulated with CpG ODN. By comparison, silencing IRF5 resulted in an 80% reduction in gene activation when compared with controls. Further analysis indicated that IRF8 and IRF5 acted on overlapping gene sets, leading us to hypothesize that there might be a direct and previously unrecognized interaction between these two IRFs. To examine this possibility, a proximity ligation assay (PLA) was performed on CAL-1 cells and freshly isolated human pDCs. PLA detects proteins that reside within 40 nM of each other. Results indicate that IRF5 and IFR8 colocalize in the cytoplasm of resting pDCs and rapidly translocate to the nucleus following CpG stimulation. These findings are consistent with the hypothesis that IRF8 physically interacts with IRF5 to modify the intensity of TLR9 signaling in human pDCs. The discovery that two transcription factors with opposing functions control TLR9 signaling contributes to our understanding of the host defense and identifies potential targets for pharmaceutical intervention.
Results
Contribution of IRF5 and IRF8 to TLR9-mediated activation of pDCs
Recent studies suggest that IRF5 plays a key role in upregulating the expression of proinflammatory cytokines and type-1 IFNs (exemplified by IL-6 and IFN-β, respectively) when human pDCs are stimulated with “K” class CpG ODN via TLR9. In the same study, IRF8 downmodulated the activation of these two genes[10]. To clarify the mechanism by which these IRFs influenced gene expression, experiments were performed on the human CAL-1 pDC cell line. CAL-1 cells share many of the phenotypic and functional properties of freshly isolated human pDCs and mirror their response to TLR9 stimulation [10].
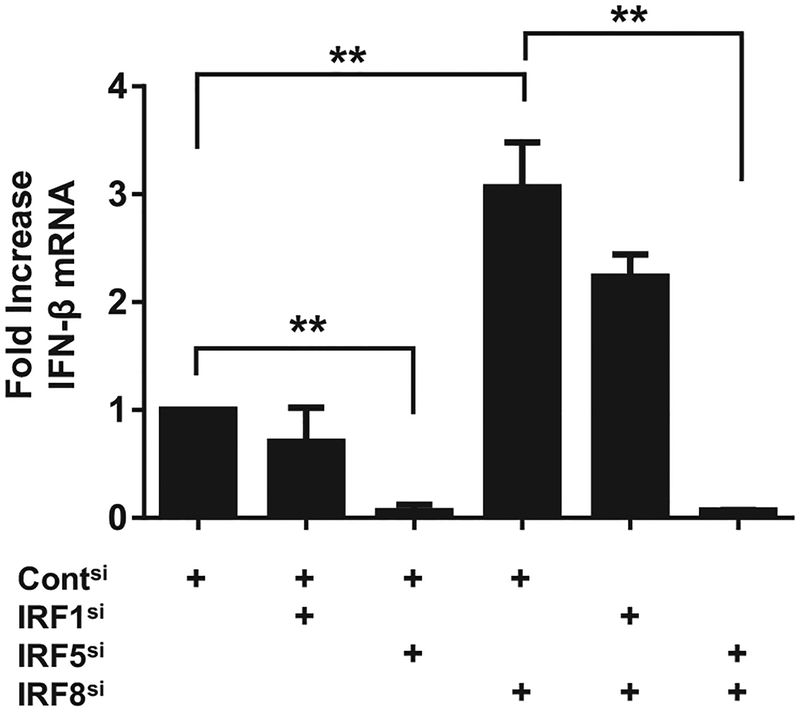
To monitor the influence of IRFs on gene expression, CAL-1 cells were transfected with siRNAs that reduced IRF expres sion levels by >70% without generating off-target effects ([10] and Supporting Information Fig. 1). Silencing IRF5 reduced IFN-β mRNA expression by >90% whereas silencing IRF8 resulted in a >300% increase in IFN-β mRNA levels (p < 0.01, Fig. 1). The specificity of these effects was established by silencing IRF-1 (which plays no role in CpG-induced immune activation) and finding no effect on gene expression (Fig. 1). When both IRF5 and IRF8 were silenced, CpG-mediated expression of IFN-β mRNA fell by 93% (p < 0.01). These findings indicate that the activity of IRF8 was contingent on the presence of IRF5. Consistent with that interpretation, the expression of IFN-β mRNA increased significantly when IRF8 and control IRF1 were both silenced (Fig. 1). IRF8 also influenced the cytokine response of CpG-stimulated CAL-1 cells. The amount of IFN-β and IL-6 secreted by IRF8-silenced CAL-1 cells rose by ≈fivefold when compared with similarly stimulated control cells (Supporting Information Fig. 1).
Figure 1.
Influence of silencing IRFs on CpG-mediated gene activation. CAL-1 cells were transfected with 0.5 nM of each indicated siRNA to silence gene expression. The siRNA transfected cells were stimulated 20 h later with 1 μM of “K” class CpG ODN and IFN-ß mRNA expression assessed by RT-PCR. GAPDH was used as an endogenous control and fold changes in mRNA level determined by comparison to identically treated cells transfected with control siRNA. Data are shown as the mean + SD from 3 independent experiments, each performed in triplicate. **p < 0.01; ANOVA one-way analysis of variance.
Effect of IRF5 and IRF8 on global gene expression following TLR9 activation of CAL-1 cells
To determine whether IRF5 and IRF8 had broad effects on TLR9-dependent gene expression, mRNAs levels were monitored by microarray. CAL-1 cells were transfected with siRNA targeting IRF5 (IRF5si) and/or IRF8 (IRF8si). These transfections reduced IRF5 and IRF8 expression levels by 67 and 85%, respectively (Supporting Information Fig. 2). The silenced cells were then stimulated with CpG ODN for 9 h. This time point was selected based on earlier studies showing gene activation peaked at that time [7]. Genes whose expression increased or decreased significantly following CpG stimulation when compared with cells transfected with control siRNA (Contsi) were identified.
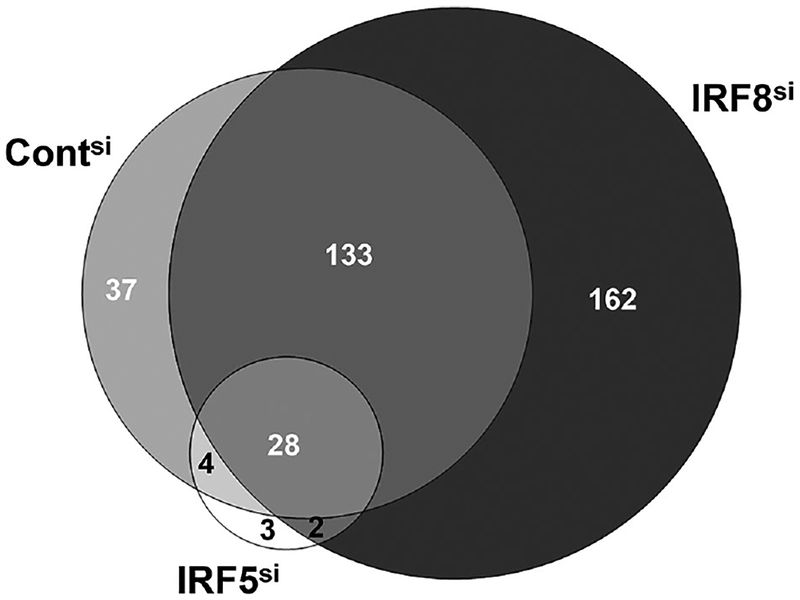
Results from four independent studies demonstrated that CpG stimulation of IRF-8 silenced CAL-1 cells upregulated 60% more genes than identically stimulated cells transfected with control siRNA (Fig. 2). Conversely, silencing IRF5 resulted in an 80% reduction in the number of genes activated by TLR9 ligation (Fig. 2). The graphical representation of these results shows that a common core of 28 genes is upregulated by CpG treatment of CAL-1 cells regardless of IRF silencing.
Figure 2.
Effect of silencing IRF5 and IRF8 on the number of genes upregulated following TLR9 activation. CAL-1 cells were transfected with 1 nM of siRNA targeting IRF5 (IRF5si), IRF8 (IRF8si) or with control siRNA (Contsi) as described in Fig. 1. Cells were then stimulated with 1 μM of CpG ODN for 9 h and gene expression monitored by microarray. The Venn diagram shows the number of genes significantly upregulated (p <0.001) in each population as determined in four independent experiments. A total of 202 genes were upregulated after CpG stimulation of Contsi cells (light gray), 325 genes in IRF8si cells (dark gray), and 37 genes in IRF5si cells (white).
IPA categorization of TLR9-activated genes
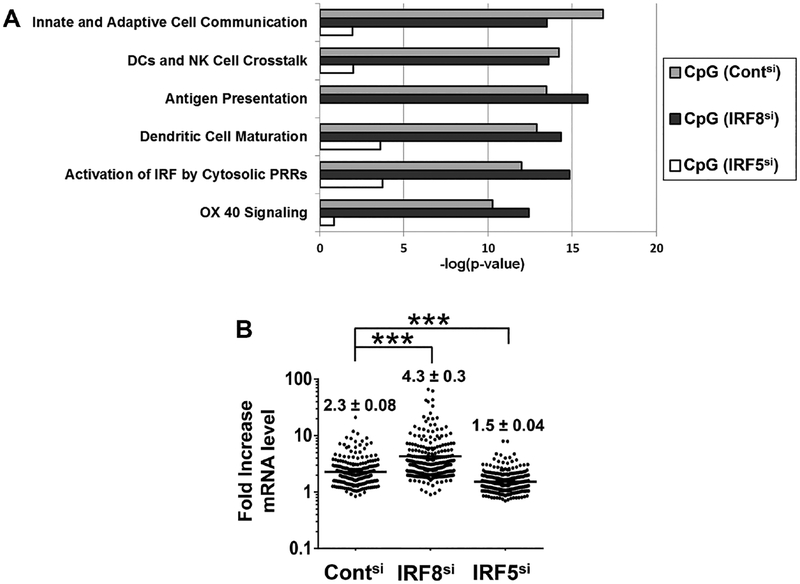
The genes activated when CAL-1 cells were stimulated via TLR9 were classified using Ingenuity Pathway Analysis (IPA). IPA char acterizes gene products based on their function and role in regulatory pathways. Six functional groups were selectively upregulated in CpG stimulated CAL-1 cells. These included cellular immune responses involving the communication/maturation of DCs and antigen presentation (Fig. 3A). When IRF5 was silenced, expression of genes utilizing these pathways fell significantly. Moreover, the same pathways were upregulated when IRF8si cells were stimulated with CpG ODN. This set of findings is consistent with the hypothesis that IRF5 and IRF8 act on the same genes and functional pathways.
Figure 3.
Analysis of CpG activated genes. (A) All genes identified in Fig. 2 were classified using the “Canonical Pathway” feature of IPA. The statistical probabilities assigned by IPA to the most strongly upregulated pathways are shown. (B) The level of expression of each gene upregulated in each population of CAL-1 cells after siRNA silencing is shown. Numbers represent the mean ± SEM expression level in each group as determined in four independent experiments. ***p < 0.001; ANOVA one-way analysis of variance.
The magnitude of gene activation in IRF-silenced CAL-1 cells was evaluated. On average, CpG stimulation induced a 2.3 ± .08-fold increase in mRNA expression in wild-type CAL-1 cells, a 4.3 ± 0.3-fold increase in IRF8 silenced cells and a 1.5 ± 0.04 increase in the IRF5 silenced population (p < 0.001, Fig. 3B and Supporting Information Table 1). These data suggest that IRF8 and IRF5 influenced both the number and magnitude of genes being upregulated.
Genes targeted by IRF8 and IRF5
There are two likely interpretations of the above findings. (1) IRF5 could be a dominant upstream mediator of CpG-induced gene activation and IRF8 may act as an independent downstream brake on gene expression by pDCs. (2) IRF8 could directly interfere with the ability of IRF5 to upregulate gene expression in TLR9-stimulated pDCs.
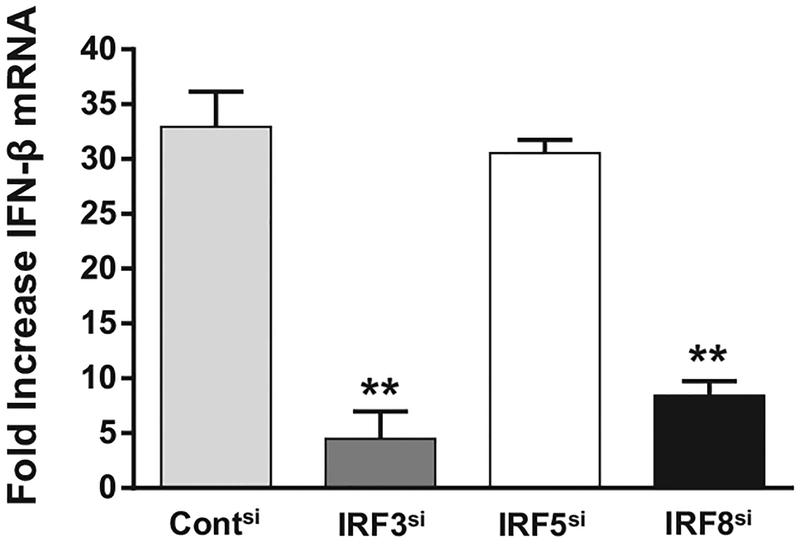
To examine whether IRF8 downregulated gene expression mediated by another IRF, CAL-1 cells were stimulated with cGAMP (a cytosolic STING activator that acts through IRF3 rather than IRF5) [17, 18]. IFN-β mRNA levels increased >30-fold following cGAMP stimulation. As expected, silencing IRF3 reduced IFN-β gene expression after cGAMP stimulation by ≈85% (Fig. 4, p < 0.05) whereas silencing IRF5 had no effect. In contrast to the effect observed on CpG activated cells, silencing IRF8 significantly suppressed gene expression in cGAMP stimulated CAL-1 cells (Fig. 4, p < 0.05). These findings indicate that IRF8 is not a universal inhibitor of gene expression following the activation of human pDCs (thereby enhancing the possibility of a specific interaction between IRF8 and IRF5).
Figure 4.
Effect of IRF silencing on cGAMP induced gene activation. CAL-1 cells were transfected with 1 nM of each siRNA to silence gene expression as described in Fig. 1. The siRNA transfected cells were stimulated 20 h later with 2 μg/mL of cGAMP (2’–5’) for 3 h and IFN-β mRNA expression assessed by RT-PCR. GAPDH was used as an endogenous control and fold changes in mRNA level determined by comparison to identically treated cells stimulated with CpG ODN in each experiment. Data are shown as mean + SD from three independent experiments, each performed in triplicate. **p < 0.01; Student’s t-test.
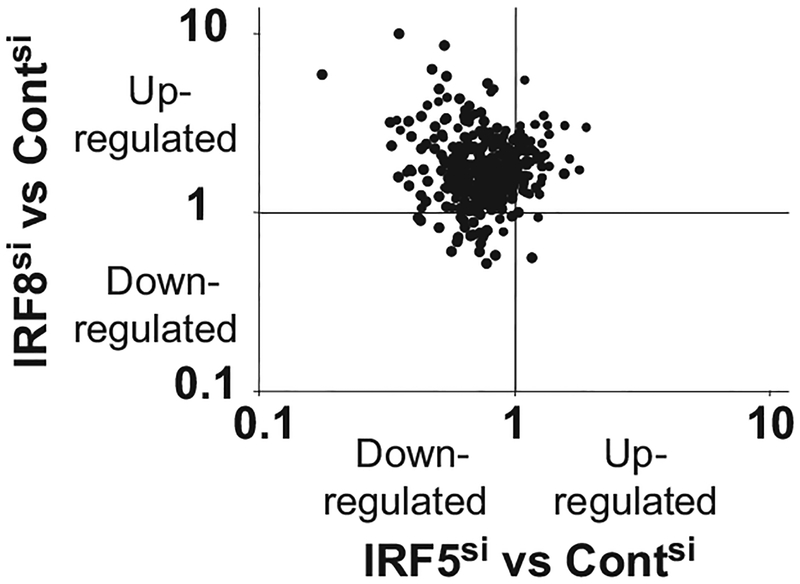
To pursue this possibility, the level of expression of all CpG activated genes was compared in IRF5 versus IRF8 silenced cells. 89% of all CpG stimulated genes shared a common characteristic: their expression increased when IRF8 was silenced and decreased when IRF5 was silenced. This set of genes can be seen in the upper left quadrant of the scatterplot shown in Fig. 5 (p < 10−5, linear regression analysis). This biased distribution of gene expression supports the interpretation that IRF8 acts on the same genes that are upregulated by IRF5, raising the possibility that these two transcription factors are interacting directly.
Figure 5.
Expression levels of CpG activated genes in IRF8 versus IRF5 silenced CAL-1 cells. The expression level of every gene activated in response to CpG stimulation in IRF5 and IRF8 silenced cells was evaluated. Each dot represents a different gene whose location reflects the magnitude with which that gene was up- or downregulated when compared to CpG stimulated cells transfected with control siRNA (Contsi cells). Note the cluster of points in the upper left quadrant representing genes whose mRNA levels rose when IRF8 was silenced but fell when IRF5 was silenced. This skewed outcome (p < 10−5, linear regression analysis) indicates that the same genes are regulated by both transcription factors. Data represent the averaged results from four independent experiments.
Colocalization of IRF8 with IRF5
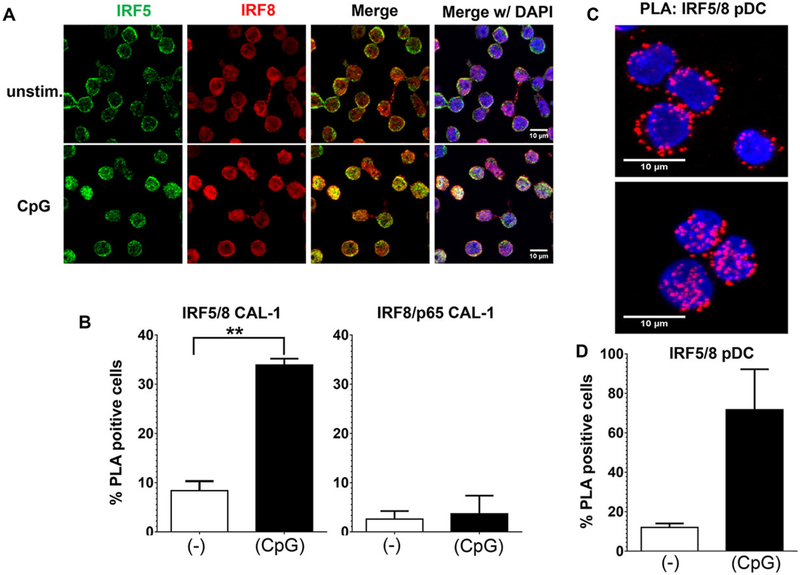
IRF8 is known to form heterodimers with a variety of transcription factors including PU.1, IRF1, and E47 [9, 19] although no association between IRF8 and IRF5 has been reported. To explore the possibility of such an interaction, the intracellular localization of these factors was examined using fluorescent probes. IRF5 was present primarily in the cytoplasm of resting CAL-1 cells and rapidly translocated to the nucleus following stimulation with “K” class CpG (Fig. 6A and [10]). IRF8 showed a similar pattern of redistribution although the magnitude of nuclear translocation was lower.
Figure 6.
Colocalization of IRF5 with IRF8 following CpG-stimulation of human pDCs. (A) CAL-1 cells were stimulated for 60 min with 1 μM of CpG ODNs. The cells were then fixed, permeabilized, and stained with antibodies to detect IRF5 (green) and IRF8 (red). Nuclei were stained with DAPI (blue). One representative image from three independent experiments is shown. (B) A proximity ligation assay was performed on the cells described in (A) using Abs against IRF5 and IRF8 (or p65 and IRF8 as a negative control). The percentage of cells with significant nuclear PLA signals, averaged from three independent experiments (mean + SD) is shown. **p < 0.01, Student’s t-test. (C) Primary pDCs from healthy volunteers were stimulated for 60 min with CpG ODN and analyzed by PLA as described in (B). PLA signals appear as red dots. One representative image from two independent experiments is shown. (D) The percentage of cells with significant nuclear PLA signals, from two independent experiments (mean + SD), was determined using ImageJ software.
A PLA was used to examine whether IRF5 and IRF8 were colocalizing. PLA generates a signal only when the proteins of interest are in close physical proximity (<40 nm, [20]) as occurs during the formation of heterodimers. After one hour of CpG ODN treatment significant nuclear colocalization of IRF5 with IRF8 was observed, exceeding background levels in unstimulated cells by >three fold (p < 0.01, Fig. 6B). This nuclear colocalization was specific since no change in the frequency of NF-κB p65 colocalizing with IRF8 in identically treated cells was detected by PLA (Fig. 6B).
A similar effect was observed when freshly isolated pDCs were analyzed by PLA. Prior to stimulation, IRF5 and IRF8 were detected primarily in the cytoplasm of human pDCs. The addition of CpG ODN led to a dramatic increase in IRF5/IRF8 colocalization with PLA signals now being detected in >70% of cells and primarily in the nucleus (Fig. 6C, D).
Discussion
pDCs make critical contributions to both the innate and adaptive arms of the immune system. Human pDCs express TLR9 that enables them to sense the unmethylated CpG motifs present in bacterial DNA. Activated pDCs excel in antigen presentation and produce IFNs required for host defense [1, 2]. They support the induction of Th1-biased immunity and their activation by “K” class CpG ODN has proven useful in the treatment/prevention of cancer, allergy, and infectious diseases in preclinical and clinical trials [6, 21–24]. Understanding the signaling cascades and patterns of gene expression elicited by the recognition of CpG DNA by human pDCs could thus have basic and therapeutic implications.
The production of IFNs by TLR9-stimulated pDCs is mediated by IFN regulatory factors. IRFs are transcription factors that influence a variety of cellular functions ranging from differentiation to proliferation to apoptosis [9, 25]. A recent study suggested that IRF5 and IRF8 might play central but opposing roles in the regulation of IFN-β mRNA expression by TLR9-stimulated human pDCs. IRF5 promoted while IFR8 inhibited the upregulation of that gene [10].
To broaden our understanding of how IRFs influence gene expression, we combined RNAi technology with microarray studies. Initial efforts to apply these techniques to study primary human pDCs failed due to the rarity of such cells (<0.5% of the PBMC pool) and their propensity to activate during the purification process [14, 26]. Instead, the human pDC-derived CAL-1 cell line was utilized as its response to CpG stimulation closely mirrors that of freshly isolated human pDC [10, 15, 16]. Several findings support the technical reliability of the knockdown experiments. First, siRNA transfection significantly reduced mRNA and protein levels of the targeted gene (Supporting Information Fig. 2 and [10]). Second, no off-target activity was detected following siRNA transfection (Supporting Information Fig. 2 and [10]). Third, transfection did not change the cytokine or gene expression pattern/response of CAL-1 cells (Supporting Information Fig. 1 and [10]). Use of CAL-1 cells does introduce the possibility of donor-specific mutations and/or abnormalities due to malignant transformation. However, sequencing established that the IRF8 gene present in CAL-1 cells contained no mutations (data not shown). Moreover, the interaction of IRF8 with IRF5 detected in CAL-1 cells was confirmed in PLA studies of human pDC (Fig. 6).
Silencing IRF5 decreased the number of genes activated following CpG ODN by >80% and the magnitude of mRNA expression by >30% (Figs. 2 and 3B). As a consequence, the functional pathways triggered by TLR9 ligation were significantly downregulated (Fig. 3A). This result is consistent with murine studies showing that CpG driven type-1 IFN and cytokine production are reduced in IRF5 KO mice [27, 28]. As the IRF5 KO strain is now known to also carry a mutation in the DOCK2 gene (which affects pDC development and type-1 IFN production), current findings support the conclusion that IRF5 (rather than DOCK2) plays a key role in modulating gene expression in TLR9 stimulated DC [29, 30].
A very different outcome was observed in studies of the IRF8 gene. Rather than reducing gene expression as observed when IRF5 was silenced, silencing IRF8 resulted in a 60% increase in the number and twofold increase in the average level of gene expression induced by CpG stimulation (Figs. 2 and 3B). These changes did not alter the regulatory pathways triggered by TLR9 ligation (Fig. 3A). To our knowledge this is the first evidence of widespread IRF8-dependent inhibition of gene expression in human cells.
IRF8 is required for the development of pDCs (and CD8α+ DCs) in humans and mice [9, 11, 12, 31]. Yet little is known concerning its ability to modulate gene expression in immune cells. Studies of human monocytes and murine cDCs found that IRF8 promoted (rather than inhibited) type-1 IFN production [11, 32]. Spleen cells from IRF8 KO mice manifest generalized defects in proinflammatory cytokine production and decreased expression of activation markers following CpG stimulation [33]. In contrast to the effect on murine cDCs, current results show that IRF8 inhibits gene expression in CpG-stimulated human pDCs. This inhibition is not a universal property of IRF8 since no such effect was observed when CAL-1 cells were stimulated with cGAMP (which utilizes IRF3 rather than IRF5, Fig. 4) [17, 18]. Further study is needed to clarify the mechanism(s) by which IRF8 mediates either activation or repression depending on cell type and stimulus.
Two hypotheses were considered to explain the antithetical effects of IRF5 and IRF8 on human pDCs. Either (1) IRF8 might act broadly as a downstream brake on IRF-mediated gene activation in human pDC or (2) IRF8 might directly interfere with the ability of IRF5 to promote gene expression. To differentiate between these alternatives we compared the effect of silencing IRF8 to that of silencing both IRF8 plus IRF5 versus IRF8 plus IRF1 (as a negative control). Gene expression rose when IRF8 (or IRF8 plus IRF1) were silenced but not when IRF5 and IRF8 were silenced (Fig. 1). These findings suggest that IRF5-mediated gene activation must be present to observe the inhibitory effect of IRF8. Moreover, silencing IRF8 did not increase gene expression mediated via IRF3 when CAL-1 cells were stimulated with cGAMP. These findings support the conclusion that IRF8 is not a general inhibitor of gene expression in human pDCs but instead selectively blocks the activity of IRF5.
To further explore this possibility, the expression of all genes stimulated by CpG treatment of CAL-1 cells was evaluated. Eighty-nine percent of these genes were upregulated by IRF5 but inhibited by IRF8 (Fig. 5, p <10−5). A direct interaction between these two transcription factors could explain their antithetical effects. IRF8 has been shown to bind a variety of transcription factors/proteins via an “IRF association domain” [19]. For example, IRF8 associates with IRF3 to cooperatively induce IFN-β mRNA expression in LPS stimulated human blood monocytes [32] and with IRF1 in U937 human monocytes to decrease gene expression [34]. Interactions between IRF8 and additional transcription factors (including AP-1 and BATF) have also been reported [35, 36]. Current results from immunofluorescence studies show that both IRF5 and IRF8 translocate from the cytoplasm to the nucleus following CpG stimulation of CAL-1 cells. PLA was used to detect IRF5/IRF8 interactions under physiologic conditions and confirmed that CpG stimulation induced the formation of such complexes in the nucleus of both CAL-1 cells and freshly isolated human pDCs (Fig. 5). These findings support the conclusion that IRF8 directly interacts with IRF5 to inhibit CpG-induced gene expression in human pDCs.
Further study is needed to precisely define how IRF8 interacts with IRF5. One possibility is that IRF8 interacts with IRF5 within a larger molecular complexes that includes MyD88 and TRAF6 under resting conditions. Such a hypothesis is consistent with evidence showing that IRF5 binds to MyD88 in the cytosol and that this interaction terminates following CpG stimulation [10]. It is suggested that IRF8 also complex with MyD88 via TRAF6 [37] although whether this is affected by TLR stimulation is currently unknown. We hope to use mass spectroscopy to detect and quantify the formation of such molecular complexes before and after CpG stimulation of human pDC.
Current results provide evidence that IRF5 plays a critical role in upregulating the vast majority of genes triggered by TLR9 engagement of human pDCs and that IRF8 acts as a brake on this stimulation via a direct interaction with IRF5. There is growing evidence that polymorphisms in IRF5, IRF8, and pDC-derived type 1 IFNs contribute to the etiopathogenesis of human autoimmune diseases including lupus, RA, and MS [12, 38–40]. The discovery that two transcription factors with opposing functions control TLR9 mediated signaling in human pDCs thus contributes to our understanding of autoimmunity as well as host defense and might help identify targets for pharmaceutical intervention. Future studies should seek to (i) clarify the mechanism by which IRF8 blocks IRF5 activity and (ii) identify whether other molecules contribute to the formation of IRF5/IRF8 complexes.
Materials and methods
Oligonucleotides
Endotoxin free ODNs were synthesized at the CBER core facility (CBER/FDA, Bethesda, MD). “K” type CpG ODNs contained an equimolar mixture of three phosphorothioate sequences: K3 (5′ ATCGACTCTCGAGCGTTCTC 3′), K23 (5′ TCGAGCGTTCTC 3 ′, and K123 (5′ TCGTTCGTTCTC 3′) and were endotoxin free.
Cell culture, preparation, and stimulation
The CAL-1 human pDC cell line was grown in complete RPMI 1640 medium (Lonza, Walkersville, MD) supplemented with 2 mM L-Glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 1 × MEM NEAA (all from Gibco, Grand Island, NY) to which 10% heat inactivated fetal bovine serum (Lonza) was added. Cells were cultured at 37°C in a CO2 in air incubator. Prior to stimulation, the CAL-1 cells were serum starved for 16 h in complete RPMI supplemented with 0.1% FBS and then treated with 1 μM “K” ODN for the indicated times as previously described [10].
Mononuclear cell enriched human buffy coats were obtained by leukopheresis (DTM, NIH, Bethesda, MD) using an IRB approved protocol. Following Ficoll Hypaque (Sigma, St. Louis, MO) and Percoll gradient (Pharmacia, Uppsala, Sweden) centrifugation of the buffy coat, pDCs were MACS sorted using a BDCA two purification kit per manufacturer’s instructions (Miltenyi Biotechm Auburn, CA). The pDCs isolated by this procedure were 93–95% pure and their viability was >95%. A total of 5 × 105 freshly isolated pDC/well were cultured in 48-well plates in complete media and then stimulated with 1 μM “K” ODN for the times indicated.
Microarray studies to detect changes in gene expression
A total of 12 × 106 transfected CAL-1 cells were transferred into 25 cm2 flasks and serum starved for 16 h in 0.1% FBS complete RPMI media prior to stimulation with 1 μM CpG (“K” ODN) for 9 h or left untreated. All experiments were independently repeated a minimum of four times.
Total RNA was extracted from CAL-1 cells using TRIzol Reagent (Invitrogen, Carlsbad, CA) as specified by the manufacturer. The RNA was quantified using an ND 1000 (NanoDrop Technologies, Wilmington, DE) and assessed for the absence of degradation by electrophoresis. Ten micrograms of total RNA was reversed transcribed as previously described [7]. For all samples, a universal reference RNA (Strategene) was processed in parallel. Both cDNA’s were purified using a MinElute PCR Purification Kit (Qiagen) and labeled with Cy5 (sample DNA) or Cy3 (reference RNA) as previously described [41]. The probes were mixed, diluted in 5 μL DMSO plus 1.7 μL of 1 M NaHCO3 and hybridized to prehybridized 36 K human 60 mer oligonucleotide array slides (Human Array Set: Hs MI Opv4 0 3 July 2007, produced by Microarray Inc., Huntsville, AL) at 42°C for 18 h in a MAUI hybridization system (BioMicro Systems, Salt Lake City, UT) followed by washing, centrifugation, and air drying.
Analysis of gene expression
Arrays were scanned using a GenePix 4000B Scanner (Axon Instruments, Union City, CA) and analyzed using the GenePix Pro 6.0 Software Tool (Axon Instruments) using GAL files provided by the manufacturer at http://madb.nci.nih.gov/. Data were uploaded to CIT/BIMAS NCI/CCR Microarray Database and formatted via export function to BRB Array Tools. Gene expression analysis was performed using BRB Array Tools Version 3.8.1 developed by R. Simon and A. P. Lam (NCI Biometric Research Branch, Bethesda, MD) as previously described [7]. Genes whose response to CpG stimulation was increased by silencing IRF5 and/or IRF8 were identified based on the following criteria: their level of expression was significantly elevated when compared with (i) unstimulated cells transfected with control siRNA (p-value cutoff of 0.001) and (ii) cells transfected with the same siRNA in the absence of CpG stimulation (p-value cutoff of 0.01). Based on these criteria differentially expressed genes were identified using a paired random variance t-test [42]. Data were further analyzed using QIAGEN’s Ingenuity7 Pathway Analysis (IPA7, QIAGEN Redwood City, www.qiagen.com/ingenuity) and BIOBASE (BIOBASE Biological Databases, Beverly, MA). Differences between gene sets (e.g. in terms of affiliation to canonical pathways) were assessed using a right-tailed Fisher Exact Test (cutoff: −log(E-12). For simple linear regression analysis of microarray data shown in the scatter plot (Fig. 5), results were log transformed to satisfy homogeneity of variance requirements.
RT-PCR expression analysis
Total RNA was extracted from CAL-1 cells per manufacturer’s instructions (Qiagen, Germantown, MD). The RNA was reverse transcribed into cDNA (QuantiTect RT Kit; Qiagen) and quantified by TaqMan-based real-time PCR (Applied Biosystems, Foster City, CA). The following TaqMan probes were used: IFNB1 (Hs02621180 s1), IRF1 (Hs00971960 m1), IRF3 (Hs01547283 m1), IRF5 (Hs00158114 m1), IRF8 (Hs0 0175238 m1), and GAPDH ((Hs02758991 g1) as an endogenous control (Applied Biosystems). GAPDH levels did not change upon stimulation or during siRNA gene silencing. Data were analyzed by StepOne Software v2.1 using GAPDH as an endogenous control.
Cell transfection
CAL-1 cells were transfected at a density of 1.5 × 106 cells/well with 1 nM of siRNA using the Amaxa 96-well shuttle nucleofector system using program DN100 (cell line SF, Lonza). siRNA to IRF1, IRF5, or Silencer Select Negative Control #1(Silencer Select, Ambion) and siRNA to IRF3 or IRF8 (Stealth RNAi, Invitrogen) were used. Transfected cells recovered in complete media supplemented with 10% FBS for 4 h and then serum starved for 16 h in 0.1% FBS complete RPMI media prior to stimulation. In some experiments CAL-1 cells were transfected with 2’3’-cGAMP (Invivogen, San Diego, CA) using Lipofectamin 2000 (Life Technologies, Carlsbad, CA) according to manufacturer’s instruction.
Proximity ligation and Immunofluorescence assays
CAL-1 cells and primary pDCs were stimulated with “K” CpG ODN for 60 min. Cells were then fixed in 2% paraformaldehyde and permeabilized with methanol. CultureWell chambered cover slips (Electron Microscopy Sciences, Hatfield PA) were treated with 0.05 μg/μL of Cell-Tak (BD Biosciences, Franklin Lakes, NJ). Cells were seeded onto the cover slips, blocked and stained with mouse anti-IRF5 (10T1) (Abcam) and rabbit anti-IRF8 (D20D8) (Cell Signaling Technology) Ab. For immunofluorescence studies, washed cells were incubated with complementary anti-mouse and anti-rabbit secondary antibodies conjugated with AlexaFluor 488 and AlexaFluor 546, respectively. For PLA studies, washed cells were incubated with anti-mouse and anti-rabbit secondary PLA probes (Olink Bioscience, Uppsalla, Sweden) and then with ligation and Red Amplification solutions per manufacturer’s instruc tions. Washed cells were sealed onto the slide using Duolink II Mounting Medium with DAPI. Image stacks were captured using an inverted Zeiss LSM 710 confocal microscope and PLA positive cells were evaluated based on nuclear signals.
Supplementary Material
Acknowledgments:
The authors would like to thank Dr. T. Maeda and Dr. S. Kamihira (Department of Island Medicine, Nagasaki University, Japan) for providing CAL-1 cells. We further like to thank Gregory W. Alvord (Director, Statistical Consultation, NCI, Frederick, MD) for statistical advice and Adelle McFarland (Molecular and Cellular Biology, University of Washington), Ram Savan (Assistant Professor, Immunology, University of Washington), and Hongchuan Li (Cancer and Inflammation Program, NCI, Frederick, MD) for discussions and advice. We also thank Kathleen Meyer (BIMAS/DCB/CIT/NIH, Bethesda, MD) for microarray data upload to Geo as well as Mario Fox (University of Bonn) for technical advice.
This research was supported by the Intramural Research Program of the NIH, NCI. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- CAL-1
human pDC-like cell line
- CpG ODN
phosphorothioate oligonucleotide expressing immunostimulatory CpG motif
- IPA
Ingenuity Pathway Analysis
- IRF
interferon regulatory factor
- ODN
oligonucleotide
- pDC
plasmacytoid dendritic cell
- PLA
proximity ligation assay
Footnotes
Published 2015. This article is a U.S. Government work and is in the public domain in the USA
Additional supporting information may be found in the online version of this article at the publisher’s web-site
Accession code
Microarray data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series Accession Number: GSE70429.
Conflict of interest: Dr. Dennis Klinman and members of his lab are co-inventors on a number of patents concerning CpG ODN and their use. All rights to these patents have been assigned to the Federal government.
References
- 1.Gilliet M, Cao W and Liu YJ, Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol 2008. 8: 594–606. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald-Bocarsly P, Dai J and Singh S, Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 2008. 19: 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T and Akira S, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol 2010. 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 4.Jarrossay D, Napolitani G, Colonna M, Sallusto F and Lanzavecchia A, Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol 2001. 31: 3388–3393. [DOI] [PubMed] [Google Scholar]
- 5.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F and Liu YJ, Subsets of human dendritic cell precursors express different Toll-like receptors and respond to different microbial antigens. J. Exp. Med 2001. 194: 863–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinman DM, Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol 2004. 4: 249–258. [DOI] [PubMed] [Google Scholar]
- 7.Steinhagen F, Meyer C, Tross D, Gursel M, Maeda T, Klaschik S and Klinman DM, Activation of type I interferon-dependent genes characterizes the “core response” induced by CpG DNA. J. Leukoc. Biol 2012. 92: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Kanzler H, Duramad O, Cao W and Liu YJ, Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood 2006. 107: 2423–2431. [DOI] [PubMed] [Google Scholar]
- 9.Tamura T, Yanai H, Savitsky D and Taniguchi T, The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol 2008. 26: 535–584. [DOI] [PubMed] [Google Scholar]
- 10.Steinhagen F, McFarland AP, Rodriguez LG, Tewary P, Jarret A, Savan R and Klinman DM, IRF-5 and NF-kappaB p50 co-regulate IFN-beta and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. Eur. J. Immunol 2013. 43: 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, Gabriele L and Ozato K, The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 2007. 27: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salem S, Langlais D, Lefebvre F, Bourque G, Bigley V, Haniffa M, Casanova JL et al. , Functional characterization of the human dendritic cell immunodeficiency associated with the IRF8(K108E) mutation. Blood 2014. 124: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S et al. , The nature of the principal type 1 interferon-producing cells in human blood. Science 1999. 284: 1835–1837. [DOI] [PubMed] [Google Scholar]
- 14.Gallucci S, Lolkema M and Matzinger P, Natural adjuvants: endogenous activators of dendritic cells. Nat. Med 1999. 11: 1249–1255. [DOI] [PubMed] [Google Scholar]
- 15.Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D et al. , Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 2008. 135: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda T, Murata K, Fukushima T, Sugahara K, Tsuruda K, Anami M, Onimaru Y et al. , A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int. J. Hematol 2005. 81: 148–154. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Sun L, Chen X, Du F, Shi H, Chen C and Chen ZJ, Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013. 339: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP et al. , cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 2013. 498: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A and Meraro D, ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J. Interferon Cytokine Res 2002. 22: 153–160. [DOI] [PubMed] [Google Scholar]
- 20.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 2006. 3: 995–1000. [DOI] [PubMed] [Google Scholar]
- 21.Haining WN, Davies J, Kanzler H, Drury L, Brenn T, Evans J, Angelosanto J et al. , CpG oligodeoxynucleotides alter lymphocyte and dendritic cell trafficking in humans. Clin. Cancer Res 2008. 14: 5626–5634. [DOI] [PubMed] [Google Scholar]
- 22.Halperin SA, Ward B, Cooper C, Predy G, az-Mitoma F, Dionne M, Embree J et al. , Comparison of safety and immunogenicity of two doses of investigational hepatitis B virus surface antigen co-administered with an immunostimulatory phosphorothioate oligodeoxyribonucleotide and three doses of a licensed hepatitis B vaccine in healthy adults 18–55 years of age. Vaccine 2012. 30: 2556–2563. [DOI] [PubMed] [Google Scholar]
- 23.Rynkiewicz D, Rathkopf M, Sim I, Waytes AT, Hopkins RJ, Giri L, DeMuria D et al. , Marked enhancement of the immune response to BioThrax(R) (Anthrax Vaccine Adsorbed) by the TLR9 agonist CPG 7909 in healthy volunteers. Vaccine 2011. 29: 6313–6320. [DOI] [PubMed] [Google Scholar]
- 24.Steinhagen F, Kinjo T, Bode C and Klinman DM, TLR-based immune adjuvants. Vaccine 2011. 29: 3341–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paun A and Pitha PM, The IRF family, revisited. Biochimie 2007. 89: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L et al. , The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci 2000. 97: 13766–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S et al. , Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 2005. 434: 243–249. [DOI] [PubMed] [Google Scholar]
- 28.Paun A, Reinert JT, Jiang Z, Medin C, Balkhi MY, Fitzgerald KA and Pitha PM, Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J. Biol. Chem 2008. 283: 14295–14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purtha WE, Swiecki M, Colonna M, Diamond MS and Bhattacharya D, Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc. Natl. Acad. Sci. USA 2012. 109: E898–E904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda K, Nundel K, Watkins AA, Dhawan T, Bonegio RG, Ubellacker JM, Marshak-Rothstein A et al. , Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int. Immunol 2013. 25: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sica A, Schioppa T, Mantovani A and Allavena P, Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur. J. Cancer 2006. 42: 717–727. [DOI] [PubMed] [Google Scholar]
- 32.Li P, Wong JJ, Sum C, Sin WX, Ng KQ, Koh MB and Chin KC, IRF8 and IRF3 cooperatively regulate rapid interferon-beta induction in human blood monocytes. Blood 2011. 117: 2847–2854. [DOI] [PubMed] [Google Scholar]
- 33.Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM and Ozato K, Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J. Immunol 2004. 172: 6820–6827. [DOI] [PubMed] [Google Scholar]
- 34.Sharf R, Azriel A, Lejbkowicz F, Winograd SS, Ehrlich R and Levi BZ, Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J. Biol. Chem 1995. 270: 13063–13069. [DOI] [PubMed] [Google Scholar]
- 35.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander LB, Khan AA et al. , A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 2012. 338: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, KC W, Albring JC, Satpathy AT, Rotondo JA et al. , Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012. 490: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kono DH, Baccala R and Theofilopoulos AN, TLRs and interferons: a central paradigm in autoimmunity. Curr. Opin. Immunol 2013. 25: 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Kong HJ, Li H, Huang B, Yang M, Zhu C, Bognunvic M et al. , IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the crosstalk between TLR and IFN-gamma signaling pathways. J. Biol. Chem 2006. 15:10073–10080. [DOI] [PubMed] [Google Scholar]
- 39.Rowland SL, Riggs JM, Gilfillan S, Bugatti M, Vermi W, Kolbeck R, Unanue ER et al. , Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med 2014. 211: 1977–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baccala R, Gonzalez-Quintial R, Blasius AL, Rimann I, Ozato K, Kono DH, Beutler B et al. , Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc. Natl. Acad. Sci. USA 2013. 110: 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaschik S, Tross D and Klinman DM, Inductive and suppressive networks regulate TLR9-dependent gene expression in vivo. J. Leuk. Biol 2009. 85: 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright GW and Simon RM, A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003. 19: 2448–2455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.