Abstract
Immune cells constitute a large fraction of the tumor microenvironment and modulate tumor progression. Clinical data indicate that chronic inflammation is present at tumor sites and that IL4 in particular is upregulated. Here, we demonstrate that T follicular helper (Tfh) cells arise in tumor-draining lymph nodes where they produce an abundance of IL4. Deletion of IL4-expressing Tfh cells improves antitumor immunity, delays tumor growth, and reduces the generation of immunosuppressive myeloid cells in the lymph nodes. These findings suggest that IL4 from Tfh cells affects antitumor immunity and constitutes an attractive therapeutic target to reduce immunosuppression in the tumor microenvironment, and thus enhance the efficacy of cancer immunotherapy.
Introduction
Lymph nodes constitute an essential element of the immune system. They contain T and B lymphocytes and antigen-presenting cells (APC) that play key roles in supporting the host response to foreign antigens (Ag) and tumors (1, 2). Dendritic cells are APCs that come in contact with tumor-associated Ags in the periphery, then migrate to draining lymph nodes where they contribute to the priming and activation of an effector T-cell response (3–5). Conversely, tumors can escape immune surveillance by supporting the generation of an immunosuppressive response in the draining lymph nodes (2, 6–8). Although draining lymph nodes are critical sites for the generation of immune responses that determine whether tumors are tolerated or eradicated, relatively few studies have analyzed the responses generated within tumor-draining lymph nodes.
CD4 T cells orchestrate a broad range of acquired immune responses and can differentiate into multiple T-cell subsets (9, 10). CD4 T cells contribute to shaping tumor-specific immunity. For example, Th1 cells can exert potent antitumor immunity by overcoming tolerance to self Ags expressed by the tumor (11–13). Harnessing these effector T cells would therefore support cancer immunotherapy. On the other hand, certain CD4 T-cell subsets, particularly regulatory T cells, suppress antitumor immunity and thus promote cancer growth (2, 14, 15). This activity reflects the importance of maintaining immune homeostasis and self-tolerance without which auto-immunity and pathologic inflammation could result (16, 17). Identifying and targeting the CD4 T cells that contribute to the inflammation and immune suppression that support tumor growth represents an important step toward improving anti-tumor immunity.
Increased IL4 is commonly detected in primary and metastatic cancers of animals and humans. Although some believe that this IL4 is produced by Th2 cells in the tumor microenvironment, its precise source and role is poorly understood. Our study initially sought to detect the changes in gene expression associated with CD4 T-cell responses in the tumor micro-environment. Consistent with earlier work, IL4 expression increased shortly after cancer cell challenge. Follicular helper CD4 T (Tfh) cells expressing IL21, BCL6, ICOS, PD-1, and CXCR5 proved to be the source of this IL4. IL4 from these Tfh cells induced myeloid cells to differentiate into M2 macrophages. Supporting the importance of this cell type, our studies using CNS2-deleted mice, in which IL4 production by Tfh cells was impaired, found enhanced antitumor immunity and delayed tumor growth. These results establish the important contribution of Tfh cells to the host’s response to tumors.
Materials and Methods
Animals and tumor cell lines
BALB/c and C57Bl/6 mice were obtained from the National Cancer Institute (Frederick, MD) or Japan SLC (Hamamatsu, Japan) and studied at 6 to 10 weeks of age. IL4/GFP–enhanced transcript (4GET; C.129-Il4tm1Lky/J), CD11c-DTR/EGFP, RAG1, and CD1d knockout (KO) mice were obtained from The Jackson Laboratory. Ja18 KO mice were provided by Cui and colleagues (18). CNS2 KO mice were provided by Harada and colleagues (19). BALB-neuT mice expressing the rat neu oncogene under the control of a chimeric mouse mammary tumor virus (MMTV) promoter were provided by Sakai and colleagues (20). All studies were approved by the NCI Frederick Animal Care and Use Committee (ACUC) or the Institutional Committee for the Use and Care of Laboratory Animals of Tohoku University. The following cell lines were purchased from the ATCC in 2011 and 2012: TC-1, which is a lung epithelial tumor cell line that expresses the E7 oncoprotein from human papillomavirus 16; 4T1, which is a breast cancer cell line; CT26, which is a colon cancer cell line. MC38, which is a colon cancer cell line, was kindly provided by G. Trinchieri (NCI, Frederick, MD) in 2012. These cell lines were used at the third or fourth passage. Authentications were not made.
In vivo tumor studies
All in vivo experiments with CNS2 KO mice were conducted using respective age- and sex-matched littermate wild-type (WT) or CNS2 heterozygous (HT) progeny as controls. Mice were injected subcutaneously with viable tumor cells (the number of cells varied with the tumor type as described in the figure legends). Tumor size was calculated by the formula: (length × width × height)/2 (21). Tumor growth curves were generated from 3 to 5 mice per group, and all results were derived by combining data from two to three independent experiments. Any animal whose tumor exceeded a diameter of 2.0 cm was immediately euthanized as per ACUC protocol. To deplete CD4+ or CD8+ cells, mice were injected intraperitoneally with 500 μg rat antibody to mouse CD8 (53.6.72) or to mouse CD4 (GK1.5) from BioXCell. These mAbs were delivered intraperitoneally on day 1 after tumor challenge.
RT2 profiler PCR array
The specific gene expression of Th cell–related inflammatory response was evaluated using a Mouse Th1 and Th2 Responses RT2 Profiler PCR Array (Qiagen), which contained primers for the detection of 84 different known Th cell–related genes. This analysis was carried out according to the manufacturer’s instructions. Briefly, total RNA was reverse transcribed to cDNA using the RT2 First Stand Kit (Qiagen) followed by combination with the RT2 qPCR Mastermix. This volume was evenly distributed among two PCR array plates. Consecutive rounds of qPCR were performed under normal thermal conditions followed by data analysis according to the abovementioned Ct method post-normalized to five independent controls.
Flow cytometry
Cells were washed with PBS, fixed in 4% paraformaldehyde for 10 minutes, and stained with fluorochrome-conjugated mAbs for 30 minutes at 4°C. Fluorochrome-conjugated CD45, CD3, CD4, CD8, and PD-1 mAbs were purchased from BD Pharmingen. Fluorochrome-conjugated CD62L, CD44, TCRβ, CXCR5, and ICOS mAbs were purchased from Biolegend.
Stained cells were washed, re-suspended in PBS/0.1% BSA plus azide, and analyzed by FACSCalibur (BD Pharmingen).
Cell preparation
Fresh lymph node cells were stained with fluorochrome-conjugated mAbs and then sorted by FACS to isolate IL4-expressing CD4 T cells (CD3+, CD4+, eGFP+), naïve CD4 T cells (CD3+, CD4+, CD62L+, CD44−). CD11b+ cells were purified from lymph nodes by magnetic cell sorting system (MACS) according to the manufacturer’s instructions (Miltenyi Biotec). The purity of CD11b+ cells by this method was 90% to 95% as determined by flow cytometry. Th2 cells were generated by stimulating with recombinant IL4 (10 ng/mL; R&D Systems), and mAbs to IFNγ (10 μg/mL; BD Pharmingen), CD3 (0.1 μg/mL; BD Pharmingen), and CD28 (0.1 μg/mL; BD Pharmingen) in vitro for 3 days. The cells were cultured in fresh medium for another 3 days (22).
Histology
Lymph node cells were flash-frozen, sectioned in a cryostat (Histoserv). Sections were fixed in 0.3% H2O2 methanol, then stained with mAb to B220 and biotinylated peanut agglutinin (PNA), followed by goat anti-rabbit IgG Alexa 594 (Life Technologies Inc.) and streptavidin Alexa 647 (BD Biosciences) or Alexa 594–conjugated anti-CD4 and biotinylated anti-B220, followed by streptavidin Alexa 647. Sections were visualized and photographed with a Zeiss LSM510 Meta confocal microscope (original magnification × 20) at room temperature, and images were acquired with Zeiss LSM image browser software (Zeiss). Germinal centers analysis was performed by immunostaining using biotinylated PNA (Vector Laboratories) plus streptavidin–HRP and diaminobenzidine substrate (DAB Substrate Kit; BD Biosciences).
RT-PCR and quantitative RT-PCR
Total RNA was extracted from target cells using TRIzol reagent (Life Technologies) as recommended by the manufacturer. One μg of total RNA was reverse-transcribed in first strand buffer (50 mmol/L Tris-HCl, pH 7.5, 75 mmol/L KCl, and 25 mmol/L MgCl2), containing 25 μg/mL oligo-(dT), 200 U Moloney leukemia virus reverse-transcriptase, 2 mmol/L dinucleotide triphosphate, and 10 mmol/L dithiothreitol. The reaction was conducted at 42°C for 1 hour. The expressions of mRNA levels were examined using the Applied Biosystems StepOne RT-PCR system, in which primers obtained from the Gene Expression Assay set (Applied Biosystems) were amplified using the TaqMan Gene Expression Master Mix Kit. All primers used for quantitative RTPCR analysis were purchased from Applied Biosystems. mRNA expression levels were then calculated by step-one software (Applied Biosystems) after correction for GAPDH expression independently for each sample.
ELISPOT assay
Single-cell suspensions were prepared from lymph node. HPV 16 E7(49–57) peptide RAHYNIVTF (H-2 Db) was used for the stimulation of CD8+ T cells. A total of 1.5–3.0 × 105cells per well were stimulated for 12 to 14 hours with 0.1 μg/mL of E7 peptide in 96-well Immulon II plates (Millipore) previously coated with monoclonal rat anti-IFNγ (R4–6A2; BD Biosciences). The plates were washed and treated with biotinylated polyclonal goat anti-IFNγ (R&D Systems) followed by streptavidin alkaline phosphatase. Spots were visualized by the addition of a 5-bromo-4-chloro-3-indolyl phosphatase solution (Sigma Aldrich) in low melt agarose (Sigma-Aldrich) and counted manually under × 40 magnification. The number of cytokine-secreting cells was determined by a single blind reader, and all data were generated by analyzing three separate wells per sample.
Statistical analysis
A two-sided unpaired Student t test was used to analyze tumor growth and cellular responses. t tests were performed in Excel software for statistical analysis with P < 0.05 considered to be statistically significant.
Results
IL4 expression is increased in tumor-draining lymph nodes
Previous studies established that the host’s immune system contributes to development of the tumor microenvironment (23–26). To evaluate changes in gene expression associated with T-cell responses after tumor inoculation, BALB/c mice were injected subcutaneously in the right flank with CT26 colon cancer cells. Tumor draining subiliac lymph nodes were removed 4 days later and the expression profile of 84 genes involved in Th cell–related inflammatory responses was monitored by PCR array. Expression of 16 genes was increased significantly (>2-fold) in this gene set, including IFNγ, IL5, IL10, IL13, IL17, and TGFβ, whereas expression of 5 genes was decreased (<0.5-fold; Table 1).
Table 1.
Increased and decreased expression of helper T-cell–related genes in tumor-draining lymph nodes compared with naïve lymph nodes
| Name of gene | Symbol | Fold change | SD | P |
|---|---|---|---|---|
| Increased expressiona | ||||
| Interleukin 4 | Il4 | 76.54 | 36.10 | 0.022267 |
| Suppressor of cytokine signaling 3 | Socs3 | 4.86 | 1.39 | 0.008650 |
| Inducible T-cell co-stimulator | Icos | 3.29 | 0.12 | 0.000004 |
| Tumor necrosis factor superfamily, member 4 | Tnfsf4 | 3.43 | 0.16 | 0.000014 |
| Interleukin 6 | Il6 | 3.20 | 1.52 | 0.040122 |
| Chemokine (C-C motif) receptor 3 | Ccr3 | 2.92 | 0.88 | 0.019473 |
| Suppressor of cytokine signaling 1 | Socs1 | 2.85 | 0.34 | 0.000689 |
| Interleukin 2 | Il2 | 2.79 | 0.54 | 0.004394 |
| Janus kinase 2 | Jak2 | 2.78 | 0.67 | 0.010155 |
| Interleukin 12b | Il12b | 2.74 | 0.50 | 0.003800 |
| Interleukin 18 | Il18 | 2.43 | 0.68 | 0.021357 |
| Chemokine (C-C motif) receptor 5 | Ccr5 | 2.40 | 0.57 | 0.013410 |
| Chemokine (C-C motif) receptor 4 | Ccr4 | 2.35 | 0.33 | 0.002088 |
| Cytotoxic T-lymphocyte-associated protein 4 | Ctla4 | 2.33 | 0.65 | 0.024373 |
| B-cell CLL/lymphoma 6 | Bcl6 | 2.26 | 0.14 | 0.000105 |
| Interleukin 2 receptor, alpha | Il2ra | 2.00 | 0.47 | 0.021042 |
| Decreased expressionb | ||||
| Inhibin alpha | Inha | 0.217 | 0.03 | 0.000004 |
| Polycomb group ring finger 2 | Pcgf2 | 0.409 | 0.11 | 0.000899 |
| Interleukin 23, alpha subunit p19 | Il23a | 0.433 | 0.34 | 0.046563 |
| Interleukin 17A | Il17a | 0.449 | 0.19 | 0.00837 |
| Chemokine (C-C motif) ligand 11 | Ccl11 | 0.456 | 0.09 | 0.000618 |
NOTE: A total of 2.5 × 105 CT26 colon cancer cells were injected subcutaneously into the right flank of syngeneic mice. Tumor-draining lymph nodes were removed on day 4 and analyzed gene expressions by RT2 Profiler PCR array. Data show the mean fold change + SD from three independent lymph nodes.
A total of 16 genes showed increased expression of 2.0-fold or more.
A total of 5 genes showed decreased expression of 0.5-fold or less.
IL4 expression in the draining subiliac lymph nodes was enhanced by 76-fold, which far exceeded changes in expression of all other genes. A similar but more modest increase was detected in the axillary draining lymph nodes (Supplementary Fig. S1A and S1B). No such effect was observed in the subiliac or axillary lymph nodes removed from control mice injected with PBS rather than tumor cells. Moreover, contralateral lymph nodes showed no change in IL4 expression (Supplementary Fig. S1B).
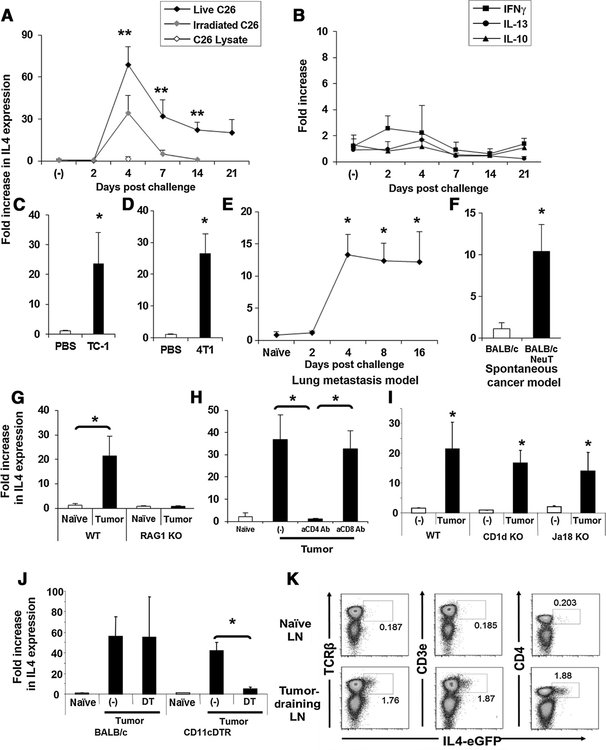
These findings led us to focus on IL4 expression in tumor-draining lymph nodes. The kinetics defining the development of the IL4 response was examined. IL4 expression remained at baseline through day 2 after subcutaneous CT26 challenge, rose rapidly to peak on day 4, and persisted at >20-fold baseline for at least for 3 weeks (Fig. 1A). Increased expression of IL4 was observed when mice were challenged with either live or apoptotic (lethally irradiated) CT26 cells, although the magnitude and persistence of IL4 production was reduced (falling to baseline at day 7) when dead cells were injected (Fig. 1A). However, when tumor cell lysates (prepared by repeated freeze/thaw and sonication of CT26 cells) were administered, no change in IL4 production was observed. Consistent with PCR array data, cytokines whose production is associated with the activation of helper T cells, including the Th2 cytokines IL10 and IL13, the Th1 cytokine IFNγ, and the Th17 cytokine IL17 were not upregulated by tumor challenge (Fig. 1B). Multiple tumor types (including TC-1, 4T1, and MC38) strongly induced IL4 expression in the tumor-draining lymph nodes by day 7 (Fig. 1C, D and G). ELISPOT assays confirmed that the number of IL4-producing cells in these lymph nodes significantly increased (Supplementary Fig. S1C).
Figure 1.
Increased IL4 expression in tumor-draining lymph nodes (LN). A and B, 2.5 × 105 live or irradiated CT26 colon cancer cells or CT26 cell lysate were injected subcutaneously. Tumor-draining lymph nodes were removed as indicated and analyzed for IL4, IFNγ, IL10, and IL13 mRNA expression. C and D, 2.5 × 105 TC-1 cervical or 4T1 breast cancer cells were injected subcutaneously into the right flank of syngeneic mice and IL4 expression examined 7 days later. E, 2.5 × 105 4T1 cells were injected intravenously and IL4 mRNA levels examined over 16 days. F, Tumor-draining lymph nodes from 4-month-old BALB-neuT mice were removed and analyzed for IL4 mRNA. A total of 2.5 × 105 MC38 (G, I) or CT26 (H, J, K) colon cancer cells were injected subcutaneously into C57BL/6, RAG1 KO, BALB/c 4GET, CD1d KO, Jα18 KO, or CD11c-DTR/EGFP mice. H, BALB/c mice were also injected intraperitoneally with 500 μg of depleting antibodies to CD4 or CD8 on day 1. Tumor-draining lymph nodes were removed on day 7 and analyzed for IL4 mRNA levels (G–J). J, CT26 colon cancer cells were injected subcutaneously into CD11-DTR or BALB/c mice that were injected the next day with 100 ng of diphtheria toxin (DT). Tumor-draining lymph nodes were removed and analyzed for IL4 mRNA (J). All results represent the mean + SD of results from 4 to 8 independent lymph nodes. Experiments were repeated two or three times with similar results. K, Naïve or tumor-draining lymph nodes from 4GET mice were removed on day 7 and analyzed for the expression of eGFP and TCRβ, CD3ε, or CD4 by flow cytometry. *, P < 0.01 when compared with PBS-treated or naïve group. **, P < 0.01 when compared with irradiated tumor cell–treated group.
To determine whether this effect was limited to inguinal lymph nodes after subcutaneous tumor delivery, 4T1 cells were injected intravenously rather than subcutaneous administration. This resulted in the development of multiple tumors in the lung. Bronchial lymph nodes from these animals also showed significantly increased IL4 expression at days 4 to 16 (Fig. 1E). Indeed, similar findings were observed in studies of spontaneous breast cancer. BALB-neuT transgenic mice express an activated HER2/neu oncogene in mammary epithelial cells (20). These animals consistently develop carcinoma in situ at approximately 4 months of age. Draining lymph nodes (axillary lymph nodes) from these animals showed significantly increased IL4 expression by 4 to 5 months of age (Fig. 1F). Thus, elevated IL4 expression was consistently found in the draining lymph nodes of tumors evaluated in a wide variety of disparate cancer models.
IL4-secreting cells in the tumor-draining lymph nodes are CD4 T cells
Multiple immune cells including T, NK, NKT, eosinophils, and mast cells are capable of producing IL4 (27). Experiments were conducted to establish which cell type was the source of IL4 in the tumor-draining lymph nodes. RAG1 KO mice lack T and B cells, and no IL4 was detected in their draining lymph nodes after tumor challenge (Fig. 1G). Consistent with the IL4-secreting cells being CD4-positive T cells, administration of anti-CD4 (but not anti-CD8) depleting Ab completely abolished IL4 expression (Fig. 1H).
Previous studies suggested that NKT cells could be an important source of IL4 and IL13 in the tumor microenvironment (28). However, CD1d and Ja18 KO mice (which lack NKT cells) showed normal IL4 production following tumor challenge (Fig. 1I). If CD4 T cells were responsible for producing IL4 in response to tumor, the process might require antigen presentation by dendritic cells. To investigate this possibility, CD11c-DTR mice, in which the diphtheria toxin receptor is under control of the CD11c promoter, were treated with DT to deplete dendritic cells. When challenged with tumor, the expression of IL4 by these animals was significantly reduced when compared with similarly challenged mice that were not treated with DT (Fig. 1J). Taken together, these findings suggest that CD4 T cells in the draining lymph nodes produce IL4 in response to tumor Ags presented by dendritic cells.
This cellular source of IL4 was confirmed in studies using IL4 reporter mice, which are IL4/GFP-enhanced transcript (4GET) mice. These mice have an enhanced GFP gene inserted at the 3′-untranslated region of their endogenous IL4 locus, such that GFP accumulates within cells that upregulate expression of IL4 mRNA (29). Consistent with previous reports, very few lymph nodes cells in naive 4GET mice are GFP+ (<0.4%; ref. 29). Following tumor cell challenge, the frequency of such cells increases >9-fold in the draining lymph nodes where the GFP/IL4–expressing cells were identified as TCRβ, CD3ε, and CD4 positive (Fig. 1K).
IL4-secreting cells in tumor-draining lymph nodes are T follicular helper cells
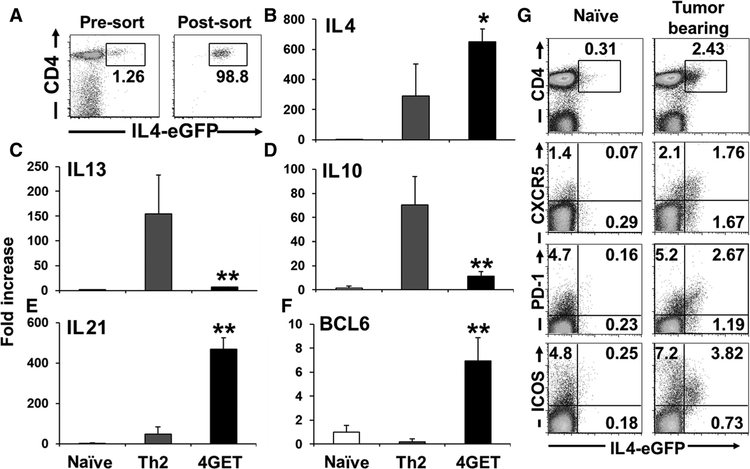
To further characterize this IL4-expressing population, 4GET cells were sorted by flow cytometry (Fig. 2A) and analyzed for gene expression by quantitative RT-PCR. They differed from both in vitro–generated Th2 cells and naïve CD4 T cells (CD62L+ and CD44−). Naïve CD4 T cells did not express IL4, whereas Th2 cells expressed IL10 and IL13 in addition to IL4. In comparison, 4GET cells expressed IL4 and IL21, but not IL10 or IL13 (Fig. 2B–E). Tfh cells produce IL4 and IL21 and express the BCL6 transcription factor (19, 30, 31). CD4 T cells isolated from the draining lymph nodes of tumor-challenged 4GET mice upregulated IL21 and BCL6 (Fig. 2E and F), whereas expression of IL21 was increased in the subcutaneous tumor model, lung metastasis model, and spontaneous tumor model (Supplementary Fig. S1D–S1F). These 4GET CD4 T cells did not express the Th2 marker GATA3 or the Treg marker FoxP3 (Supplementary Fig. S2A and S2B).
Figure 2.
IL4-producing cells in the draining lymph nodes (LN) had a Tfh phenotype. A, Tumor-draining lymph nodes of 4GET mice were removed on day 7 and FACS sorted on the basis of their expression of CD3ε, CD4, and IL4-eGFP. B–F, Sorted 4GET cells were analyzed for mRNA expression. Naïve CD4 T cells were sorted on the basis of their expression of CD62L+ and CD44−. Th2 cells were generated by stimulating with IL4, anti-IFNγ and anti-CD3 and anti-CD28 in vitro for 4 days. Results represent the mean + SD of results from 5 to 9 independent sorted cell populations. *, P < 0.05 when compared with Th2 cells. **, P < 0.01 when compared with Th2 cells. G, Naïve or tumor-draining lymph nodes cells isolated from 4GET mice were removed 7 days after tumor inoculation. The expression of eGFP and CD4 in CD3+ cells, or eGFP and CXCR5, PD-1, or ICOS in CD4+ cells were analyzed by flow cytometry. Experiments were repeated three times with similar results.
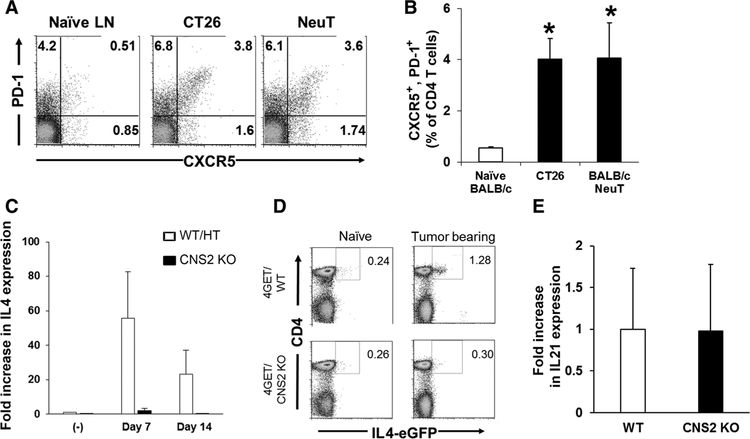
To confirm that the IL4-secreting cells isolated from tumor-draining lymph nodes were Tfh cells, we examined their expression of ICOS, PD-1, and CXCR5 (30, 31). IL4+/CD4+ T cells isolated from tumor-draining lymph nodes expressed all of these Tfh surface markers (Fig. 2G). Kinetic studies showed that the number of IL4+/CD4+ T cells expressing these markers was elevated by day 4 and peaked on day 7 after tumor challenge (Supplementary Fig. S3A). Consistent with this finding, PD-1+/CXCR5+ Tfh cells increased significantly in tumor-draining lymph nodes over this period in the subcutaneous tumor model and spontaneous tumor model (Fig. 3A and B). These results indicate that Tfh cells are increased in tumor-draining lymph nodes and a majority of the CD4 T cells expressing IL4 in lymph nodes are phenotypically Tfh cells.
Figure 3.
Increased Tfh cells and IL4 expression in tumor-draining lymph nodes (LN) of WT and CNS2 KO mice. A and B, lymph nodes were isolated as described in Fig. 1A and F and analyzed for the expression of PD-1 and CXCR5 in CD4+ cells. A, Representative results from one mouse per group and (B) mean + SD from 5 to 8 independently analyzed mice per group, showing PD-1+ and CXCR5+ cells of CD4+ cells. *, P < 0.01 when compared with naïve group. C and D,+2.5 × 105 C26 colon cancer cells were injected subcutaneously into 4GET/CNS2 KO, wild-type (WT) or CNS2 heterozygous (HT) control litter mates. Tumor-draining lymph nodes were removed and analyzed for IL4 mRNA expression at the times indicated (C) and IL21 mRNA expression on day 7 (E). Results represent the mean + SD of five independent lymph nodes. D, Naïve or tumor-draining lymph nodes were removed from 4GET or 4GET/CNS2 KO mice and analyzed for expression of CD4 and eGFP in CD3 gated cells by flow cytometry. Experiments were repeated five times with similar results.
As further evidence that IL4 expression in tumor-draining lymph nodes is derived from Tfh cells, conserved noncoding sequence 2 (CNS2) KO mice were examined. CNS2 is an essential enhancer element for IL4 expression in Tfh, but not Th2, cells (19). Studies of CNS2 KO mice demonstrated that Tfh cells were normal in terms of development and expression of BCL6 and IL21, but that their production of IL4 was impaired (19). We therefore injected CNS2 KO mice with tumor cells and examined the response in their draining lymph nodes. Consistent with expectations, IL4 expression in tumor-draining lymph nodes of CNS2 KO mice was severely impaired (Fig. 3C and D), whereas the frequency of Tfh (CD4+, PD-1+, and CXCR5+) cells and IL21 expression in tumor-draining lymph nodes of CNS2 KO mice were similar (Fig. 3E). These findings all support the conclusion that Tfh cells are primarily responsible for the expression of IL4 in tumor-draining lymph nodes.
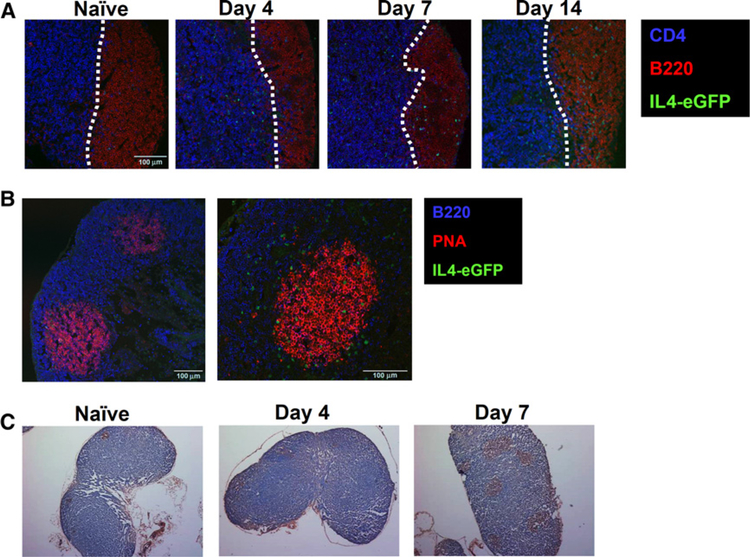
Tfh cells typically localize to B-cell follicles within the lymph nodes as this is a prerequisite for their interaction with B cells (30, 31). To explore whether the IL4+/CD4+ T cells in tumor-draining lymph nodes showed such localization, lymph nodes from tumor-challenged 4GET mice were analyzed immunohistochemically. IL4+/GFP+ T cells were virtually undetectable in naïve mice (Fig. 4A). IL4-secreting cells could not be detected in B-cell follicles until day 7 where they persisted through day 14 (Fig. 4A and B).
Figure 4.
IL4-expressing cells localized to the germinal center of tumor-draining lymph nodes (LN). Confocal imaging of tumor-draining lymph nodes from 4GET mice. A, Time course analysis showing CD4 (blue), B220 (red) and IL4-eGFP (green). B, Analysis on day 14 showing B220 (blue), PNA (red), and IL4-eGFP (green). C, Section of tumor-draining lymph nodes at indicated days stained with PNA (brown). Experiments were repeated five times with similar results.
A principal function of Tfh cells is to interact with B cells and promote germinal center formation (30, 31). Sections stained with peanut agglutinin (PNA) showed that it took until day 7 post-challenge for the number and size of germinal centers in the tumor-draining lymph nodes to increase, concomitant with an influx of IL4-expressing cells (Fig. 4B and C). Consistent with this finding, the frequency of PNA+ B cells increased significantly in tumor-draining lymph nodes over this period. These data suggest that Tfh cells activated in the tumor-draining lymph nodes induce B-cell expansion and activation/maturation.
Effect of IL4 from Tfh cells on tumor growth and the tumor microenvironment
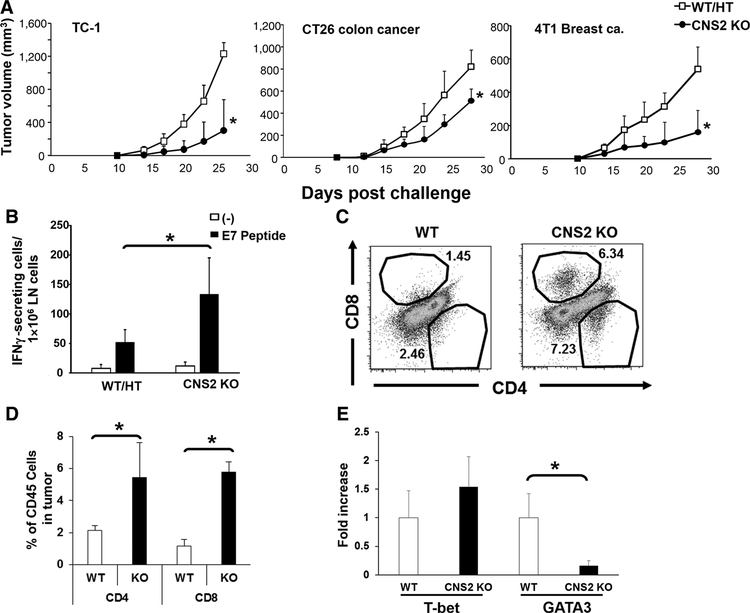
To examine the role of IL4-expressing Tfh cells on tumor growth, various tumor cell lines were inoculated into either CNS2 KO or control litter mates (WT/HT mice). The growth of TC-1, CT26, and 4T1 tumor cells was significantly reduced in the CNS2 KO mice (Fig. 5A). To clarify the mechanism underlying this effect, single-cell suspensions were prepared from the draining lymph nodes of TC-1 and CT26 tumor-bearing CNS2 KO mice. These cells were then stimulated ex vivo with their cognate tumor Ag (human papillomavirus E7 protein for TC-1 and AH1 tumor Ag for CT26), and analyzed by ELISPOT for IFNγ production. The number of IFNγ-producing cells in CNS2 KO mice was significantly higher than in controls after challenge with TC-1 or CT26 tumors (P < 0.05, Fig. 5B and unpublished observation). Consistent with the enhancement in antitumor immunity observed in CNS2 KO mice, the fraction of CD4 and CD8 T cells infiltrating their tumors was significantly increased, although the total number of CD45+ cells infiltrating the tumor bed was similar (Fig. 5C and D and unpublished observation). Tumor-infiltrating CD4 T cells from these animals were sorted by flow cytometry and analyzed for gene expression by quantitative RT-PCR. Expression of the Th1 cell marker T-bet did not change, whereas the Th2 cell marker GATA3 expression fell significantly in CNS2 KO mice (Fig. 5E), such that the Th1/Th2 balance was shifted in favor of Th1 immunity. Tumor-infiltrated CD4 T cells did not express CXCR5 demonstrating that they were not Tfh cells (Supplementary Fig. S3B). Thus, tumor specific IFNγ responses and tumor infiltrate T cells were increased and tumor growth delayed in the absence of IL4 producing Tfh cells.
Figure 5.
Tumor growth and tumor-infiltrating T cells in CNS2 KO mice. A, 105 TC-1, CT26, or 4T1 cancer cells were injected subcutaneously into WT/CNS2 HT or CNS2 KO mice. Data represent the combined means + SD of 10–16 mice/group from two to three independent experiments. *, P < 0.01 when compared with WT/HT mice. B, 105 TC-1 cells were injected subcutaneously into WT/CNS2 HT or CNS2 KO mice. Tumor draining lymph nodes (LN) were removed on day 21 and the cells stimulated in vitro with 0.1 μg of E7 peptide. Results show the number of IFNγ-producing cells quantified by ELISPOT. Results were evaluated independently in each mouse, and data represent the mean + SD of 5 mice per group. *, P < 0.05. C and D, CT26 tumors were removed at day 21 and the number of tumor-infiltrating CD45+ cells that expressed CD4 and CD8 determined by FACS. C, Representative results from one mouse per group and (D) mean + SD from 5 independently analyzed mice per group, showing CD4- and CD8-expressing cells as a percentage of all tumor-infiltrating CD45+ lymphocytes. E, CD4 or CD8 T cells were sorted by FACS and analyzed for T-bet and GATA3 mRNA expression. Results represent the mean + SD of results from four independent sorted cell populations; *, P < 0.05.
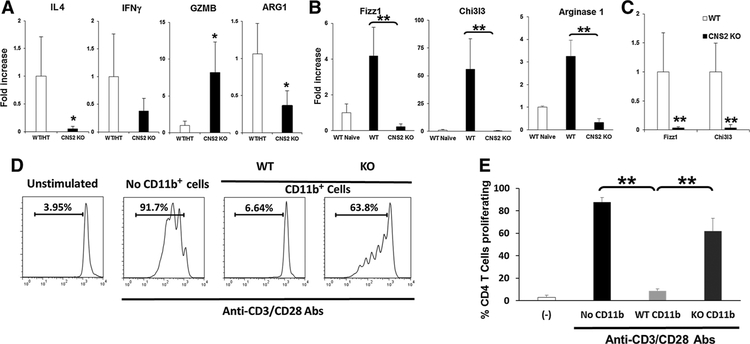
Increased IL4 has been reported at the tumor sites of patients with various cancers, such as colon cancer, renal cell cancer, lung cancer, breast cancer, melanoma, gastric cancer, and other type of tumor (32–38). To examine the effect of IL4 from Tfh cells in the tumor microenvironment, the pattern of cytokine production (expression of IL4 and IFNγ) was evaluated by quantitative RT-PCR. Increased IL4 expression was detected in tumors of WT mice but was severely impaired in tumors from CNS2 KO mice (Fig. 6A). Although IFNγ was decreased in the CNS2 tumor microenvironment, that difference did not reach statistical significance. However, granzyme B (GZMB) was significantly increased in tumors from CNS2 KO mice (Fig. 6A).
Figure 6.
Altered gene expression and suppressive function of myeloid cells in CNS2 KO mice. A, 105 CT26 cells were injected subcutaneously into WT/CNS2 HT or CNS2 KO mice. CT26 tumors were removed at day 21 and analyzed for indicated mRNA by real time qPCR. B, CD11b+ myeloid cells from lymph nodes (LN) of CT26 tumor bearing mice were isolated by MACS and analyzed for Fizz1, Chi3l3, and ARG-1 mRNA by real time qPCR. Mean SD, from 6 to 8 independently analyzed mice per group, are shown. C, Tumors were removed on day 25 and sorted by FACS based on their expression of CD45+, CD11b+, Ly6c−, and Ly6g−. Sorted macrophages were analyzed for mRNA expression. D and E, CD11b+ cells from the spleen of CT26 tumor-bearing WT or CNS2 KO mice were isolated on day 25 post-tumor challenge by MACS (final purity, 90%–95%). A total of 5 × 105 cells were cultured for 4 days with 2.5 × 105 CFSE-labeled syngeneic CD4 T cells in the presence or absence of 0.1 mg/mL anti-CD3/anti-CD28. T-cell proliferation was monitored by CFSE dilution. D, Representative example and (E) mean SD (n = 4 independent CD11b+ cells preparations in three independent experiments) are shown; *, P < 0.05. **, P < 0.01.
IL4 has been reported to affect CD11b+ myeloid cells in the tumor microenvironment, such as monocytic MDSC (mMDSC) and TAMs (23, 39–41). They exert their immunosuppressive effects via arginase-1 production (23, 39, 40). Indeed, arginase-1 was reduced in tumors from CNS2 KO mice (Fig. 6A). TAMs express markers similar to M2 macrophages [including Fizz-1 (Retnla) and Chi3l3; ref. 39, 40)]. When CD11b+ cells from lymph nodes were sorted by MACS and the expression of these markers evaluated by quantitative RT-PCR, Fizz1, Chi3l3, and arginase-1 were significantly increased in tumor-bearing WT mice compared with naïve mice and severely reduced in CNS2 KO mice (Fig. 6B, P < 0.01). Tumor-infiltrating macrophages (CD45+, CD11b+, Ly6c−, Ly6g−) were sorted by flow cytometry and analyzed for gene expression by quantitative RT-PCR. Consistent with the above findings, Fizz1, Chi3l3, and arginase-1 mRNA in macrophages was significantly reduced in CNS2 KO mice (Fig. 6C).
To examine the effect of IL4 from Tfh cells on macrophage, CD11b+ cells were isolated from the spleens of tumor-bearing WT or CNS2 KO mice (sufficient numbers of CD11b+ cells could not be obtained from tumor-draining lymph nodes). These CD11b+ cells were mixed with CD8+ T cells isolated from tumor-free syngeneic mice that had been stimulated to proliferate using anti-CD3/CD28 Abs (Fig. 6D and E). Their proliferation was severely reduced when CD11b+ cells from tumor-bearing WT mice were added to the culture whereas CD11b+ cells from CNS2 KO mice did not suppress T-cell proliferation (Fig. 6D and E). These findings suggest that the generation of immunosuppressive myeloid cells is significantly reduced when IL4 production from Tfh cells are depleted.
Discussion
Draining lymph nodes are the site where APC-carrying tumor Ags initiate an adaptive immune response. We found that this was associated with a dramatic elevation in IL4 mRNA. IL4-expressing cells exhibited a phenotype characteristic of Tfh cells: they expressed IL21, BCL6, CXCR5, PD-1, and ICOS and localize to germinal centers. The number of Tfh cells increased in tumor-draining lymph nodes and these localized to germinal centers where they drive the proliferation of B-cell follicles. CNS2 KO mice are selectively deficient in IL4 expression by Tfh cells, but not Th2 cells (19). When we challenged them with tumor cells, these animals showed severely impaired IL4 expression in tumor-draining lymph nodes, enhanced antitumor immunity, and delayed tumor growth. The number of M2 macrophage (assessed by expression of Fizz1 and Chi3l3) and the ability of CD11b+ myeloid cells to suppress T-cell proliferation were also significantly reduced in CNS2 KO when compared with WT mice. This constellation of findings suggested that Tfh cells were triggered by tumor Ags reaching the draining lymph nodes and that these cells contribute to the immunosuppressive microenvironment that supports tumor growth.
Increased IL4 is commonly detected in the tumors of animals and patients with cancer (32–38, 42, 43). IL4 is a Th2 cytokine produced primarily by T cells, basophils, and mast cells (27). We found that IL4 expression increased significantly in the draining lymph nodes within 4 days of tumor cell inoculation. Dogma holds that IL4 in the tumor microenvironment derives primarily from Th2 cells (32–38), but our findings suggest that Tfh cells were a major source of IL4 in the draining lymph nodes. These cells expressed archetypal Tfh surface markers including CXCR5, IL21, PD-1, ICOS, and BCL6, and localize to the germinal centers. Moreover, IL4 in the draining lymph nodes was markedly reduced in CNS2 KO mice. In this context, two reports showed tumor-infiltrating Tfh cells in human cancers. Gu-Trantien and colleagues found that Tfh cells localize primarily to peritumoral tertiary lymphoid structures and are a major source of CXCL-13 in patients with breast cancer (44). Bindea and colleagues reported that CXCL-13 production is provoked by highly mutated tumor cells in patients with colon cancer (45). Unlike patient samples, the murine tumor models used in the current report were lethal in a matter of weeks. This provided insufficient time for the development of tertiary lymphoid structures, the accumulation of genetic alterations, or increased production of CXCL-13.
In comparison, increased IL4 expression was detected in the draining lymph nodes by day 4 after tumor challenge. Several findings suggest that this IL4 was produced by naïve T cells, because (i) BCL6 expression by IL4+, CD4+ T cells did not increase until day 7, (ii) IL4-expressing cells were not present in B-cell areas until day 7, (iii) the expression of PD-1 and CXCR5 by IL4+, CD4+ T cells peaked on day 7, and (iv) whereas both live and dead tumor cells induced IL4 production through day 4, only live tumors maintained this response at later time points. The possibility that T follicular regulatory cells, which reportedly also express Bcl6 and CXCR5 (46), might be involved seems unlikely as cells in the draining lymph nodes did not express Foxp3.
Optimal induction of IL4 in the draining lymph nodes requires that live tumor cells be present: we found that tumor cell lysates were ineffective and irradiated/apoptotic tumor cells triggered only transient IL4 expression. This finding suggests that persistent exposure to tumor Ag is critical to maintaining the adaptive response. We postulate that APCs are exposed to Ag at the tumor site and then process and present these tumor Ags to CD4 T cells in the draining lymph nodes, which then preferentially differentiate into Tfh cells.
IL4 plays a crucial role in tumor immunology (47). IL4 was initially believed to be a potent antitumor cytokine, as IL4-secreting tumor cells could induce long-lasting antitumor immunity (48). However, clinical evidence suggests that IL4 can also act as a tumor-promoting molecule, as it is found at high levels in multiples types of human primary and metastatic cancers (32–38). This is consistent with studies in animal models showing that IL4 can promote tumor growth and that eliminating IL4 (by use of IL4R- or STAT6-deficient mice or treatment with depleting anti-IL4 Ab) can significant delay cancer cell proliferation (42, 43, 49, 50).
IL4 can suppress tumor immunity in several ways. This cytokine can downregulate the development of Th1 immunity and acts on CD8 T cells to render them noncytotoxic (51). Our results extend the latter observation by showing that the induction of tumor-specific CTLs was enhanced in CNS2 KO mice, whose expression of IL4 by Tfh cells was markedly deficient. IL4 also supports the generation of immunosuppressive cells of the CD11b+ myeloid lineage such as TAMs and MDSCs (39–41). They promote tumor growth by supporting angiogenesis and creating an immunosuppressive milieu that inhibits the lytic activity of CTLs and NK cells (39–41).
In this study, IL4 in Tfh cells induced a dramatic change in the tumor immune environment. Tumor-infiltrating CD4 and CD8 T cells and granzyme B expression in the tumor were significantly increased. On the other hand, suppressive cytokines such as arginase-1 were significantly reduced in the tumor. In this context, the frequency of M2 macrophage (based on marker expression) and the suppressive activity of CD11b+ myeloid cells from tumor-draining lymph nodes was significantly reduced in CNS2 KO mice. The production of IL21 by Tfh cells in the tumor microenvironment is sometimes associated with improved survival and enhanced antitumor immunity (44, 45). Our results are consistent with that finding, as the expression of IL21 in tumor-draining lymph nodes was comparable in WT and CNS2 KO mice.
Tfh cells can express cytokines typically associated with Th1, Th17, and Th2 cells (including IFNγ, IL17, and IL4, respectively). We found that the production of IL4 and IL21, but not IFNγ or IL17, increased in tumor-draining lymph nodes. Others have found evidence that B cells can play a supporting role in tumor development (52). Our data showed that the number of B cells in the germinal center of draining lymph nodes increased after tumor cell inoculation, an effect associated with the expansion of Tfh cells. It seems unlikely, however, that antitumor Abs play an important role in the studies described herein, as tumor progression is very rapid in these murine models.
In summary, this work demonstrates that Tfh cells are a major producer of IL4 in tumor-draining lymph nodes and that Tfh cells influences the tumor microenvironment, affecting antitumor immunity, and macrophage polarization. These findings suggest that Tfh cells and the IL4 these produce could be important targets for cancer immunotherapy.
Supplementary Material
Grant Support
This work was supported by the Intramural Research Program of the National Cancer Institute of the National Institutes of Health, USA and the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI, Japan.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.von Andrian UH, Mempel TR. Homing and cellular traffic in lymph nodes. Nat Rev Immunol 2003;11:867–78. [DOI] [PubMed] [Google Scholar]
- 2.Munn DH, Mellor AL. The tumor-draining lymph node as an immune-privileged site. Immunol Rev 2006;213:146–58. [DOI] [PubMed] [Google Scholar]
- 3.Shu S, Cochran AJ, Huang RR, Morton DL, Maecker HT. Immune responses in the draining lymph nodes against cancer: implications for immunotherapy. Cancer Metastasis Rev 2006;25:233–42. [DOI] [PubMed] [Google Scholar]
- 4.Fransen MF, Arens R, Melief CJ. Local targets for immune therapy to cancer: tumor-draining lymph nodes and tumor microenvironment. Int J Cancer 2013;132:1971–6. [DOI] [PubMed] [Google Scholar]
- 5.Jeanbart L, Ballester M, de Titta A, Corth esy P, Romero P, Hubbell JA, et al. Enhancing efficacy of anticancer vaccines by targeted delivery to tumor-draining lymph nodes. Cancer Immunol Res 2014;2:436–47. [DOI] [PubMed] [Google Scholar]
- 6.Okita Y, Ohira M, Tanaka H, Tokumoto M, Go Y, Sakurai K, et al. Alteration of CD4 T cell subsets in metastatic lymph nodes of human gastric cancer. Oncol Rep 2015;34:639–47. [DOI] [PubMed] [Google Scholar]
- 7.Swartz MA. Immunomodulatory roles of lymphatic vessels in cancer progression. Cancer Immunol Res 2014;2:701–7. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura S, Yaguchi T, Kawamura N, Kobayashi A, Sakurai T, Higuchi H, et al. TGF-β1 in tumor microenvironments induces immunosuppression in the tumors and sentinel lymph nodes and promotes tumorprogression. J Immunother 2014;37:63–72. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt N, Ueno H. Regulation of human helper T cell subset differentiation by cytokines. Curr Opin Immunol 2015; 34:130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, et al. Plasticity of human CD4 T cell subsets. Front Immunol 2014;5:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, Akpinarli A, Maris C, Hipkiss EL, Lane M, Kwon EK, et al. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J Exp Med 2010;207:651–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 2010;207:637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344:641–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willimsky G, Blankenstein T. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature 2005;437:141–6 [DOI] [PubMed] [Google Scholar]
- 15.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res 2014;2:91–8 [DOI] [PubMed] [Google Scholar]
- 16.Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol 2010;11:7–13 [DOI] [PubMed] [Google Scholar]
- 17.Toomer KH, Chen Z. Autoimmunity as a double agent in tumor killing and cancer promotion. Front Immunol 2014;5:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, et al. Requirement for Va14 NKT cells in IL12-mediated rejection of tumors. Science 1997; 278:1623–6 [DOI] [PubMed] [Google Scholar]
- 19.Harada Y, Tanaka S, Motomura Y, Harada Y, Ohno S, Ohno S, et al. The 3′ enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity 2012;36: 188–200. [DOI] [PubMed] [Google Scholar]
- 20.Sakai Y, Morrison BJ, Burke JD, Park JM, Terabe M, Janik JE, et al. Vaccination by genetically modified dendritic cells expressing a truncated neu oncogene prevents development of breast cancer in transgenic mice. Cancer Res 2004;64:8022–8. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N, Hong C, Klinman DM, Shirota H. Oligodeoxynucleotides expressing polyguanosine motifs promote antitumor activity through the upregulation of IL2. J Immunol 2013;190:1882–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirota H, Sano K, Hirasawa N, Terui T, Ohuchi K, Hattori T, et al. Novel roles of CpG oligodeoxynucleotides as a leader for the sampling and presentation of CpG-tagged antigen by dendritic cells. J Immunol 2001;167:66–74. [DOI] [PubMed] [Google Scholar]
- 23.Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. Adv Cancer Res 2015;128:95–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci 2015;22:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira ER, Jones D, Jung K, Padera TP. The lymph node microenvironment and its role in the progression of metastatic cancer. Semin Cell Dev Biol 2015;38:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol 2015;15:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May RD, Fung M. Strategies targeting the IL4/IL13 axes in disease. Cytokine 2015;75:89–116. [DOI] [PubMed] [Google Scholar]
- 28.Iwamura C, Nakayama T. Role of NKT cells in allergic asthma. Curr Opin Immunol 2010;22:807–13. [DOI] [PubMed] [Google Scholar]
- 29.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL4 reporter. Immunity 2001;15:303–11. [DOI] [PubMed] [Google Scholar]
- 30.Crotty S T follicular helper cell differentiation, function, and roles in disease. Immunity 2014;41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat Immunol 2015;16:142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedroza-Gonzalez A, Xu K, Wu TC, Aspord C, Tindle S, Marches F, et al. Thymic stromal lymphopoietin fosters human breast tumor growth by promoting type 2 inflammation. J Exp Med 2011;208: 479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevala WK, Vachon CM, Leontovich AA, Scott CG, Thompson MA, Markovic SN, et al. Evidence of systemic Th2-driven chronic inflammation in patients with metastatic melanoma. Clin Cancer Res 2009;15:1931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J, Li Y, Liu X, Xu X, Zhao Y. Relationship between cytokine levels and clinical classification of gastric cancer. Asian Pac J Cancer Prev 2011;12: 1803–6. [PubMed] [Google Scholar]
- 35.Li J, Wang Z, Mao K, Guo X. Clinical significance of serum T helper 1/T helper 2 cytokine shift in patients with non-small cell lung cancer. Oncol Lett 2014;8:1682–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onishi T, Ohishi Y, Imagawa K, Ohmoto Y, Murata K. An assessment of the immunological environment based on intratumoral cytokine production in renal cell carcinoma. BJU Int 1999;83:488–92. [DOI] [PubMed] [Google Scholar]
- 37.Gao J, Wu Y, Su Z, Amoah Barnie P, Jiao Z, Bie Q, et al. Infiltration of alternatively activated macrophages in cancer tissue is associated with MDSC and Th2 polarization in patients with esophageal cancer. PLoS ONE 2014;9:e104453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baier PK, Wolff-Vorbeck G, Eggstein S, Baumgartner U, Hopt UT. Cytokine expression in colon carcinoma. Anticancer Res 2005;25:2135–9. [PubMed] [Google Scholar]
- 39.Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol 2013;120: 163–84. [DOI] [PubMed] [Google Scholar]
- 40.Allavena P, Mantovani A. Immunology in the clinic review series; focus on cancer: tumour-associated macrophages: undisputed stars of the inflammatory tumour microenvironment. Clin Exp Immunol 2012; 167:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol 2009;182:4499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi M, Kobayashi H, Pollard RB, Suzuki F. A pathogenic role of Th2 cells and their cytokine products on the pulmonary metastasis of murine B16 melanoma. J Immunol 1998;160:5869–73. [PubMed] [Google Scholar]
- 43.Li Z, Jiang J, Wang Z, Zhang J, Xiao M, Wang C, et al. Endogenous interleukin-4 promotes tumor development by increasing tumor cell resistance to apoptosis. Cancer Res 2008;68:8687–94. [DOI] [PubMed] [Google Scholar]
- 44.Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, et al. CD4+ follicular helper T-cell infiltration predicts breast cancer survival. J Clin Invest 2013;123:2873–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 46.Sage PT, Sharpe AH. T follicular regulatory cells in the regulation of B-cell responses. Trends Immunol 2015;36:410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Chen L, Qin Z. Paradoxical roles of IL4 in tumor immunity. Cell Mol Immunol 2009;6:415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tepper RI, Pattengale PK, Leder P. Murine interleukin-4 displays potent anti-tumor activity in vivo. Cell 1989;57:503–12. [DOI] [PubMed] [Google Scholar]
- 49.Terabe M, Matsui S, Noben-Trauth N, Chen H, Watson C, Donaldson DD, et al. NKT cell-mediated repression of tumor immunosurveillance by IL13 and the IL4R-STAT6 pathway. Nat Immunol 2000;1:515–20. [DOI] [PubMed] [Google Scholar]
- 50.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009;16:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villacres MC, Bergmann CC. Enhanced cytotoxic T cell activity in IL4-deficient mice. J Immunol 1999;162:2663–70. [PubMed] [Google Scholar]
- 52.Affara NI, Ruffell B, Medler TR, Gunderson AJ, Johansson M, Bornstein S, et al. B cells regulate macrophage phenotype and response to chemotherapy in squamous carcinomas. Cancer Cell 2014;25:809–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.