Abstract
Flow cytometry has the potential to facilitate understanding the heterogenous responses of diverse brain cell populations to a variety of stimuli. However, existing methods of applying flow cytometry to brain tissues are each limited in certain ways. They either require genetically labeled cells to achieve separation of specific populations, are not applicable to previously fixed tissue, or are not compatible with downstream mRNA analysis. Here, we describe a group of related methods that overcome many previous limitations and allow robust sorting and downstream molecular analysis of highly enriched populations of specific neuronal and non-neuronal cells from any mammalian brain. We illustrate these techniques, which are compatible with antibodies for both nuclear and non-nuclear epitopes and do not require transgenic animals, with three examples. First, we describe the separation and downstream mRNA analysis of four types of cortical interneurons (somatostatin, parvalbumin, calretinin, calbindin) from paraformaldehyde-fixed rat brain sections. Second, we demonstrate separation of neurons and non-neurons from zinc-fixed mouse brain cortical sections followed by analysis of enzymatic activity (ACE2 activity) and mRNA expression. Third, we show that routinely fixed post-mortem human brain can be analyzed by isolating parvalbumin-containing neurons from cortical samples that were fixed for periods of up to 8 weeks in formalin. In each case, sorted cell identity was confirmed with mRNA analysis. The neurocytometry methodology described here has the potential to significantly expand studies to analyze the effects of drugs, environmental manipulations, and disease states on the nucleic acid and protein content of specific brain cell populations.
Keywords: FACS, flow cytometry, brain, neuron, interneuron, neurocytometry
Introduction
Flow cytometry is a widespread technique used to identify, analyze, and separate complex mixtures of cells in suspension. Brain tissue is comprised of exceedingly diverse cellular populations, and flow cytometric methods have the potential to facilitate studies of individual cell types found within close proximity throughout the brain matrix. Fluorescence activated cell sorting (FACS) of brain tissue was first used to segregate a small population of rat neurons based on intracellular markers three decades ago1. Since that report, however, neuroscientists have been relatively slow to widely employ flow cytometry. This was, perhaps, due to difficulties in achieving single cell suspensions from brain tissue. Recently, a number of reports have been published that demonstrate the power of neurocytometry – flow cytometry applied to brain tissue - for analyzing a variety of neuronal and non-neuronal brain cell populations.
Previous work has shown that a combination of enzymatic and mechanical techniques of dissociating brain cells allows for the separation of live, GFP-tagged neurons from the brains of genetically modified mice2. More recently, neurons were separated from glia using FACS without requiring transgenic animals that express fluorescent proteins in the cells of interest3. These researchers also isolated activated neurons from non-activated neurons using cFos immunofluorescence4, and later refined the technique by separating neurons from a single rat striatum in sufficient quantity for downstream RNA analysis of sorted populations5. These methods involve dissociating fresh brain tissue, with subsequent fixation of the dissociated cells in ethanol before proceeding to immunostaining and FACS3–5. Other researchers have described the use of a zinc-based fixative treatment of cells isolated from dissociated fresh brain tissue to achieve the simultaneous segregation of neurons, astrocytes, and microglia with FACS6. Most recently, chromatin precipitation from dopamine 1 receptor-expressing cells of the nucleus accumbens was performed following purification by FACS using transgenic animals expressing the fluorescent protein dTomato in D1R expressing cells7.
Although existing neurocytometric methods are useful, they each differ in application and have certain limitations. For example, some methods isolate relatively intact cell bodies that retain cytoplasm from brain without fixation prior to sorting, but these require the relevant cells to be genetically labeled with a fluorescent protein to perform FACS2, 7, 9. There has been one report describing FACS of neurons from brain fixed using Zamboni’s fixative prior to dissociation1, but this method is not compatible with downstream nucleic acid or enzymatic analyses. A series of more recently described protocols incorporate fixation of brain cells after dissociation from fresh tissue, and are useful for sorting neurons using a variety of antigens, including Fos and NeuN, and for downstream nucleic acid analysis3–6. Many cytosolic proteins, however, are lost from neurons using these procedures, resulting in small, rounded cell bodies that consist primarily of nuclei (i.e.,3, 6, 8), necessitating the use of nuclear antigens for maximum sorting efficiency.
A significant limitation of current flow cytometry methodology is in the analysis of human brain tissue. The vast majority of brain tissues available from various sources have been fixed in formalin, or have been frozen for an indeterminate time prior to processing. It is difficult to obtain fresh samples that have not been frozen or fixed and therefore compatible with existing FACS methodology. Although a technique to isolate human neuronal nuclei by FACS has been recently developed and used for RNA-sequencing10, cytoplasm, intracelluar proteins, and extracellular membranes are lost, and this technique requires tissue that has never been fixed. Currently, to our knowledge, the only practical method to analyze DNA and RNA from specific subtypes of neurons or other cells from routinely available human brain tissue requires immunohistochemical labeling of specific cells followed by laser capture microdissection (e.g.,11). This is a time consuming and laborious technique that is relatively inefficient for producing large numbers of purified cells relative to FACS.
In order to overcome the limitations of requiring fresh tissue, genetically labeled cells, or the necessity to use primarily nuclear antigens, we developed new methodology that allows for the separation of neurons and cells from previously fixed brain tissue using non-nuclear antigens and maintains the ability to perform downstream analysis of mRNA expression. Here, we describe our methods in a group of related protocols and demonstrate that they allow for the separation of specific types of neurons and other brain cells following fixation in a variety of agents, including solutions of zinc salts, formalin, and paraformaldehyde. Fixed neurons and cells remain largely intact during dissociation, with preservation of cytoplasmic proteins. This enables the robust use of non-nuclear epitope markers, in addition to nuclear, for FACS. Importantly, the purified cells retain RNA suitable for downstream analysis. We demonstrate the effectiveness of our methodology by describing three experiments. In the first we separate four distinct types of cortical interneurons (somatostatin, parvalbumin, caltretinin, and calbindin) from paraformaldehyde-fixed rat brain. In the second example, we demonstrate sorting and mRNA analysis of highly purified parvalbumin-expressing interneurons from routinely prepared, formalin-fixed adult human brain cortex. In a third example, we show that zinc-based fixation of frozen mouse brain, followed by dissociation and FACS, allows for the analysis of enzyme activity (angiotensin converting enzyme 2 (ACE2) activity) in neurons compared with non-neurons. In general, the methodologies we describe here can potentially be used to more easily obtain large numbers of highly purified populations of specific cells from any mammalian brain to analyze the effects of drugs, genetic manipulations, or disease states. In particular, the application of neurocytometry to human brain tissue will allow for purification of large populations of specific cell types from fresh, frozen, or routinely banked fixed brain tissues that until now were inaccessible to sorting and subsequent molecular analysis.
Results
Paraformaldehyde-Based Fixation
Whole neuronal somata, including cytoplasmic proteins that differentiate many neuronal cell types, can be segregated by FACS following fixation with paraformaldehyde. To demonstrate this technique, 4 major classes of GABAergic interneurons were isolated from single cortical sections of individual rats. Somatostatin (SOM) and calretinin (CR) neurons were sorted simultaneously, and consist of mutually exclusive populations of interneurons (Figure 1A–E), as has been reported previously16, 17. Parvalbumin (PV(+)) and calbindin (CB(+)) neurons from the same brain sections were also sorted simultaneously, however, some overlap was found between these populations (Figure 2A–E), as expected from previous immunohistochemical studies16, 17. Two distinct populations of CB(+) cells can be seen; a large group of neurons with low to moderate levels of CB immunoreactivity relative to background, and a smaller group of interneurons with very high levels of CB immunoreactivity (Figure 2D–E). The sorting strategy in these experiments (see Methods) was such that PV(+) cells were sorted first, without regard for CB immunoreactivity, and CB(+) cells were sorted as the top ~0.8% of the remaining PV(−) population. Because PV(+) cells were sorted without regard for CB immunoreactivity, some PV(+) cells can be seen to display CB staining (Figure 2E). The fraction of PV(+) cells displaying very high levels of CB immunoreactivity was small (~3%), however, a much larger fraction (~40%) can be seen to express moderate, above-background, levels of CB immunoreactivity.
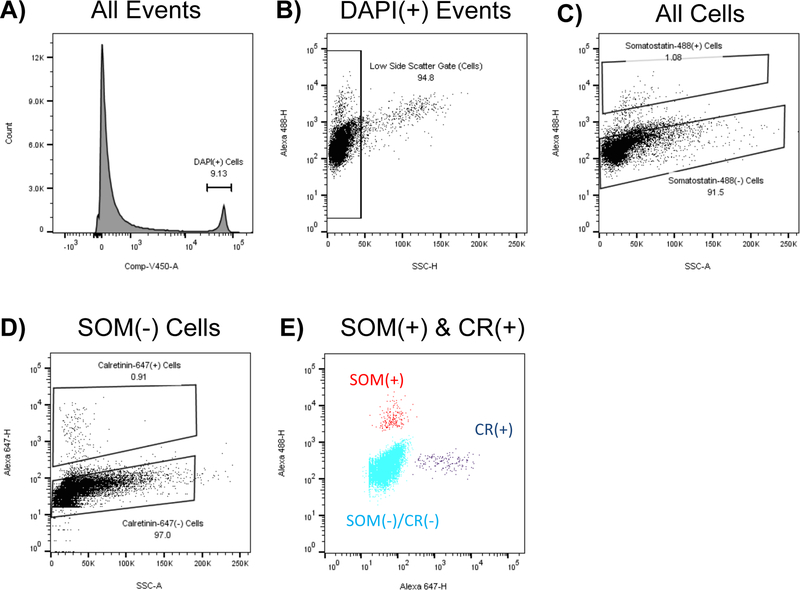
Figure 1: FACS allows the simultaneous separation of somatostatin and calretinin interneurons:
Rat motor and somatosensory cortex was fixed in paraformaldehyde, dissociated, and stained. Sorting strategy for separating somatostatin (SOM) and calretinin (CR) expressing interneurons is shown from a representative sort. A) All Events are gated based on DAPI. B) The top 5% of SSC-H events are excluded to avoid debris C) SOM(+) cells are gated. D) CR(+) cells are gated. E) Plot of SOM(+), CR(+), and SOM/CR(−) cells. Note the nearly 100% mutual exclusivity between these SOM(+) and CR(+) populations.
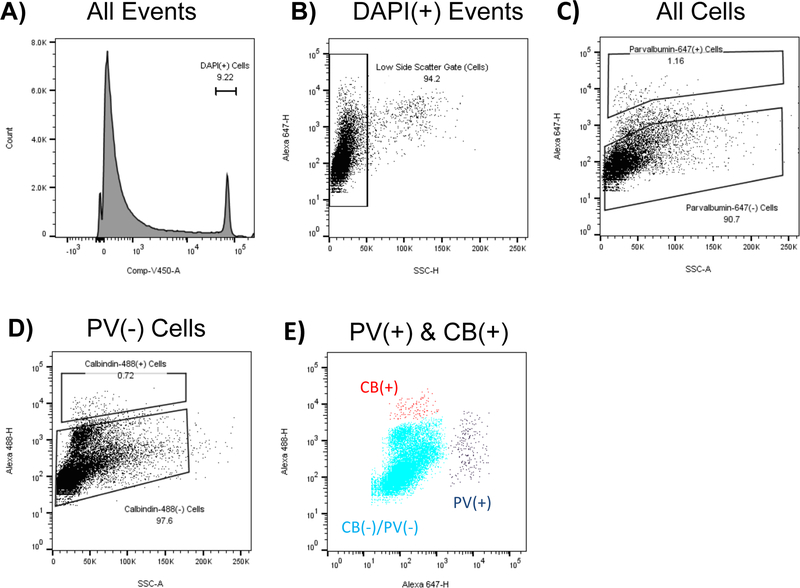
Figure 2: FACS allows the simultaneous separation of parvalbumin and calbindin interneurons:
Rat motor and somatosensory cortex was fixed in paraformaldehyde, dissociated, and stained. Sorting strategy for separating parvalbumin (PV) and calbindin (CB) expressing interneurons is shown from a representative sort. A) All Events are gated based on DAPI. B) The top 5% of SSC-H events are excluded to avoid debris C) PV(+) cells are gated. D) CB(+) cells are gated. E) Plot of PV(+), CB(+), and PV(−)/CB(−) cells. Note that some cells expressing high levels of parvalbumin also express moderate or high levels of calbindin, and that a large group of cells expresses moderate, but above background levels of calbindin.
In order to demonstrate that the staining observed is specific to neurons, we performed separate control experiments on rat somatosensory cortex fixed in paraformaldehyde. In these experiments, dissociated tissue was stained for either somatostatin or parvalbumin in combination with NeuN, a neuronal marker (Figure 3). Analysis of neuronal populations reveals that neurons exhibit bimodal populations with respect to PV and SOM staining, in which a small population of neurons exhibits above background signal (Figure 3C, 3G). In contrast, non-neuronal populations do not exhibit a bimodal distribution and only a few scattered events contaminate the positive gate (Figure 3D, 3H). This data illustrates that non-neurons exposed to identical dissociation and staining conditions do not stain for the proteins parvalbumin or somatostatin.
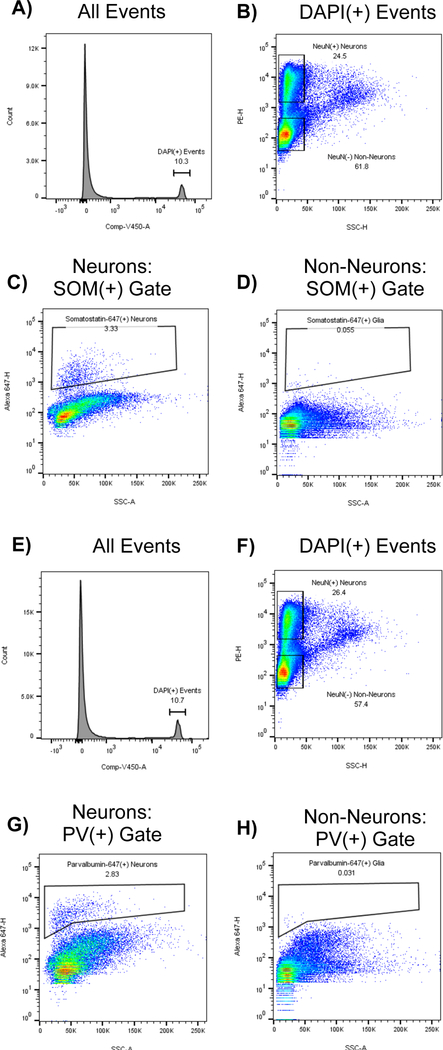
Figure 3: Specific staining for interneuron markers somatostatin and parvalbumin is largely absent from non-neuronal populations:
As a control, somatosensory cortex was fixed in paraformaldehyde, dissociated, split into two aliquots, and stained with DAPI, NeuN-PE, and either somatostatin-647 (A-D) or parvalbumin-647 (E-H). Top 4 panels (A-D) show the gating strategy for tissue stained for somatostatin: A) DAPI(+) events are gated B) Neurons and Non-Neurons are gated based on NeuN-PE signal, but excluding events with high SSC-H. C) Somatostatin(+) gate drawn on the NeuN(+) population. D) Somatostatin(+) gate drawn on NeuN(−) population. Note that neurons contain a distinct subpopulation of somatostatin(+) events, while nonneurons do not. Botttom 4 panels (E-H) show the gating strategy for tissue stained for parvalbumin: E) DAPI(+) events are gated F) Neurons and Non-Neurons are gated based on NeuN-PE signal, but excluding events with high SSC-H. G) Parvalbumin(+) gate drawn on the NeuN(+) population. H) Parvalbumin(+) gate drawn on the NeuN(−) population. Note that neurons contain a distinct subpopulation of parvalbumin(+) events, while nonneurons do not.
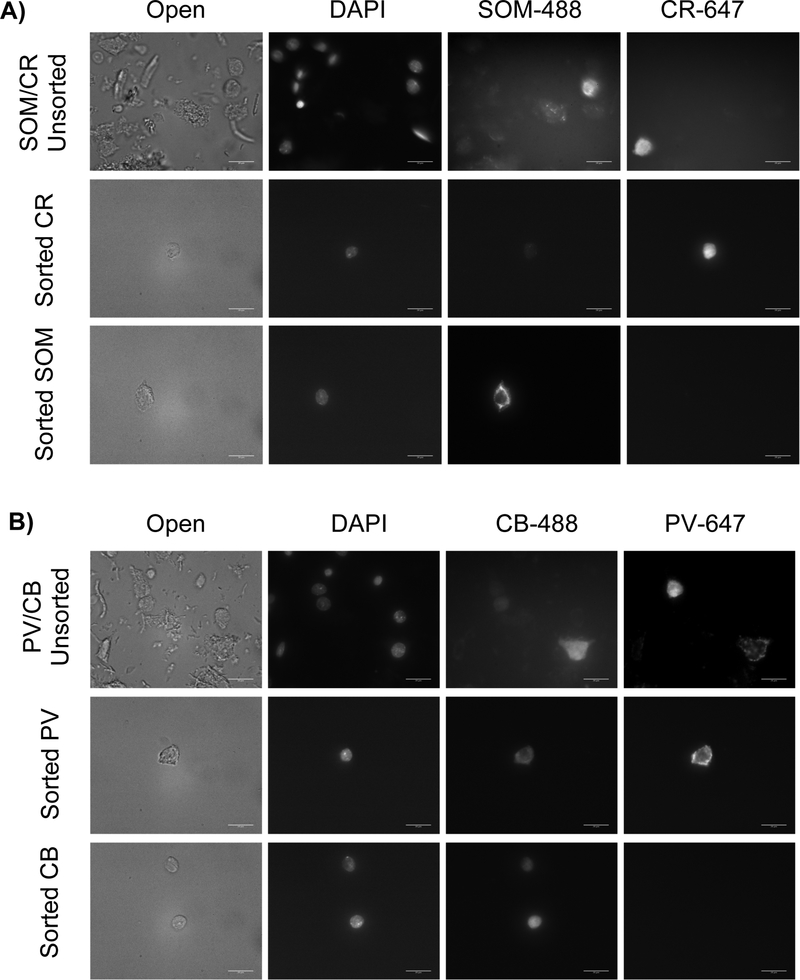
The appearances of stained and dissociated but unsorted tissues, along with sorted cellular populations, are shown from representative photomicrographs in Figure 4. Unsorted tissue from CR/SOM experiments contain a mixture of DAPI(+) cells and debris, along with scattered individual cells immunoreactive for either CR or SOM (Figure 4A, top row). Sorted cells from CR/SOM experiments appropriately display either CR or SOM immunoreactivity (Figure 4A, bottom two rows). Similarly, unsorted tissue from PV/CB experiments contain a mixture of DAPI(+) cells and debris, along with scattered individual cells immunoreactive for either PV, CB, or both (Figure 4B, top row). PV(+) sorted cells from PV/CB experiments display either PV staining only, or PV and some CB staining (Figure 4B, middle row). CB(+) sorted cells from PV/CB experiments display only CB staining (Figure 4C, bottom row), as the CB(+) population was gated from the PV(−) population.
Figure 4: Photomicrographs of unsorted tissue and sorted interneurons:
Dissociated tissue and sorted sorted cells were collected and viewed under a microscope at 63× and imaged with 4 different filters: Open/brightfield (left), DAPI (middle left), 488 (middle right), and 647 (right). Top Panel (A): Dissociated tissue and cells from SOM/CR sorts. Bottom Panel (B): Dissociated tissue and cells from PV/CB sorts. Note that the imaged PV(+) cell does contain some CB-488 immunoreactivity. Scale bar = 20 microns
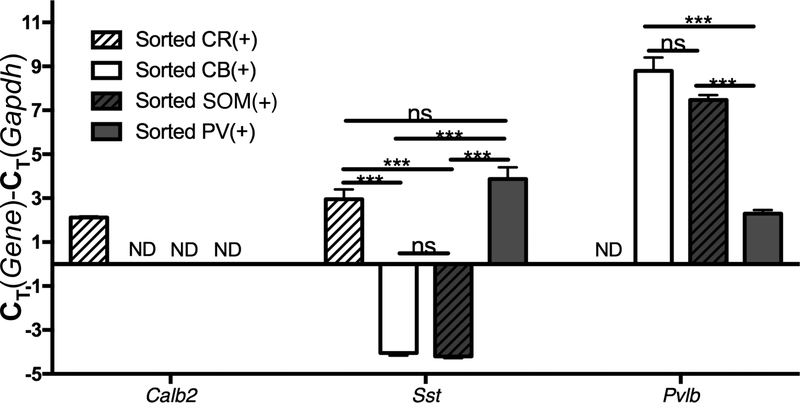
Between 700–2,500 neurons/subtype/sample from each of the four groups of interneurons were sorted from each sample, and then compared for mRNA composition. CR(+) neurons were the only group in which calb2 was detected (Figure 5), as expected, given that calretinin is the protein product of calb2. Both SOM(+) and CB(+) interneurons displayed similar sst mRNA levels about 7 cycles (~128 fold) higher than PV(+) and CR(+) cells (Figure 5), indicating these populations share considerable cellular overlap, as previously reported16. PV(+) cells displayed much higher levels of pvlb expression relative to CB(+) and SOM(+) cells, as expected, and pvlb was not detected at all in CR(+) cells. In other experiments in which PV/SOM and PV/CR cells were stained simultaneously and sorted by FACS, very little overlap was found between PV and SOM staining (not shown), and between PV and CR staining (not shown) as expected, given that PV(+) interneurons rarely express these interneuronal markers16, 17.
Figure 5: QPCR validation of sorted interneuron identity:
FACS-purified parvalbumin (PV+), somatostatin (SOM+), calretinin (CR+), and calbindin (CB+) neurons were collected and analyzed for mRNA content with QPCR. Note that only CR(+) interneurons contain calb2. Also note that both SOM(+) and CB(+) expressing cells contain high levels of sst, while only PV(+) cells contain high levels of pvlb. “***” and “ns” indicate p<0.001 and nonsignificant, One-way ANOVA. N=3/group, Tukey post-hoc.
Zinc Fixation
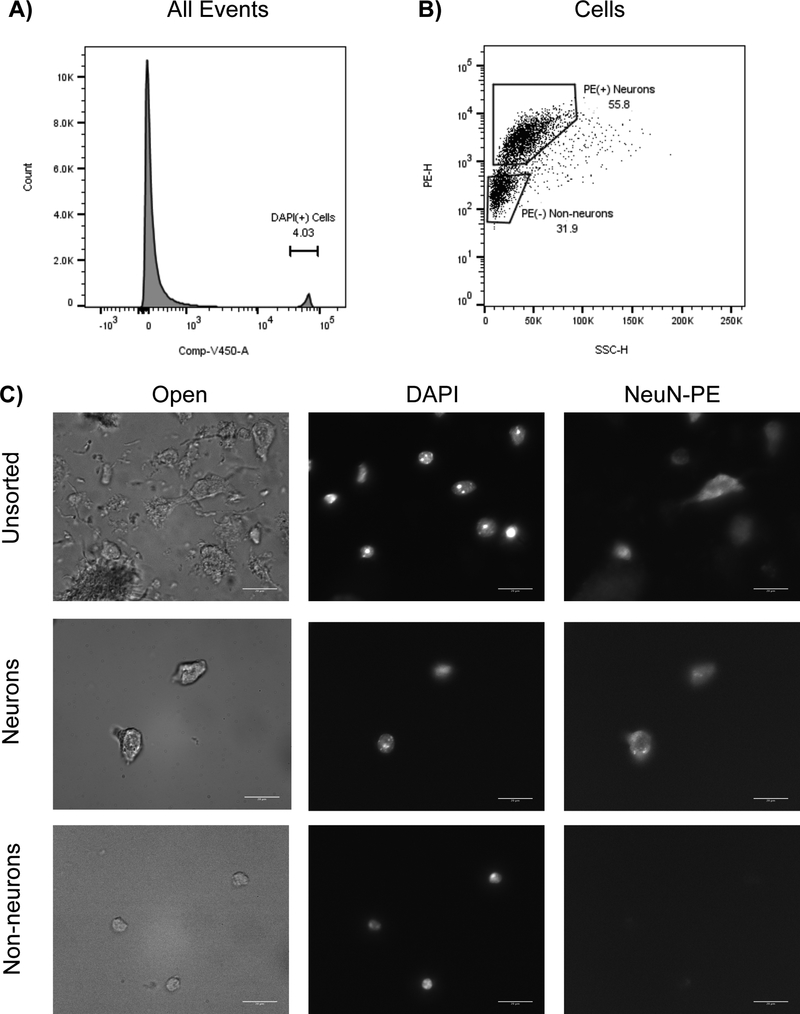
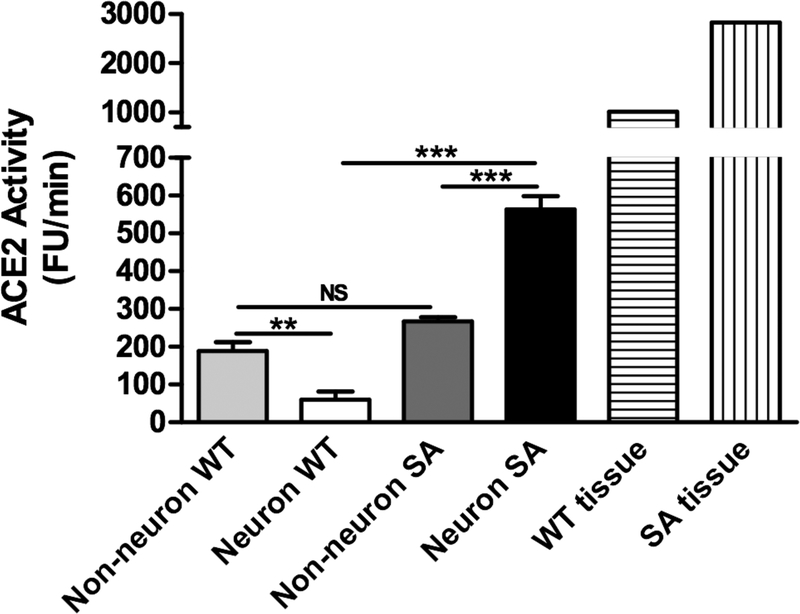
Zinc-based fixation followed by dissociation results in brain cells that retain cell bodies and some proximal processes. The gating strategy for isolating neurons and non-neurons prepared with this method is shown in Figure 6. Illustrative photomicrographs of Zn-fixed brain tissue stained with DAPI and NeuN-PE is shown in Figure 6C. Unsorted tissue clearly displays debris and cells (DAPI(+) objects), including neurons (NeuN(+) cells) and non-neurons (NeuN(−) cells); sorted cell populations are exclusively either NeuN(+) (neurons) or NeuN(−) (non-neurons). Sorted neurons and non-neurons were collected from wild-type (WT) mice and from mice overexpressing human ACE2 under control of the synapsin promoter (SA mice). Synapsin is expressed primarily in neurons, and SA mice have been previously demonstrated to exhibit high levels of ACE2 activity in the brain relative to WT mice12. Enzymatic activity was subsequently assayed in the sorted neurons and non-neurons from the cortices of both strains (Figure 7). ACE2 activity assays demonstrate that SA neurons exhibit much higher levels of ACE2 activity than WT neurons, while SA non-neurons exhibit similar levels of activity relative to WT non-neurons (Figure 7). These results demonstrate that zinc-based fixation methods preserve cell bodies and associated ACE2 enzymatic activity contained in the cells.
Figure 6: FACS sorting strategy and cell appearance following zinc-based fixation and dissociation:
Frozen cortex from mouse was fixed in zinc solution, dissociated, stained, and FACS was performed. A) DAPI(+) cells are gated from DAPI(−) debris. B) Neurons are gated from non-neurons based on the expression of NeuN-PE. C) Dissociated tissue was collected and viewed under a microscope at 63×. Unsorted (top row), post-sort neurons (middle row), and post-sort non-neurons (bottom row) are shown, imaged with 3 different filters: Open/brightfield (left column), DAPI (middle column), and NeuN-PE/TRITC (right column). Note that intact cellular bodies survive FACS. Also, note the lack of NeuN-PE sorting in the non-neuronal population. Scale bar = 20 microns.
Figure 7: Zinc-based fixation preserves ACE2 enzymatic activity through cell sorting:
Sorted neurons and non-neurons were analyzed for ACE2 activity following cell sorting. SA mice exhibit high levels of ACE2 activity in neurons relative to WT neurons, whereas non-neuronal populations exhibit similar levels of ACE2 activity between WT and SA mice. Data from unsorted tissue (N=1) is shown for comparison on the right. Data for sorted samples is normalized to the number of cells collected for each sorted sample (N=3 mice/group). “**”, “***”, and “ns” indicate p<0.01, p<0.001, and non-significant, One-way ANOVA, Tukey post-hoc.
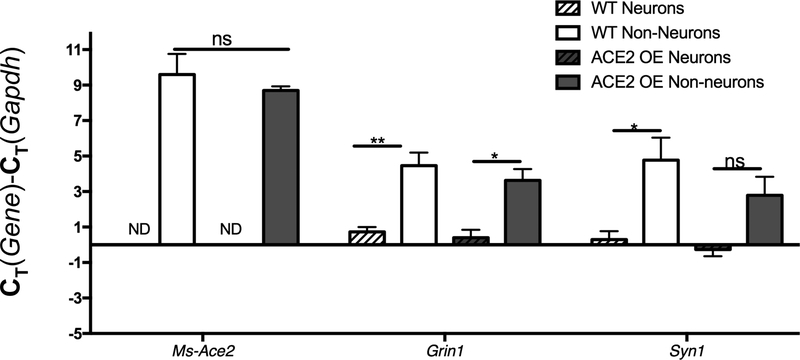
Zinc-based fixation also allows for the recovery of mRNA from sorted cells (Figure 8). Non-neurons from both WT and SA mice displayed detectable, albeit very low, levels of mouse-ace2 relative to gapdh, whereas neurons from SA and WT mice displayed non-detectable levels of mouse-ace2 (Figure 8). This data correlates with levels of ACE2 activity detected in sorted WT cells, where non-neurons displayed higher activity than neurons (Figure 7). In our analysis of mRNA levels of the neuron-specific genes grin1 and syn1 (relative to gapdh), neurons exhibit higher levels of these genes compared with non-neurons in WT and SA mice, as expected (Figure 8). However, the increase in syn1 expression did not reach statistical significance in SA mice (Figure 8).
Figure 8: Zinc-based fixation preserves RNA:
Sorted neurons and non-neurons from wild-type WT and SA mice were compared for gene expression with QPCR. Mouse ace2 was detected only in non-neurons of both strains, while neuronal genes grin1 and syn1 were found at higher levels in neurons. Data is shown relative to gapdh expression. “*”, “**”, and “ns” indicate p<0.05, p<0.01, and non-significant, One-way ANOVA, Tukey post-hoc.
Formalin-Based Fixation of Human Tissue
Human frontal cortical samples were fixed and processed as described in the methods. PV(+) cells were successfully segregated from other brain cells in all samples (N=4) processed. The gating strategy for sorting and qPCR analysis of sorted populations is shown in Figure 9. Note that the NeuN epitope is lost by the excessive formalin fixation applied to these human samples, and that fluorescence in the NeuN-PE channel therefore provides a convenient estimate of background in the channel of interest (PV-647). The sorted populations were highly pure, as illustrated by the large increase in pvlb expression in PV(+) cells (>8 cycles; >256 fold) relative to PV(−) cells, as well as the finding that slc17a7, a transcript found primarily in excitatory neurons, was not detectable in PV(+) cells.
Figure 9: Pure populations of human neuronal subtypes can be sorted using FACS:
Human frontal cortex was fixed, dissociated, stained, sorted, and compared via QPCR. FACS gating shown in top row from representative sort (A-C): A) DAPI(+) events are gated from the majority of debris events. B) DAPI(+) events with high SSC-H are gated out. C) Parvalbumin cells (PV+) are separated from all non-parvalbumin cells (PV-) based on PV-647 and NeuN-PE fluorescence. D) QPCR analysis of sorted cells: Expression of pvlb is much higher in PV(+) neurons than PV(−) neurons, while slc17a7 and htr2a is not detectable in PV(+) neurons.
Discussion
In this report, we demonstrate neurocytometric techniques suitable for the purification of highly enriched cellular populations from mammalian brain, and downstream analysis of nucleic acids and ACE2 enzyme activity. These techniques have the potential to facilitate a variety of studies investigating the effects of disease, drugs, aging, or the environment on particular types of cells in the brain. Our methods differ from most previously described techniques for cell sorting and analysis of brain tissue preparations because we fix brains prior to cellular dissociation. This maintains the integrity of cytoplasmic and extracellular membrane constituents in addition to nucleic acids. Another feature of our methodology is that it allows for the use of previously frozen brain tissue, permitting an investigator to bank brains at −80° C until they are ready for processing. Significantly, this is the first report to demonstrate preservation of measurable enzymatic activity in brain cells following cell sorting, and the first to sort specific neuronal subtypes from routinely fixed human brain tissue.
The utility of paraformaldehyde-based fixation prior to dissociation and FACS is exemplified by the sorting and mRNA analysis of four distinct types of neurons from a single rat cortical section. Our results here are consistent with previous reports, indicating PV(+) cells comprise a group of GABAergic interneurons distinct from somatostatin and calretinin interneurons, though containing occasional highly immunoreactive calbindin(+) cells and abundant moderately stained calbindin cells. Our results corroborate previous immunohistochemical studies of cortical calbindin distribution, indicating two subpopulations, a small subpopulation with high calbindin levels, and a large subpopulation with moderate calbindin levels17. We also find an abundance of sst mRNA in high-expressing calbindin interneurons, indicating extensive overlap between SOM(+) and CB(+) populations, in agreement with previous immunohistochemical studies that found the majority of CB(+) and SOM(+) cells co-express both peptides16,17. We provide several lines of evidence that the populations we sort are specifically stained and highly pure. First, the proportions of positive cells are approximately as expected from known distributions of cell types, including the fractions of overlap between markers, and the observation of two distinct levels of CB staining. Second, non-neuronal cells do not exhibit appreciable staining for somatostatin or parvalbumin, as expected. Third, dissociated and sorted cells have the expected pattern of staining when viewed microscopically. Fourth, the qPCR analyses indicate a relative abundance of expected transcripts and a relative lack of unexpected transcripts.
We have previously used a similar paraformaldehyde-based method to study the effects of psychedelics on gene expression, specifically in parvalbumin and somatostatin neurons8. In that report, we estimated the purity of sorted interneurons to be greater than 94% based on low levels of slc17a7 mRNA in sorted interneurons; it is conceivable that actual sort purity is higher, as rat interneurons are known to occasionally contain this transcript18. One may also estimate sensitivity or yield of this method by comparing the total amount of a given transcript (eg., pvlb) between negative and positive populations (eg., PV(+) vs PV(−)). In other experiments, we found that >75% of the total detectable pvlb transcripts are in the top 2% of PV immunoreactive cells (~top 4% PV(+) neurons) indicating that the majority of cells of a given type are collected within the positive population (data not shown). However, we note that not all cells of a given subtype are successfully sorted using this methodology. A fraction of cells that are truly positive are discarded in the negative population to ensure that the sorted population is relatively pure. As in many FACS experiments, a compromise between purity and yield can be decided upon through gate placement.
Considerations for using paraformaldehyde as a fixative include known modifications that occur on nucleic acids and proteins19. While useful mRNA can be recovered from such tissues with extended heat treatment to reverse chemical modifications of nucleic acids (see Methods), enzymatic activity can be permanently lost. Also, paraformaldehyde fixation leads to relatively poorly dissociating material. However, yields of several thousand pure parvalbumin interneurons are readily achievable from a 1–2 mm thick section of rat cortex, and this number is enough to reliably measure mRNA expression for many genes of moderately low abundance. The amount of tissue used can be readily scaled up to increase yield. We also note that permeabilization of the membrane occurs with this methodology, allowing antibody penetration to intracellular epitopes. It is conceivable that some leakage of cellular contents may occur during dissociation, and many neuronal processes are lost. However, we have successfully sorted using each cytosolic epitope we have tested. Human tissues fixed for long periods in formalin are also poorly dissociating, however, adequate yields for downstream qPCR analysis can still be achieved with extended heat treatment of sorted cells. Whether nucleic acids recovered from these cells are suitable for sequencing remains untested
A general pitfall of using routinely fixed human brain samples is that these tissues are stored at room temperature and in formalin for extended periods. This can lead to overfixation of certain epitopes, like NeuN14. This also results in lower amounts of quantifiable RNA yields than can be achieved if the brains are handled as described according to the paraformaldehyde-based fixation method. Therefore, for sensitive human studies of lowly expressed genes, brain tissue should be collected with short post-mortem intervals, fixed quickly, and kept at 4°C until dissociation. Alternately, frozen brain sections can be thawed in fixative, allowing the preservation of some epitopes (i.e., somatostatin), but not others (i.e., parvalbumin), which may leak from ruptured cell membranes upon thawing13.
Importantly, adequate staining of cellular populations must be achieved for accurate sorting. Antigens that are uniformly and abundantly distributed throughout the cytoplasm or nucleus are ideal, however we also have had success sorting cells using antibodies directed against extracellular membrane GPCR proteins (not shown). The protein somatostatin is often only present in sections of cytoplasm and usually not found in the nucleus (see Figure 4), yet it is sufficiently abundant in the cell to easily sort SOM(+) cells of high purity with FACS. Consideration of background fluorescence is critical when determining placement of sort gates in order to select true antigen-positive populations. Background fluorescence in all channels tends to correlate with the scatter properties of particles, and gates are often best drawn on distributions of cells against the channel of interest and SSC-H to separate positive populations. Another option is to use a control antibody that will only bind non-specifically to provide an estimate of background in the channel of interest (Figure 9). Utilizing stricter, more conservative sort gates will facilitate increased sort purity although at the expense of decreased cell yield. It is important when validating any FACS gating methodology to verify purity of the sorted populations via microscopy and qPCR.
Dissociation is a critical factor in performing FACS on brain tissue. Too little dissociation leads to decreased cellular yield, large autofluorescent debris, and aggregates - events consisting of two or more cells stuck together, visible as multiple DAPI(+) peaks (Supplemental Figure 1). Too much dissociation can strip cells of their cytoplasm, leaving behind only nuclei and rendering sorting based on cytoplasmic makers impossible. Consistency is important in all dissociation procedures to achieve comparable and meaningful results between experiments.
Zinc-based fixation has certain advantages over paraformaldehyde, as it is less toxic and leaves at least one tested enzyme (ACE2) in a functional state. Whereas some antigens are preserved using this method (i.e., NeuN), others (i.e., parvalbumin) are not, so it may not be as versatile as paraformaldehyde-based fixation in terms of cell-type selection. Desired antigens must be individually tested for compatibility with zinc fixation. However, tissue dissociation is facilitated using zinc solutions as a fixative, hence larger numbers of cells can be sorted using this method. RNA can be readily extracted from zinc-fixed cells without special protocols. Note that we were unable to detect mouse-ace2 mRNA in WT or SA neurons, yet we were able to detect (very low) levels of ACE2 activity in WT neurons. This may indicate that genes with low levels of mRNA may be difficult to detect with the zinc protocol or that mRNA is degraded to some degree using this protocol. However, we note that all other expected mRNA species tested for were detected using zinc protocols.
In summary, the novel neurocytometry methodologies we describe here can be used to sort highly purified populations of individual types of mammalian brain cells that can be subsequently analyzed for gene expression and enzyme activity. Because cellular integrity is maintained, one can sort brain cells regardless of the location of the identifying antigen within the cell, as long as there is a specific antibody that can recognize the antigen. These methods expand the applicability of neurocytometry to tissues from non-genetically modified animals that are freshly prepared, frozen, or fixed. Significantly, routinely fixed human postmortem brain tissue can be sorted and analyzed.
Methods
Animals used in these studies were maintained in accordance with the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals, and all protocols were approved by the Institutional Animal Care and Use Committee of LSUHSC.
Sprague-Dawley rats (Harlan, Indianapolis, IN) weighing between 250–300 g (n=3) were singly housed in translucent home cages, kept on a 12:12 hr light/dark cycle, and had access to food and water ad libitum. Rats were anesthetized with isofluorane and decapitated. Brains were isolated within 3 minutes, and 2 mm coronal sections from a single hemisphere, from +3.2 mm to +1.2 mm (relative to Bregma), were sliced on a cold brain block, and immediately immersion fixed in ice-cold 4% paraformaldehyde in PBS. Control experiments shown in Figure 3 utilized 1.0 mm coronal sections from +1.2 mm to +0.2 mm.
Wild type C57BL/6 and transgenic synapsin-hACE2 (SA) mice12 were used (n=3). Adult mice were anesthetized and decapitated. Brains were isolated within 3 minutes and frozen in isopentane cooled by dry ice and stored at −80°C. The frontal cortices of these mice were dissected on a cold plate and thawed in an ice-cold solution of zinc salts.
Human brain tissue was supplied by the Louisiana State University Health Science Center Pathology Department, and all studies using this tissue met Institutional Review Board approval. Entire brains (n=4) were removed during autopsy and immersed in 10% formalin according to standard procedures. Postmortem interval between death and autopsy ranged from a minimum of 6 hours to a maximum of 36 hours.
Fixation methods:
A) Paraformaldehyde-based fixation: 2 mm thick coronal sections from one hemisphere of rat brain (freshly removed or previously frozen and stored at −80° C) were fixed in freshly prepared, ice-cold 4% paraformaldehyde in PBS for 12 hours. The anterior cingulate, motor, and somatosensory cortices were dissected from fixed sections and together removed to ice-cold PBS supplemented with 0.1% Tween-20 (PBS-T), and washed 4× for 10 minutes each. No white matter was included in dissections. Note: Previously frozen tissue may be thawed in fixative to achieve separation based on some proteins (eg., somatostatin) but not others (eg., parvalbumin), where non-previously frozen tissues must be used13.
B) Zinc-based fixation: The zinc fixative (0.1 M Tris-HCl pH=6.5, 0.5% ZnCl2, 0.5% Zn Acetate, 0.05% CaCl2, final pH=6.3) is kept at 4° C until use. The frontal cortices of previously flash-frozen mouse brains, stored at −80° C, were immersed in at least 10× volume of zinc-fixative solution at 4° C overnight. Tissues were then removed to ice-cold PBS before washing 3 times in PBS-T (20 min/wash).
C) Formalin-based fixation: Human brain tissues were collected at autopsy and stored in 10% Formalin at room temperature for a minimum of 30 days and a maximum of 51 days in prior to our obtaining them. A 2 mm thick section of frontal cortex was removed to cold PBS and stored at 4°C until processing. For processing, frontal cortex samples (~2 mm × 2 mm × 3 mm), containing all layers of grey matter and no white matter, were removed to ice-cold PBS-T overnight. Tissues were subsequently washed 4 times for 10 min in PBS-T prior to dissociation.
Dissociation techniques
Note: All steps are performed at 4° C or on ice unless otherwise noted. Centrifugation steps must be carried out using a swinging-bucket rotor centrifuge to avoid cell loss.
A) Paraformaldehyde fixation: Fixed and washed tissue is minced very finely on a cold, flat glass plate for at least 5 minutes in a drop of PBS-T using two new sharp flat razor blades. The sample is transferred with a 1 ml syringe (with no needle) into a low-protein binding microfuge tube. The volume is brought to 650 μl with PBS-T and the suspension is passed through a 19 Gauge needle back and forth 12×. The suspension is centrifuged at 250 g for 3 min, and the supernatant discarded. One ml of Accutase enzyme solution (Innovative Cell Technologies Inc, San Diego, CA) is added to the tissue pellet, and the mixture is shaken at room temperature for 1 hour (microfuge tube on its side on a rotary shaker at 200 rpm). The partially digested tissue is centrifuged at 250 g for 3 min, the supernatant discarded, and the tissue washed an additional 1× in cold PBS-T to remove excess enzyme. Four rounds of trituration are completed in 600 μl cold PBS-T, starting with 12 passes of the tissue through a 21 G needle, avoiding bubbles, followed by 5 min of settling on ice. The cloudy supernatant (~500 μl) is then removed to a separate low-protein binding tube, and additional PBS-T (500 ul) is added to remaining tissue for another round of trituration. The next round of trituration repeats the above steps using a 23 G needle, and the final two rounds use a 25 G needle. All supernatants are combined and filtered through a 70 um mesh prior to antibody staining and FACS. Filtered cells are spun at 250 g for 3 minutes in a swinging bucket centrifuge at 4° C. Supernatant is removed, and the cells resuspended in 200 μl PBS-T and stored at 4° C until staining.
B) Zinc Fixation: Fixed and washed tissue is minced very finely on a cold, flat glass plate for 3 minutes in a drop of PBS-T with two sharp flat razor blades as described above. The sample is transferred with a 1 ml syringe with no needle into a low-protein binding tube. Four rounds of trituration are completed in 600 μl cold PBS-T, starting with 12 passes of the tissue through a 21 G needle, avoiding bubbles, followed by 5 min of settling. The cloudy supernatant (~500 ul) is removed, following settling, to a separate low-protein binding tube, and additional PBS-T (500 ul) is added to remaining tissue for another round of trituration. The next round of trituration uses a 23 G needle and the final two rounds use a 25 G needle. All supernatants are combined and filtered through a 70 um mesh prior to antibody staining and FACS. Filtered cells are spun at 250 g for 3 minutes in a swinging bucket centrifuge at 4° C. Supernatant is removed, and the cells resuspended in 200 μl PBS-T and stored at 4° C until staining.
C) Human brain tissue (subject to prolonged formalin fixation after autopsy) – The dissociation method used was identical to that described for paraformaldehyde fixation, above. Filtered cells are spun at 250 g for 3 minutes in a swinging bucket centrifuge at 4° C. Supernatant is removed, and the cells resuspended in 200 μl PBS-T and stored at 4° C until staining.
Staining Methods
Cell suspensions can be divided into the appropriate number of tubes for a given experiment, or used in total, and were stained in PBS-T supplemented with 1 mM dithiothreitol and an RNAse inhibitor (RNAsin, Promega) at 1 unit/μl. Staining was carried out in low-protein binding microfuge tubes shaking vigorously (rotator platform at ~200 RPM) and protected from light. Centrifugation steps were carried out in a swinging bucket centrifuge.
A) Paraformaldehyde-fixed rat tissue from a single brain was split into two aliquots (100 μl each), and stained with one of the following combinations of antibodies for 1.5 hours at room temperature: 1) goat-anti-somatostatin (sc-7819, 1:100, Santa Cruz) and mouse-anti-calretinin (1:200, Swant); 2) rabbit-anti-parvalbumin (PV-25, 1:10,000, Swant) and mouse-anti-calbindin (1:400, Sigma). Tissues were then washed 2× for 5 min in cold PBS-T before a second-round of staining using the appropriate secondary antibody: anti-goat-AlexaFluor647 (1:750, Jackson), anti-mouse-AlexaFluor488 (1:750 Jackson), anti-rabbit-AlexaFluor647 (1:750, Molecular Probes). All secondary reagents were highly cross-adsorbed. Samples also received 4’,6-diamidino-2-phenylindole (DAPI) at a final concentration of 1.43 μM. Secondary antibodies were incubated for 30 min at 4° C with shaking and protected from light. Samples were washed 2× prior to sorting in PBS-T. (Alternatively, fluorophore-labeled primary antibodies can be used in all three methods. In this case, only a single incubation step and wash are necessary prior to FACS). Control experiments in Figure 3 on rat somatosensory cortex used either anti-somatostatin or anti-parvalbumin (at the above concentrations) in combination with anti-NeuN-PE (1:200, Millipore) and DAPI.
B) Filtered zinc-fixed cells from a single mouse frontal cortex were stained with anti-NeuN-PE (1:200, Millipore) and DAPI (1.43μM) in 1 ml of PBS-T for 1.5 hrs at 4°C on a rotating platform ~200 RPM. Cells were washed 1× before sorting in PBS-T.
C) Formalin-fixed human cells were stained at room temperature in 100 ul for 1.5 hours. Solutions contained rabbit-anti-parvalbumin (PV-25, 1:5,000, Swant) and anti-NeuN-PE (1:250, Millipore). Tissues were then washed 2× for 5 min and incubated with anti-rabbit-AlexaFluor647 (1:750, Molecular Probes) and DAPI (1.43 μM) for 30 min at 4°C on a rotating platform ~200 RPM. Cells were washed 2× and were sorted in PBS-T.
Flow Cytometric Analysis and Sorting
All samples were sorted on a FacsAria (Becton-Dickinson, San Jose, CA) at 45 psi using a 70 um nozzle. Sorted cells were collected into empty 1.7 ml microfuge tubes. The sample loader and collection tubes were kept at 4°C. Compensation for spectral overlap between PE and AlexaFluor-488 was calculated by FACSDiva software using single stained samples. 50,000–100,000 events from each sample were acquired to set gates prior to cell sorting.
A) Paraformaldehyde-fixed rat cells were first gated on DAPI-Area (DAPI-A) to exclude debris and select the nucleated cell population (average of all runs: 10.4% of all measured events) (Figure 1A, 2A). Another gate was drawn to exclude the top 5% of DAPI(+) events for side-scatter-height, as these events were found to contain high levels of autofluorescent debris (Figure 1B, 2B) (also see Supplementary Figure 1). Cells co-stained with somatostatin and calretinin were first gated according to somatostatin-488-height (488-H) vs. side-scatter-area (SSC-A), and cells clearly above background fluorescence levels in 488-H (which increases with SSC-A) were sorted as somatostatin(+) (average: 1.4%) (Figure 1C). The lowest ~90% of 488-H cells, somatostatin(−) cells, were gated for calretinin according to calretinin-647-height (647-H) vs SSC-A. Cells that were clearly above background (average: 0.7%) were sorted as calretinin(+) (Figure 1D). In separate tubes, cells co-stained for parvalbumin and calbindin were first gated according to parvalbumin-647-height vs SSC-A, and cells clearly above background levels were sorted as parvalbumin(+) (average: 1.2%) (Figure 2C). The lowest ~90% of cells, with respect to 647-H relative to SSC-A, parvalbumin(−) cells, were further gated based on calbindin-488-H. There are two populations of calbindin(+) cells (see Figure 2D); the population displaying the highest calbindin-488-H intensity (average: 0.8%) was sorted for downstream analysis (Figure 2D).
B) Zinc-fixed mouse cells were first gated on DAPI-A to exclude debris and include nucleated cells (average of all runs: 4.0% of all events) (Figure 6). Cells were then sorted on NeuN-PE-H vs SSC-H to separate neurons (PE(+): average 56% of cells) from non-neurons (PE(−): average 36%). Events with high SSC-H levels are contaminated with highly autofluorescent debris and were excluded from collection, Cells with intermediate levels of PE-H were also excluded from collection (Figure 6).
C) Formalin-fixed human cells were first gated on DAPI-A to exclude debris and include nucleated cells (Figure 9). 5% of DAPI(+) events exhibiting highest side-scatter were also gated out. The epitope for NeuN is destroyed by excessive formalin fixation, as noted elsewhere14. Background fluorescence intensity in the channel of interest correlates well with fluorescence in the PE channel, and therefore provides a convenient gating strategy for true positives (Figure 9). In parvalbumin(−) cells, 647-H increases with PE-A, and parvalbumin(+) cells (average of all runs: 1.2%) were selected as those cells that exhibit 647-H intensity clearly above background, relative to PE-A (Figure 9). Parvalbumin(−) cells were gated to include cells with the same range of PE-A intensity as parvalbumin(+) cells, but which displayed background levels of 647-H (average: 48%).
RNA Extraction
A) Paraformaldehyde-fixed sorted cells were pelleted at 13,000 g and resuspended in 70 μl RNA extraction buffer (15mM Tris-HCL, pH=8.0, 1% SDS, and 4 mg/ml Proteinase K) before incubation overnight (16 hours) at 60°C. This solution was then extracted with 150 μl of Trizol, phase separated with 7.5 μl bromo-anisole, and spun at 13,000 g for 15 minutes. The supernatant was added to an equal volume of isopropanol, supplemented with 1 μg of glycogen to aid RNA precipitation. RNA was washed in ethanol and dissolved in 10 μl nuclease free water for cDNA synthesis.
B) Zinc-fixed sorted cells were extracted with 100 μl of Trizol, phase separated with 5.0 μl bromo-anisole, and spun at 13,000 g for 15 minutes. The supernatant was added to an equal volume of isopropanol, supplemented with 1 μg of glycogen to aid RNA precipitation. RNA was washed in ethanol and dissolved in 10 ul nuclease free water for cDNA synthesis.
C) Formalin-fixed human neurons were extracted as paraformaldehyde-fixed rat cells were, except neurons were incubated for 40 hours in RNA extraction buffer before Trizol extraction.
QPCR
First strand cDNA was synthesized using the ImPromp-II kit from Promega (Madison, WI) using all RNA extracted from each sample. For quantitative RTPCR (qPCR) experiments, the Universal ProbeLibrary system (Roche Indianapolis, IN) was used to design primer/probe pairs. Primers were synthesized by IDT (Coralville, IA) (sequences provided in Table 1). Duplicate amplification reactions were performed on a Roche 480 LightCycler II using the Roche Light Cycler Master Mix following the manufacturer’s directions. For rat and mouse tissues, amplification of gapdh, a housekeeping gene, was performed simultaneously in every well to normalize amplification thresholds to input cDNA quantity. Note that gapdh levels may vary between cell type, therefore care must be taken in interpreting gene levels across cell types. For human tissues, gapdh was amplified in separate wells containing the same quantity of input cDNA, for normalization. Relative expression for each gene of interest (GOI) was determined by subtracting the gapdh amplification threshold from the GOI threshold for each sample, using the average point of 2nd derivative maximum, as estimated by the Roche software package. If no amplification was detected before cycle 38, a gene was considered not detectable. Statistical analyses of qPCR relative expression data were performed using a one-way analysis of variance (ANOVA) between compared groups for each GOI, or a t-test, as appropriate. Tukey’s multiple comparisons testing was used for ANOVA testing, and significance levels were set at p < 0.05.
Table 1:
Primer sequences used for the qPCR experiments to examine mRNA expression levels
| Gene | Left Primer | Right Primer |
|---|---|---|
| Rat gapdh | tgatggcatggactgtgg | ctgcaccaccaactgcttag |
| Rat ssf | agcccaaccagacagagaac | cctcatctcgtcctgctca |
| Rat pvlb | ttctggacaaagacaaaagtgg | ctgaggagaagcccttcaga |
| Rat calb2 | gccaaccgaagagaatttcc | gtgtcatacttccgccaagc |
| Mouse gapdh | atggtgaaggtcggtgtga | aatctccactttgccactgc |
| Mouse ace2 | gacatctcaaattccactgaagc | tccagggctctgaatttcc |
| Mouse grini | gcttttgcagccgtgaac | cgggctctgctctaccact |
| Mouse syn1 | cacagctggcccagaaac | ttggtcagagactgggatttg |
| Human gapdh | ag ccacatcgctcag acac | gcccaatacgaccaaatcc |
| Human pvlb | caaaattggggttgacgaa | ggggcagtcagtgcttctta |
| Human slc17a7 | ccaggaggagtttcggaag | cactcagctccagcgtctc |
| Human htr2a | tgatgtcacttgccatagctg | caggtaaatccagactgcacaa |
ACE2 Activity Assay
Cells were suspended in ACE2 reaction buffer (1M NaCl, 0.5mM ZnCl2, and 75 mM Tris-HCl, pH 6.5) with 0.5% (v/v) Triton X-100. ACE2 activity was determined as the difference in the hydrolysis rates of fluorogenic ACE2 substrate Mca-APK(Dnp)15. Assays were run at 37°C with 10 ul sample in 90 ul ACE2 reaction buffer containing 10 M Mca-APK(Dnp) in the absence or presence of 10 μM ACE2 inhibitor DX600 (Phoenix Pharmaceuticals, Inc., Burlingame, CA). The fluorescence emission at 405 nm, after excitation at 320 nm, was measured in a Spectramax M2 microplate reader from Molecular Devices (Sunnyvale, CA). Slope of fluorescence development between 10 and 120 min of incubation was calculated. The data are presented in fluorescence units (FU/min), as amounts of fluorescence substrate converted to product per minute and normalized by the number of cells used per well.
Microscopy
A drop of unsorted or sorted cells was placed onto a microscope slide and viewed with a Leica (DMRA2) epifluorescence microscope and photographed with a Cooke Sensi-cam camera at 63× and Slide Book software (Intelligent Imaging Innovations, Denver CO). For comparison of cell suspensions and sorted cells using a given fixative, all images were taken during one session. Exposure and display settings were identical between samples.
Supplementary Material
Acknowledgements
We would like to thank Dr. Bruce Hope and his laboratory at NIDA for assistance with training in dissociation procedures and FACS.
This work was partially supported by NIH grant P30GM106392 and P60AA009803.
References
- [1].Paden CM BD, Hapner SJ, Welsh CJ. (1986) A flow cytometric method for intracellular labeling and purification of rare neuronal populations: isolation of fixed neurophysin neurons., Brain Reseach, 310–319. [DOI] [PubMed] [Google Scholar]
- [2].Lobo MK, Karsten SL, Gray M, Geschwind DH, and Yang XW (2006) FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains, Nat Neurosci 9, 443–452. [DOI] [PubMed] [Google Scholar]
- [3].Guez-Barber D, Fanous S, Harvey BK, Zhang Y, Lehrmann E, Becker KG, Picciotto MR, and Hope BT (2012) FACS purification of immunolabeled cell types from adult rat brain, J Neurosci Methods 203, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guez-Barber D, Fanous S, Golden SA, Schrama R, Koya E, Stern AL, Bossert JM, Harvey BK, Picciotto MR, and Hope BT (2011) FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons, J Neurosci 31, 4251–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu QR, Rubio FJ, Bossert JM, Marchant NJ, Fanous S, Hou X, Shaham Y, and Hope BT (2014) Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS), J Neurochem 128, 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Smith SM, Kimyon RS, and Watters JJ (2014) Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method, ASN Neuro 6, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Finegersh A, and Homanics GE (2016) Chromatin immunoprecipitation and gene expression analysis of neuronal subtypes after fluorescence activated cell sorting, J Neurosci Methods 263, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martin DA, and Nichols CD (2016) Psychedelics Recruit Multiple Cellular Types and Produce Complex Transcriptional Responses Within the Brain, EBioMedicine 11, 262–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Saxena A, Wagatsuma A, Noro Y, Kuji T, Asaka-Oba A, Watahiki A, Gurnot C, Fagiolini M, Hensch TK, and Carninci P (2012) Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain, Biotechniques 52, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, Hodge R, McCarthy JK, Kelder M, McCorrison J, Aevermann BD, Fuertes FD, Scheuermann RH, Lee J, Lein ES, Schork N, McConnell MJ, Gage FH, and Lasken RS (2016) Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons, Nat Protoc 11, 499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herai RR, Stefanacci L, Hrvoj-Mihic B, Chailangkarn T, Hanson K, Semendeferi K, and Muotri AR (2014) Micro RNA detection in long-term fixed tissue of cortical glutamatergic pyramidal neurons after targeted laser-capture neuroanatomical microdissection, J Neurosci Methods 235, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feng Y, Xia H, Cai Y, Halabi CM, Becker LK, Santos RA, Speth RC, Sigmund CD, and Lazartigues E (2010) Brain-selective overexpression of human Angiotensin-converting enzyme type 2 attenuates neurogenic hypertension, Circ Res 106, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fuchtbauer EM, Rowlerson AM, Gotz K, Friedrich G, Mabuchi K, Gergely J, and Jockusch H (1991) Direct correlation of parvalbumin levels with myosin isoforms and succinate dehydrogenase activity on frozen sections of rodent muscle, J Histochem Cytochem 39, 355–361. [DOI] [PubMed] [Google Scholar]
- [14].Korzhevskii DE, Gilerovich EG, Zin’kova NN, Grigor’ev IP, and Otellin VA (2006) Immunocytochemical detection of brain neurons using the selective marker NeuN, Neurosci Behav Physiol 36, 857–859. [DOI] [PubMed] [Google Scholar]
- [15].Pedersen KB, Sriramula S, Chhabra KH, Xia H, and Lazartigues E (2011) Species-specific inhibitor sensitivity of angiotensin-converting enzyme 2 (ACE2) and its implication for ACE2 activity assays, Am J Physiol Regul Integr Comp Physiol 301, R1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gonchar Y, and Burkhalter A (1997) Three distinct families of GABAergic neurons in rat visual cortex, Cereb Cortex 7, 347–358. [DOI] [PubMed] [Google Scholar]
- [17].Kubota Y, Hattori R, and Yui Y (1994) Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex, Brain Res 649, 159–173. [DOI] [PubMed] [Google Scholar]
- [18].Rossier J, Bernard A, Cabungcal JH, Perrenoud Q, Savoye A, Gallopin T, Hawrylycz M, Cuenod M, Do K, Urban A, and Lein ES (2015) Cortical fast-spiking parvalbumin interneurons enwrapped in the perineuronal net express the metallopeptidases Adamts8, Adamts15 and Neprilysin, Mol Psychiatry 20, 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Evers DL, Fowler CB, Cunningham BR, Mason JT, and O’Leary TJ (2011) The effect of formaldehyde fixation on RNA: optimization of formaldehyde adduct removal, J Mol Diagn 13, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.