Abstract
Geriatric fractures take longer to heal and heal with more complications than those of younger patients; however, the mechanistic basis for this difference in healing is not well understood. To improve this understanding, we investigated cell and molecular differences in fracture healing between 5 month-old (young adult) and 25 month-old (geriatric) mice healing utilizing high-throughput analysis of gene expression. Mice underwent bilateral tibial fractures and fracture calluses were harvested at 5, 10, and 20 days post fracture (DPF) for analysis. Global gene expression analysis was performed using Affymetrix MoGene 1.0 ST microarrays. After normalization, data were compared using ANOVA and evaluated using Principal Component Analysis (PCA), CTen, heatmap, and Incromaps analysis. PCA and cross-sectional heatmap analysis demonstrated that DPF followed by age had pronounced effects on changes in gene expression. Both un-fractured and 20 DPF aged mice showed increased expression of immune associated genes (CXCL8, CCL8, and CCL5) and at 10 DPF, aged mice showed increased expression of matrix-associated genes, (Matn1, Ucma, Scube1, Col9a1, and Col9a3). Cten analysis suggested an enrichment of CD8+ cells and macrophages in old mice relative to young adult mice and, conversely, a greater prevalence of mast cells in young adult mice relative to old. Finally, consistent with the PCA data, the classic bone healing pathways of BMP, Indian Hedgehog, Notch and Wnt clustered according to the time post-fracture first and age second.
Keywords: Geriatric fracture healing, Bone regeneration, Molecular basis for fracture healing, Microarray, inflammation and fracture healing, Mouse model of fracture healing
Introduction
When long bone healing proceeds normally, the end result is restoration of normal bone morphology and near normal limb function. Failures in bone healing (such as nonunions), however, can result in persistent loss of function(1). While delayed unions and non-unions have proven difficult to accurately predict, there are factors recognized to increase risk; among them, the currently unmodifiable variable of advanced age, stands out as a particular concern(1,2). Older patients, when they do heal, can heal with insufficient mineral density and are also prone to re-fracture(3–5).
In secondary bone healing, the fracture site is initially filled by a hematoma marked by high levels of inflammatory cytokines and infiltrating innate immune cells(6,7). Following establishment of the hematoma, mesenchymal progenitors lead to a chondrogenic phase to form a soft callus(8–10), then hypertrophy and calcification occurs,(11,12) and, finally, ossification of the soft callus results in a bony callus that is remodeled into functional bone(13). Our current understanding of how these processes are altered at cellular and molecular levels during geriatric fracture healing are not well-understood.
Murine models of fracture healing have become a standard method for the assessment of cellular and molecular influences on bone repair, and, increasingly, fracture healing models in aged mice have been used to better understand alterations and failures in geriatric human populations. Previous gene studies in bone healing suggest the molecular complexity of the repair process, with almost 600 known genes and over 100 novel genes(14). And while specific elements of classic canonical bone formation pathways such as Wnt/beta-catenin(15,16) and BMP(17,18) have been studied as a function of aging fracture healing, they have not been evaluated together within the context of global gene expression nor have they been well studied in very old (rather than moderately aged) mice. Moreover, there has not been a transcriptome-level analysis of fracture healing in aged mice.
Therefore, one of our goals was to characterize the differential dysregulation of classical canonical signaling pathways, such as Wnt, BMP and Notch (within an overarching goal of identifying patterns of differences for further study), as well as to identify as of yet unknown gene regulatory pathways that fundamentally impair healing in our aged fracture model. Based on our previous tissue and cell based analysis(19), we expected that very old mice would lag temporally behind young adult mice in the expression of genes important for regeneration when compared to young fracture healing. Within that context, we also hypothesized that evidence of increase in inhibitory processes, such as sustained inflammation(20,21) would characterize the delay in aged fracture healing.
Improved understanding of the biological differences in fracture healing between young adult and geriatric populations will offer a basis for targeted therapeutic intervention. We have previously characterized altered healing patterns in a mouse model of geriatric fracture healing that reflects what is observed in humans(19). To further our understanding beyond the tissue and cellular levels and to begin the identification of signaling pathways and genetic networks for potential therapeutic manipulation, we have investigated the molecular differences between old and young adult fracture healing by characterizing the gene expression profile of fracture calluses in young adult and old mice. Better understanding of differential gene expression between young and old mice as it relates to the stages of fracture healing will enable more rationally designed studies of functionally significant genes and pathways that may explain differences in healing outcomes between young and aged cohorts.
Methods
Animal Experimentation and Surgical Model
All animal procedures were approved by our Institutional Animal Care and Use Committee. 25-month-old (m/o) C57BL/6 mice represent 50 to 75% survival (hypothesized to be consistent with humans of advanced age) whereas 5 m/o mice represent young adult status; all mice were obtained from the NIH aged rodent colony (https://www.nia.nih.gov/research/dab/aged-rodent-colonies-handbook) and underwent transverse bilateral tibial fractures as previously described(19,20). Briefly, mice were administered buprenorphine and isoflurane, their legs were prepared aseptically, prestabilized with an intramedullary pin (the same sized pin was used for all animals), and a traumatic closed fracture was created with a blunt guillotine (three-point bend mechanism). The mice were allowed to move freely and given buprenorphine/Nutella ® (Ferrero USA, Inc, Somerset, NJ, USA) for 3 additional days and euthanized via CO2 inhalation and cervical dislocation.
Mouse tibial fractures were gathered at 4 time points (pre-fracture, 5 days post fracture, 10 days post fracture, 20 days post fracture) and 2 age groups (5 months old, 25 months old). Five biological replicates, 1 each from 5 distinct mice, were gathered at each time/age point except for 25 months old, pre-fracture, where 6 replicates were gathered. Based on previous fracture and gene expression studies (and in consultation with the Microarray Core Facility), we estimated that 5 independent animal samples would provide appropriate power for statistical validation of microarray data accounting for variations between animals. In total the experiment used 21 mice at 25-months-of-age and 20 mice at 5-months-of-age. Partek genomics suite was used for quality control, normalization, comparisons, and analysis. For the time point 0, we used whole diaphyseal bone because no fracture was created and this would provide a baseline gene expression profile against which to compare the fracture healing time points. For all subsequent time points, the callus was carefully dissected from the remaining bone ends. For RNA extraction, after the mouse was euthanized the samples (either whole bone or just callus) were lysed in TriZOL (ThermoFisher, Waltham, MA) then homogenized via tissue tearor, and, finally, frozen to release RNA. RNA quality was assed via Agilent Bioanalyzer 2100.
Microarray assay
Microarray services were provided by the University of Pennsylvania Molecular Profiling Facility, including quality control tests of the total RNA samples by Agilent Bioanalyzer and Nanodrop spectrophotometry. All protocols were conducted as described in the NuGEN Ovation Pico WTA system v2 user guide and the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, 50ng of total RNA was converted to first-strand cDNA using reverse transcriptase primed by poly(T) and random oligomers that incorporated an RNA priming region. Second-strand cDNA synthesis was followed by ribo-SPIA linear amplification of each transcript using an isothermal reaction with RNase, RNA primer and DNA polymerase, and the resulting ssDNA was assessed by Bioanalyzer, fragmented and biotinylated by terminal transferase end labeling. Five and a half micrograms of labeled cDNA were added to Affymetrix hybridization cocktails, heated at 99°C for 5 min and hybridized for 16 h at 45°C to Mouse Gene 1.0 ST GeneChips (Affymetrix Inc., Santa Clara CA) using the GeneChip Hybridization oven 645.
The microarrays were then washed at low (6× SSPE) and high (100mM MES, 0.1M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A GeneChip 3000 7G scanner was used to collect fluorescence signal. Affymetrix Command Console and Expression Console were used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters.
Five biological replicates were captured at each time point and age group, with a sixth replicate for 25-month-old, 0 DPF mice, yielding a total of 41 samples across eight conditions. Affymetrix probe intensities (.cel files) were imported into Partek Genomics Suite (v6.6, Partek Inc., St. Louis, MO) were log2-transformed expression intensities were calculated with RMA. To correct for batch effects, ComBat(22) was applied. Partek was subsequently used to perform an ANOVA analysis on age-DPF interaction, to create comparisons between age groups and DPF time points, and to generate of a principal component analysis (PCA). Partek’s comparisons are based on two-group “contrast” comparison as part of the ANOVA analysis; this method leverages better statistical power in variance estimation versus Student’s t test. Because each tibial sample was from a different mouse, comparisons were unpaired.
Top 10 table
Two group comparison results were ranked by fold-change, taking the top 10 largest magnitude fold-change genes, across increases and decreases in gene expression, respectively. Genes secondarily required a p-value less than 0.005 for inclusion on the list. Whereas comparisons across DPF time points yielded many statistically significant genes after multiple test correction, comparisons across age within the same DPF time point did not always produce significant genes after multiple test correction. For this reason, and due to the small sample size and nuanced expression differences between age groups, a fold-change rank based approach was taken.
Removal of muscle associated genes
Due to the sensitivity of RNA-based expression assays to heterogeneous cellular composition, all genes associated with muscle tissue were removed from the top 10 tables and heatmaps. The muscle contamination gene list was taken from an empirically produced, publicly available gene list from Ayturk et al.(23) Other visualizations (i.e. cell type associated gene heat map, InCroMap visualizations) did not omit muscle contamination associated genes as those gene lists were predefined and therefore not subject to change. For completeness, the same heatmaps and top 10 tables as shown were also created without removing muscle contained associated genes (Supp. Fig. 1, Supp Table 1, and Supp Table 2).
Heatmap generation
R was used to visualize heatmaps via the built-in “heatmap” function. The top 5 greatest fold-change genes, subject to p<0.005, from each two group comparison, after removing muscle contamination associated genes, were used to make a cross-sectional heatmap. Subsequent heatmaps related to signaling pathways and cell type markers were also generated via R's heatmap function, but without removal of muscle contamination genes or filtered based on p-value (Supp. Fig. 1). Signaling and cell type indicator genes were chosen via manual curation from the literature.
CTen analysis
At each time point, genes differentially expressed between young and old mice were loaded into CTen(24) for a two group (young vs. old) comparison, requiring p<0.01 in Partek's two group comparison and a fold-change greater than 1.3 for inclusion in the gene list. The typical list size used by CTen was between 100–500 genes, which is slightly smaller than CTen's reference gene lists. Whereas CTen was designed to derive cell type from absolute gene expression values, here it was given differential genes in an effort to deduce which cell types were present in different amounts between the two groups.
InCroMap visualization
Biological pathway figures were generated using InCroMap(25), which images quantitative values overlayed atop Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Fold-change measures from Partek's two group comparisons were used as input to InCroMap's visualization tool. InCroMap's internal pathway statistical significance tool was used to calculate p-values on differential expression within each pathway by selecting genes with fold change greater than 1.5 between age groups at each time point.
Results
Fracture callus gene expression patterns clustered by healing time and animal age
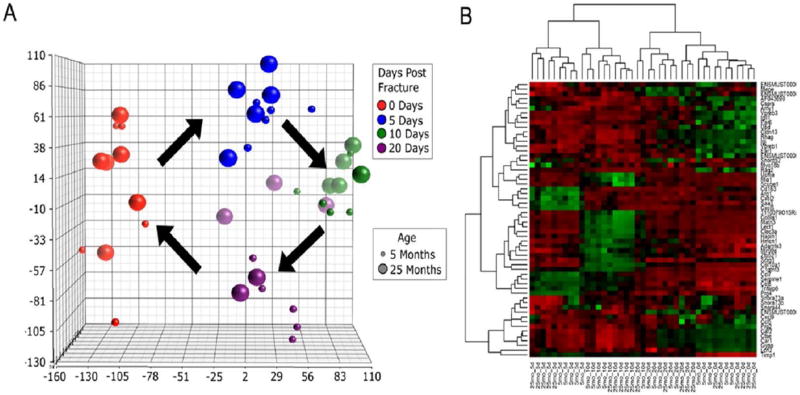
Principal component analysis (PCA) indicated a cyclical progression of overall gene expression following fracture (Fig. 1A). As time post-fracture progressed, differential gene expression was greatest at 10 days post fracture (DPF) and moved closer to baseline by 20 DPF. The analysis also indicated that gene expression clustering was secondarily determined by age of animals. While aged mice show a cyclical progression in gene expression similar to young mice, the older mice show greater heterogeneity at day 20 DPF with some aged individuals appearing to show a pattern more similar to 10 DPF (Fig. 1A).
Fig. 1. Principle component and hierarchical clustering analysis of gene expression.
(A) Principal component analysis indicates a cyclical progression of gene expression during fracture healing. As time progresses, differential gene expression is greatest at 10 days and moves closer to baseline by 20 days. Gene expression profiles clustered primarily on the basis of DPF, then by age. (B) Hierarchal clustering analysis of top 5 most differentially expressed genes at each DPF. Gene expression clustered initially on days post fracture and secondarily on animal age. Red indicates higher level of expression, whereas green indicates lower level of expression.
To further investigate the components governing gene expression during fracture healing, hierarchical clustering analysis was performed on the top five most differentially expressed genes from each of time point. As with principal component analysis, gene expression clustered initially on DPF and secondarily on animal age (Fig. 1B) with 20 DPF clustering closer to 0 DPF relative to 10 DPF.
Top ten differentially expressed genes during fracture healing progression were identified using a size-adjusted step-up p-value of less than 0.005 and then sorted according to fold-change comparing 5 versus 0, 10 vs 5, and 20 vs 10 DPF in each age group (Table 1). At 5 DPF, there were seven common upregulated genes and four common downregulated genes comparing 5-month-old to 25-month-old mice (55% of the top regulated genes were common between 5- and 25-month-old mice at day 5). At 10 DPF six genes were upregulated in common, and five were downregulated (55% of the top regulated genes are common between 5- and 25-month-old mice at day 10), while at 20 DPF, three genes were upregulated in common, and five were in common downregulated (40% of the top regulated genes are common between 5- and 25-month-old mice at day 20). Patterns of expression from 0 to 5, 5 to 10, and 10 to 20 DPF were consistent, regardless of age, with inflammation followed by mesenchymal activation, inflammatory clearing, and then chondrogenesis and bone formation.
Table 1.
Top ten up- and down-regulated genes during fracture healing between time points post fracture.
| 5DPF vs Unfractured | 10DPF vs 5 DPF | 20 DPF v 10 DPF | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5-Months Old | 25-Months Old | 5-Months Old | 25-Months Old | 5-Months Old | 25-Months Old | |||||||||||||
| Gene Name |
Fold Change |
q-value | Gene Name |
Fold Change |
q-value | Gene Name |
Fold Change |
q-value | Gene Name | Fold Change |
q-value | Gene Name | Fold Change |
q-value | Gene Name | Fold Change |
q-value | |
| Upregulated Genes | Ccl8 | 51.7068 | 5.46E-13 | Timp1 | 16.319 | 8.89E-12 | Clec3a | 32.8468 | 2.10E-07 | Clec3a | 72.6445 | 1.80E-09 | Car1 | 7.60468 | 0.00347466 | Car1 | 25.0829 | 1.85E-05 |
| Ccl7 | 23.0152 | 2.70E-10 | Tnfaip6 | 15.7304 | 1.02E-10 | Col10a1 | 25.5779 | 2.87E-09 | Col10a1 | 33.574 | 2.17E-10 | Ctse | 4.21029 | 0.0215834 | Gypa | 17.9 | 0.000184688 | |
| Timp1 | 19.4469 | 1.83E-12 | Ccl7 | 15.6649 | 3.29E-09 | Matn3 | 11.3424 | 1.12E-06 | Matn3 | 27.0598 | 2.36E-09 | Car2 | 4.02812 | 0.0117012 | Rhag | 17.677 | 0.000119596 | |
| Serpine1 | 18.7733 | 8.26E-13 | Prg4 | 14.4678 | 2.87E-08 | Lect1 | 11.2742 | 2.19E-07 | Lect1 | 22.9154 | 7.47E-10 | Gdpd2 | 4.01189 | 4.90E-05 | Ctse | 15.9103 | 4.13E-05 | |
| Tnfaip6 | 18.532 | 3.26E-11 | Serpine1 | 14.4383 | 8.89E-12 | Scrg1 | 11.1695 | 4.76E-06 | Hapln1 | 19.6306 | 1.42E-06 | Cldn13 | 3.87113 | 0.0223476 | Car2 | 12.9331 | 2.11E-05 | |
| Sfrp1 | 18.0681 | 4.70E-10 | Cthrc1 | 14.1115 | 2.96E-09 | Hapln1 | 10.8943 | 5.57E-05 | Acan | 16.6014 | 1.51E-06 | Stab2 | 3.75166 | 1.10E-05 | Slc4a1 | 12.1416 | 0.000588406 | |
| Saa3 | 17.6519 | 1.26E-10 | Vcan | 13.6722 | 8.89E-12 | Zim1 | 7.14084 | 2.87E-09 | Ucma | 15.5522 | 5.89E-10 | Spna1 | 3.55237 | 0.0166092 | Cldn13 | 11.0874 | 0.000127954 | |
| Ms4a6d | 17.2707 | 1.44E-11 | Ccl8 | 12.3968 | 9.52E-10 | Agtr2 | 6.4351 | 4.05E-06 | 3110079O15Rik | 15.2108 | 4.91E-08 | Snca | 3.26757 | 0.00742677 | Epb4.2 | 10.0517 | 0.000131137 | |
| Cxcl5 | 17.0611 | 2.31E-08 | Sfrp1 | 11.2241 | 1.35E-08 | Chad | 6.35486 | 0.000321803 | Scrg1 | 14.6 | 5.74E-07 | Gal | 3.23887 | 0.000977659 | Igk-V1 | 9.9375 | 7.79E-05 | |
| Ccl12 | 16.0445 | 5.79E-13 | Saa3 | 10.7828 | 4.84E-09 | Lipi | 6.19965 | 9.35E-09 | Col9a1 | 13.4898 | 6.44E-11 | Ms4a1 | 3.13358 | 0.00492443 | Kel | 9.14906 | 0.000165466 | |
| Downregulated Genes | Gypa | −36.7476 | 1.16E-06 | Ear1 | −20.2929 | 7.23E-07 | Cxcl5 | −14.468 | 9.92E-07 | Cxcl5 | −11.0661 | 2.37E-06 | C1qtnf3 | −19.9331 | 1.54E-07 | Matn3 | −32.2182 | 1.52E-09 |
| Rhag | −31.0394 | 1.28E-06 | Ear6 | −14.6878 | 1.02E-07 | Arg1 | −10.049 | 2.87E-09 | Cxcl2 | −10.2895 | 8.44E-06 | Scrg1 | −15.8569 | 5.18E-07 | Hapln1 | −23.8115 | 9.30E-07 | |
| Car1 | −29.982 | 1.07E-06 | Vpreb1 | −11.9426 | 1.62E-09 | Saa3 | −9.149 | 3.36E-07 | Saa3 | −6.66003 | 1.82E-06 | Matn3 | −14.8953 | 1.92E-07 | Lect1 | −22.1161 | 1.52E-09 | |
| Cldn13 | −24.9896 | 2.09E-07 | Prg2 | −11.7774 | 2.52E-06 | Cxcl2 | −8.7526 | 4.17E-05 | Car1 | −6.11081 | 0.00622303 | Lect1 | −11.9316 | 1.55E-07 | Scrg1 | −21.246 | 9.87E-08 | |
| Car2 | −22.3932 | 1.11E-07 | Vpreb3 | −11.4993 | 1.05E-09 | Prg4 | −8.4259 | 1.39E-05 | Cd163 | −5.7244 | 5.75E-07 | Hapln1 | −11.3573 | 3.71E-05 | Clec3a | −20.9901 | 1.41E-06 | |
| Ctse | −22.3846 | 9.61E-07 | Ly6g | −10.0661 | 1.36E-05 | Il1rl1 | −6.0817 | 2.03E-06 | Prg4 | −5.33996 | 0.000171637 | AF357355 | −8.56187 | 5.63E-06 | C1qtnf3 | −19.9725 | 9.87E-08 | |
| Kel | −21.8589 | 1.50E-07 | Rhag | −9.62421 | 0.000333338 | Il6 | −5.8171 | 0.000249328 | Gypa | −5.20701 | 0.0188113 | 1500015O10Rik | −8.54579 | 3.60E-07 | Ucma | −17.4602 | 3.72E-10 | |
| Rhd | −21.5165 | 3.67E-07 | Igk-V1 | −9.49088 | 1.30E-05 | Ccl2 | −5.6197 | 1.30E-05 | Arg1 | −5.02311 | 7.91E-07 | Ccl8 | −8.1571 | 4.23E-07 | Acan | −16.2878 | 2.73E-06 | |
| Ly6g | −20.394 | 2.50E-07 | Mcpt8 | −8.78738 | 9.34E-08 | Ms4a4a | −4.8786 | 2.05E-07 | Tspan8 | −5.01726 | 0.000373912 | Mfap4 | −7.54815 | 5.80E-08 | Col9a1 | −12.442 | 2.55E-10 | |
| Ear1 | −20.3285 | 8.70E-07 | Gypa | −8.77516 | 0.00083786 | Cd163 | −4.8702 | 4.33E-06 | Serpina3n | −4.88816 | 7.54E-07 | Zim1 | −7.41162 | 3.88E-09 | 3110079O15Rik | −10.0848 | 1.23E-06 | |
Bold genes are present in both 5 and 25 month
10 differentially up-regulated and down-regulated genes for old vs. young mice at each time point post fracture were also identified (Table 2). Consistent with PCA and hierarchical clustering analyses, comparisons between time points (Table 1) produced greater differential gene expression than did comparisons between age groups (Table 2). Both un-fractured and 20 DPF aged mice showed increases in immune associated genes, in particular CXCL8, CCL8, and CCL5 were more highly expressed than in 5-month-old mice. At 10 DPF, the aged mice show increases in a number of matrix associated genes, in particular Matn1, Ucma, Scube1, Col9a1, and Col9a3. Genes that were increased in the 5-month-old mice relative to 25-month-old mice, did not show clear patterns, except for increases in Mir99a and Mr15a at day 5, several genes that encode for small nucleolar RNAs at days 5, 10, and 20, and a set of Mup genes that are classified as major urinary proteins.
Table 2.
Top 10 up- and down-regulated between old versus young mice at each time point post-fracture.
| 25-month vs 5 month | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unfractured | 5 Days Post Fracture | 10 Days Post Fracture | 20 Days Post Fracture | |||||||||
| Gene Name | Fold Change | p-value | Gene Name |
Fold Change | p-value | Gene Name | Fold Change | p-value | Gene Name |
Fold Change | p-value | |
| Higher Expression at 25-Months | Cxcl9 | 3.9176 | 0.0003 | Igj | 3.88103 | 0.0006 | Ucma | 3.21115 | 1.47E-05 | Ccl8 | 3.72603 | 5.58E-06 |
| Ccl8 | 3.82189 | 1.99E-06 | Amy1 | 3.54147 | 1.56E-05 | Mia1 | 2.77897 | 4.05E-05 | Cxcl9 | 3.66272 | 0.0009 | |
| Ubd | 3.54092 | 4.31E-05 | Tspan8 | 2.92372 | 0.0019 | 3110079O15Rik | 2.42711 | 0.004 | Ccl5 | 3.52583 | 0.0003 | |
| Ccl5 | 2.98618 | 0.0007 | Krt18 | 2.89207 | 6.70E-05 | Scube1 | 2.13944 | 4.99E-05 | Lcn2 | 3.15219 | 0.0015 | |
| Csprs | 2.61911 | 3.39E-08 | Nrap | 2.74706 | 0.0022 | Col9a1 | 2.13406 | 0.0004 | AW112010 | 2.94374 | 4.93E-05 | |
| Rtp4 | 2.55427 | 6.54E-05 | Camk2b | 2.63232 | 8.52E-06 | Moxd1 | 2.07754 | 0.0029 | Pglyrp1 | 2.72714 | 0.0012 | |
| Oasl2 | 2.52529 | 6.00E-05 | Cmya5 | 2.62953 | 0.0013 | Col9a3 | 2.00654 | 0.0001 | Apol10a | 2.66505 | 0.0016 | |
| Gm7609 | 2.50707 | 6.22E-08 | Trdn | 2.56702 | 0.0049 | Matn1 | 1.95358 | 1.54E-05 | Ms4a4b | 2.64142 | 0.0001 | |
| Nlrc5 | 2.4735 | 4.46E-06 | Fsd2 | 2.53774 | 0.001 | Cspg4 | 1.8078 | 0.004 | Klrc1 | 2.63664 | 0.0032 | |
| Gm7609 | 2.45712 | 2.48E-08 | Gm10674 | 2.51765 | 0.0015 | Mfge8 | 1.80739 | 0.0003 | Lrg1 | 2.61073 | 0.0008 | |
| Lower Expression at 25-Months | Adamts3 | −1.66205 | 0.001 | Mir99a | −3.10473 | 0.0003 | Mup7 | −4.62095 | 0.0032 | Vpreb1 | −2.29738 | 0.0019 |
| Rag2 | −1.62362 | 0.0003 | Snora73a | −3.06453 | 0.0002 | Mup11 | −4.4348 | 0.0028 | Hmcn1 | −2.1022 | 0.0003 | |
| Mepe | −1.59314 | 0.004 | Snora73b | −3.03144 | 0.0002 | Mup19 | −4.23417 | 0.0031 | Igll1 | −1.85577 | 0.0015 | |
| Zfp521 | −1.51664 | 0.004 | Snora44 | −2.74157 | 0.001 | Mup2 | −4.11126 | 0.0031 | Snord52 | −1.82012 | 0.0002 | |
| Eif2b3 | −1.40897 | 0.0002 | Crabp1 | −2.69092 | 1.55E-05 | Mup1 | −3.26439 | 0.0031 | Hmcn1 | −1.70504 | 0.0038 | |
| 2310047B19Rik | −1.3452 | 0.0006 | Npm3 | −2.04601 | 1.56E-05 | Snord118 | −2.74323 | 0.0005 | Syne2 | −1.61705 | 0.0015 | |
| 2700097O09Rik | −1.33643 | 0.0032 | Fcrls | −2.03889 | 2.11E-05 | Adipoq | −2.49978 | 0.0009 | Hmcn1 | −1.58635 | 0.0033 | |
| Etv1 | −1.33367 | 0.0001 | Mir15a | −1.99839 | 0.0029 | Mcpt4 | −2.38792 | 3.50E-06 | Prss48 | −1.57496 | 0.0001 | |
| Epha4 | −1.31656 | 0.0022 | Mir15a | −1.98857 | 0.0002 | Snora23 | −2.26599 | 0.0035 | Arhgef5 | −1.5678 | 0.0005 | |
| Mrpl21 | −1.28749 | 0.0005 | Arsi | −1.92852 | 0.0007 | Vpreb1 | −2.17515 | 0.0034 | Fign | −1.55236 | 0.0004 | |
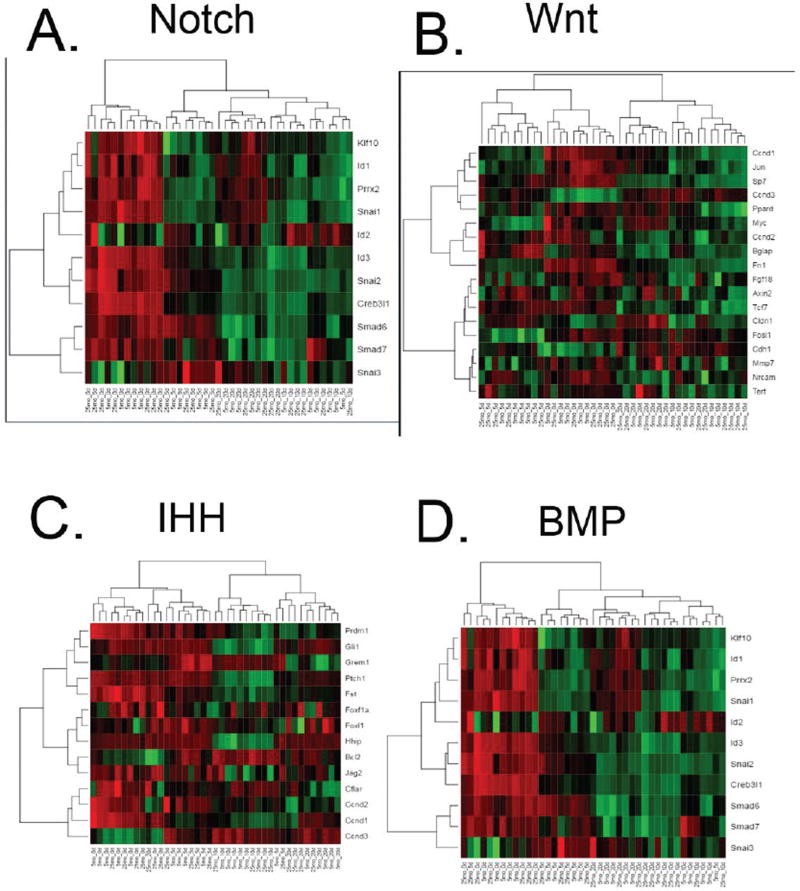
To examine activation of signal pathways classically relevant to bone formation, target genes representing activation of bone morphogenetic protein (BMP), Indian hedgehog (IHH), Notch, and Wnt pathways were subjected to hierarchical clustering analysis. As with overall gene expression, clustering of all signaling pathways was first on the basis of time post fracture, then by age group (Fig 2). The Notch pathway was expressed higher at earlier time points, 0 and 5 DPF, compared to later time points for both young and old mice (Fig 2C). The highest level of Wnt and BMP signaling associated genes was at 0 DFP for both young and old mice, with moderate expression at 5 and 10 DFP (Fig 2A, D).
Fig. 2. Hierarchical clustering of signal pathway target genes.
Analysis of (A) BMP, (B) IHH, (C) Notch, and (D) Wnt target genes cluster first on the basis of time post fracture, then by age group.
Cell type prevalence and proliferation differs based on animal age
Differential enrichment of specific cell types between old and young mice at individual time points was assessed using CTen (http://www.influenza-x.org/~jshoemaker/cten/)(24). Data were consistent with greater prevalence of immune cells in geriatric mice relative to young mice (Fig 3) including the saturation of CD8+ cells and macrophages in old mice relative to young mice. Conversely, there was evidence of mast cells in young mice relative to old mice.
Fig. 3. Cell Type Enrichment (CTen) analysis of gene expression.
Differential enrichment of specific cell types between old and young adult mice at individual time points was assessed using CTen. Aged mice showed a greater tendency towards CD8+ T cell and activated macrophage enrichment, where younger cells showed a greater tendency towards mast cell and osteoblast enrichment.
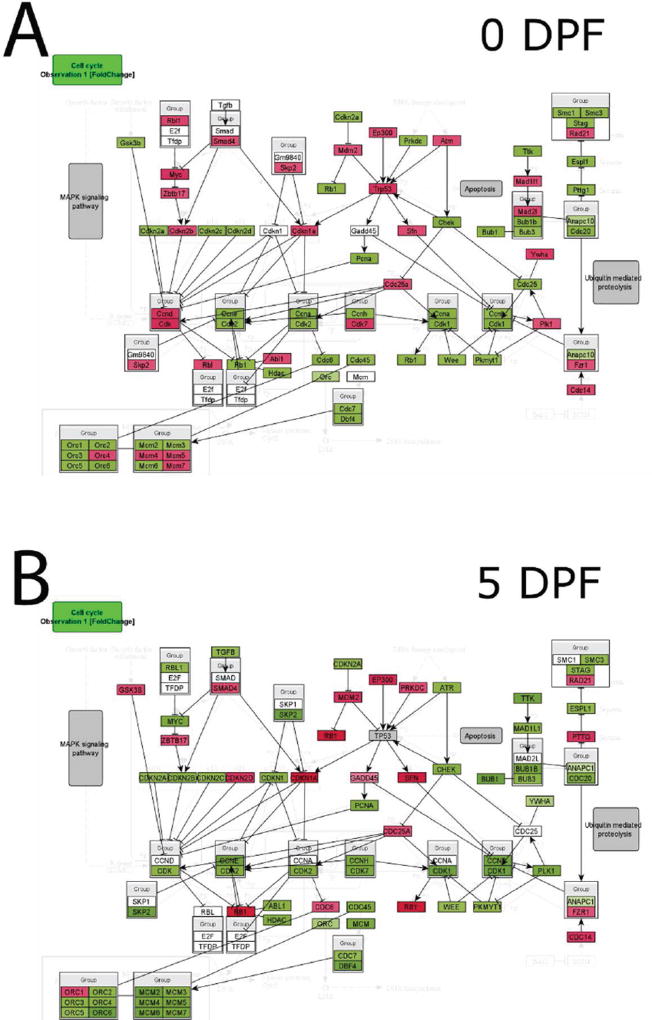
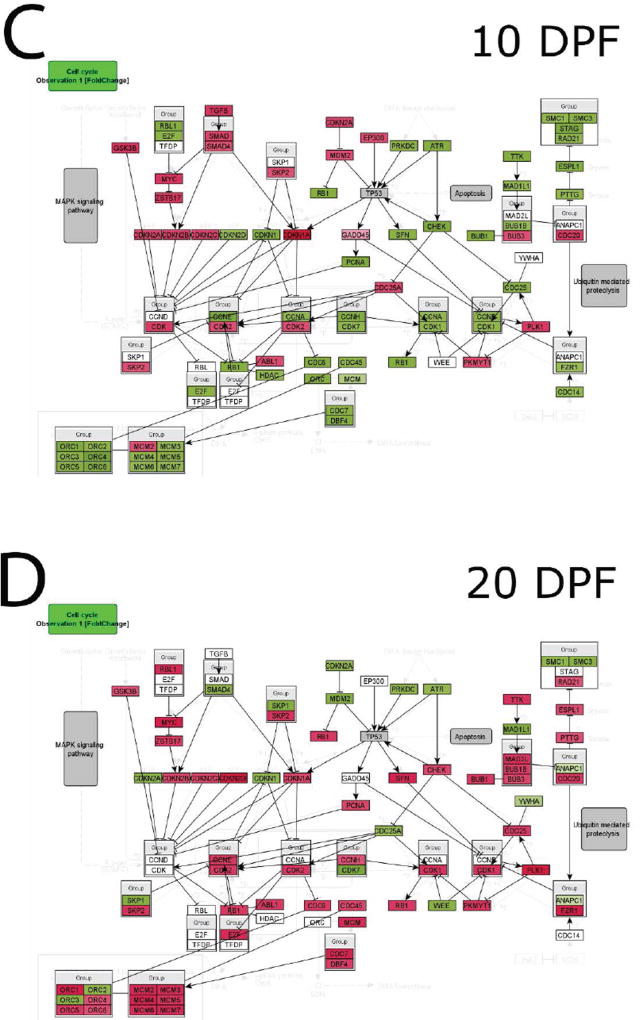
To investigate potential alterations in cell cycle progression during fracture healing in old versus young mice, InCroMap was used to visualize cell cycle regulatory genes comparing expression in old and young mice at each time point following fracture (Fig. 4). At 0, 5, and 10 DPF, young mice demonstrated higher expression of cell cycle progression genes (Fig. 4 A–C). Old mice, however, showed higher expression of cell cycle progression genes at 20 DPF (Fig. 4 D).
Fig. 4. Cell cycle marker gene expression analysis.
InCroMap was used to visualize cell cycle regulatory genes comparing expression in old and young adult mice at (A) 0, (B) 5, (C) 10, and (D) 20DPF. Green represents higher expression in young adult mice, whereas red indicates higher expression in old mice. At 0, 5, and 10 DPF, young adult mice demonstrated higher expression of cell cycle progression genes. Old mice showed higher expression of cell cycle progression genes at 20 DPF.
Discussion
It is well-accepted that fracture healing in aged patients occurs less robustly than in young individuals(19,26). The molecular mechanisms underpinning this poor healing, however, are not well defined. In order to identify aged-based divergence in the cellular milieu and variations in signaling pathway activation, we performed gene expression analyses comparing young adult (5 months) and geriatric (25 months) mice throughout the fracture healing process.
Principal component analysis of global gene expression and hierarchical clustering showed clustering primarily on the basis of time post-fracture (Fig 1 A, B) and secondarily on age. Gene expression from aged mice at 20 DPF were closer to 10 DPF young adult potentially reflecting the lag in the fracture healing morphologically of aged mice compared to young adult. This is graphically represented in Figure 1 where the 10 day clustering of both ages (in all 3 dimensions) appears to proceed to continued clustering in the young cohort (small purple circles) but less so in the aged (large purple) where 2 animals cluster closer to the young but 3 cluster closer to the 10 day in the Y, X, and Z axes (1, 2 and 3 animals, respectively).
Cell Type Enrichment (CTen) analyses(24) indicated that cells of both innate and adaptive immunity were more highly enriched in aged mice (Fig 3) throughout the healing process, particularly at 0 DPF and 20 DPF. Furthermore, the enrichment of gene expression, in aged mice, approximating that of LPS stimulated macrophages is consistent with an underlying basal inflammatory state often associated with aging. Similarly, in aged 20 DPF fractures, we detected enrichment of CD8+ T cells, which has been shown to negatively impact bone regeneration(21,27). We also found differential immunological pathway activation between age groups at 0 DPF and 20 DPF, which further supports the difference in activity of immune cells between young adult and old mice (Supp Table 3). Conversely, we found mast cells, which have been identified as necessary for proper bone healing, were enriched in young adult fractures at the midpoint of the healing process (10 DPF)(28–30) but not in aged fracture healing. Taken together, these cell type enrichment data suggest an “immune” cellular environment that represses healing in the aged mice relative to the young adult mice.
Examination of individual genes during the fracture healing process further supported a basal pro-inflammatory state and an increased variation in bone formation in aged mice. Prior to fracture and late in the fracture healing process, aged mice demonstrated higher expression of the proinflammatory cytokine CCL8, the T cell chemoattractant CXCL9, and the T cell-secreted cytokine CCL5 (Table 2)(31–33). Aged mice showed decreased expression of Matrix Extracellular Phosphoglycoprotein (MEPE), which is a component of mineralizing bone matrix and Ephrin receptor A4 (Epha4), which has been implicated in osteoblast maturation (Table 2)(34,35). Furthermore, aged mice show lower 5 DPF expression of Mir-15a (Table 2), whose pathologic decreased expression in Myasthenia Gravis has been shown to promote proinflammatory cytokines production(36). Finally, Mup1, a regulator for glucose and lipid metabolism in mice(37,38), and other Mup genes are down regulated at 10 DPF in aged mice, possibly to conserve energy to promote fracture healing (Table 2).
We also analyzed classic canonical signaling pathways (BMP, Wnt, Notch, Hedgehog) that are known to be important to both bone formation and healing. Clustering of signaling pathways demonstrated that days post-fracture followed by age were drivers of differential gene expression (Fig 2). The higher earlier levels of Notch signaling in old and young adult fractures which decreases with time may suggest that sustaining signaling could better sustain the proliferative stages of healing. In combination with our previous study which indicated that low basal-levels of Notch signaling in mesenchymal progenitor cells of geriatric mice were still inducible(19), these results support the further investigation of the Notch pathway as a therapeutic target. On the other hand, the lack of substantial age-based differential relative expression in classical osteogenic pathways suggests that investigation of potential therapeutic targets for improving age-associated fracture healing should be expanded.
Finally, we compared the expression levels of cell cycle regulatory genes. Minimal differences are observed in the unfractured bone at 0 DPF (Fig 4A). At 5 DPF, however, there was an increased activation of cell cycle genes in young adult mice in comparison to old mice (Fig 4B). By 20 DPF, there was an increase in activation of cell cycle genes in geriatric mice (Fig 4D). These data suggest that old mice, while capable of initiating regeneration, lag behind young adult mice during the fracture healing process, in part secondary to sustained inflammation and delayed cell proliferation.
The primary goal of this study was to probe the cellular and molecular basis for our previous observation of reduced magnitude of tissue-based aged healing(19). Therefore, our study focused on microarray gene expression analysis patterns. While our data demonstrated that the fundamental molecular machinery and pathways governing the bone healing process are not lost with advanced age, underlying immune dysfunction and delayed cellular proliferation likely contribute to the substantially reduced capacity to heal observed with age. Future experiments will explore the function importance of changes in the activation of specific pathways and critical genes within them.
Supplementary Material
Supplemental Figure 1. Hierarchal clustering analysis of top 5 most differentially expressed genes at each DPF with the muscle cells included. Gene expression clustered initially on days post fracture and secondarily on animal age. Red indicates higher level of expression, whereas green indicates lower level of expression.
Clinical Significance.
Greater understanding of age-dependent molecular changes with healing will help form a mechanistic basis for therapies to improve patient outcomes.
Acknowledgments
One or more of the authors certify that they have received, during the study period, funding from NIH R03AG040670 (JA, KDH), American Geriatric Society Jahnigan Award (JA), McCabe Foundation Pilot Award (JA, KDH), and Penn Center for Musculoskeletal Disorders Pilot Award (NIH P30AR050950) (JA, KDH). Two of the authors (KDH, JA) certifies that he or she, or a member of his or her immediate family, has received or may receive payments or benefits, during the study period, an amount of less than USD 10,000 from Skelegen (Philadelphia, PA, USA). One of the authors (JA) certifies that he or she, or a member of his or her immediate family, has received or may receive payments or benefits, during the study period, an amount less than USD 10,000 from Synthes Inc (West Chester, PA, USA), an amount of USD 10,000 to USD 100,000 from Merck & Co, Inc (Whitehouse Station, NJ, USA). JWA was supported by the University of Pennsylvania Postdoctoral Opportunities in Research and Teaching (PENN-PORT) fellowship funded by the National Institute of General Medical Sciences Institutional Research and Career Development Award (IRACDA; 5 K12 GM081259-08).
Footnotes
Author contributions: Ahn J – designed experiments, interpreted data, and edited manuscript; Hankenson KD – designed experiments, interpreted data, and edited manuscript; Lopas L – designed and performed experiments, analyzed and interpreted data, and drafted manuscript; Ashley JA analyzed data and edited manuscript. Daniels L – analyzed data and edited manuscript. Hebb JH – analyzed data and edited manuscript. All authors have read and approved the final manuscript
References
- 1.Hak DJ, Fitzpatrick D, Bishop JA, Marsh JL, Tilp S, Schnettler R, et al. Delayed union and nonunions: epidemiology, clinical issues, and financial aspects. Injury. 2014 Jun;45(Suppl 2):S3–7. doi: 10.1016/j.injury.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Nandra R, Grover L, Porter K. Fracture non-union epidemiology and treatment. Trauma. 2016 Jan 1;18(1):3–11. 2016. [Google Scholar]
- 3.Robinson CM, Royds M, Abraham A, McQueen MM, Court-Brown CM, Christie J. Refractures in patients at least forty-five years old. a prospective analysis of twenty-two thousand and sixty patients. The Journal of bone and joint surgery American volume. 2002 Sep;84-A(9):1528–33. doi: 10.2106/00004623-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Posen J, Beaton DE, Sale J, Bogoch ER. Bone mineral density testing after fragility fracture: Informative test results likely. Canadian family physician Medecin de famille canadien. 2013 Dec;59(12):e564–71. [PMC free article] [PubMed] [Google Scholar]
- 5.Giannotti S, Bottai V, Dell'osso G, Pini E, De Paola G, Bugelli G, et al. Current medical treatment strategies concerning fracture healing. Clinical cases in mineral and bone metabolism : the official journal of the Italian Society of Osteoporosis, Mineral Metabolism, and Skeletal Diseases. 2013 May;10(2):116–20. [PMC free article] [PubMed] [Google Scholar]
- 6.Horst K, Eschbach D, Pfeifer R, Hubenthal S, Sassen M, Steinfeldt T, et al. Local inflammation in fracture hematoma: results from a combined trauma model in pigs. Mediators of inflammation. 2015;2015:126060. doi: 10.1155/2015/126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolar P, Gaber T, Perka C, Duda GN, Buttgereit F. Human early fracture hematoma is characterized by inflammation and hypoxia. Clinical orthopaedics and related research. 2011 Nov;469(11):3118–26. doi: 10.1007/s11999-011-1865-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki A, Tsunoda M, Kinoshita S, Saura R. Role of fracture hematoma and periosteum during fracture healing in rats: interaction of fracture hematoma and the periosteum in the initial step of the healing process. Journal of orthopaedic science : official journal of the Japanese Orthopaedic Association. 2000;5(1):64–70. doi: 10.1007/s007760050010. [DOI] [PubMed] [Google Scholar]
- 9.Yoo JU, Johnstone B. The role of osteochondral progenitor cells in fracture repair. Clinical orthopaedics and related research. 1998 Oct;(355 Suppl):S73–81. doi: 10.1097/00003086-199810001-00009. [DOI] [PubMed] [Google Scholar]
- 10.D'Amelio P, Cristofaro MA, Grimaldi A, Ravazzoli M, Pluviano F, Grosso E, et al. The role of circulating bone cell precursors in fracture healing. Calcified tissue international. 2010 Jun;86(6):463–9. doi: 10.1007/s00223-010-9362-3. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson C, Alpern E, Miclau T, Helms JA. Does adult fracture repair recapitulate embryonic skeletal formation? Mechanisms of development. 1999 Sep;87(1–2):57–66. doi: 10.1016/s0925-4773(99)00142-2. [DOI] [PubMed] [Google Scholar]
- 12.Bahney CS, Hu DP, Miclau T, 3rd, Marcucio RS. The multifaceted role of the vasculature in endochondral fracture repair. Frontiers in endocrinology. 2015;6:4. doi: 10.3389/fendo.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindeler A, McDonald MM, Bokko P, Little DG. Bone remodeling during fracture repair: The cellular picture. Seminars in cell & developmental biology. 2008 Oct;19(5):459–66. doi: 10.1016/j.semcdb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Dimitriou R, Giannoudis PV. The genetic profile of bone repair. Clin Cases Miner Bone Metab. 2013 Jan;10(1):19–21. doi: 10.11138/ccmbm/2013.10.1.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leucht P, Jiang J, Cheng D, Liu B, Dhamdhere G, Fang MY, et al. Wnt3a reestablishes osteogenic capacity to bone grafts from aged animals. J Bone Joint Surg Am. 2013 Jul 17;95(14):1278–88. doi: 10.2106/JBJS.L.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baht GS, Silkstone D, Vi L, Nadesan P, Amani Y, Whetstone H, et al. Exposure to a youthful circulaton rejuvenates bone repair through modulation of beta-catenin. Nat Commun. 2015 May 19;6:7131. doi: 10.1038/ncomms8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Histing T, Heerschop K, Klein M, Scheuer C, Stenger D, Herath SC, et al. Effect of Stabilization on the Healing Process of Femur Fractures in Aged Mice. J Invest Surg. 2016 Aug;29(4):202–8. doi: 10.3109/08941939.2015.1127448. [DOI] [PubMed] [Google Scholar]
- 18.Naik AA, Xie C, Zuscik MJ, Kingsley P, Schwarz EM, Awad H, et al. Reduced COX-2 expression in aged mice is associated with impaired fracture healing. J Bone Miner Res. 2009 Feb;24(2):251–64. doi: 10.1359/jbmr.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopas LA, Belkin NS, Mutyaba PL, Gray CF, Hankenson KD, Ahn J. Fractures in geriatric mice show decreased callus expansion and bone volume. Clin Orthop Relat Res. 2014;472(11):3523–32. doi: 10.1007/s11999-014-3829-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dishowitz MI, Terkhorn SP, Bostic SA, Hankenson KD. Notch signaling components are upregulated during both endochondral and intramembranous bone regeneration. J Orthop Res. 2012 Feb;30(2):296–303. doi: 10.1002/jor.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinke S, Geissler S, Taylor WR, Schmidt-Bleek K, Juelke K, Schwachmeyer V, et al. Terminally differentiated CD8(+) T cells negatively affect bone regeneration in humans. Sci Transl Med. 2013 Mar 20;5(177):177ra36. doi: 10.1126/scitranslmed.3004754. [DOI] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007 Jan;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.Ayturk UM, Jacobsen CM, Christodoulou DC, Gorham J, Seidman JG, Seidman CE, et al. An RNA-seq protocol to identify mRNA expression changes in mouse diaphyseal bone: applications in mice with bone property altering Lrp5 mutations. J Bone Miner Res. 2013 Oct;28(10):2081–93. doi: 10.1002/jbmr.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker JE, Lopes TJ, Ghosh S, Matsuoka Y, Kawaoka Y, Kitano H. CTen: a web-based platform for identifying enriched cell types from heterogeneous microarray data. BMC Genomics. 2012;13:460. doi: 10.1186/1471-2164-13-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrzodek C, Eichner J, Buchel F, Zell A. InCroMAP: integrated analysis of cross-platform microarray and pathway data. Bioinformatics. 2013 Feb 15;29(4):506–8. doi: 10.1093/bioinformatics/bts709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber R, Koch H, Doll BA, Tegtmeier F, Einhorn TA, Hollinger JO. Fracture healing in the elderly patient. Experimental gerontology. 2006 Nov;41(11):1080–93. doi: 10.1016/j.exger.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Toben D, Schroeder I, El Khassawna T, Mehta M, Hoffmann JE, Frisch JT, et al. Fracture healing is accelerated in the absence of the adaptive immune system. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011 Jan;26(1):113–24. doi: 10.1002/jbmr.185. [DOI] [PubMed] [Google Scholar]
- 28.Behrends DA, Cheng L, Sullivan MB, Wang MH, Roby GB, Zayed N, et al. Defective bone repair in mast cell deficient mice with c-Kit loss of function. European cells & materials. 2014;28:209–21. doi: 10.22203/ecm.v028a14. discussion 21-2. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi H. Mast cells in fracture healing: an experimental study using rat model. Nihon Seikeigeka Gakkai zasshi. 1990 Oct;64(10):949–57. [PubMed] [Google Scholar]
- 30.Banovac K, Renfree K, Makowski AL, Latta LL, Altman RD. Fracture healing and mast cells. J Orthop Trauma. 1995;9(6):482–90. doi: 10.1097/00005131-199509060-00005. [DOI] [PubMed] [Google Scholar]
- 31.Marques RE, Guabiraba R, Russo RC, Teixeira MM. Targeting CCL5 in inflammation. Expert opinion on therapeutic targets. 2013 Dec;17(12):1439–60. doi: 10.1517/14728222.2013.837886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing Z, Lu C, Hu D, Yu YY, Wang X, Colnot C, et al. Multiple roles for CCR2 during fracture healing. Disease models & mechanisms. 2010 Jul-Aug;3(7–8):451–8. doi: 10.1242/dmm.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunology and cell biology. 2011 Feb;89(2):207–15. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staines KA, MacRae VE, Farquharson C. The importance of the SIBLING family of proteins on skeletal mineralisation and bone remodelling. The Journal of endocrinology. 2012 Sep;214(3):241–55. doi: 10.1530/JOE-12-0143. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda C, Kubota S, Kawata K, Aoyama E, Sumiyoshi K, Oka M, et al. Distribution, gene expression, and functional role of EphA4 during ossification. Biochemical and biophysical research communications. 2008 Sep 12;374(1):22–7. doi: 10.1016/j.bbrc.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 36.Liu XF, Wang RQ, Hu B, Luo MC, Zeng QM, Zhou H, et al. MiR-15a contributes abnormal immune response in myasthenia gravis by targeting CXCL10. Clin Immunol. 2016 Mar;164:106–13. doi: 10.1016/j.clim.2015.12.009. Epub 2016/02/01. [DOI] [PubMed] [Google Scholar]
- 37.Hui X, Zhu W, Wang Y, Lam KS, Zhang J, Wu D, et al. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem. 2009 May;284(21):14050–7. doi: 10.1074/jbc.M109.001107. Epub 2009/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem. 2009 Apr;284(17):11152–9. doi: 10.1074/jbc.M900754200. Epub 2009/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Hierarchal clustering analysis of top 5 most differentially expressed genes at each DPF with the muscle cells included. Gene expression clustered initially on days post fracture and secondarily on animal age. Red indicates higher level of expression, whereas green indicates lower level of expression.