Abstract
Two-component systems (TCSs) are critical to the pathogenesis of Xanthomonas oryzae pv. oryzae (Xoo). We mutated 55 of 62 genes annotated as responsive regulators (RRs) of TCSs in the genome of Xoo strain PXO99A and identified 9 genes involved in Xoo virulence. Four (rpfG, hrpG, stoS, and detR) of the 9 genes were previously reported as key regulators of Xoo virulence and the other 5 have not been characterized. Lesion lengths on rice leaves inoculated with the mutants were shorter than those of the wild type and were significantly restored with gene complementation. The population density of the 5 mutants in planta was smaller than that of PXO99A at 14 days after inoculation, but the growth curves of the mutants in rich medium were similar to those of the wild type. These newly reported RR genes will facilitate studies on the function of TCSs and of the integrated regulation of TCSs for Xoo pathogenesis.
Keywords: response regulator, two-component regulatory system, Xanthomonas
Living organisms have well-developed signal transduction pathways that rapidly respond to various external changes. In both prokaryotes and eukaryotes, these systems separately or co-operatively regulate the expression of genes associated with most biological processes. The two-component system (TCS) is a dominant signaling pathway in bacteria and is typically composed of two proteins; a membrane-bound histidine kinase (HK), which functions as a sensor, and the other is a cytoplasmic responsive regulator (RR), which typically functions as a transcription factor. The perception of extracellular stimuli by HKs activates RRs via phosphorylation and alternate gene expression (Stock et al., 2000). Therefore, mechanism of the TCS signaling in bacteria has been considered relatively simple compared to the complex signaling by multiple proteins in eukaryotes. However, recent findings acknowledge that more complex mechanisms exist owing to the diverse phosphotransmission pathways and cross or joint regulation by TCSs (Buelow and Raivio, 2010). It might be an evolutionary consequence that bacteria are able to respond to diverse environmental challenges with a limited number of genes for TCSs. Successful reprogramming through simple and more complex pathways involving TCSs surprisingly guarantees the survival of bacteria in diverse conditions. The role of TCSs is also crucial in pathogenic bacteria, which immediately face exclusive conditions in hosts after infection and cannot survive the competition without rapid reprogramming of gene expression by signal pathways such as TCSs. Study of pathogenic bacterial TCSs, therefore, provides critical information to understand microbe-host interactions, but few are functionally defined. In this study, we attempted to identify all RR genes contributing to the virulence of a phytopathogenic bacterium, Xanthomonas oryzae pv. oryzae (Xoo), using 55 RR gene knock-out mutants. We believe that the new RR genes identified from our experiments will extend the knowledge of the TCSs function in Xoo pathogenesis and provide essential clues in the study of the signaling network of TCSs to understand the integrated systems for full fitness of the bacteria in host conditions.
Xoo is a causal agent of a bacterial leaf blight disease in the staple crop plant rice. The pathovar is commonly responsible for loss of 10 to 20% of rice production. To date, genome sequencing of 44 Xoo strains has been completed, and the information is available in the National Center for Biotechnology Information (NCBI) genome database (https://www.ncbi.nlm.nih.gov/). According to the database, the Xoo genome is generally predicted to possess just over 4,000 genes encoding proteins; around 100 of them are annotated as HK and RR genes for TCSs. First, we chose a representative Xoo strain isolated from the Philippines (PXO99A) and selected 62 RR genes in the genome for generation of knock-out mutant strains based on the NCBI and another database, P2CS (Prokaryotic Two-Component System, http://www.p2cs.org/) (Table 1). PXO99A has more RR genes in the genome than the other two representative Xoo strains (MAFF 311018 and KACC 10331). According to P2CS, PXO99A has 58 RRs with 4 mispredicted TCS proteins; MAFF 311018, isolated in Japan, has 52 RRs with 4 mispredicted TCS proteins; and KACC 10331, isolated in Korea, has 53 RRs with 5 mispredicted TCS proteins. However, 53 is the actual number of RR proteins that PXO99A can produce from 58 RR genes because 10 of the 58 RR genes are duplicates (PXO_RS12150 and PXO_RS13080, PXO_RS11975 and PXO_RS12905, PXO_RS11965 and PXO_RS12895, PXO_RS11880 and PXO_RS12810, PXO_RS11880 and PXO_RS12785) with 100% sequence identity (Table 1). We then selected an additional 9 genes (RR8, RR17, RR23, RR24, RR25, RR30, RR36, RR43, and RR51) annotated as hybrids in the NCBI genome database, but not in the P2CS. With these 62 RR gene (including hybrids) candidates, we successfully generated 55 mutant strains, covering 88.7% of all predicted RR genes in the PXO99A genome (Table 1) using marker-exchange mutagenesis (Lee and Ronald, 2007). We suspect that three (RR7, RR18, and RR30) of the 7 genes that we could not generate knock-out mutants are presumably lethal, while another 4 genes [CitB (RR14), ColR (RR28), PhoP (RR37), and RaxR (RR55)] have been previously characterized with knock-out mutants in Xoo (Burdman et al., 2004; Lee et al., 2008; Sahebi et al., 2015; Subramoni et al., 2012). CitB, ColR, and PhoP deficiency fully or mildly weakens virulence in Xoo, whereas RaxR is a required gene for a-virulence activity by rice XA21 protein; however, mutation of the genes failed in the present study despite many attempts. We secured more than three independent replicates for each mutant, and three of the strains for each mutant were used in the experiments of this study. The primer sets used for cloning of each gene and restriction enzymes used for insertion of the kanamycin cassette to carry out marker-exchange mutagenesis are listed in Supplementary Table 1.
Table 1.
Sixty-two candidate RR genes in the PXO99A genome
| Strains | Locus tag No. of mutated gene | Family | P2CS description | References or source |
|---|---|---|---|---|
| RR1 | PXO_RS 08445 | CheY | Response Regulator | in this study |
| RR2 | PXO_RS 08330 | CheB | Response Regulator | in this study |
| RR3 | PXO_RS 08270 | RpfG | Response Regulator | RpfG (He et al., 2010), in this study |
| RR4 | PXO_RS 07450 | OmpR | Response Regulator | in this study |
| RR5 | PXO_RS 09090 | PleD-VieA | Response Regulator | in this study |
| RR6 | PXO_RS 08775 | PleD | Response Regulator | in this study |
| RR7 | PXO_RS 08715 | RpfG | Response Regulator | |
| RR8 | PXO_RS 10565 | Hybrid | in this study | |
| RR9 | PXO_RS 10155 | NarL | Response Regulator | in this study |
| RR10 | PXO_RS 11120 | PrrA | Response Regulator | in this study |
| RR11 | PXO_RS 11220 | unclassified | Response Regulator | in this study |
| RR12 | PXO_RS 12150/PXO_RS 13080 | CheY | Response Regulator | in this study |
| RR13 | PXO_RS 11975/PXO_RS 12905 | CheY | Response Regulator | VemR(Xcc) (Tao and He, 2010), in this study |
| RR14 | PXO_RS 11965/PXO_RS 12895 | NarL | Response Regulator | CitB (Sahebi et al., 2015) |
| RR15 | PXO_RS 11880/PXO_RS 12810 | CheV | Response Regulator | in this study |
| RR16 | PXO_RS 11855/PXO_RS 12785 | PleD-VieA | Response Regulator | PdeR (Yang et al., 2012), RavR(Xcc) (He et al., 2009), in this study |
| RR17 | PXO_RS 15740 | Hybrid | in this study | |
| RR18 | PXO_RS 15515 | CheY | Response Regulator | |
| RR19 | PXO_RS 15395 | NarL | Response Regulator | in this study |
| RR20 | PXO_RS 15100 | CheY | Response Regulator | in this study |
| RR21 | PXO_RS 14975 | CheY | Response Regulator | in this study |
| RR22 | PXO_RS 14970 | CheY | Response Regulator | in this study |
| RR23 | PXO_RS 14605 | Hybrid | in this study | |
| RR24 | PXO_RS 14600 | Hybrid | in this study | |
| RR25 | PXO_RS 14595 | Hybrid | in this study | |
| RR26 | PXO_RS 14460 | unclassified | Response Regulator | in this study |
| RR27 | PXO_RS 18055 | OmpR | Response Regulator | HrpG (Tsuge et al., 2006), in this study |
| RR28 | PXO_RS 18925 | OmpR | Response Regulator | ColR (Subramoni et al., 2012) |
| RR29 | PXO_RS 19035 | OmpR | Response Regulator | in this study |
| RR30 | PXO_RS 19370 | Hybrid | ||
| RR31 | PXO_RS 19400 | NarL | Response Regulator | in this study |
| RR32 | PXO_RS 19415 | OmpR | Response Regulator | TctD(Xcv) (Tamir-Ariel et al., 2011), in this study |
| RR33 | PXO_RS 19870 | OmpR | Response Regulator | PhoB, in this study |
| RR34 | PXO_RS 20355 | RpfG | Response Regulator | in this study |
| RR35 | PXO_RS 20535 | CheY | Response Regulator | in this study |
| RR36 | PXO_RS 23080 | Hybrid | in this study | |
| RR37 | PXO_RS 22695 | OmpR | Response Regulator | PhoP (Lee et al., 2008) |
| RR38 | PXO_RS 22160 | VieA | Response Regulator | in this study |
| RR39 | PXO_RS 21800 | NtrC | Response Regulator | XibR(Xcc) (Pandey et al., 2016), in this study |
| RR40 | PXO_RS 21790 | unclassified | Response Regulator | in this study |
| RR41 | PXO_RS 21740 | CheY | Response Regulator | in this study |
| RR42 | PXO_RS 21605 | OmpR | Response Regulator | in this study |
| RR43 | PXO_RS 21535 | Hybrid | StoS (Ikawa et al., 2014; Zheng et al., 2016), in this study | |
| RR44 | PXO_RS 23555 | NarL | Response Regulator | in this study |
| RR45 | PXO_RS 01340 | NtrC | Response Regulator | in this study |
| RR46 | PXO_RS 02255 | FrzZ | Response Regulator | in this study |
| RR47 | PXO_RS 02040 | OmpR | Response Regulator | in this study |
| RR48 | PXO_RS 02690 | NtrC | Response Regulator | in this study |
| RR49 | PXO_RS 03980 | LytTR | Response Regulator | in this study |
| RR50 | PXO_RS 04300 | CheY | Response Regulator | SreR (Zheng et al., 2016), in this study |
| RR51 | PXO_RS 04305 | Hybrid | SreS (Zheng et al., 2016), in this study | |
| RR52 | PXO_RS 04760 | OmpR | Response Regulator | in this study |
| RR53 | PXO_RS 04595 | OmpR | Response Regulator | in this study |
| RR54 | PXO_RS 06090 | NarL | Response Regulator | in this study |
| RR55 | PXO_RS 06050 | OmpR | Response Regulator | |
| RR56 | PXO_RS 05400 | unclassified | Response Regulator | in this study |
| RR57 | PXO_RS 05395 | CheY | Response Regulator | in this study |
| RR58 | PXO_RS 05125 | OmpR | Response Regulator | DetR (Nguyen et al., 2016), in this study |
| RR59 | PXO_RS 06365 | VieA | Response Regulator | in this study |
| RR60 | PXO_RS 06355 | CheB | Response Regulator | in this study |
| RR61 | PXO_RS 06785 | OmpR | Response Regulator | in this study |
| RR62 | PXO_RS 06935 | NtrC | Response Regulator | in this study |
Gray cells indicate that the knock-out mutant strain of the RR gene reduced Xoo virulence in the present study.
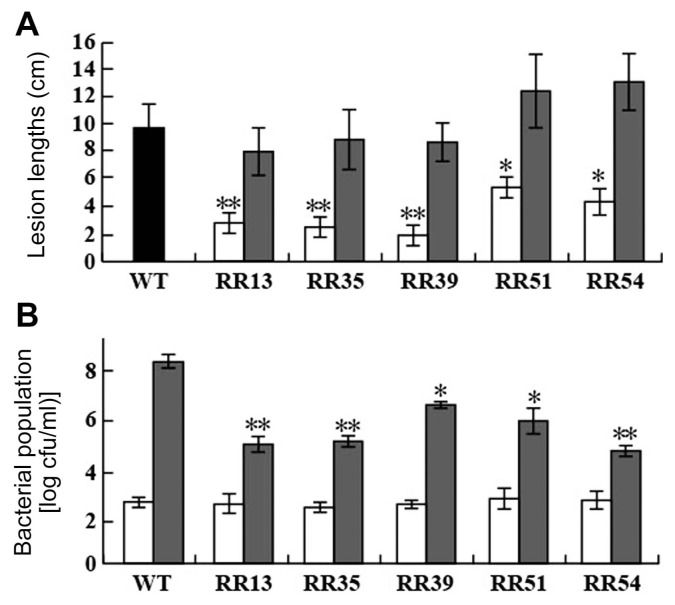
Virulence of the 55 mutant strains was examined after inoculation on leaves of Dong-jin rice plants (a japonica variety, compatible with PXO99A). For the inoculation test, plants were inoculated with a large population (107 cfu/ml) of mutant strain using a clipping method (Kauffman et al., 1973). Lesion lengths on leaves inoculated by the 9 mutant strains (RR3, RR13, RR27, RR35, RR39, RR43, RR51, RR54, and RR58) were significantly reduced compared to those on leaves inoculated by other mutants and PXO99A (Fig. 1, Supplementary Fig. 1, 2). Among them, RR3, RR27, RR43, and RR58 are deficient mutants of RpfG, HrpG, StoS, and DetR proteins, respectively. A TCS mediated by RpfC/G is involved in signal sensing and the transduction of diffusible signal factor and is important for Xoo virulence (Jeong et al., 2008). Both rpfC and rpfG knockout mutants decrease production of several virulence determinants including exopolysaccharide, lipopolysaccharide, and extracellular enzymes except cellulase in Xoo. RR3, a knock-out mutant strain of the rpfG gene (Table 1), also showed reduced virulence in our test (Supplementary Fig. 2). HrpG was first identified in Xanthomonas campestris pv. vesicatoria (Wengelnik et al., 1996) and is an RR protein well-known as a representative virulence factor in Xanthomonas spp. HrpG belongs to the OmpR family, is the key regulator of pathogenicity (hrp) genes in susceptible rice plants, and is responsible for hypersensitive responses in resistant plants. HrpG regulates all genes required for the type III secretion system (T3SS), genes encoding T3SS effectors, and the genes encoding type II secretion system substrate with a transcriptional activator, HrpX (Guo et al., 2011). The critical role of HrpG and regulation of gene expression in Xoo pathogenicity were also investigated (Tsuge et al., 2006). The PXO_RS18055 (hrpG) knock-out mutant (RR27) also reduced lesion length in our virulence test (Supplementary Fig. 2). PXO_RS21535 encodes a hybrid protein containing PAS domain sensor HK, which was previously published as StoS (stress tolerance-related oxygen sensor) in Xoo (Ikawa et al., 2014). The knock-out mutant has decreased stress tolerance to high osmolality, sodium, and H2O2 and shows mildly reduced virulence. The RR43 strain in our inoculation results also showed mild attenuation in lesion length on inoculated leaves compared to PXO99A (Supplementary Fig. 2). DetR, recently published by our group as a key regulator of Xoo virulence, controls exopolysaccharide synthesis, ROS detoxification, and iron homeostasis (Nguyen et al., 2015). Defects in the DetR of Xoo strongly reduced virulence (Supplementary Fig. 2).
Fig. 1.
Virulence phenotypes of PXO99A (WT) and RR mutant strains. (A) Lesion lengths caused by PXO99A (WT, black), RR mutant (white), and the complementary (gray) strains on Dongjin rice leaves at 14 days after infection (DAI). Rice leaves were infected with 107 cfu/ml of bacterial suspension by the scissor clipping method. Bars are mean ± SD (n = 20). * or ** indicate that the lesion length of each mutant was significantly different from that of PXO99A by Duncan’s test (P < 0.05). (B) Bacterial population at 0 (white) and 14 (gray) DAI. Immediately after infection (0 DAI) and at 14 DAI, 1-cm2 and 25-cm2 samples from the infected site of three leaves were used to extract the bacteria population at 0 and 14 dpi, respectively. Bars are mean ± SD (n = 3). * and ** indicate that the population of mutants was significantly smaller than that of PXO99A by Duncan’s test (P < 0.05). C) All experiments were repeated three times with high consistency, and the results from one experiment are shown.
The other 5 genes (RR13, RR35, RR39, RR51, and RR54) are new RRs that have not yet been investigated in Xoo. The RR13 mutant has no RR protein encoded in PXO_RS11975 or PXO_RS12905. Lesion lengths of the RR13 mutant strain (2.6 ± 1.4 cm) were reduced compared with those of PXO99A (9.8 ± 1.9 cm) (Fig. 1). The RR13 gene is a homolog of vemR identified in Xanthomonas campestris pv. campestris (Xcc) ATCC33913. The amino acid sequence of the Xoo RR13 protein has 100% identity with 96% query coverage. Only 5 amino acids at the C-terminus are different between the full-length Xoo and Xcc proteins. VemR is involved in exopolysaccharide production and mobility to regulate Xcc pathogenicity. Our results indicated that the homolog of Xcc VemR also regulates virulence in Xoo. PXO_RS20535 (RR35) encodes an orphan RR protein belonging to the CheY family. The lesion length on leaves inoculated with RR35 mutant strain was 2.6 ± 1.5 cm (Fig. 1), while that of PXO99A was 9.8 ± 1.9 cm (Fig. 1). The RR35 gene is well-conserved in all Xanthomonas spp., but has not yet been characterized. Suppression of PXO_RS21800 gene expression (RR39) exhibited a reduction of lesion lengths on inoculated leaves (Fig. 1). RR39 is a homolog of Xcc XibR (Xanthomonas iron binding regulator) with a 93% identity in a 99% query coverage. XibR is required for fine-tuning and regulating the expression of iron-regulated genes and virulence-associated function (Pandey et al., 2016). Our results indicated that signaling by the homolog of the Xcc XibR is also important in Xoo virulence. With regard to StoS (RR43 in this study, Supplementary Fig. 2), there is another report concluding that Xoo StoS and SreKRS (salt response kinase, regulator, sensor) orchestrate the behavior of Xoo in rice by regulating virulence factor expression (Zheng et al., 2016). However, the absence of StoS or SreK is not reported to weaken the virulence of Xoo (Zheng et al., 2016). The sreKRS genes in an operon are annotated in NCBI as a PAS-domain S-box protein, a response regulator, and a hybrid HK/RR, respectively. Although SreK was not included in our candidates, SreR and SreS correspond to RR50 and RR51, respectively. In our inoculation test, RR50 did not show any significant change in lesion length. However, the lesion lengths of RR51 were reduced by half compared to those of PXO99A (Fig. 1), which is similar to the results of RR43 shown in Supplementary Fig. 2. It is not obvious if Zheng et al. (2016) tested the SreS mutant for Xoo virulence; however, our results strongly indicate that the hybrid (SreS) contributes to Xoo virulence. PXO_RS06090 (RR54), located with a cognate HK gene (PXO_RS06085) in an operon of the PXO99A genome, encodes an RR protein belonging to the NarL family. It is well-conserved in all Xanthomonas spp. The lengths of lesions on leaves inoculated by RR51 were reduced (4.1 ± 1.7 cm) compared to those of PXO99A, indicating that TCS mediated by the RR is involved in Xoo virulence (Fig. 1). Except these 5, we have another questionable RR, published as PdeR (Yang et al., 2012). Yang et al. (2012) reported that deletion of pdeR and the cognate HK gene, pdeK, attenuates virulence and decreases secretion of exopolysaccharide in Xoo (Yang et al., 2012). Indeed the homologs of Xoo pdeR and pdeK were published previously in Xcc with different names, RavR and RavS (He et al., 2009). Deletion of RavR and RavS similarly reduces Xcc virulence. However, RR16 (pdeR deletion mutant) in our study did not show a conclusive change in virulence (Supplementary Fig. 2). The PXO99A genome has two copies of the gene at PXO_RS11855 and PXO_RS12785, with 100% identity. One possible explanation is that our mutant (RR16) still has one of these two without a disruption.
We next generated complementary strains for the new 5 gene knock-out mutants (Supplementary Table 2). It is to test if the reduced virulence of the 5 mutant strains was caused by unexpected expression changes of the neighboring genes, not caused by the target genes. To generate complementary strains, the full-lengths of RR13, RR35, RR39, RR51, and RR54 genes followed by the sequence for 6 His residues was cloned into a pBBR1MCS-5 vector and introduced into the 5 knock-out mutant strains (RR13, RR35, RR39, RR51, and RR54), respectively. After confirmation of protein expression with western blot analysis (Supplementary Fig. 3), the complementary strains were inoculated on the leaves of Dong-jin rice plants. We then observed a significant restoration in virulence of the complementary strains (Fig. 1A). This result revealed that the observed decrease in virulence of the 5 knock-out mutants was not due to a polar effect. Furthermore, the bacterial population in inoculated rice leaves was also monitored at 0 and 14 days after inoculation (DAI, Fig. 1B). We harvested rice leaves immediately (0 days) after inoculation and 14 DAI and extracted the bacteria by the 1 h shaking-incubation of chopped rice leaves. Whereas the population density of PXO99A at 14 DAI reached 8.2 ± 0.5 cfu/ml in the log phase, the population of mutants only reached 6.4 ± 0.3 cfu/ml even in the most virulent strain RR39 (Fig. 1B). Finally, we established growth curves of the 5 knock-out mutant strains to determine if the reduced virulence was caused by a defect of bacterial growth in normal conditions and was not directly associated with pathogenicity in host conditions (Supplementary Fig. 4). The population of the mutant strains was measured in peptone sucrose broth (PSB) medium every 12 h for 4 days using colony counting. We found no difference in growth between PXO99A and the 5 mutants. This result suggests that the RRs and/or TCSs mediated by the RRs are most likely specific to Xoo virulence.
Judging from our results and from previous reports, Xoo possesses at least 12 RRs (or 13 if PedR is included) that are closely associated with pathogenicity. Seven (RpfG, RR3; CitB, RR14; PdeR, RR16; HrpG, RR27; ColR, RR28; PhoP, RR37; DetR, RR58) have been previously characterized, and we identified 5 novel RRs (RR13; Xcc VemR homolog; RR35, PXO_RS20535; RR39, Xcc XibR homolog; RR51, sreS; RR54, PXO_RS06090) in the present study. Although the 12 (or 13) RR genes cannot be asserted to be all that is required for Xoo virulence, we suspect it is likely. Our findings extend the knowledge of RRs (or TCSs mediated by the new RRs) required for Xoo virulence and will facilitate further studies to elucidate the network between RRs for full fitness of Xoo in host conditions. We are now in the progress of transcriptome analysis with all 12 RR knock-out mutants.
Supplementary Information
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2017R1A2B4004035).
References
- Buelow DR, Raivio TL. Three (and more) component regulatory systems – auxiliary regulators of bacterial histidine kinases. Mol Microbiol. 2010;75:547–566. doi: 10.1111/j.1365-2958.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- Burdman S, Shen Y, Lee SW, Xue Q, Ronald P. RaxH/RaxR: A two-component regulatory system in Xanthomonas oryzae pv oryzae required for AvrXa21 activity. Mol Plant-Microbe Interact. 2004;17:602–612. doi: 10.1094/MPMI.2004.17.6.602. [DOI] [PubMed] [Google Scholar]
- Guo Y, Figueiredo F, Jones J, Wang N. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Mol Plant-Microbe Interact. 2011;24:649–661. doi: 10.1094/MPMI-09-10-0209. [DOI] [PubMed] [Google Scholar]
- He YW, Boon C, Zhou L, Zhang LH. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol Microbiol. 2009;71:1464–1476. doi: 10.1111/j.1365-2958.2009.06617.x. [DOI] [PubMed] [Google Scholar]
- He YW, Wu J, Cha JS, Zhang LH. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol. 2010;10:187. doi: 10.1186/1471-2180-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y, Furutani A, Ochiai H, Tsuge S. StoS, a hybrid histidine kinase sensor of Xanthomonas oryzae pv. oryzae, is activated by sensing low O2 concentration and is involved in stress tolerance and virulence. Mol Plant-Microbe Interact. 2014;27:537–545. doi: 10.1094/MPMI-09-13-0263-R. [DOI] [PubMed] [Google Scholar]
- Jeong KS, Lee SE, Han JW, Yang SU, Lee BM, Noh TH, Cha JS. Virulence reduction and differing regulation of virulence genes in rpf mutants of Xanthomonas oryzae pvoryzae. Plant Pathol J. 2008;24:143–151. doi: 10.5423/PPJ.2008.24.2.143. [DOI] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Lee SW, Ronald PC. Marker-exchange mutagenesis and complementation strategies for the Gram-negative bacteria Xanthomonas oryzae pvoryzae. Methods Mol Biol. 2007;354:11–18. doi: 10.1385/1-59259-966-4:11. [DOI] [PubMed] [Google Scholar]
- Lee S-W, Jeong K-S, Han S-W, Lee S-E, Phee B-K, Hahn T-R, Ronald P. The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J Bacteriol. 2008;190:2183–2197. doi: 10.1128/JB.01406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MP, Yoon JM, Cho MH, Lee SW. Prokaryotic 2-component systems and the OmpR/PhoB superfamily. Can J Microbiol. 2015;61:799–810. doi: 10.1139/cjm-2015-0345. [DOI] [PubMed] [Google Scholar]
- Nguyen MP, Park J, Cho MH, Lee SW. Role of DetR in defence is critical for virulence of Xanthomonas oryzae pv. oryzae. Mol Plant Pathol. 2016;17:601–613. doi: 10.1111/mpp.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SS, Patnana PK, Lomada SK, Tomar A, Chatterjee S. Co-regulation of iron metabolism and virulence associated functions by iron and XibR, a novel iron binding transcription factor, in the plant pathogen Xanthomonas. PLoS Pathog. 2016;12:e1006019. doi: 10.1371/journal.ppat.1006019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahebi M, Taheri E, Tarighi S. CitB is required for full virulence of Xanthomonas oryzae pvoryzae. World J Microbiol Biotechnol. 2015;31:1619–1627. doi: 10.1007/s11274-015-1914-2. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Subramoni S, Pandey A, Vishnu Priya MR, Patel HK, Sonti RV. The ColRS system of Xanthomonas oryzae pv. oryzae is required for virulence and growth in iron-limiting conditions. Mol Plant Pathol. 2012;13:690–703. doi: 10.1111/j.1364-3703.2011.00777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir-Ariel D, Rosenberg T, Burdman S. The Xanthomonas campestris pv. vesicatoria citH gene is expressed early in the infection process of tomato and is positively regulated by the TctDE two-component regulatory system. Mol Plant Pathol. 2011;12:57–71. doi: 10.1111/j.1364-3703.2010.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, He C. Response regulator, VemR, positively regulates the virulence and adaptation of Xanthomonas campestris pv. campestris. FEMS Microbiol Lett. 2010;304:20–28. doi: 10.1111/j.1574-6968.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- Tsuge S, Nakayama T, Terashima S, Ochiai H, Furutani A, Oku T, Tsuno K, Kubo Y, Kaku H. Gene involved in transcriptional activation of the hrp regulatory gene hrpG in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2006;188:4158–4162. doi: 10.1128/JB.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik K, Van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant-Microbe Interact. 1996;9:704–712. doi: 10.1094/MPMI-9-0704. [DOI] [PubMed] [Google Scholar]
- Yang F, Tian F, Sun L, Chen H, Wu M, Yang CH, He C. A novel two-component system PdeK/PdeR regulates c-di-GMP turnover and virulence of Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 2012;25:1361–1369. doi: 10.1094/MPMI-01-12-0014-R. [DOI] [PubMed] [Google Scholar]
- Zheng D, Yao X, Duan M, Luo Y, Liu B, Qi P, Sun M, Ruan L. Two overlapping two-component systems in Xanthomonas oryzae pv oryzae contribute to full fitness in rice by regulating virulence factors expression. Sci Rep. 2016;6:22768. doi: 10.1038/srep22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.