Abstract
Ethylene regulates plant abiotic stress responses and tolerances, and ethylene-insensitive3 (EIN3)/EIN3-like (EIL) proteins are the key components of ethylene signal transduction. Although the functions of EIN3/EIL proteins in response to abiotic stresses have been investigated in model plants, little is known in non-model plants, including mulberry (Morus L.), which is an economically important perennial woody plant. We functionally characterized a gene encoding an EIN3-like protein from mulberry, designated as MnEIL3. A quantitative real-time PCR analysis demonstrated that the expression of MnEIL3 could be induced in roots and shoot by salt and drought stresses. Arabidopsis overexpressing MnEIL3 exhibited an enhanced tolerance to salt and drought stresses. MnEIL3 overexpression in Arabidopsis significantly upregulated the transcript abundances of ethylene biosynthetic genes. Furthermore, MnEIL3 enhanced the activities of the MnACO1 and MnACS1 promoters, which respond to salt and drought stresses. Thus, MnEIL3 may play important roles in tolerance to abiotic stresses and the expression of ethylene biosynthetic genes.
Keywords: MnEIL3, Mulberry, Arabidopsis, Abiotic stresses, Salt stress, Drought
Introduction
Ethylene is gaseous hormone that regulates many physiological processes, including seed germination, seedling growth, leaf expansion, flower opening, senescence, and fruit development. Ethylene is synthesized from methionine by a catalysis mediated by S-adenosyl-L-methionine synthetase, 1-aminocyclopropane-1-carboxylic acid synthase (ACS), and 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) (Kende, 1993; Yang & Hoffman, 1984). ACS and ACO are the rate-limiting enzymes that regulate ethylene biosynthesis, and several regulators influence ethylene production by changing the activities and gene expression levels of ACS and ACO.
The ethylene perception and signal transduction pathway have been well studied in model plants, like Arabidopsis thaliana, tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum). In the presence of ethylene, the endoplasmic reticulum-localized receptors (ethylene receptors, ETRs) perceive ethylene, resulting in the inactivation of constitutive triple response 1, which is the negative regulatory factor of ethylene responses (Clark et al., 1998; Hua & Meyerowitz, 1998; Rodríguez et al., 1999). Then, ethylene insensitive 2 (EIN2), an essential positive regulator of ethylene signaling, is dephosphorylated, and its carboxyl terminus is cleaved and enters into the nucleus, where it binds to ethylene insensitive 3/EIN3-like proteins (EIN3/EILs) (Chao et al., 1997; Qiao et al., 2012). Eventually, the activated EIN3/EILs regulate the transcription of ethylene-responsive factors (ERFs) and other downstream genes (Alonso & Stepanova, 2004; Wang, Li & Ecker, 2002).
EIN3/EILs are the key elements that initiate the ethylene-mediated downstream transcriptional cascade (An et al., 2018). The mutation of EIN3/EILs genes, AtEIN3 (AT3G20770) and AtEIL1 (AT2G27050), result in ethylene-insensitive performance, and plants overexpressing AtEIN3 and AtEIL1 show enhanced ethylene production and triple responses in Arabidopsis. The ein3-1 eil1-1 double mutant completely abolishes the ethylene response in etiolated Arabidopsis seedlings (Alonso et al., 2003; Chao et al., 1997). The stabilities of EIN3/EILs are regulated by EIN3-binding F-box proteins (EBF1 and EBF2) in the EBF1- and EBF2-mediated ubiquitin-proteasome degradation pathway, and mutations of EBF1 and EBF2 accumulate EIN3/EIL proteins and display constitutive ethylene responses (Potuschak et al., 2003). Ethylene quickly stabilizes EIN3/EIL1 by promoting EBF1 and EBF2 proteasomal degradation, which contributes to the ethylene responses (An et al., 2010). In addition, the MKK9-MPK3/MPK6 cascades promote EIN3-mediated transcription in ethylene signaling by regulating the phosphorylation and protein stability of EIN3 (Yoo et al., 2008).
The EIN3/EILs family are plant-specific transcription factors (TFs) and bind to primary ethylene response elements (PEREs) and EIL conserved binding sequences (ECBSs) in the promoters of downstream genes involved in the response to ethylene (Yin et al., 2010). Thus, EIN3/EILs regulate many physiological processes, including apical hook formation, hormone responses, fruit development, abiotic stress responses, seedling photomorphogenesis, and light perception, by activating the expression of a wide range of downstream genes (An et al., 2012; He et al., 2011; Peng et al., 2014; Shan et al., 2012; Shi et al., 2012; Shi et al., 2018; Zhu et al., 2011). Recently, studies have focused on the functions of EIN3/EILs in abiotic stress tolerances. Peng et al. (2014) demonstrated that EIN3/EIL1 are essential for the enhanced ethylene-induced salt tolerance in Arabidopsis, and salt stress stabilizes EIN3/EIL1 proteins by promoting EBF1/EBF2 proteasomal degradation in an EIN2 independent manner. In addition, a large number of EIN3/EIL1-regulated genes that participate in salt stress responses have been identified using whole-genome transcriptome analyses, including many genes encoding reactive oxygen species scavengers. An AP2 domain-containing gene, ESE1, is an ethylene-modulated gene downstream of EIN3/EIL1 in the salt response (Zhang et al., 2011). Mutations of EIN3 increase the sensitivity in response to water stress stimulated by polyethylene glycol (PEG) 6,000 (Cui et al., 2015). Genetic and biochemical analyses revealed that EIN3 proteins act as negative factors against freezing stress by repressing the expression of C-repeat binding factors and type-A Arabidopsis response regulator (ARR) 5, ARR7, and ARR15 (Shi et al., 2012). The functions of the EIN3/EIL1 proteins in response to heavy metal stresses have also been studied. Kong et al. (2018) found that cadmium (Cd) inhibits EIN3 protein degradation in Arabidopsis, and the ein3-1 eil1-1 double mutant plants display an increased tolerance to Cd. EIN3 enhances root growth inhibition under Cd stress by regulating the expression of the xyloglucan endotransglucosylase/hydrolase 33 and response to low sulfur 1 genes, which are involved in cell wall modification and sulfur metabolic processes, respectively (Kong et al., 2018).
Mulberry (Morus L.) is an economically important perennial woody plant belonging to Moraceae of Rosales, which have multiple uses in silkworm rearing, ecology, pharmaceuticals, and traditional Chinese medicines (He et al., 2013). Mulberry adapts well to drought, salinity, waterlogging, and other abiotic stress conditions, but little is known regarding the molecular mechanisms of the tolerance. In our previous studies, the elements involved in mulberry ethylene biosynthesis and signal transduction were identified and its functions in fruit development were clarified (Liu et al., 2014, 2015). However, the functions of mulberry ethylene biosynthesis and signal pathway related genes in other aspects of the lifecycle remain unclear, especially in abiotic stress responses and tolerances. In this study, we investigated the physiological functions of a mulberry gene encoding EIN3-like proteins, MnEIL3, in salt and drought tolerances by analyzing its expression patterns and its heterologous overexpression in Arabidopsis. MnEIL3’s expression was significantly upregulated by salt and drought stresses, and its overexpression in Arabidopsis led to enhanced salt and drought stress tolerances and the upregulated expression of ethylene biosynthetic genes. Furthermore, MnEIL3 significantly enhanced the activities of MnACO1 and MnACS1 promoters. Thus, a working model for MnEIL3 in plant tolerance to abiotic stresses was suggested.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 and the ein3-1 eil1-1 mutant were used as plant materials and were grown at 24 °C/22 °C under a 16-h light/8-h dark photoperiod.
A mulberry (M. notabilis Schneid) tree, which was used for genome sequencing, is an isolated wild mulberry species with a chromosome number of 14. The seedlings of M. notabilis were used in this study and grown in a PQX-type plant incubator with artificial intelligence capability (Ningbo Southeast Instrument Corporation, China) under a 16-h light/8-h dark photoperiod at 26 °C/22 °C (day/night). For stress treatments, the one-month-old seedlings were subjected to salt [0.6% (m/v) NaCl] and drought [20% (m/v) PEG6000]. The roots and shoot of the treated seedlings were sampled at 0, 1, 3, 6, 12, and 24 h post-treatment. The 14-day-old seedlings were treated independently with 200 mM NaCl and 200 mM mannitol, and the treated seedlings were sampled at 0, 1, 3, 6, and 12 h post-treatment. The harvested materials were frozen immediately in liquid nitrogen for total RNA extraction.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA extraction, first-strand of cDNA synthesis, and qRT-PCR analysis were performed as described in our previous study (Wei et al., 2014). The ACTIN3 and β-actin2 genes were used as internal controls for mulberry and Arabidopsis, respectively, and the relative expression was defined as 2−[Ct(target gene) Ct(control gene)]. All qRT-PCRs were performed with three independent biological replicates. The primers used are specified in Table S2.
Plasmid construction
The full-length coding sequence of MnEIL1 (GenBank accession number: XM_010107825) and MnEIL3 (XM_010093690) were cloned into the NcoI and BglII restriction sites of the pCAMBIA1302 expression vector under the control of the CaMV35S promoter, and MnEIL2 (XM_010107826) was cloned into the BglII and SpeI restriction sites of the pCAMBIA1302 expression vector. Finally, the CaMV35S::MnEIL1, CaMV35S::MnEIL2, and CaMV35S::MnEIL3 recombinant plasmids were generated.
The 5′ upstream regions of the MnACS1 and MnACS3 genes were independently inserted into the EcoRI and NcoI restriction sites of the pCAMBIA1301 expression vector, producing the MnACS1pro::GUS and MnACS3pro::GUS recombinant plasmids, respectively. The primers used are specified in Table S2.
Plant transformation
The recombinant plant expression vectors were transformed into Agrobacterium tumefaciens strain GV3101. MnEIL3, MnACS1pro::GUS and MnACS3pro::GUS were eventually independently transformed into Arabidopsis thaliana (Columbia-0) using the floral dip method (Clough & Bent, 1998). The transgenic Arabidopsis lines were evaluated by GUS staining, genomic PCR, inverse PCR, and qRT-PCR analyses. The homozygous lines of the T3 generation were used for further research.
Stress treatments of transgenic Arabidopsis
Wild type, ein3-1 eil1-1, and MnEIL3 transgenic seeds were germinated on 1/2 Murashige and Skoog (MS) agar medium. The 7-day-old seedlings were transferred into pots containing the soil supplemented with normal nutrients were grown at 24 °C/22 °C under a 16-h light/8-h dark photoperiod. The 21- and 14-day-old seedlings were treated with salt [1.2% (m/v) NaCl] and drought (watering treatments withheld), respectively, and the proline, hydrogen peroxide (H2O2), and malondialdehyde (MDA) contents, were measured using their respective test kits (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. Each treatment was replicated three times.
The seeds of the transgenic plants that contained MnACS1pro::GUS and MnACS3pro::GUS were germinated on 1/2 MS agar medium. The 10-day-old seedlings were exposed to salt (200 mM NaCl) and drought (200 mM mannitol) treatments. The samples were subjected to GUS staining after 0, 1, 3, 6, and 12 h of exposure.
Transient expression assays and GUS activity detection
CaMV35S::MnEIL1, CaMV35S::MnEIL2, and CaMV35S::MnEIL3 recombinant plasmids were used as effector plasmids, and MnACO1pro::GUS, MnACO2pro::GUS, MnACS1pro::GUS, and MnACS3pro::GUS recombinant plasmids were used as reporter plasmids. Of these vectors, the MnACO1pro::GUS and MnACO2pro::GUS recombinant plasmids have been reported in a previous study (Yu et al., 2017). The reporter and effector plasmids were transformed into Agrobacterium tumefaciens strain GV3101. The bacteria were mixed and co-injected into the strawberry fruit as described in a previous study (Spolaore, Trainotti & Casadoro, 2001). The injected tissues were sampled and used for the GUS staining analysis. Meanwhile, the GUS activity of the injected tissues was detected by 4-nitrophenyl-β-D-glucopyranoside methods (Jefferson, 1987).
Statistical analyses
The statistical analyses methods were as described in a previous study (Liu et al., 2017a). All data were conducted using SPSS statistical software 17.0 (SPSS Inc., Chicago, IL, USA) and Excel 2013 (Microsoft, Redmond, CA, USA). The results are presented as mean values ± SEs. The significant differences between samples were analyzed using a one-way ANOVA in SPSS Statistics 17.0. The analyses of significant differences (P < 0.05) were measured by Student’s t-test analysis.
Results
Expression patterns of MnEIL genes under salt and drought stresses
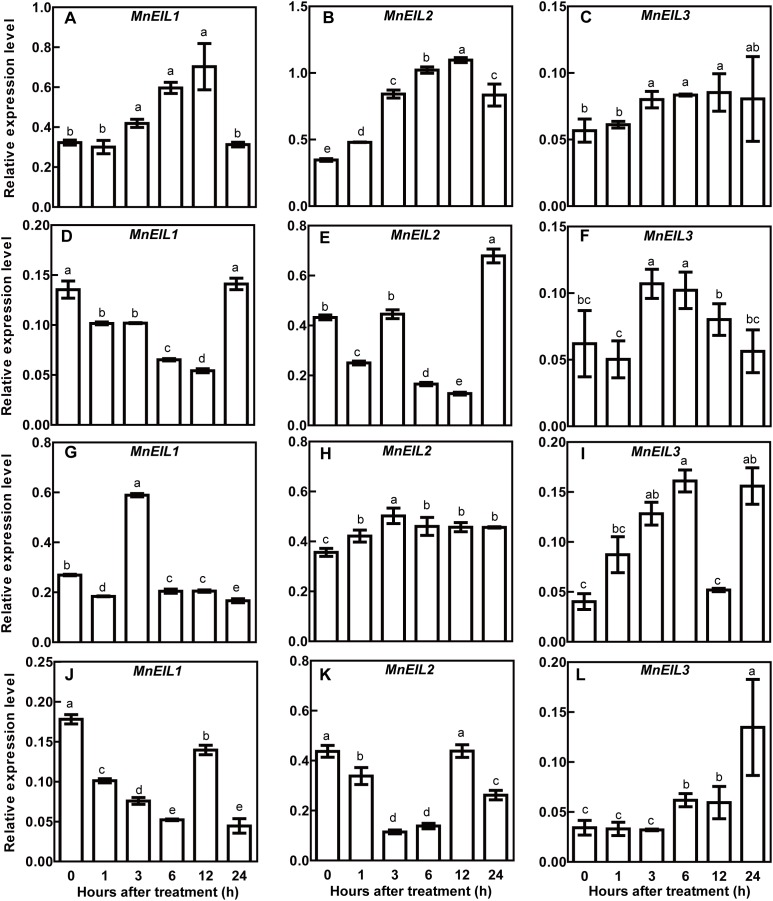
The expression levels of MnEIL genes under NaCl and PEG treatments were assessed by qRT-PCR. Under salt stress, the transcript abundance of MnEIL1 was significantly upregulated and downregulated in roots and shoots, respectively, 3 h after the NaCl treatment, but the expression levels after 24 h were not different than those at 0 h (Figs. 1A and 1B). The expression of MnEIL2 in roots was significantly upregulated after 1 h of NaCl treatment. MnEIL2’s expression in shoots was downregulated at 1, 6, and 12 h, but its expression was upregulated at 24 h after the NaCl treatment (Figs. 1A and 1B). The transcript abundances of MnEIL3 in roots and shoot were significantly upregulated 3 h after the NaCl treatment, but the expression of MnEIL3 in shoots after 24 h showed no difference with that at 0 h (Figs. 1A and 1B). Under drought stress conditions, MnEIL1’s expression levels in roots and shoot were downregulated by the NaCl treatment, although MnEIL1’s expression in roots was upregulated at 3 h (Figs. 1C and 1D). The transcript abundance of MnEIL2 was significantly upregulated and downregulated after 3 h of NaCl treatment in roots and shoot, respectively (Figs. 1C and 1D). The expression of MnEIL3 in roots was significantly upregulated after a PEG treatment, although its expression showed no response to PEG at 12 h. MnEIL3’s expression in shoots was significantly upregulated 6 h after the PEG treatment and exhibited a strong expression peak at 24 h (Figs. 1C and 1D).
Figure 1. Expression profiles of MnEIL genes in response to salt and drought stresses.
Mulberry seedlings were subjected to salt [0.6 % (m/v) NaCl] and drought [20% (m/v) PEG6000]. (A–C) MnEIL gene expression levels in roots under salt stress. (D–F) MnEIL gene expression levels in shoots under salt stress. (G–I) MnEIL gene expression levels in roots under drought stress. (J–L) MnEIL gene expression levels in shoots under drought stress. Data are means ± SEs (n = 3). Means within a column with different letters are significantly different (P < 0.05). Means within a column with the same letters are not significantly different (P > 0.05).
The overexpression of MnEIL3 in Arabidopsis enhances salt and drought tolerances
The MnEIL3 gene was selected for further investigation base on its responses to salt and drought stresses. The full-length sequence of the MnEIL3 gene was inserted into the pCAMBIA1302 vector under the control of the CaMV35S promoter and transformed into wild type Arabidopsis plants. Transgenic lines were obtained using hygromycin resistance and confirmed by genomic PCR and qRT-PCR analyses (Figs. S1A and S1B). In addition, the insertion site of the transgene construct was determined using inverse PCR. The CaMV35S::MnEIL3 recombinant plasmid was inserted into chromosome 2 of the Arabidopsis genome (Fig. S1C).
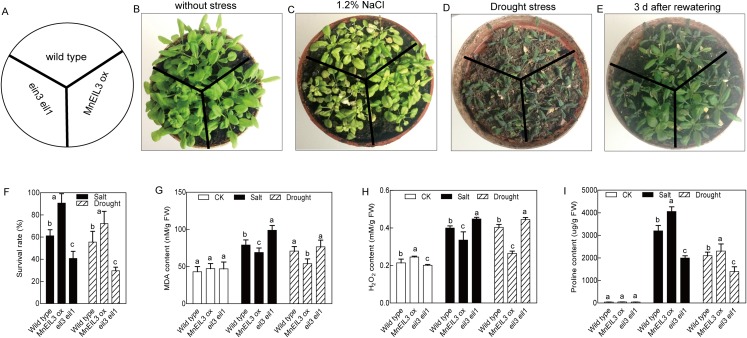
To evaluate the tolerance of MnEIL3-overexpressing (MnEIL3ox) plants against salt stress, the 21-day-old wild type, MnEIL3ox, and ein3-1 eil1-1 Arabidopsis seedlings were treated with 1.2% (m/v) NaCl. After treatment, MnEIL3ox plants showed relatively greater growth rates than wild type and ein3-1 eil1-1 Arabidopsis. Additionally, the ein3-1 eil1-1 seedlings showed a decreased salt tolerance compared with wild type plants (Figs. 2A and 2B). To characterize the performance of MnEIL3ox plants under drought stress, the 14-day-old seedlings of wild type, MnEIL3ox, and ein3-1 eil1-1 Arabidopsis plants were treated with drought stress. The growth of wild type, MnEIL3ox, and ein3-1 eil1-1 plants showed no difference under drought stress conditions. However, MnEIL3ox showed a greater capability to survive than wild type and ein3-1 eil1-1 plants when the treated plants were re-watered (Figs. 2A and 2B).
Figure 2. Stress tolerance analyses of wild type, MnEIL3ox, and ein3-1 eil1-1 Arabidopsis.
(A–E) The growth of wild type, MnEIL3ox, and ein3-1 eil1-1 Arabidopsis plants under normal, salt and drought conditions. (F–I) The survival rates (F), MDA content (G), H2O2 content (H), and proline content (I) of wild type, MnEIL3ox, and ein3-1 eil1-1 Arabidopsi plants under normal and stress conditions. Data are means ± SEs (n = 3). Means within a column with different letters are significantly different (P < 0.05). Means within a column with the same letters are not significantly different (P > 0.05).
To understand the mechanism behind the enhanced sensitivity to drought and salt stresses caused by MnEIL3’s overexpression, the accumulated levels of H2O2, MDA, and proline were analyzed. The MDA and H2O2 contents in MnEIL3ox and ein3-1 eil1-1 plants were lower and higher than in wild type Arabidopsis, respectively (Figs. 2C and 2D). The proline contents in MnEIL3ox and ein3-1 eil1-1 plants were higher and lower than in wild type Arabidopsis, respectively (Figs. 2C and 2D). Thus, MnEIL3 may negatively regulate drought and salt stress tolerances.
The enhanced expressions of ACS and ACO genes in MnEIL3ox plants
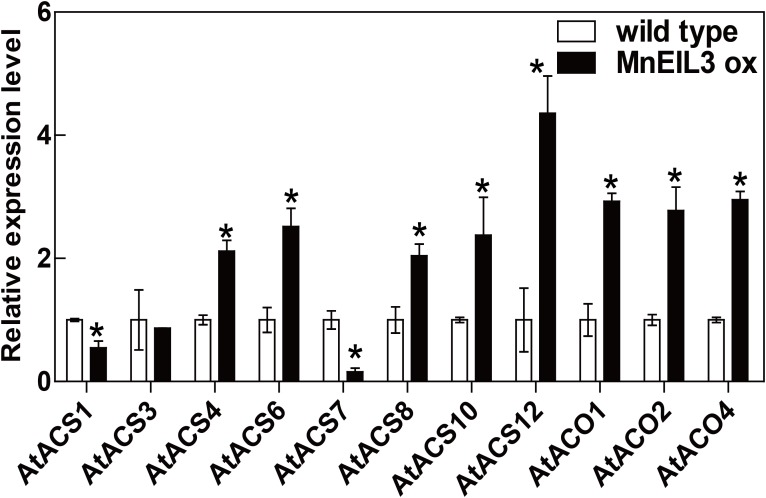
In this study, the expression levels of ACS- and ACO-encoding genes were detected in MnEIL3ox plants. The transcript abundances of AtACS4/6/8/10/12 genes in MnEIL3ox plants were higher than in wild type Arabidopsis, while the expression levels of AtACS1/7 genes showed were lower in MnEIL3ox plants. Moreover, there was no difference in the expression of AtACS3 between MnEIL3ox and wild type plants (Fig. 3). All of the detected AtACO genes, AtACO1/2/4, showed higher expression levels in MnEIL3ox plants than in wild type Arabidopsis (Fig. 3).
Figure 3. qRT-PCR analysis of changes in AtACS and AtACO genes in MnEIL3ox plants.
Data are means ± SEs (n = 3). Significant differences (P < 0.05) are marked with asterisks. The gene expression in wild type was set as 1.
Mulberry EIL proteins modulate MnACO1, MnACO2, MnACS1, and MnACS3 promoter activities
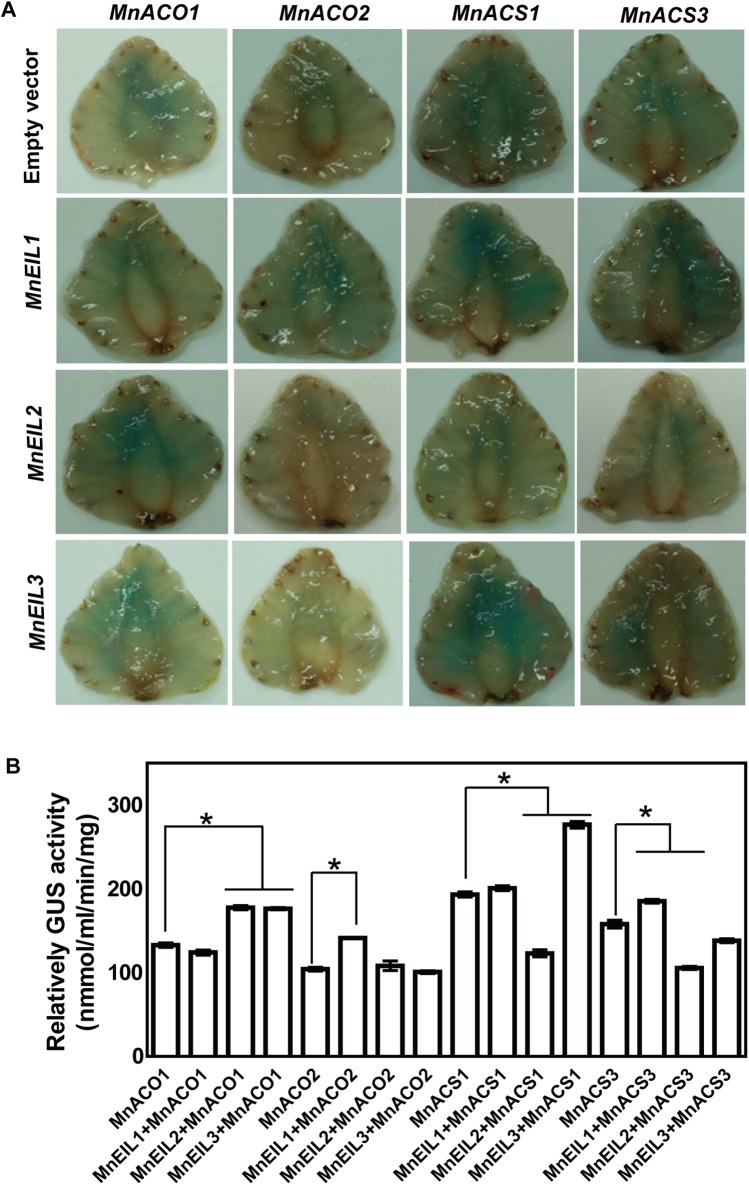
To explore the correlations between the expression of MnEIL3 and ethylene biosynthetic genes, MnACO1, MnACO2, MnACS1, and MnACS3 were selected for promoter isolation. All these genes have been determined as the key genes involved in ethylene biosynthesis in mulberry (Liu et al., 2014, 2015). The gene’ promoters were downloaded from the Morus genome database (http://morus.swu.edu.cn/morusdb/) and isolated from mulberry (M. notabilis). By searching for cis-acting regulatory elements, PERE- and ECBS-binding sites were identified in the promoters of MnACO1, MnACO2, MnACS1, and MnACS3 (Table S1). In vivo interactions between MnEIL3 and these promoters were estimated by transient analyses in strawberry fruit. MnEIL3 significantly enhanced the activities of MnACO1 and MnACS1 promoters, while no significant effects on the activities of MnACO2 and MnACS3 promoters were found (Fig. 4). Thus, EIL proteins may act as the transcriptional activators of ethylene biosynthetic genes. We also detected correlations between the other two MnEIL genes, MnEIL1/2, and ethylene biosynthetic genes. Thus, MnEIL1 acted as the activator of MnACO2 and MnACS3 promoters, while MnEIL2 regulated the activities of the MnACO1 and MnACS3 promoters (Fig. 4).
Figure 4. In vivo interactions of MnEIL3 with ethylene biosythetic genes promoters.
(A) GUS staining of the injected strawberry fruit. (B) The detection of GUS activities. Data are means ± SEs (n = 3). Significant differences (P < 0.05) are marked with asterisks.
The activities of MnACS1 and MnACS3 promoters were regulated by salt and drought stresses
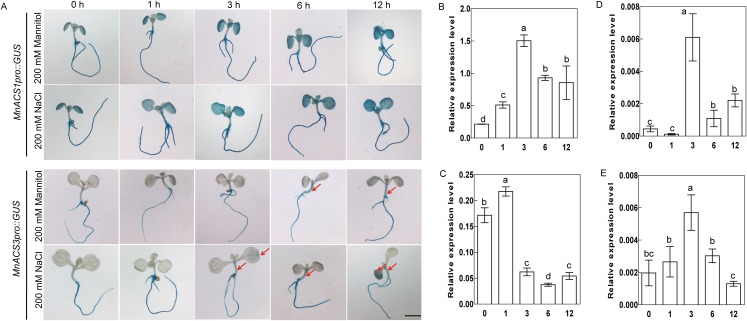
To examine the responsiveness of the MnACS1 and MnACS3 genes under salt and drought stresses, transgenic Arabidopsis were generated by introducing MnACS1pro::GUS- and MnACS3pro::GUS-fused genes, and then the 10-day-old seedlings were exposed to stresses (Fig. 5A). The MnACS1 promoter in leaves responded to NaCl and mannitol treatments (Fig. 5A). The GUS reporter in the MnACS3pro::GUS transgenic Arabidopsis was mainly expressed in roots, while little GUS accumulation levels in stems and leaves were detected. Under salt stress, the GUS accumulation levels in the stems of MnACS3pro::GUS transgenic Arabidopsis were enhanced after 6 h of NaCl treatment, while the GUS levels in stems and leaves were enhanced after 3 h of mannitol treatment (Fig. 5A).
Figure 5. Analyses of promoter activities and expression levels of MnACS1 and MaACS3 genes in response to salt and drought treatments.
(A) Histochemical GUS staining of the MnACS1pro::GUS and MnACS3pro::GUS transgenic Arabidopsis under salt [200 mM NaCl] and drought (200 mM mannitol) treatments. Scale bar, two mm. (B) The expression level of MnACS1 in response to salt treatment. (C) The expression level of MnACS1 in response to drought treatment. (D) The expression level of MnACS3 in response to salt treatment. (E) The expression level of MnACS3 in response to drought treatment. Data are means ± SEs (n = 3). Means within a column with different letters are significantly different (P < 0.05). Means within a column with the same letters are not significantly different (P > 0.05).
The expression levels of MnACS1 and MnACS3 are regulated by salt and drought stresses
The 14-day-old seedlings were treated with NaCl and mannitol, and then used to detect the expression levels of MnACS1 and MnACS3. MnACS1’s expression level was significantly upregulated after 1 h of NaCl treatment, and its expression was transiently upregulated after 1 h of mannitol treatment and then was downregulated (Figs. 5B and 5C). The transcript abundance of MnACS3 was significantly upregulated after 3 h of NaCl treatment, and its expression was upregulated after 3 h of mannitol treatment, but it showed no response at any other time point (Figs. 5D and 5E).
Discussion
EIN3/EILs proteins are positive factors in ethylene signal transduction. In model plants, EIN3/EILs are involved in many aspects of the life cycle, including seed germination, soil emergence, seedling development, leaf senescence, pigments biosynthesis, light perception, and abiotic stress responses (An et al., 2018; Kim, Cho & Yoo, 2017; Yu et al., 2013; Yu et al., 2016; Zhong et al., 2014). Additionally, the regulatory functions of EIN3/EILs in response to abiotic stresses have also attracted considerable attention (Cui et al., 2015; Kong et al., 2018; Peng et al., 2014; Shi et al., 2012; Zhang et al., 2011). However, there are no reports on the functions of EIN3/EILs in the abiotic stress tolerance of woody plants, including mulberry. In the present study, the expression levels of mulberry MnEIL genes under salt and drought stresses were revealed, and they showed different patterns. Among these genes, the expression of MnEIL3 was significantly upregulated by salt and drought stresses in roots and shoots (Fig. 1), which is similar to the expression patterns of Arabidopsis AtEIN3 and AtEIL1 genes (Fig. S2). The full-length coding sequence of MnEIL3 was transformed into Arabidopsis for stress tolerance analysis. The expression levels of MnEIL3 were significantly upregulated in roots and shoots by NaCl treatments (Fig. 1), and the overexpression of this gene in Arabidopsis enhanced salt stress tolerance (Fig. 2). MnEIL3’s overexpression decreased the MDA and H2O2 contents and enhanced the proline content under salt stress. Thus, MnEIL3 positively regulated plant salt tolerance, which was similar to the results described by Peng et al. (2014). In this study, ein3-1 eil1-1 plants showed decreased tolerances to drought stress compared with wild type plants (Fig. 2), and mutation of EIN3/EILs decreased the MDA and H2O2 contents and enhanced the proline content under drought stress. This was similar to the results of a previous study which reported that the ein3-1 mutant exhibited a decreased tolerance to drought stress stimulated by PEG6000 (Cui et al., 2015). In addition, MnEIL3ox plants showed a greater ability to survive drought stress than wild type and ein3-1 eil1-1 plants. Thus, MnEIL3 may play a positive role in abiotic stress tolerances, and it indicates that the functions of EIL3/EILs in response to abiotic stresses are relatively conserved in plants.
The overexpression of kiwifruit (Actinidia deliciosa) EIN3-like TFs, AdEIL2 and AdEIL3, increased ethylene production by upregulating the expression of ACS and ACO genes in transgenic Arabidopsis (Yin et al., 2010). Based on the data reported by Liu et al. (2017b), several genes that were involved in ethylene biosynthesis and signal transduction showed lower expression levels in ein3-1 eil1-1 plants compared with wild type Arabidopsis. Here, the transcript abundances of AtACS and AtACO genes were mainly upregulated in MnEIL3ox plants. The analysis suggested the positive feedback regulation of EIN3/EILs in ethylene production.
When plants receive the ethylene signal, EIN3/EILs are activated, and then, they regulate the transcriptional expression of downstream responsive genes, including ERF, ACO, xyloglucan endo-transglycosylase, and cell wall-modifying genes (Huang et al., 2010; Ireland et al., 2014; Solano et al., 1998; Yin et al., 2010). The PERE and ECBS motifs in promoters have been identified as EIN3-interactive motifs. MnACO1, MnACO2, MnACS1, and MnACS3 promoters contain PERE and ECBS motifs, which suggests that ethylene biosynthetic genes can be regulated by EIN3/EILs. In the present study, we found that MnEIL3 and two other MnEILs (MnEIL1 and MnEIL2) modulate the activities of MnACO1 and MnACO2 promoters as assessed by transient analysis in strawberry (Fragaria × ananassa Duch.) fruit. This result was similar to those reported in kiwifruit and melon (Cucumis melo L. cv. Andes) (Huang et al., 2010; Yin et al., 2010). MnACS1 and MnACS3 promoter’ activities were regulated by MnEIL1/2/3 (Fig. 4). Thus, MnEIL proteins provide positive feedback regulation during ethylene production by directly regulating the transcription of ethylene biosynthetic genes. We also constructed MnACS1pro::GUS and MnACS3pro::GUS vectors and independently introduced them into Arabidopsis. The GUS activities in the stems and leaves of the transgenic Arabidopsis seedlings were enhanced by salt and drought stresses (Fig. 5A). Additionally, the expression levels of MnACS1 and MnACS3 significantly responded to salt and drought stresses (Figs. 5B–5E). The promoter’ activities and gene expression levels of MnACO1 and MnACO2 were also enhanced by abiotic stresses (Yu et al., 2017).
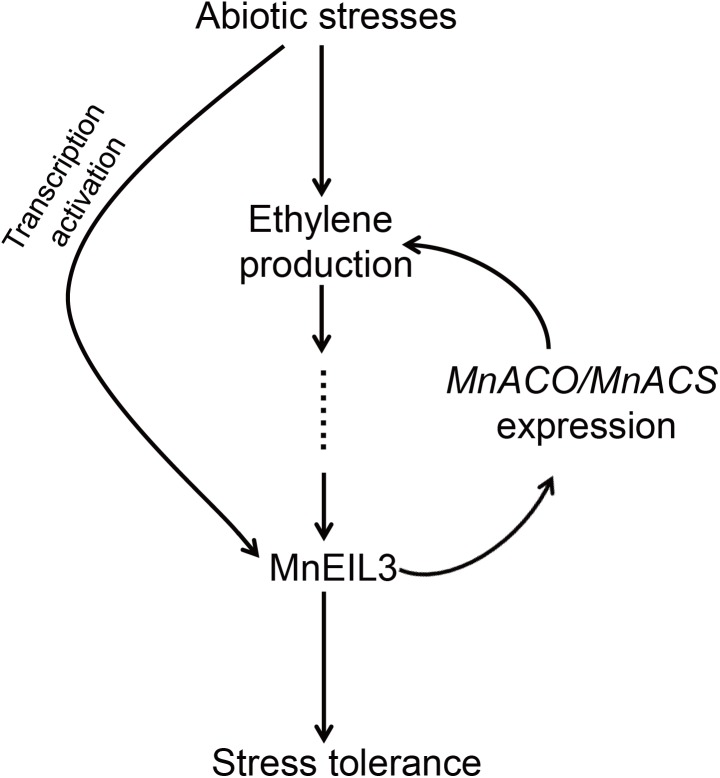
On the basis of our results, we proposed a working model for the regulatory network of mulberry MnEIL3 in response to abiotic stresses (Fig. 6). When plants are exposed to abiotic stresses, the stress signals are perceived by plant cells, leading to the enhanced accumulation of ethylene. Then, the ethylene signal transduction pathway was induced and the nucleus-localized MnEIL3 proteins’ accumulation and stability were enhanced. MnEIL3’s expression was also significantly induced by stresses. MnEIL3 eventually positively regulates abiotic stress tolerances by activating downstream stress-responsive genes. In addition, MnEIL3 binds to the target regions in the promoters of ACO and ACS genes, and activates gene expression, which contributes to the accumulation of ethylene. MnEIL1 and MnEIL2 may function in other processes, such as fruit development and maturation, by modulating ethylene responses (Liu et al., 2015). However, more work is needed to investigate the roles of the ethylene–EIN3/EILs–ACO/ACS regulatory loop in abiotic stress tolerances.
Figure 6. A possible model of MnEIL3 in responses to abiotic stresses.
Conclusions
In summary, our results explored the functions of a gene encoding an EIN3-like protein from mulberry, MnEIL3. The expression level of MnEIL3 significantly increased in response to salt and drought stresses in roots and shoot. Transgenic Arabidopsis overexpressing MnEIL3 exhibited an enhanced tolerance to salt and drought stresses. The overexpression of MnEIL3 significantly upregulated the expression levels of ethylene biosynthetic genes in Arabidopsis. Moreover, MnEIL3 could enhance the activities of MnACO1 and MnACS1 promoters, which suggested an ethylene–EIN3/EILs–ACO/ACS regulatory loop in abiotic stress tolerance. This research provides insights into the functions of MnEIL3 in abiotic stress tolerance and their influence on the expression levels of ethylene biosynthetic genes.
Supplemental Information
(A) Genomic PCR analysis of transgenic lines. (B) Quantitative real-time PCR. (C) Inverse PCR.
(A–B) AtEIN3 and AtEIL1 genes’ expression in roots under salt stress. (C–D) AtEIN3 and AtEIL1 genes’ expression in shoot under salt stress. (E–F) AtEIN3 and AtEIL1 genes’ expression in roots under drought stress. (G–H) AtEIN3 and AtEIL1 genes’ expression in shoot under drought stress. These data were downloaded from the ePlant database (http://bar.utoronto.ca/eplant/).
This file contains:
– Raw data for Figure 1: MnEIL genes expression level under salt and drought stresses.xlsx.
– Raw data for Figure 2: MDA, H2O2, proline contents and survival rate of Arabidopsis.xlsx.
– Raw data for Figure 3: Expression of AtACS and AtACO genes in MnEIL3ox plants.xlsx.
– Raw data for Figure 4: The detection of GUS activities.xlsx.
– Raw data for Figure 5: MnACS1 and MnACS3 genes expression level under salt and drought stresses.xlsx.
– Raw data for Figure S1: MnEIL3 expression level in transgenic plants.xlsx.
– Raw data for Figure S2: AtEIN3 and AtEIL1 genes expression level under salt and drought stresses.xlsx.
– Raw data for Table S1: The sequences and EIL-binding sites of MnACO and MnACS gene promoters.docx.
Acknowledgments
We thank Dr. Hongwei Guo (Institute of Plant and Food Science, Department of Biology, Southern University of Science and Technology, Shenzhen 518055, P.R. China) for providing the seeds of ein3-1 eil1-1 Arabidopsis.
Funding Statement
This work was supported by the China Agriculture Research System (No. CARS-18–ZJ0201), the Special Fund for Agro-scientific Research in the Public Interest of China (No. 201403064), the Fundamental Research Funds for the Central Universities (No. XDJK2018C008), and the National Natural Sciences Foundation of China (Grant No. 31801126). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Changying Liu conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jun Li performed the experiments.
Panpan Zhu performed the experiments.
Jian Yu performed the experiments.
Jiamin Hou performed the experiments.
Chuanhong Wang performed the experiments.
Dingpei Long analyzed the data.
Maode Yu conceived and designed the experiments.
Aichun Zhao conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data are available in Data S1.
References
- Alonso & Stepanova (2004).Alonso JM, Stepanova AN. The ethylene signaling pathway. Science. 2004;306(5701):1513–1515. doi: 10.1126/science.1104812. [DOI] [PubMed] [Google Scholar]
- Alonso et al. (2003).Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An et al. (2012).An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Research. 2012;22(5):915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An et al. (2018).An J-P, Wang X-F, Li Y-Y, Song L-Q, Zhao L-L, You C-X, Hao Y-J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiology. 2018;178(2):808–823. doi: 10.1104/pp.18.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An et al. (2010).An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, Zhang S, Ecker JR, Guo H. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22(7):2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao et al. (1997).Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89(7):1133–1144. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Clark et al. (1998).Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough & Bent (1998).Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui et al. (2015).Cui M, Lin Y, Zu Y, Efferth T, Li D, Tang Z. Ethylene increases accumulation of compatible solutes and decreases oxidative stress to improve plant tolerance to water stress in Arabidopsis. Journal of Plant Biology. 2015;58(3):193–201. doi: 10.1007/s12374-014-0302-z. [DOI] [Google Scholar]
- He et al. (2011).He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, Wen X, Li P, Chu J, Sun X, Yan C, Yan N, Xie D-Y, Raikhel N, Yang Z, Stepanova AN, Alonso JM, Guo H. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23(11):3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2013).He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, Lee T-H, Wang X, Cai Q, Li D, Lu M, Liao S, Luo G, He R, Tan X, Xu Y, Li T, Zhao A, Jia L, Fu Q, Zeng Q, Gao C, Ma B, Liang J, Wang X, Shang J, Song P, Wu H, Fan L, Wang Q, Shuai Q, Zhu J, Wei C, Zhu-Salzman K, Jin D, Wang J, Liu T, Yu M, Tang C, Wang Z, Dai F, Chen J, Liu Y, Zhao S, Lin T, Zhang S, Wang J, Wang J, Yang H, Yang G, Wang J, Paterson AH, Xia Q, Ji D, Xiang Z. Draft genome sequence of the mulberry tree Morus notabilis. Nature Communications. 2013;4(1):2445. doi: 10.1038/ncomms3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua & Meyerowitz (1998).Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94(2):261–271. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2010).Huang S, Sawaki T, Takahashi A, Mizuno S, Takezawa K, Matsumura A, Yokotsuka M, Hirasawa Y, Sonoda M, Nakagawa H, Sato T. Melon EIN3-like transcription factors (CmEIL1 and CmEIL2) are positive regulators of an ethylene- and ripening-induced 1-aminocyclopropane-1-carboxylic acid oxidase gene (CM-ACO1) Plant Science. 2010;178(3):251–257. doi: 10.1016/j.plantsci.2010.01.005. [DOI] [Google Scholar]
- Ireland et al. (2014).Ireland HS, Gunaseelan K, Muddumage R, Tacken EJ, Putterill J, Johnston JW, Schaffer RJ. Ethylene regulates apple (Malus x domestica) fruit softening through a dose x time-dependent mechanism and through differential sensitivities and dependencies of cell wall-modifying genes. Plant and Cell Physiology. 2014;55(5):1005–1016. doi: 10.1093/pcp/pcu034. [DOI] [PubMed] [Google Scholar]
- Jefferson (1987).Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter. 1987;5(4):387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- Kende (1993).Kende H. Ethylene biosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology. 1993;44(1):283–307. doi: 10.1146/annurev.pp.44.060193.001435. [DOI] [Google Scholar]
- Kim, Cho & Yoo (2017).Kim GD, Cho Y-H, Yoo S-D. Regulatory functions of cellular energy sensor SNF1-related kinase1 for leaf senescence delay through ETHYLENE-INSENSITIVE3 repression. Scientific Reports. 2017;7(1):3193. doi: 10.1038/s41598-017-03506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong et al. (2018).Kong X, Li C, Zhang F, Yu Q, Gao S, Zhang M, Tian H, Zhang J, Yuan X, Ding Z. Ethylene promotes cadmium-induced root growth inhibition through EIN3 controlled XTH33 and LSU1 expression in Arabidopsis. Plant, Cell & Environment. 2018;41(10):2449–2462. doi: 10.1111/pce.13361. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2017b).Liu X, Liu R, Li Y, Shen X, Zhong S, Shi H. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell. 2017b;29(12):3051–3067. doi: 10.1105/tpc.17.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2014).Liu CY, Lu RH, Li J, Zhao AC, Wang XL, Diane U, Wang XH, Wang CH, Yu YS, Han SM, Lu C, Yu MD. Characterization and expression profiles of MaACS and MaACO genes from mulberry (Morus alba L.) Journal of Zhejiang University-Science B. 2014;15(7):611–623. doi: 10.1631/jzus.B1300320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2017a).Liu C, Wei C, Zhang M, Xu Y, Xiang Z, Zhao A. Mulberry MnMAPK1, a group C mitogen-activated protein kinase gene, endowed transgenic Arabidopsis with novel responses to various abiotic stresses. Plant Cell, Tissue and Organ Culture. 2017a;131(1):151–162. doi: 10.1007/s11240-017-1272-x. [DOI] [Google Scholar]
- Liu et al. (2015).Liu C, Zhao A, Zhu P, Li J, Han L, Wang X, Fan W, Lü R, Wang C, Li Z, Lu C, Yu M, Xu C. Characterization and expression of genes involved in the ethylene biosynthesis and signal transduction during ripening of mulberry fruit. PLOS ONE. 2015;10(3):e0122081. doi: 10.1371/journal.pone.0122081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng et al. (2014).Peng JY, Li ZH, Wen X, Li WY, Shi H, Yang LS, Zhu HQ, Guo HW. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLOS Genetics. 2014;10(10):e1004664. doi: 10.1371/journal.pgen.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak et al. (2003).Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115(6):679–689. doi: 10.1016/S0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Qiao et al. (2012).Qiao H, Shen Z, Huang S-sC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338(6105):390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez et al. (1999).Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283(5404):996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Shan et al. (2012).Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY, Lu WJ. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. Journal of Experimental Botany. 2012;63(14):5171–5187. doi: 10.1093/jxb/ers178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2018).Shi H, Lyu M, Luo YW, Liu SC, Li Y, He H, Wei N, Deng XW, Zhong S. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(25):6482–6487. doi: 10.1073/pnas.1803861115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2012).Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 2012;24(6):2578–2595. doi: 10.1105/tpc.112.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano et al. (1998).Solano R, Stepanova A, Chao QM, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Development. 1998;12(23):3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolaore, Trainotti & Casadoro (2001).Spolaore S, Trainotti L, Casadoro G. A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. Journal of Experimental Botany. 2001;52(357):845–850. doi: 10.1093/jexbot/52.357.845. [DOI] [PubMed] [Google Scholar]
- Wang, Li & Ecker (2002).Wang KL-C, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14(Suppl 1):S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2014).Wei C, Liu X, Long D, Guo Q, Fang Y, Bian C, Zhang D, Zeng Q, Xiang Z, Zhao A. Molecular cloning and expression analysis of mulberry MAPK gene family. Plant Physiology and Biochemistry. 2014;77:108–116. doi: 10.1016/j.plaphy.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Yang & Hoffman (1984).Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology. 1984;35(1):155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- Yin et al. (2010).Yin XR, Allan AC, Chen KS, Ferguson IB. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology. 2010;153(3):1280–1292. doi: 10.1104/pp.110.157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo et al. (2008).Yoo S-D, Cho Y-H, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451(7180):789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2017).Yu J, Liu C-Y, Zhao A-C, Wang C-H, Cai Y-X, Yu M-D. Functional analysis of 1-aminocyclopropane-1-carboxylate oxidase gene’s promoter in mulberry. Acta Agronomica Sinica. 2017;43(6):839–848. doi: 10.3724/SP.J.1006.2017.00839. [DOI] [Google Scholar]
- Yu et al. (2016).Yu Y, Wang J, Shi H, Gu J, Dong J, Deng XW, Huang R. Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiology. 2016;170(4):2340–2350. doi: 10.1104/pp.15.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2013).Yu Y, Wang J, Zhang Z, Quan R, Zhang H, Deng XW, Ma L, Huang R. Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLOS Genetics. 2013;9(12):e1004025. doi: 10.1371/journal.pgen.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2011).Zhang L, Li Z, Quan R, Li G, Wang R, Huang R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiology. 2011;157(2):854–865. doi: 10.1104/pp.111.179028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong et al. (2014).Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. Ethylene-orchestrated circuitry coordinates a seedling's response to soil cover and etiolated growth. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(11):3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2011).Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim J-M, To TK, Li W, Zhang X, Yu Q, Dong Z, Chen W-Q, Seki M, Zhou J-M, Guo H. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(30):12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Genomic PCR analysis of transgenic lines. (B) Quantitative real-time PCR. (C) Inverse PCR.
(A–B) AtEIN3 and AtEIL1 genes’ expression in roots under salt stress. (C–D) AtEIN3 and AtEIL1 genes’ expression in shoot under salt stress. (E–F) AtEIN3 and AtEIL1 genes’ expression in roots under drought stress. (G–H) AtEIN3 and AtEIL1 genes’ expression in shoot under drought stress. These data were downloaded from the ePlant database (http://bar.utoronto.ca/eplant/).
This file contains:
– Raw data for Figure 1: MnEIL genes expression level under salt and drought stresses.xlsx.
– Raw data for Figure 2: MDA, H2O2, proline contents and survival rate of Arabidopsis.xlsx.
– Raw data for Figure 3: Expression of AtACS and AtACO genes in MnEIL3ox plants.xlsx.
– Raw data for Figure 4: The detection of GUS activities.xlsx.
– Raw data for Figure 5: MnACS1 and MnACS3 genes expression level under salt and drought stresses.xlsx.
– Raw data for Figure S1: MnEIL3 expression level in transgenic plants.xlsx.
– Raw data for Figure S2: AtEIN3 and AtEIL1 genes expression level under salt and drought stresses.xlsx.
– Raw data for Table S1: The sequences and EIL-binding sites of MnACO and MnACS gene promoters.docx.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in Data S1.