Abstract
Insulin deficiency and uncontrolled diabetes lead to a catabolic state with decreased muscle strength, contributing to disease-related morbidity. FoxO transcription factors are suppressed by insulin and thus are key mediators of insulin action. To study their role in diabetic muscle wasting, we created mice with muscle-specific triple knockout of FoxO1/3/4 and induced diabetes in these M-FoxO-TKO mice with streptozotocin (STZ). Muscle mass and myofiber area were decreased 20–30% in STZ-Diabetes mice due to increased ubiquitin-proteasome degradation and autophagy alterations, characterized by increased LC3-containing vesicles, and elevated levels of phosphorylated ULK1 and LC3-II. Both the muscle loss and markers of increased degradation/autophagy were completely prevented in STZ FoxO-TKO mice. Transcriptomic analyses revealed FoxO-dependent increases in ubiquitin-mediated proteolysis pathways in STZ-Diabetes, including regulation of Fbxo32 (Atrogin1), Trim63 (MuRF1), Bnip3L, and Gabarapl. These same genes were increased 1.4- to 3.3-fold in muscle from humans with type 1 diabetes after short-term insulin deprivation. Thus, FoxO-regulated genes play a rate-limiting role in increased protein degradation and muscle atrophy in insulin-deficient diabetes.
Introduction
Uncontrolled diabetes causes decreased muscle strength and aerobic capacity, both of which contribute to disability and mortality in patients with the disease. Likewise, older individuals with type 2 diabetes lose muscle strength faster than those without diabetes, and this loss of muscle strength is highly correlated with death rates in these populations (1,2). Patients with newly diagnosed or poorly controlled type 1 diabetes also exhibit loss of lean mass (3,4). Additionally, physical fitness in patients with type 1 diabetes is directly correlated with hemoglobin A1c levels, indicating that poor control of glycemia is associated with poor fitness in type 1 diabetes (5). Uncontrolled diabetes is accompanied by numerous metabolic abnormalities, including insufficient insulin levels, hyperglycemia, lipid abnormalities, hyperglucagonemia, and glucocorticoid elevations, each of which may contribute to abnormalities in muscle function. However, the exact molecular mechanisms that account for muscle loss with loss of insulin action in diabetes are still not understood.
Insulin action induces profound effects on fuel metabolism and protein turnover. Insulin and amino acids (AAs) coordinate protein synthesis and degradation in muscle (6). In patients with type 1 diabetes, insulin deprivation leads to a state of high protein turnover with a net loss of muscle mass. This is due to increased rates of protein degradation that exceed the increased synthesis rate at the organismal level (7,8). Conversely, insulin treatment suppresses protein degradation in muscle. Insulin also stimulates muscle protein synthesis but only when coinfused with AAs (9), mimicking the conditions of the postprandial state. Thus, insulin’s role in protein degradation is more important than in synthesis in muscle. The effect of insulin to suppress protein degradation has been demonstrated in vitro and in rodent models (10,11). Defining the rate-limiting steps in the pathways by which insulin regulates protein degradation could provide an important target in treatment of diabetes that would help limit disability related to muscle atrophy.
Insulin acts via its tyrosine kinase receptor to mediate metabolic changes and cellular growth. Insulin receptor (IR) and the closely related IGF-1 receptor (IGF1R) have overlapping roles in mediating muscle growth and glucose homeostasis (12). IR and IGF1R act via the phosphoinositide 3-kinase/Akt and the MAPK/ERK pathways to influence a broad range of cellular functions, including glucose uptake, growth, proliferation, and protein turnover. Part of insulin’s action in muscle is to modulate transcription (13,14). Activation of Akt in response to insulin or IGF-1 induces phosphorylation of FoxO transcription factors, which suppresses their transcriptional activity. FoxO proteins are metabolic and stress-responsive transcription factors that are ubiquitously expressed and conserved in the animal kingdom (15). Insulin’s ability to suppress FoxO1-mediated transcription is central to insulin action in the liver, and deletion of FoxO1 in liver can rescue many of the gene expression changes observed in mice with a liver-specific IR knockout (16,17).
FoxOs control a broad range of atrophy-related genes in muscle, including Fbxo32 (Atrogin1) and Trim63 (MuRF1), and autophagy genes (18–21). Indeed, deletion of FoxOs in muscle prevents muscle atrophy in response to starvation and denervation (22). Likewise, we have found that deletion of the three FoxO isoforms in muscle (FoxO1, FoxO3, and FoxO4) is able to rescue the profound muscle atrophy from muscle-specific deletion of IR and IGF1R, demonstrating the critical role of FoxOs in muscle protein degradation and proteostasis (23), but the degree to which FoxOs control muscle proteostasis in response to insulin-deficient diabetes, where numerous metabolic abnormalities are present (including hyperglycemia, lipid abnormalities, glucocorticoid elevations, and ketosis), has not been determined.
In the current study, we have explored the role of FoxO transcription factors in the muscle atrophy of insulin-deficient diabetes in both mice and humans. We show that muscle atrophy in streptozotocin (STZ) diabetes mice is prevented in STZ FoxO triple knockout mice (STZ FoxO-TKO). This occurs without any effect on hyperglycemia, hypoinsulinemia, or suppression of muscle protein synthesis by STZ treatment. Muscle mass is maintained in STZ FoxO-TKO mice owing to suppression of autophagy-lysosome and ubiquitin-proteasome degradation without changes in myocyte number or oxidative/glycolytic distribution. Transcriptomic analysis reveals that induction of transcripts involved in protein degradation pathways in STZ-Diabetes mice is prevented in muscle from STZ FoxO-TKO mice. Finally, we show that transcripts of the ubiquitin-proteasome and autophagy-lysosome systems are increased in muscle biopsies from people with type 1 diabetes who were deprived of insulin for as little as 8 h. Thus, we demonstrate that muscle atrophy in response to insulin-deficient diabetes is mediated by FoxO-driven protein degradation and that blocking this pathway can provide protection from this complication of diabetes.
Research Design and Methods
Animal Care and Use
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee at both the Joslin Diabetes Center and the University of Iowa. Male mice were used for studies unless otherwise indicated. Muscle-specific FoxO1, FoxO3, and FoxO4 triple knockout (M-FoxO-TKO) mice were generated using ACTA1-Cre (stock number 006149; The Jackson Laboratory) and FoxO1/3/4 triple floxed mice, provided by Dr. Domenico Accili, Colombia University Medical Center, as previously described (23). Littermate controls were used for all experiments, as the mice are on a mixed background containing C57Blk6, C57Blk6J, and 129 strains.
Diets and Treatments
Animals were maintained on a standard chow diet (Mouse Diet 9F, 5020; LabDiet). Fed mice were allowed ad libitum access to food and were sacrificed at 9:00 a.m. For STZ treatments, mice were fasted overnight and then injected intraperitoneally with a single high dose of STZ (150 mg/kg) (S0130; Sigma) dissolved in 100 mmol/L citrate buffer (pH 4.5) or injected with citrate buffer alone as a control, then immediately given access to food. Female mice were given a 150 mg/kg dose followed by a 75 mg/kg dose of STZ on day 3, since female mice are less responsive to STZ (24). Mice were monitored for hyperglycemia on days 3, 7, 12, and 15, and only mice in which random blood glucose remained >300 mg/dL for the following 9–12 days were used for experiments. For colchicine treatments, mice were injected intraperitoneally with colchicine (0.4 mg/kg/day) (C9754; Sigma) or saline daily for 2 days as previously described (23,25). All mice were sacrificed between 12 and 15 days after STZ injection.
Proteolysis Assay
Proteolysis was measured as previously described (11,23), with the following modifications. Soleus and extensor digitorum longus (EDL) muscles were isolated from 8-week-old mice and preincubated in 1 mL Kreb’s-Ringer buffer (in mmol/L: 117 NaCl, 4.7 KCl, 2.5 CaCl2, 1.2 KH2PO4, 1.2 MgSO4, 24.6 NaHCO3, and 5 glucose) for 30 min with constant bubbling of 95% O2/5% CO2 prior to transfer to fresh 1 mL Kreb’s-Ringer buffer containing 0.5 mmol/L cycloheximide to inhibit protein synthesis for 2 h. Incubation buffer was collected and tyrosine concentration was determined in 0.5 mL of incubation buffer as previously described (26) and was normalized to muscle weight.
Protein Synthesis Assay
Muscle protein synthesis was measured using the SUnSET (surface sensing of translation) method to determine puromycin incorporation into newly synthesized proteins in quadriceps (Quad) tissue as previously described (27). Briefly, mice were injected intraperitoneally with puromycin at a dose of 0.04 μmol/g body wt, anesthetized 20 min later, and sacrificed 30 min after puromycin injection. Western blot analysis of puromycin-incorporated proteins was performed using antibodies in Supplementary Table 3. Fed and 24-h fasted mice were injected or not injected with puromycin to validate the method.
Proteasome Activity Assays
Proteasome activity was determined from muscle homogenates using substrates for trypsin-like activity of the 26S proteasome as previously described (23,28). Briefly, frozen powdered muscle was homogenized in 50 mmol/L Tris-HCl, 5 mmol/L MgCl2, 250 mmol/L sucrose, 2 mmol/L ATP, 1 mmol/L dithiothreitol, and 0.5 mmol/L EDTA, pH 7.5, and then centrifuged (600g for 20 min) at 4°C to pellet contractile proteins and membranes. The supernatant was removed and centrifuged (16,000g for 10 min at 4°C) to yield the final supernatant for proteasome activity. Twenty micrograms of protein from the proteasome supernatant were mixed with Boc-Leu-Ser-Thr-Arg-7-amido-4-methylcoumarin (LSTR) (300 μmol/L final concentration) (B4636; Sigma) in assay buffer (50 mmol/L Tris-HCl, pH 7.5, and 5 mmol/L MgCl2, 40 mmol/L KCl, 2 mmol/L ATP, 1 mmol/L dithiothreitol, and 0.5 mg/mL BSA) in a 96-well black plate, and fluorescence was monitored (360 nm excitation and 460 nm emission) every 3 min for 1.5 h at 37°C. Enzyme activity (Vmax) was determined as the change in fluorescence during the linear phase of the reaction without MG132 (carbobenzoxy-Leu-Leu-leucinal) minus the activity with 200 μmol/L MG132, each converted to nanomoles per minute per milligram using a standard curve of 7-amido-4-methylcoumarin (A9891; Sigma-Aldrich).
Physiologic Measurements
Whole blood glucose levels were measured using a glucose meter. Serum insulin levels were measured using an insulin ELISA according to the manufacturer’s protocol (Crystal Chem). Muscle tissue fluid AA levels were measured in mixed Quad and gastrocnemius muscle at the Mayo Clinic Metabolomics Resource Core using time-of-flight mass spectrometry as previously described (12). Please note that muscle tissue fluid AA levels from the control group and M-FoxO-TKO group were incorporated in the previous publication (23) and are also presented as important control groups in the current study. Muscle grip strength was measured as previously described (23).
Histology
Frozen cross sections of tibialis anterior (TA) muscle were stained for succinate dehydrogenase by immersion of slides in staining buffer containing PBS with 0.5 mol/L disodium succinate, 20 mmol/L MgCl2, and 0.5 mg/mL nitro blue tetrazolium for 15 min at 37°C and quantified as previously described (12). For cross-sectional area (CSA), immunofluorescence staining of TA sections for laminin was used. For LC3 vesicle density, TA sections were costained with LC3A and myosin IIa and quantified as previously described (23). See Supplementary Table 3 for antibodies used.
Western Analysis
Muscle tissue was homogenized in radioimmunoprecipitation assay buffer (Millipore) with protease and phosphatase A and B inhibitors (Bimake). Lysates were subjected to SDS-PAGE and blotted using antibodies as detailed in Supplementary Table 3.
Transcriptomic Analysis and Quantitative RT-PCR
For RNA-Seq experiments, RNA was extracted from a mixture of powdered Quad and gastrocnemius muscles using Trizol reagent (Invitrogen). Each sample was verified for quality and then enriched for polyadenylated RNAs used for library construction. All samples were run once for counts of reads and then twice for verification after adjustment to obtain >1 million reads per sample. Each sample was aligned and annotated using STAR (29) to the mouse genome. In the alignment, noncanonical and unannotated splice junctions were removed. Mapped reads were counted for gene features using featureCounts (30). In this process, overlap features and multimapping reads were not counted. One sample was removed, as it had almost empty reads. Filtering was performed, and we only kept genes having counts per million (cpm) >1 in at least two samples. STZ-Diabetes mice versus controls and STZ FoxO-TKO versus M-FoxO-TKO were compared using linear modeling in the limma package (31). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was performed using the sigPathway package (32). Heat maps were normalized by row (i.e., by gene) to compute z scores and were plotted with the gplots package. Volcano plots were plotted with the ggplot2 package (33). All statistical analysis was done in the R software.
For RT-PCR, RNA was reverse transcribed into cDNA (Applied Biosystems) according to the manufacturer’s protocol. RT-PCR was carried out using SYBR green (Bio-Rad or Bimake) with primers as detailed in Supplementary Table 1 (mouse) and Supplementary Table 2 (human), and normalized to TBP, except for proteasome subunits, which were normalized to GAPDH, as TBP was different between groups for that experimental run.
Muscle cDNA from Insulin-Treated and Insulin-Deprived Patients With Type 1 Diabetes
Muscle biopsy cDNA was obtained in collaboration with Dr. K.S.N. The details of the study have previously been published (34,35). Briefly, participants with type 1 diabetes underwent two studies: one with insulin treatment and one with insulin deprivation for an average of 8.6 h, separated by 1–2 weeks. Informed written consent was obtained after a detailed review of the protocol, which had been approved by the institutional review board of the Mayo Clinic and Mayo Clinic Foundation. Participants taking long-acting insulin were instructed to discontinue the long-acting insulin for 3 days prior to the study day, and volunteers on insulin pumps were instructed to continue using ultra rapid-acting insulin until admission to the Clinical Research Unit. Volunteers were admitted to the Clinical Research Unit at 1700 h on the evening before each study day and given a standard dinner at 1800 h, after which subjects remained fasting until the completion of the study the next day. On the insulin treatment study day, regular human insulin was infused into a forearm vein to maintain blood glucose between 4.44 and 5.56 mmol/L overnight until 1200 h the next day. The dose of insulin was adjusted based on plasma glucose levels every 30–60 min. On the insulin deprivation study day, the insulin infusion was discontinued for 8.6 ± 0.6 h. Vastus lateralis muscle samples were obtained at the end of the study (8.6 ± 0.6 h after insulin withdrawal or at a similar time point on the insulin treatment day) under local anesthesia (lidocaine [2%]) with a percutaneous needle as described previously (34). RNA was isolated and cDNA was synthesized as mentioned above.
Statistical Analyses
All data are presented as mean ± SEM. Two-way ANOVA was performed for comparison of STZ-treated groups to determine significance. For RT-PCR analyses of muscle cDNA from insulin-treated and insulin-deprived patients with type 1 diabetes, paired t tests were done, since each patient contributed to both the insulin-treated and the insulin-deprived groups.
Results
Deletion of FoxO Isoforms in Muscle Prevents Loss of Muscle Mass in Insulin-Deficient Diabetes
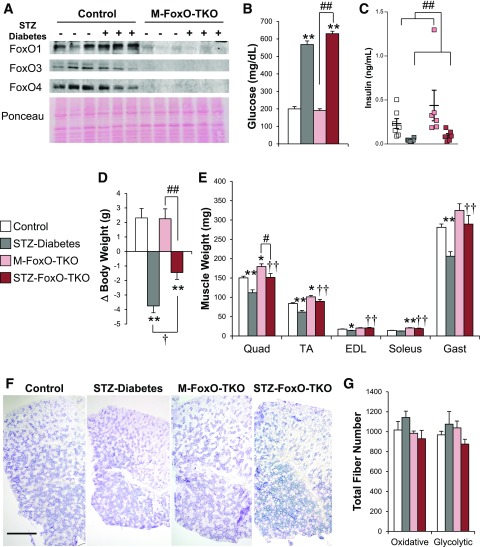
We have previously shown that skeletal muscle expresses FoxO1, -3, and -4 and that deletion of all three isoforms of FoxOs in muscle can prevent atrophy from muscle-specific deletion of IR and IGF1R (23). To determine the impact of FoxO deletion on muscle proteostasis in the context of insulin-deficient diabetes, we used M-FoxO-TKO mice and control mice rendered diabetic with STZ (23). These mice displayed a >95% depletion of FoxO proteins in Quad muscle (Fig. 1A), without changes in FoxO protein expression in heart or liver from M-FoxO-TKO mice (Supplementary Fig. 1A and B).
Figure 1.
FoxO deletion in muscle prevents muscle loss after STZ diabetes without altering oxidative fiber type or number. Western blot analysis (A) of FoxO isoforms 1, 3, and 4 in Quad. Blood glucose (B) and serum insulin (C) levels in control and M-FoxO-TKO mice 12–14 days after high-dose STZ treatment to induce insulin-deficient diabetes (n = 10–12 for glucose and 5–7 for insulin levels). Total change (Δ) in body weight 12–14 days after STZ (D) and dissected muscle weights (E) from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (n = 10–12). Succinate dehydrogenase staining (F) and quantification (G) of total numbers of purple oxidative and gray glycolytic fibers from whole TA muscle. Bar = 500 μm. *P < 0.05, **P < 0.01 vs. control; †P < 0.05, ††P < 0.01 STZ-Diabetes vs. STZ FoxO-TKO; #P < 0.05, ##P < 0.01 as indicated (two-way ANOVA). Gast, gastrocnemius. Blots are from parallel samples run on separate gels.
Insulin deficiency is the primary defect in type 1 diabetes but is also accompanied by multiple abnormalities in circulating hormones and metabolites that could affect muscle including increases in glucagon, inflammatory cytokines, and glucocorticoids as well as alterations of AAs and lipid metabolites in muscle. To determine whether muscle atrophy in response to insulin-deficient diabetes is mediated by FoxOs, we injected control and M-FoxO-TKO mice with STZ (single dose of 150 mg/kg) to induce diabetes and quantified muscle loss 12–14 days later. STZ-Diabetes induced marked hyperglycemia and insulin deficiency in both control and M-FoxO-TKO mice relative to vehicle-injected littermate mice within 14 days (Fig. 1B and C). During the 12–14 days after STZ treatment, body weight decreased in the STZ-Diabetes group by 3.75 g (−15%), while control and M-FoxO-TKO mice each gained 2.3 g (9%); STZ FoxO-TKO mice also lost weight, but this was attenuated to 1.45 g (−6%) (Fig. 1D and Supplementary Fig. 1C). Cardiac and adipose tissues atrophied to similar degrees in diabetic control and diabetic M-FoxO-TKO mice, while liver weights tended to increase (Supplementary Fig. 1D and E). However, STZ-Diabetes induced a dramatic 20–25% decrease in muscle size, and deletion of FoxOs in STZ FoxO-TKO mice prevented loss of muscle mass in response to diabetes (Fig. 1E), accounting for the significant preservation of body weight even with uncontrolled diabetes.
FoxOs are critical regulators of cell cycle control and modulate muscle differentiation depending on the stage of myogenesis (36). Cell culture models have demonstrated that repression of FoxO4 increased muscle progenitor cell proliferation (37), but FoxO1 has been implicated in both the suppression of and activation of myogenic differentiation by influencing expression of components of the mTOR pathway and myostatin (38–40). To determine whether our muscle-specific deletion of FoxO isoforms or insulin-deficient diabetes impacts expression of these genes and myocyte numbers in M-FoxO-TKO mice, we determined mRNA expression of myogenic factors in Quad and counted total oxidative and glycolytic fibers in TA muscles. STZ-Diabetes tended to increase myogenin and IGF-2 (Supplementary Fig. 1F). In M-FoxO-TKO and STZ FoxO-TKO Quad, MyoD and myogenin were significantly increased and IGF-2 tended to increase, while IGF-1 and myostatin remained unchanged (Supplementary Fig. 1F). However, quantitation of total oxidative and glycolytic fibers in TA cross sections revealed no changes in total fiber numbers or fiber types in STZ-Diabetes, M-FoxO-TKO, or STZ FoxO-TKO mice compared with controls (Fig. 1F and G). Thus, deletion of FoxO isoforms in skeletal muscle prevents diabetes-induced muscle loss but does not change myofiber number or oxidative type.
Deletion of FoxOs Prevents Muscle Atrophy in Response to Insulin-Deficient Diabetes
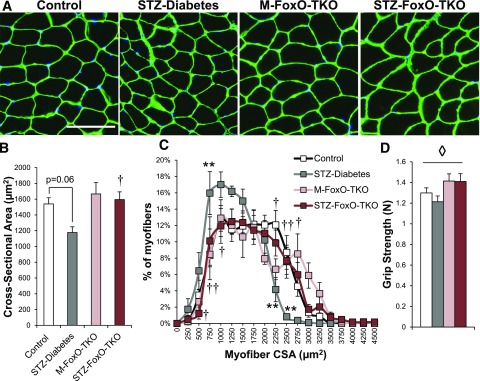
STZ-Diabetes mice showed a 24% decrease in the average myofiber CSA of TA muscle compared with controls, and this was completely prevented in STZ FoxO-TKO (Fig. 2A and B), thus confirming that the maintenance of muscle mass in STZ FoxO-TKO mice was due to prevention of myofiber atrophy. STZ-Diabetes mice showed an increased percentage of fibers <1,000 μm2 compared with controls and decreased the percentage of fibers >2,250 μm2 (Fig. 2C). This shift toward smaller fibers in STZ-Diabetes was reversed in STZ FoxO-TKO mice. Finally, muscle grip strength was improved in M-FoxO-TKO and STZ FoxO-TKO relative to controls and STZ-Diabetes mice (Fig. 2D). These data indicate that insulin-deficient diabetes induces significant muscle atrophy, which is dependent on FoxO transcription factors.
Figure 2.
Deletion of FoxOs prevents muscle atrophy in response to insulin-deficient diabetes. Immunofluorescent staining of laminin in TA muscle cross sections (A) from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (bar = 100 μm) (n = 5–6). Quantitation of CSA (B) and myofiber distribution (C) from images in A. Grip strength (D) measured 12–15 days after STZ injection (n = 8–11). **P < 0.01 vs. control; †P < 0.05, ††P < 0.01 STZ-Diabetes vs. STZ FoxO-TKO; ◊P < 0.05 genotype main effect (two-way ANOVA).
Induction of Autophagy-Lysosomal Markers Is Prevented in STZ FoxO-TKO Muscle
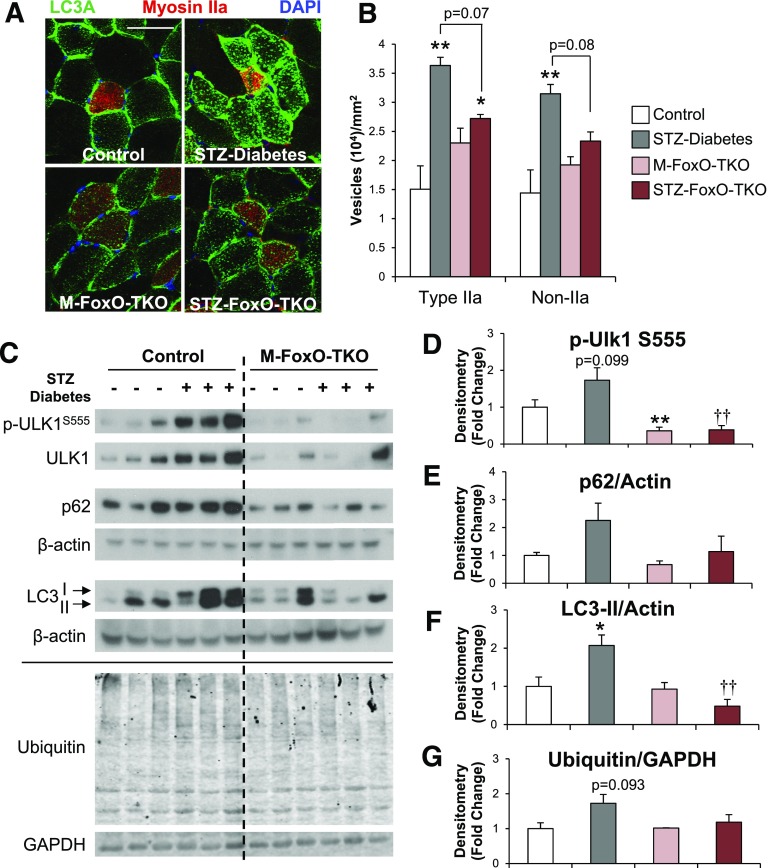
FoxOs are critical regulators of protein degradation pathways in skeletal muscle (18–22). During states of energy deprivation, such as starvation or uncontrolled diabetes, bulk autophagy (macroautophagy) is activated and can be monitored by accumulation of the autophagy protein LC3 into puncta (or vesicles) (41). We found that in both type IIa oxidative fibers and non–type IIa fibers from TA muscle, LC3 vesicles per area increased more than twofold in STZ-Diabetes compared with controls (Fig. 3A and B). This was due primarily to an increase in LC3 vesicles coupled with a small decrease in myofiber CSA in STZ-Diabetes (11 and 15% in type IIa and non-IIa, respectively) (Supplementary Fig. 2A and B). The number of vesicles per muscle fiber also increased in M-FoxO-TKO, but this was not significant, since the CSA of the fibers also increased (27 and 44% in type IIa and non-IIa, respectively) (Supplementary Fig. 2A and B). Importantly, when compared with M-FoxO-TKO, STZ FoxO-TKO mice showed no increase in LC3 vesicles per area (Fig. 3B).
Figure 3.
Induction of autophagy-lysosomal markers is prevented in STZ FoxO-TKO muscle. Immunofluorescent staining of LC3A and myosin IIa in TA muscle cross sections (A) from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (bar = 50 μm) (n = 5–6). Quantitation of LC3A-positive vesicles per CSA (B) in type IIa–positive and non-IIA fibers. Western blot analysis (C) and densitometric quantification of p-Ulk1S555 (D), p62 (E), LC3-II (F), and ubiquitin (G) levels in control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (n = 5–6, other than ubiquitin, where n = 3 per group). *P < 0.05, **P < 0.01 vs. control; ††P < 0.01 STZ-Diabetes vs. STZ-FoxO-TKO (two-way ANOVA). Blots are from parallel samples run on separate gels.
Phosphorylation of Ulk1 at serine 555 (p-Ulk1S555), a marker of autophagy induction, tended to increase in STZ-Diabetes Quad compared with controls (Fig. 3C and D). Total Ulk1 levels also tended to increase in STZ-Diabetes compared with controls. By contrast, M-FoxO-TKO mice showed decreased Ulk1 total protein levels (P < 0.05 for genotype main effect by ANOVA) and prevented the increase in STZ FoxO-TKO (Fig. 3C). Levels of LC3-II induced in STZ-Diabetes and p62/SQSTM also tended to increase but not in STZ FoxO-TKO muscle (Fig. 3E and F). For determination of the impact on autophagy flux, STZ-Diabetes and STZ FoxO-TKO mice were treated with or without colchicine for 2 days prior to sacrifice. Autophagy markers p-Ulk1S555, p62, and LC3-II tended to increase in colchicine-treated STZ-Diabetes Quad muscle compared with saline-treated STZ-Diabetes (Supplementary Fig. 2C and D). However, these same autophagy markers were significantly reduced in quad from saline-treated STZ FoxO-TKO muscle and remained low in STZ FoxO-TKO colchicine muscle. In contrast, LC3-II increased equally in the heart of STZ diabetes and STZ FoxO-TKO mice compared with their respective controls, whereas no changes were observed between any of the groups in the liver (Supplementary Fig. 2E and F). Taken together, these data indicate that STZ diabetes causes an increase in autophagy markers in muscle relative to nondiabetic mice, perhaps due to increased flux or decreased degradation of autophagosomes. However, the increase in autophagy markers is not seen when FoxO transcription factors are absent.
Total ubiquitination levels, which play a role in both proteasomal degradation and autophagy, tended to increase with STZ-Diabetes but were unchanged in STZ FoxO-TKO muscle (Fig. 3G). However, proteasome activity was unchanged and a proteasomal subunit was increased in M-FoxO-TKO and STZ FoxO-TKO relative to controls (Supplementary Fig. 3A and 3B). This may not be surprising, since the primary regulator of ubiquitin-mediated proteolysis is ubiquitination of the target and not proteasome activity. To determine how these alterations of protein degradation pathways influence total proteolysis, we measured tyrosine release from EDL and soleus muscles incubated ex vivo as previously described (23). Interestingly, we did not observe significant increases in proteolysis of STZ-Diabetes muscle compared with control in this ex vivo experiment with EDL and soleus (Supplementary Fig. 3C and D), which display more mild or no atrophy in response to STZ treatment (Fig. 1E). However, in soleus muscle, deletion of FoxOs did suppress proteolysis (Supplementary Fig. 3D), indicating that the regulation of autophagy and ubiquitin proteasome pathways by FoxOs controls muscle protein degradation.
FoxO Deletion Does Not Change a Majority of AA Levels in Muscle in Response to STZ Diabetes
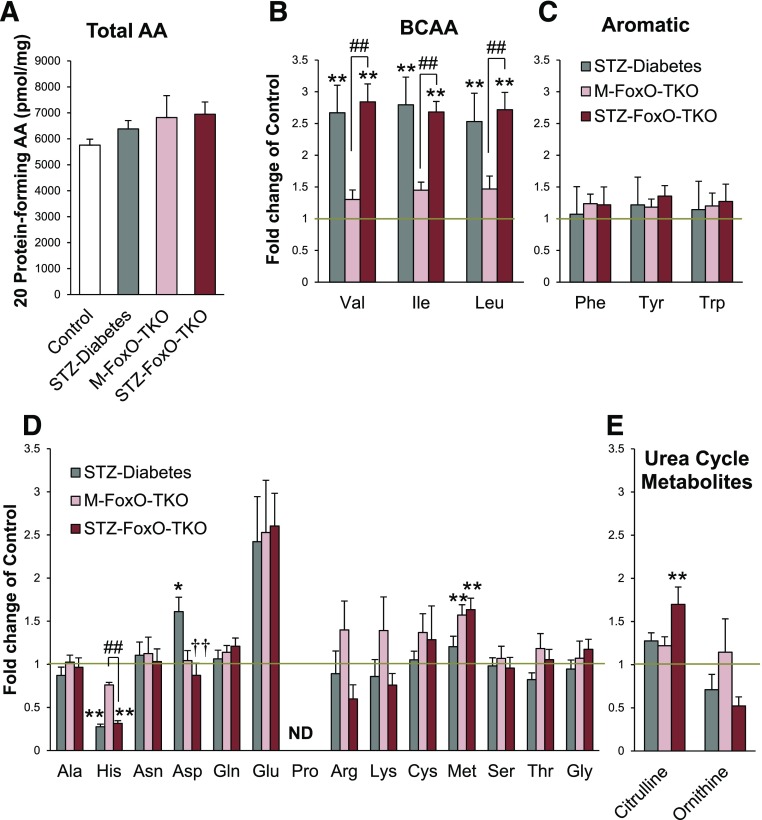
Previous reports indicate that marked alterations in plasma (35) and muscle (23) AA levels occur in mice with insulin-deficient diabetes or mice with knockout of IR/IGF1R in muscle. To determine the role of increased protein degradation and muscle atrophy in STZ diabetes and the contribution of FoxO proteins to these changes, we measured AA levels in muscle lysates from control and M-FoxO-TKO mice rendered diabetic with STZ. Total AA levels in muscle were unchanged between all four groups (Fig. 4A). Branch-chain AAs (BCAAs) increased in muscle from STZ-Diabetes mice (Fig. 4B), mimicking changes in plasma of humans with type 1 diabetes during insulin withdrawal (35). Interestingly, FoxO deletion did not prevent this rise in BCAAs, indicating that the suppression of protein degradation pathways by FoxO deletion in muscle does not contribute to the changing tissue BCAA levels. Histidine levels, on the other hand, decreased in muscle tissue from STZ diabetic mice, and this change was not dependent on FoxO signaling, while levels of other aromatic AAs were unchanged by diabetes or FoxO deletion (Fig. 4C). By contrast, methionine levels in muscle were increased in FoxO triple knockout and were not further affected by STZ-Diabetes (Fig. 4D). Aspartate levels also increased with STZ-Diabetes, and this change was prevented by FoxO deletion (Fig. 4D). Aspartate has many fates including as a degradation product in the urea cycle. Citrulline combines with aspartate to initiate the urea cycle, and citrulline levels increased in STZ FoxO-TKO muscle, perhaps indicating an increased capacity for urea cycle flux, but arginine and ornithine levels were unchanged (Fig. 4D and E). Thus, STZ diabetes induces a rise in BCAAs and decreases histidine levels in muscle, but these changes are not mediated by FoxO signaling. However, FoxO deletion in muscle does raise methionine and citrulline levels and prevents the increase in aspartate with diabetes, indicating a role for FoxOs in muscle AA metabolism.
Figure 4.
FoxO deletion does not prevent changes in most muscle tissue AA levels in response to STZ diabetes. Total tissue levels of 20 protein-forming AAs in hind-limb muscle from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (A). Fold change of BCAAs (B), aromatic AAs (C), other essential AAs (D), and urea cycle metabolites (E) relative to controls represented by gold line (n = 4–6 per group). *P < 0.05, **P < 0.01 vs. control; ††P < 0.01 STZ-Diabetes vs. STZ FoxO-TKO; ##P < 0.01 as indicated (two-way ANOVA). Note that control and M-FoxO-TKO groups contain data from our prior publication (23) and are presented as important controls in the current study. ND, not detected.
Increases in Ubiquitin-Proteasome Pathway Transcripts in STZ Diabetic Muscle Depend on FoxOs, but Abnormalities in Fatty Acid Metabolism Transcripts Are FoxO Independent
To determine the roles of FoxOs in transcriptional control of muscle protein turnover and metabolism, we performed RNA-Seq transcriptomic analyses on control and M-FoxO-TKO mice rendered diabetic with STZ. With use of a false discovery rate of <0.1, a total of 1,340 transcripts differed between STZ-Diabetes and control, with 550 increased and 790 decreased (Fig. 5A). In contrast, only 193 of these transcripts remained different between STZ FoxO-TKO and M-FoxO-TKO, with 78 increased and 115 decreased, indicating that >85% of the transcriptional changes in STZ diabetic muscle are reversed by the deletion of FoxOs (Fig. 5A). Volcano plots of these comparisons confirm that far more transcripts were altered in STZ-Diabetes versus control compared with STZ FoxO-TKO versus M-FoxO-TKO (Supplementary Fig. 5). These data indicate that FoxO transcription factors mediate most of the transcriptional changes in muscle due to diabetes.
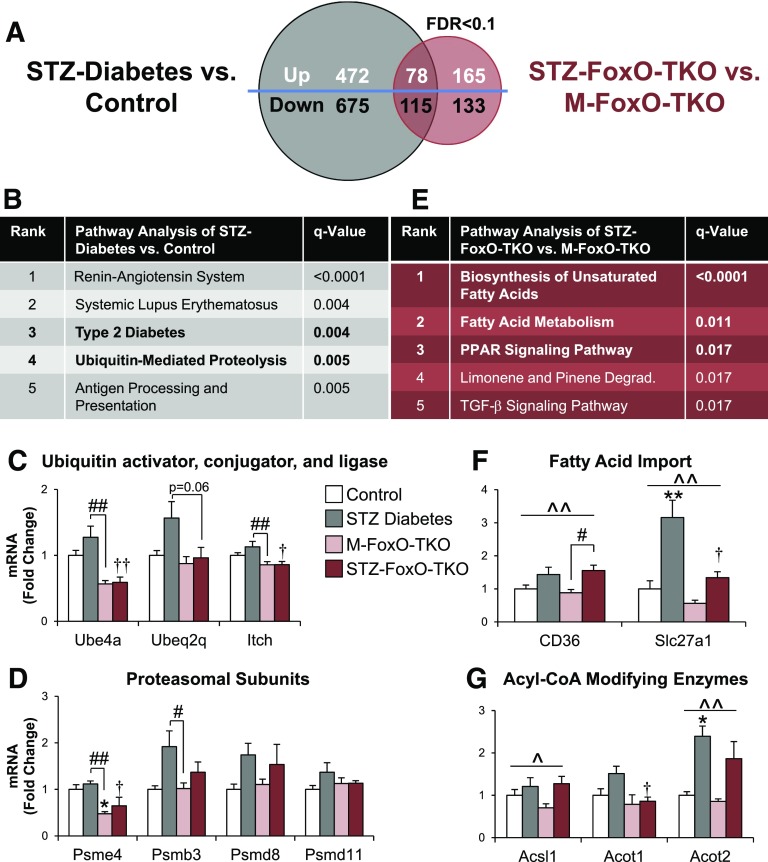
Figure 5.
Increases in ubiquitin-proteasome pathway transcripts in STZ diabetic muscle depend on FoxOs, but abnormalities in fatty acid metabolism transcripts are FoxO independent. Venn diagram of genes differentially regulated in STZ diabetes vs. control (gray circle) and STZ FoxO-TKO vs. M-FoxO-TKO (red circle) by RNA-Seq analysis of mixed Quad and gastrocnemius using a false discovery rate (FDR) of <0.1 (A). KEGG pathway analysis (B) of RNA-Seq data from STZ-Diabetes vs. control from panel A. Quantitative RT-PCR was performed to confirm mRNA levels of a ubiquitin activator (Ube4a), modifier (Ube2q2), and ligase (Itch) from the ubiquitin-mediated proteolysis pathway in Quad (C). Quantitative RT-PCR was performed for proteasomal subunit genes (D). KEGG pathway analysis (E) of RNA-Seq data from STZ FoxO-TKO vs. M-FoxO-TKO from panel A. Degrad., degradation pathway. Quantitative RT-PCR of fatty acid import genes (F), and acyl-CoA–modifying enzymes (G) in Quad from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO mice (n = 4–6 per group). *P < 0.05, **P < 0.01 vs. control; †P < 0.05, ††P < 0.01 STZ-Diabetes vs. STZ-FoxO-TKO; #P < 0.05, ##P < 0.01 as indicated; ^P < 0.05, ^^P < 0.01 STZ main effect (two-way ANOVA).
Among the top five most significantly regulated KEGG pathways between STZ-Diabetes and controls were pathways related to type 2 diabetes and the “ubiquitin-mediated proteolysis” pathway (Fig. 5B). By contrast, in STZ FoxO-TKO compared with M-FoxO-TKO, the ubiquitin-mediated proteolysis pathway was no longer significant, indicating that regulation of these pathways in diabetes requires FoxOs. A heat map of all 125 genes within the ubiquitin-mediated proteolysis pathway demonstrated a large cluster of genes that increased in STZ-Diabetes compared with controls, and this regulation was lost when FoxOs were deleted (Supplementary Fig. 6). We confirmed mRNA levels of three genes within this cluster, including a ubiquitin-activating enzyme (Ube4a), an E2-ubiquitin conjugating enzyme (Ubeq2q), and an E3-ubiquitin ligase (Itch). The results show that Ube4a, Ube2q2, and Itch tended to increase with STZ diabetes but were significantly reduced in STZ FoxO-TKO mice (Fig. 5C). Proteasomal subunit mRNAs were mildly altered by STZ diabetes and FoxO deletion. Psme4 significantly decreased in M-FoxO-TKO regardless of STZ treatment, while STZ diabetes tended to increase Psmb3 and Psmd8, with no effect of FoxO deletion (Fig. 5D).
To determine the pathways that are diabetes dependent but FoxO independent, we compared STZ FoxO-TKO with M-FoxO-TKO and found that fatty acid metabolism pathways ranked at the top (Fig. 5E). Indeed, a heat map of the transcripts in the top three pathways from Fig. 5E reveals a subset of genes that are highly induced in both STZ-Diabetes compared with control and STZ FoxO-TKO compared with M-FoxO-TKO (Supplementary Fig. 7). Fatty acid transport genes CD36 and Slc27a1 increased in muscle in STZ-Diabetes, and this increase was not blocked by loss of FoxOs in STZ FoxO-TKO muscle, although baseline levels of Slc27a1 were decreased in M-FoxO-TKO (Fig. 5F). Likewise, STZ diabetes increased Acsl1 and Acot2 (acyl-CoA–modifying enzymes) expression independent of FoxO deletion (Fig. 5G). This transcriptomic analysis reveals that insulin-deficient diabetes induces proteolysis pathways in a FoxO-dependent manner, including the ubiquitin-proteasome system, but also increases fatty acid metabolism genes, which are largely independent of FoxO regulation.
Induction of Ubiquitin-Proteasome and Autophagy-Lysosomal mRNAs in Muscle Is Dependent on FoxOs
Figure 6A shows a heat map of 49 atrogenes that are commonly regulated in all forms of muscle atrophy (42). These cluster into three main groups. Cluster 1 contained transcripts that increased with STZ diabetes and were rescued by FoxO deletion, i.e., are FoxO dependent. These include many ubiquitin-proteasome genes, such as Fbxo32 (aka Atrogin1/MAFbx), Ubc, Uba52, and proteasomal subunits. Cluster 2 was composed of transcripts that were downregulated by STZ diabetes and not rescued by FoxO deletion. These included many metabolic genes, such as oxidative phosphorylation subunits, Mdh1, Dlat (a component of pyruvate dehydrogenase complex), and Ckmt2 (the mitochondrial creatine kinase). The third cluster contained genes that showed little regulation in response to either STZ diabetes or FoxO deletion.
Figure 6.
Induction of ubiquitin-proteasome and autophagy-lysosomal mRNAs in muscle is dependent on FoxOs. A heat map of transcripts of 49 atrophy-related genes from RNA-Seq analysis of mixed Quad and gastrocnemius (A). Quantitative RT-PCR of FoxO target genes (B), E3-ubiquitin ligases (C), and autophagy genes (D) from control, STZ-Diabetes, M-FoxO-TKO, and STZ FoxO-TKO Quad muscle (n = 4–6 per group). *P < 0.05, **P < 0.01 vs. control; †P < 0.05, ††P < 0.01 STZ-Diabetes vs. STZ FoxO-TKO; ##P < 0.01 as indicated (two-way ANOVA).
We confirmed mRNA levels of several genes in cluster 1 along with other FoxO target and autophagy genes in Quad muscle from a separate cohort of control and M-FoxO-TKO mice rendered diabetic with STZ. In agreement with the RNA-Seq data, mRNA levels of FoxO target genes Gadd45a, p27 Kip1, and Eif4ebp1 were significantly increased by STZ diabetes but not in STZ-FoxO-TKO muscle (Fig. 6B). Gadd45a is of particular interest since it can directly mediate skeletal muscle atrophy (43). 4EBP1 (the enzyme product of the Eif4ebp1 gene) is a negative regulator of protein synthesis that is inactivated upon phosphorylation by mTORC1, and we observed an increase in 4EBP1 protein levels with a decrease in the p-4EBP–to–4EBP1 ratio in Quad from STZ diabetes, which was reversed in STZ FoxO-TKO mice (Supplementary Fig. 4A and B). Diabetes is known to decrease protein synthesis rates in muscle (44). To determine whether this regulation of 4EBP1 occurred simultaneously with alterations of protein synthesis rates in our mouse models, we determined protein synthesis by puromycin incorporation into muscle protein (27). Surprisingly, these data reveal that protein synthesis in Quad is decreased equally in STZ-Diabetes and STZ FoxO-TKO groups compared with nondiabetic controls (Supplementary Fig. 4C and D). These data indicate that while FoxOs can control mRNA levels of 4EBP proteins, loss of FoxOs is not sufficient to prevent the decrease in muscle protein synthesis due to diabetes.
Muscle-specific E3-ubiquitin ligases Fbxo32 and Trim63 (also known as MuRF1) were upregulated in STZ-Diabetes but were decreased in M-FoxO-TKO and remained suppressed in STZ FoxO-TKO (Fig. 6C). However, other E3-ubiquitin ligases implicated in muscle atrophy, Fbxo21 and Fbxo30 (aka SMART and MUSA1) (22,45), tended to increase in M-FoxO-TKO and were not modified by STZ treatment (Fig. 6C). Some autophagy genes, such as LC3A and LC3B, did not change significantly at the mRNA level with STZ diabetes. However, total mRNA levels of Bnip3, Bnip3L, Gabarapl, and Ulk1 were induced in STZ-Diabetes compared with controls, and this was blocked by FoxO deletion (Fig. 6D). These changes in ubiquitin-proteasome and autophagy-lysosome genes did show some specificity based on muscle type. EDL, a glycolytic muscle, showed diabetes-induced increases in both autophagy genes and E3-ubiquitin ligases that were FoxO dependent (Supplementary Fig. 4E), similar to Quad. However, oxidative soleus muscle showed very little change in autophagy genes other than an upregulation of Lamp2 and decreases in Ulk1 expression that were independent of FoxOs (Supplementary Fig. 4F). E3 ligases increased in soleus with STZ diabetes in a FoxO-regulated fashion. Together these results show that FoxO transcription factors control the induction of a broad range of atrophy-related protein degradation genes in response to uncontrolled diabetes.
Transcripts of FoxO-Dependent Ubiquitin-Proteasomal and Autophagy-Lysosomal Genes Increase in Muscle From Patients With Type 1 Diabetes After Insulin Withdrawal
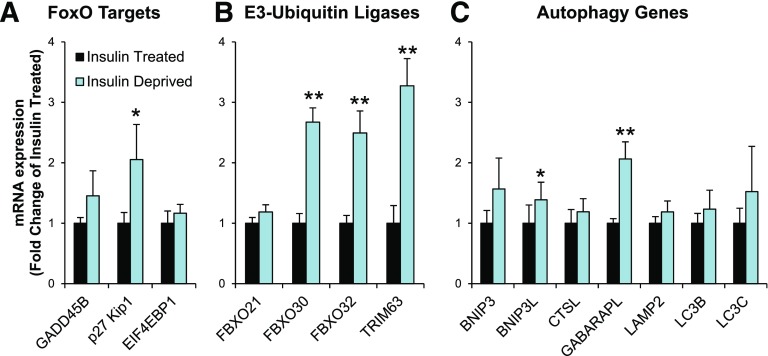
To determine whether these FoxO-dependent atrophy and autophagy genes that increase in STZ-Diabetes mice were also increased in humans with type 1 diabetes, we obtained cDNA from muscle biopsies of patients with type 1 diabetes both before and after 8 h of insulin deprivation (34). Following insulin deprivation, p27 Kip1 and three E3-ubiquitin ligases, FBXO30, FBXO32, and TRIM63, significantly increased by two- to threefold compared with samples obtained from the same patients while insulin treated (Fig. 7A and B). In addition, two of seven autophagy-related mRNAs, BNIP3L and GABARAPL, were increased significantly in the insulin-deprived group (Fig. 7C), and mRNA levels of other atrophy and autophagy genes also tended to increase after insulin deprivation. Thus, FoxO-driven muscle atrophy–related pathways and protein degradation pathways are transcriptionally upregulated in mice with STZ diabetes and in humans with type 1 diabetes in as little as 8 h after insulin withdrawal.
Figure 7.
Transcripts of FoxO-dependent ubiquitin-proteasomal and autophagy-lysosomal genes increase in muscle from patients with type 1 diabetes after insulin withdrawal. Quantitative RT-PCR of FoxO target genes (A), E3-ubiquitin ligases (B), and autophagy genes (C) from vastus lateralus muscle biopsies from patients with type 1 diabetes either treated normally with insulin or deprived of insulin for 8 h as previously reported (34). (n = 9 per group.) *P < 0.05, **P < 0.01 vs. insulin treated (paired t test).
Discussion
While much attention has focused on the role of diabetes in risk of cardiovascular disease, blindness, and renal failure, diabetes affects virtually all tissues of the body, often in more subtle, but very important, ways. For example, patients with type 1 and type 2 diabetes have decreased muscle size and strength, and this contributes to poor physical fitness and increased risks of disability (3–5). Perhaps more striking is the strong association of decreased muscle strength with mortality. In the Health, Aging and Body Composition Study (Health ABC) Study, individuals age 70–80 years showed a near 20–30% mortality rate over 6 years if they were in the bottom quartile of muscle strength, and patients with uncontrolled diabetes not only had decreased initial strength but also actually lost strength faster than individuals without diabetes (1,2,46). Furthermore, patients with diabetes have decreased strength after major surgery, such as coronary artery bypass grafting (47). This would lead to delayed recovery, increased risk of postsurgical complications, increased falls, and disability, thus supporting the critical need to identify the factors that contribute to muscle loss in diabetes. Identifying the intracellular targets that are perturbed in diabetes and that lead to decreases in muscle fitness is of critical importance, as it is not clear that these effects on protein synthesis and degradation are simply linked to uncontrolled hyperglycemia.
In the current study, we have investigated the role of FoxO transcription factors in diabetic muscle disease by creating muscle-specific FoxO triple knockout mice and rendering them diabetic with STZ. Previous studies showed that overexpression of FoxO1 was sufficient to induce muscle atrophy (21,48), and we now demonstrate that deletion of FoxOs specifically in muscle prevented diabetes-induced muscle atrophy without affecting glucose homeostasis. STZ diabetes induced a broad upregulation of both ubiquitin-proteasome and autophagy-lysosome pathways, and this was blocked by FoxO deletion. A similar upregulation of these protein degradation pathways occurs in muscle biopsies from people with type 1 diabetes after only 8 h of insulin withdrawal, indicating the exquisite control of insulin over this process in humans. Thus, insulin deficiency induces muscle protein degradation via a FoxO-dependent pathway, providing new insight and potential for therapeutic investigations to prevent diabetes-induced muscle disease.
FoxOs target several genes that regulate muscle atrophy, and our study demonstrates that many of these genes are also upregulated in uncontrolled diabetes. The E3-ubiqutin ligases Fbxo32 and Trim63 are FoxO target genes known to modulate muscle atrophy in response to starvation, immobilization, and denervation (18–21,49). Interestingly, of the proteolysis genes tested in humans with type 1 diabetes, FBXO32 and TRIM63 showed the largest induction after insulin withdrawal. Gadd45a, another known FoxO target, can promote muscle atrophy in response to various forms of stress by activating MEKK4 (43,50). Diabetes also leads to a decrease in protein synthetic rates in muscle from rodents (44). The induction of Eif4ebp1, a negative regulator of protein synthesis, in our STZ diabetic mice was prevented in STZ FoxO-TKO, allowing FoxOs to regulate aspects of both synthesis and degradation of proteins. However, our direct measurements of puromycin incorporation into muscle protein revealed that loss of FoxOs does not prevent the decrease in protein synthetic rates in diabetes. This further strengthens our conclusions that FoxOs regulate protein degradation pathways to induce muscle atrophy in response to diabetes. Indeed, autophagy markers of LC3-II and p-Ulk1S555 accumulate in STZ diabetic muscle along with LC3-positive vesicles, indicating an alteration of autophagy but not necessarily increased autophagy flux. Importantly, inhibition of autophagy in muscle can also lead to atrophy (51). Likely a combination of enhanced autophagy with impaired lysosomal degradation occurs with diabetes, but these alterations are prevented in STZ FoxO-TKO, indicating the necessity of FoxOs for this regulation. We provide evidence that FoxOs control autophagy by transcriptional mechanisms, since a number of autophagy-lysosomal genes are upregulated in a FoxO-dependent manner in mice (Bnip3, Bnip3L, Gabarapl, and Ulk1). But these genes were induced to a lesser degree in humans after insulin withdrawal. Whether this indicates that the two proteolytic pathways (ubiquitin-proteasome vs. autophagy-lysosome) contribute differentially to diabetes-induced muscle atrophy in humans and mice or whether this is simply the difference in duration of insulin deficiency in mice (which is chronic) and humans (which was only 8 h) is unknown. Further studies on the time course of induction of ubiquitin-proteasome versus autophagy intermediates may clarify whether the response to insulin withdrawal involves acute upregulation of E3-ubiquitin ligase followed by a broad activation of autophagy pathways after chronic insulin deprivation.
The diabetes-induced upregulation in protolysis pathways did not lead to a broad increase in tissue AAs. Rather, selective AA pools were affected, such as increases in BCAAs and decreased histidine, that were independent of FoxOs. Furthermore, STZ diabetes increased aspartate that was normalized with FoxO deletion, which could signify that FoxOs control synthesis or degradation of aspartate through regulation of enzymes in the urea cycle. Citrulline levels were also increased in STZ FoxO-TKO muscle, which may indicate an increased capacity for urea cycle flux, but these static metabolite measurements cannot be used to quantify flux. Aspartate can also be converted to other AAs and even TCA cycle intermediates, and the enzymes that control these processes may be regulated by FoxOs. These data indicate that diabetes and FoxOs influence AA metabolism by mechanisms independent of proteostasis.
Transcriptomic analysis revealed that FoxOs are necessary for a majority of the transcriptional changes in response to STZ diabetes. Indeed, of the 1,340 significantly changed genes in muscle from STZ-Diabetes mice, only 193 genes (14%) remained significantly changed in STZ FoxO-TKO. However, FoxOs do not mediate all aspects of muscle gene regulation in response to insulin-deficient diabetes. For example, proteolysis pathways were highly upregulated in muscle from STZ-Diabetes mice, and these were prevented by deletion of FoxOs, whereas upregulation of fatty acid import and biosynthesis genes that occurs in STZ diabetes was not mediated by FoxOs. Interestingly, of the 49 atrophy-related genes presented in the heat map, 19 were downregulated with STZ diabetes, and 10 of the 19 genes are involved in metabolic pathways that were not rescued by FoxO deletion. Thus, while FoxOs do regulate some metabolism-related genes in muscle, such as the upregulation of PDK4 in the fasted state (52), FoxOs do not regulate genes related to fatty acid import and biosynthesis. FoxOs do have an important role in insulin-mediated metabolic regulation in the liver, controlling gluconeogenesis by gene regulation of G6Pase and PEPCK (17), and recent studies suggest that FoxOs may control both lipid and glucose metabolism in the liver (16,53). Lastly, FoxOs have been implicated in mitochondrial dynamics related to muscle atrophy (54). How this might interact with the changes in protein degradation and autophagy remains to be determined.
Uncontrolled type 1 diabetes is caused by inadequate insulin levels. However, other hormonal and metabolite abnormalities also occur with insulin deficiency in uncontrolled diabetes including hyperglycemia, elevated cortisol levels, and increased inflammation. The degree to which each of these contributes to diabetes-induced muscle atrophy and whether they all impinge on FoxO transcription factors are unknown. Previous reports have suggested that insulin signaling predominates over IGF-1 to control muscle size (23,55). Other studies suggest that glucocorticoid signaling contributes significantly to muscle atrophy in the context of diabetes (56), and glucocorticoid receptors act in a synergistic way with FoxO1 to induce the ubiquitin ligase Trim63 (MuRF1) (57). The increase in inflammation and cytokine signaling during diabetes may also activate nuclear factor-κB signaling to promote muscle protein degradation (58). While these changes in circulating hormones, cytokines, and metabolites could contribute to diabetes-induced atrophy, the fact that deletion of insulin and IGF-1 receptors induces marked muscle atrophy that is entirely reversed by triple FoxO deletion (6,23) strongly supports the current data and points to a model where decreased insulin action in diabetes activates FoxO transcription, leading to muscle atrophy. Indeed, regardless of the upstream cause, our study demonstrates that in the context of uncontrolled diabetes, FoxOs are necessary for the atrophic response.
In summary, insulin deficiency and uncontrolled diabetes lead to muscle atrophy and decreased muscle strength, which can contribute to decreased health and disability. Our study demonstrates that, in muscle, FoxO transcription factors mediate the majority of transcriptional changes in response to STZ diabetes. FoxOs control the increases in proteolytic pathways that occur upon insulin deficiency, and these changes occur within 8 h after insulin deprivation in humans with type 1 diabetes. Thus, FoxOs are critical regulators of diabetes-induced muscle atrophy and represent potential therapeutic targets to prevent muscle loss in patients with diabetes.
Supplementary Material
Article Information
Funding. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health (NIH), grants R01-DK-031036 and R01-DK-033201 (to C.R.K.) and R01-DK-41973 and U24-DK-100469 (to K.S.N.). B.T.O. was funded by a K08 training award from the NIDDK (K08-DK-100543); an R03 award from the NIDDK (R03-DK-112003); a Mayo Clinic Metabolomics Resource Core grant, U24-DK-100469, from the NIDDK, which originates from the NIH Director’s Common Fund; and Mayo Clinic Clinical and Translational Science Awards grant UL1-TR-000135 from National Center for Advancing Translational Sciences of the NIH. The Joslin Diabetes and Endocrinology Research Center core facility was used for part of this work (P30-DK-36836).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. B.T.O. designed the study, researched data, and wrote the manuscript. G.B., C.M.P., P.A.S.B., and K.P. researched data, helped design experiments, and helped write the manuscript. M.T.K., K.K., and M.L. researched data and helped design experiments. H.P. and J.M.D. performed bioinformatic analyses. K.S.N. provided reagents, helped design experiments, and helped write the manuscript. C.R.K. designed the study and helped write the manuscript. C.R.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0416/-/DC1.
References
- 1.Park SW, Goodpaster BH, Strotmeyer ES, et al. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes 2006;55:1813–1818 [DOI] [PubMed] [Google Scholar]
- 2.Park SW, Goodpaster BH, Strotmeyer ES, et al.; Health, Aging, and Body Composition Study . Accelerated loss of skeletal muscle strength in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes Care 2007;30:1507–1512 [DOI] [PubMed] [Google Scholar]
- 3.Sinha A, Formica C, Tsalamandris C, et al. Effects of insulin on body composition in patients with insulin-dependent and non-insulin-dependent diabetes. Diabet Med 1996;13:40–46 [DOI] [PubMed] [Google Scholar]
- 4.Rosenfalck AM, Almdal T, Hilsted J, Madsbad S. Body composition in adults with Type 1 diabetes at onset and during the first year of insulin therapy. Diabet Med 2002;19:417–423 [DOI] [PubMed] [Google Scholar]
- 5.Nguyen T, Obeid J, Walker RG, et al. Fitness and physical activity in youth with type 1 diabetes mellitus in good or poor glycemic control. Pediatr Diabetes 2015;16:48–57 [DOI] [PubMed] [Google Scholar]
- 6.James HA, O’Neill BT, Nair KS. Insulin regulation of proteostasis and clinical implications. Cell Metab 2017;26:310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meek SE, Persson M, Ford GC, Nair KS. Differential regulation of amino acid exchange and protein dynamics across splanchnic and skeletal muscle beds by insulin in healthy human subjects. Diabetes 1998;47:1824–1835 [DOI] [PubMed] [Google Scholar]
- 8.Nair KS, Garrow JS, Ford C, Mahler RF, Halliday D. Effect of poor diabetic control and obesity on whole body protein metabolism in man. Diabetologia 1983;25:400–403 [DOI] [PubMed] [Google Scholar]
- 9.Barazzoni R, Short KR, Asmann Y, Coenen-Schimke JM, Robinson MM, Nair KS. Insulin fails to enhance mTOR phosphorylation, mitochondrial protein synthesis, and ATP production in human skeletal muscle without amino acid replacement. Am J Physiol Endocrinol Metab 2012;303:E1117–E1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulks RM, Li JB, Goldberg AL. Effects of insulin, glucose, and amino acids on protein turnover in rat diaphragm. J Biol Chem 1975;250:290–298 [PubMed] [Google Scholar]
- 11.Price SR, Bailey JL, Wang X, et al. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest 1996;98:1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, Kahn CR. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Rep 2015;11:1220–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yechoor VK, Patti ME, Ueki K, et al. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A 2004;101:16525–16530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai W, Sakaguchi M, Kleinridders A, et al. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat Commun 2017;8:14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 2013;14:83–97 [DOI] [PubMed] [Google Scholar]
- 16.O-Sullivan I, Zhang W, Wasserman DH, et al. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat Commun 2015;6:7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 2007;6:208–216 [DOI] [PubMed] [Google Scholar]
- 18.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472–483 [DOI] [PubMed] [Google Scholar]
- 19.Stitt TN, Drujan D, Clarke BA, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 2004;14:395–403 [DOI] [PubMed] [Google Scholar]
- 20.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007;6:458–471 [DOI] [PubMed] [Google Scholar]
- 21.Sandri M, Sandri C, Gilbert A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004;117:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milan G, Romanello V, Pescatore F, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun 2015;6:6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Neill BT, Lee KY, Klaus K, et al. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J Clin Invest 2016;126:3433–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deeds MC, Anderson JM, Armstrong AS, et al. Single dose streptozotocin-induced diabetes: considerations for study design in islet transplantation models. Lab Anim 2011;45:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju JS, Varadhachary AS, Miller SE, Weihl CC. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy 2010;6:929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waalkes TP, Udenfriend S. A fluorometric method for the estimation of tyrosine in plasma and tissues. J Lab Clin Med 1957;50:733–736 [PubMed] [Google Scholar]
- 27.Goodman CA, Hornberger TA. Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc Sport Sci Rev 2013;41:107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 2005;398:364–378 [DOI] [PubMed] [Google Scholar]
- 29.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–930 [DOI] [PubMed] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A 2005;102:13544–13549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham H. A layered grammar of graphics. J Comput Graph Stat 2010;19:3–28 [Google Scholar]
- 34.Karakelides H, Asmann YW, Bigelow ML, et al. Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects. Diabetes 2007;56:2683–2689 [DOI] [PubMed] [Google Scholar]
- 35.Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS One 2010;5:e10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci 2014;71:1657–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X, Wallis AM, Gerard RD, et al. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci 2012;125:5329–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu AL, Kim JH, Zhang C, Unterman TG, Chen J. Forkhead box protein O1 negatively regulates skeletal myocyte differentiation through degradation of mammalian target of rapamycin pathway components. Endocrinology 2008;149:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bois PR, Grosveld GC. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J 2003;22:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol 2007;292:C188–C199 [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacheck JM, Hyatt JP, Raffaello A, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 2007;21:140–155 [DOI] [PubMed] [Google Scholar]
- 43.Ebert SM, Dyle MC, Kunkel SD, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem 2012;287:27290–27301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wool IG, Cavicchi P. Protein synthesis by skeletal muscle ribosomes. Effect of diabetes and insulin. Biochemistry 1967;6:1231–1242 [Google Scholar]
- 45.Sartori R, Schirwis E, Blaauw B, et al. BMP signaling controls muscle mass. Nat Genet 2013;45:1309–1318 [DOI] [PubMed] [Google Scholar]
- 46.Newman AB, Kupelian V, Visser M, et al. Strength, but not muscle mass, is associated with mortality in the health, aging and body composition study cohort. J Gerontol A Biol Sci Med Sci 2006;61:72–77 [DOI] [PubMed] [Google Scholar]
- 47.Boban M, Barisic M, Persic V, et al. Muscle strength differ between patients with diabetes and controls following heart surgery. J Diabetes Complications 2016;30:1287–1292 [DOI] [PubMed] [Google Scholar]
- 48.Kamei Y, Miura S, Suzuki M, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 2004;279:41114–41123 [DOI] [PubMed] [Google Scholar]
- 49.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 2014;307:E469–E484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bullard SA, Seo S, Schilling B, et al. Gadd45a protein promotes skeletal muscle atrophy by forming a complex with the protein kinase MEKK4. J Biol Chem 2016;291:17496–17509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass. Cell Metab 2009;10:507–515 [DOI] [PubMed] [Google Scholar]
- 52.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J 2003;375:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haeusler RA, Hartil K, Vaitheesvaran B, et al. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun 2014;5:5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romanello V, Guadagnin E, Gomes L, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 2010;29:1774–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neill ED, Wilding JP, Kahn CR, et al. Absence of insulin signalling in skeletal muscle is associated with reduced muscle mass and function: evidence for decreased protein synthesis and not increased degradation. Age (Dordr) 2010;32:209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson ML, Baehr LM, Reichardt HM, Tuckermann JP, Bodine SC, Furlow JD. A cell-autonomous role for the glucocorticoid receptor in skeletal muscle atrophy induced by systemic glucocorticoid exposure. Am J Physiol Endocrinol Metab 2012;302:E1210–E1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waddell DS, Baehr LM, van den Brandt J, et al. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab 2008;295:E785–E797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai D, Frantz JD, Tawa NE Jr., et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 2004;119:285–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.