Watch a video presentation of this article
Watch the interview with the author
Abbreviations
- ALT
alanine aminotransferase
- anti‐HBc
hepatitis core antibody
- anti‐HBs
hepatitis surface antibody
- BCP
basal core promoter
- ccc
covalently closed circular
- CHB
chronic hepatitis B
- CTLA4
cytotoxic T lymphocyte antigen 4
- dlDNA
double‐stranded linear/duplex‐linear DNA
- HBcAg
hepatitis B core antigen
- HBcrAg
hepatitis B core‐related antigen
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- IL
interleukin
- mAb
monoclonal antibody
- NA
nucleos(t)ide analogue
- OBI
occult HBV infection
- pgRNA
pregenomic RNA
- Pol
polymerase
- spDNA/RNA
spliced DNA/RNA
- SVP
subviral particle
- TLR7
toll‐like receptor 7
Hepatitis B virus (HBV) plays a significant role in global morbidity and mortality with around 257 million chronically infected people worldwide.1 In 2016, the World Health Organization released a global strategy for eliminating viral hepatitis as a major public health threat by 2030. Chronic hepatitis B (CHB) has become a key focus because it contributes to 50% of all viral hepatitis‐related deaths. Even though global testing, vaccination, and treatment are the keys to achieving elimination, this must be preceded by reliable and accurate diagnosis of infection through serological and virological testing.
A sterilizing cure, which involves eradication of detectable HBsAg and all HBV DNA including covalently closed circular (ccc) and integrated DNA, is the ultimate goal but remains elusive. A functional cure, defined by Lok and colleagues2 as sustained loss of hepatitis B surface antigen (HBsAg) with or without hepatitis B surface antibody seroconversion, is more feasible. Because there are currently multiple direct‐acting antivirals and immune modulators in the pipeline designed to promote this (see Table 1), biomarkers for detecting and monitoring intrahepatic activity and viral persistence will be crucial.
Table 1.
Potential Hepatitis B Virus Therapies in the Pipeline Including Direct‐Acting Antivirals and Immune Modulators.
| Site of Action | Drug | Company | Status |
|---|---|---|---|
| Antiviral | |||
| Entry inhibitor | Myrcludex B | Hepatera/MYR GmbH | Phase 2 |
| cccDNA inhibitor | Preclinical | ||
| RNA silencers | AROHBV | Arrowhead | Phase 1/2a |
| ARB‐1467 | Arbutus | Phase 2 | |
| ARB‐1740 | Arbutus | Preclinical | |
| ALN‐HBV | Alnylam | Phase 1/2 | |
| GSK 3228836 | GlaxoSmithKline/Ionis | Phase 1 | |
| Core protein/capsid inhibitors | AL‐3778 | Alios/Johnson & Johnson | Phase 1/2 |
| ABI‐H0731 | Assembly Bioscience | Phase 1 | |
| BAY 41‐4109 | AiCuris (Germany) | Phase 1 | |
| GLS4 | HEC | Phase 1 | |
| JNJ‐56136379 | Johnson & Johnson | Phase 1 | |
| AB‐423 | Arbutus | Preclinical | |
| HBsAg release inhibitors | REP 2139 | Replicor | Phase 2 |
| REP 2165 | Replicor | Phase 2 | |
| Immune modulators | |||
| TLR7 agonist | GS‐9620 | Gilead Sciences | Phase 2 |
| Therapeutic vaccines | GS‐4774 | Gilead Sciences | Complete phase 2 |
| ABX 203 | ABIVAX (France) | Phase 2/3 | |
| AIC649 | AiCuris (Germany) | Phase 1 | |
| FB‐02.2 | Altimmune | Phase 1 | |
| INO‐1800 | Inovio | Phase 1 | |
| TG1050 | Transgene (France) | Phase 1 | |
| Small molecules* | |||
| RIG‐1 NOD2 activator* | SB 9200 | SpringBank Pharmaceuticals | Phase 2 |
| SMAC inhibitor* | Birinapant | TetraLogic | Terminated |
| Checkpoint inhibitors* | PD‐1/PD‐2 mAb | Merck Sharp and Dohme; Bristol‐Myers Squibb | |
| CTLA4 mAb |
Abbreviations: CTLA4, cytotoxic T lymphocyte antigen 4; mAb, monoclonal antibody; NOD2, nucleotide‐binding oligomerization domain‐containing protein 2; PD‐1, Programmed cell death protein 1; RIG‐I, retinoic acid‐inducible gene I; SMAC, second mitochondria‐derived activator of caspases; TLR7, toll‐like receptor 7.
Adapted with permission from Gastroenterology & Hepatology.13 Copyright 2017, Millennium Medical Publishing.
HBV Biomarkers
Hepatitis B Surface Antigen
The presence of HBsAg is the prime biomarker for diagnosing acute or chronic infection (see Table 2 and Fig. 1). HBsAg is the surface protein that envelops infectious virions (Dane particles) and is produced in excess as noninfectious subviral particles (SVPs). HBsAg is produced from episomal cccDNA and integrated HBV DNA. Decline and ultimate loss of HBsAg is the current goal of therapy, and quantification of HBsAg during therapy is important in patient management. HBsAg levels may: (1) predict the phase of infection, with the lowest levels seen in HBeAg‐negative chronic infection; (2) predict HBsAg loss in HBeAg‐negative individuals; and (3) assist with response‐guided pegylated interferon therapy.3
Table 2.
Description of Virological and Serological Markers of Acute and Chronic HBV: Their Role and Use in Diagnosis.
| Serum Biomarker | Description/Source | Role | Predictor/Outcome |
|---|---|---|---|
| HBsAg (essential) | Surface/envelope protein derived from cccDNA and/or integrated DNA | Required for infectivity, excessive quantities may inhibit immune response |
Initial diagnosis of: acute HB—transient HBs antigenemia; CHB—persistent HBs antigenemia (>6 months) Loss/seroconversion results in functional cure, quantification is used to monitor therapeutic outcomes |
| HBV DNA (essential) | Found in infectious virions | Required for replication | Used to monitor therapy and assess risk to pregnant women to avoid perinatal transmission; HBV DNA >2000 IU/mL is a key predictor of HCC risk |
| HBeAg (essential) | Secreted nonstructural protein derived from precore | Tolerogenicity, immune regulation, persistence | Defines phase CHB; seroconversion is one of the main therapeutic endpoints; quantification is used to predict seroconversion and therapeutic response |
| HBcrAg (investigational) | Composed of viral HBcAg, HBeAg, and p22cr | Combines both structural and nonstructural proteins forming empty capsids and virions | Monitoring therapy and predictor of: treatment response; HBeAg seroconversion and HBV reactivation in occult infection during immunosuppression; and risk for HCC5 |
| HBV pgRNA (investigational) | Derived directly from the TA of intrahepatic cccDNA/minichromosome | Transcriptional template for Pol protein production and HBV replication | Assesses intrahepatic cccDNA activity, used for monitoring current and novel therapies particularly when HBV DNA is suppressed, predictor of HBeAg seroconversion6 |
| Anti‐HBc (optional) | Reliable marker of host response to HBV exposure | Immune response to HBcAg | Indication of HBV exposure, especially useful in diagnosis of occult infection |
| Anti‐HBs (optional) | Host response to hepatitis B vaccination and/or after decline/loss of HBsAg | Immune response to HBsAg | Presence confirms efficacy of vaccination or achievement of functional cure in CHB |
| dlDNA (future) | Genomic form of DNA | Involved in integration | May have the potential to reflect intrahepatic activity and in particular risk for HCC11 |
| spDNA/RNA (investigational/future) | Truncated variants arising from spDNA/RNA during replication | May be involved in liver disease, including HCC, and impaired response to interferon | PCR assays for detection of spDNA/RNA may reflect increased risk for HCC10 |
Figure 1.
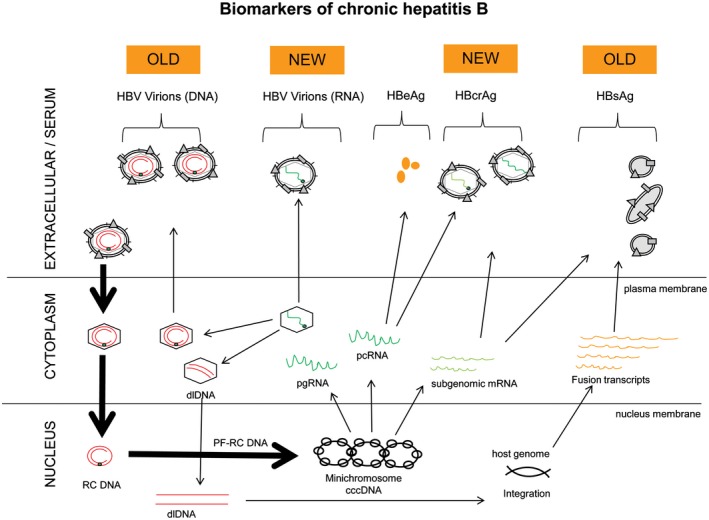
Biomarkers of chronic hepatitis B. Traditional (old) markers of CHB include HBsAg, HBV DNA, and HBeAg. Recent (new) experimental markers undergoing validation include HBV RNA and HBcrAg. Future markers include dlDNA. [Colour figure can be viewed at wileyonlinelibrary.com]
HBV DNA
HBV DNA in the peripheral component reflects the production of actively replicating virus within the hepatocytes. Quantification of serum HBV DNA is essential in staging the phase of CHB (Fig. 2) and also in monitoring effectiveness of nucleos(t)ide analogue (NA) therapy. Although suppressed HBV DNA to undetectable levels is ideal, it is currently achieved only through long‐term NA treatment, which does not target the intrahepatic cccDNA (minichromosome).
Figure 2.
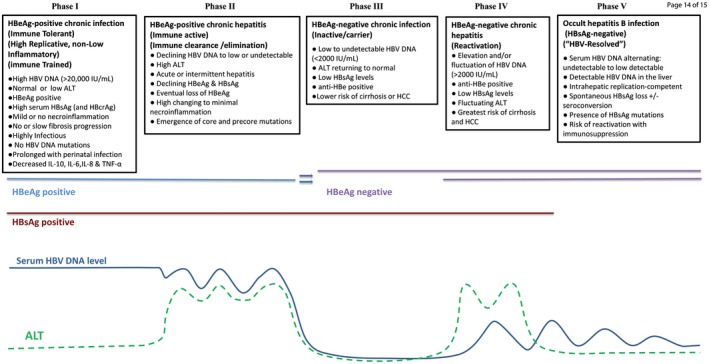
Five phases of chronic hepatitis B infection in a natural history setting. Abbreviation: IL, interleukin. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 3.
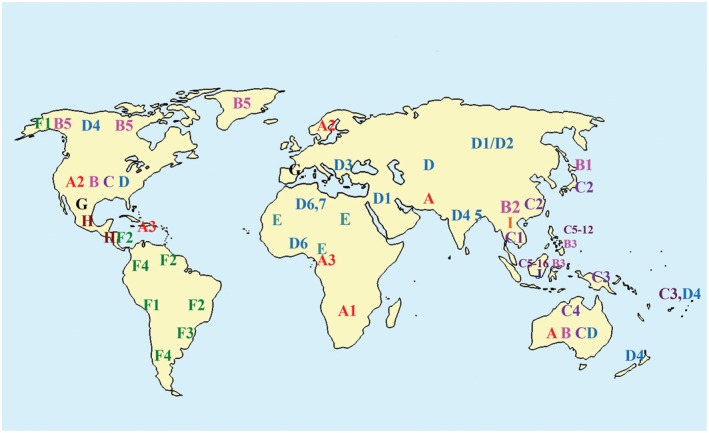
Geographic distribution of genotypes and subgenotypes. [Colour figure can be viewed at wileyonlinelibrary.com]
Hepatitis B e Antigen
Hepatitis B e antigen (HBeAg) is a nonstructural secreted protein that has tolerogenic properties and immune‐modulating activity, and plays an essential role in persistence. Its presence is used to distinguish the phase of CHB (Fig. 2). During the natural course of infection, seroconversion from HBeAg positivity to negativity often occurs. HBeAg may also be lost or reduced with the emergence of precore and/or basal core promoter (BCP) mutations. Regardless, HBeAg seroconversion is an established therapeutic endpoint associated with reduced morbidity and mortality provided HBV DNA is low or undetectable.4 Quantification of HBeAg is available clinically in many countries and is part of all HBV drug development roadmaps.
Hepatitis B Core Antigen and the Hepatitis B Core‐Related Antigen
The hepatitis B core‐related antigen (HBcrAg) assay is composed of antibodies that can detect three proteins that share a common 149‐amino acid sequence: hepatitis B core antigen (HBcAg), the structural nucleocapsid protein that cannot be easily measured directly; HBeAg; and a putative small core‐related protein, p22cr, found in DNA‐free virion‐like (empty) particles. HBcrAg is a potentially useful marker reflecting intrahepatic replication, particularly in NA‐suppressed and HBeAg‐seroconverted cases, but appears less robust for assessing cccDNA transcriptional activity (TA).5
HBV RNA
HBV pregenomic RNA (pgRNA) produced in virions is a key replicative intermediate that provides the template for protein translation of the polymerase (Pol) and genomic replication through reverse transcription. It is generally unaffected by NA therapy which targets the HBV DNA Pol. Hence, HBV RNA quantification is proving a useful investigational biomarker for monitoring therapy when HBV DNA is suppressed because its detection in serum6 may reflect intrahepatic TA.7
Hepatitis Core Antibody (Anti‐HBc)
Antibody to HBcAg, anti‐HBc, remains the most useful marker of past HBV exposure or infection. It becomes increasingly important in the diagnosis of occult HBV infection (OBI) where HBsAg is not detectable. Titers of anti‐HBc may be useful for assessing natural history, reactivation, and for predicting therapeutic response. It also indicates a lack of need for HBV vaccination.
Hepatitis Surface Antibody (Anti‐HBs)
Anti‐HBs develops as a response to prophylactic vaccination, or as a result of HBsAg seroconversion during the natural course of HBV infection or during therapy. Current diagnostic assays only detect free circulating anti‐HBs that are not complexed with the large quantities of HBsAg associated with virions or SVPs. Further studies are warranted to determine the true viral neutralizing capacity of these circulating anti‐HBs.
Genotype and HBV Mutants
There are currently 10 genotypes of HBV, A to J, that differ by 8% to 10% at the nucleotide level across the whole genome, and more than 40 subgenotypes (4% to 8% nucleotide divergence) within most genotypes (see Table 3). HBV genotype and subtype can influence disease progression and response to antiviral therapy,8, 9 as can the presence of mutations. BCP variants may increase the risk for hepatocellular carcinoma (HCC), and a deletion in preS may be a predictor of cirrhosis.9 Sequencing for drug resistance mutations may also be needed if viral loads persist and/or increase on therapy.
Table 3.
Characteristics of Hepatitis B Virus Genotypes.
| Genotypes | Subtypes | Origin/Geographic Distribution | Clinical Notes | Predominant Route of Transmission | Mutations/Recombinations | Response to Antiviral Therapy |
|---|---|---|---|---|---|---|
| A | A1 | Africa |
More likely to lose HBsAg than C and D A1 is associated with aggressive liver disease |
Parenteral or sexual | Higher frequency of BCP mutations than B and D | Responds better to interferon therapy than C and D |
| A2 | Europe/North America | |||||
| A3 | Africa/Haiti | |||||
| B | B1 | Japan |
Lower HBsAg than A and D More likely to lose HBsAg than C and D B1 is associated with fulminant hepatitis |
Perinatal | 93% of strains have recombination | Responds better to interferon therapy than C and D |
| B2 | China | |||||
| B3 | Indonesia, China, Philippines | |||||
| B4 | Vietnam, Cambodia, France | |||||
| B5 | Eskimos, Inuits | |||||
| C | C1 | Thailand, Myanmar, Vietnam |
High replication capacity can result in more severe disease Slower progression to HBeAg seroconversion Lower HBsAg levels than A and D |
Perinatal | Higher frequency of BCP mutations than B and D | |
| C2 | Japan, China, Korea | |||||
| C3 | New Caledonia, Polynesia | |||||
| C4 | Australian aborigines | |||||
| C5‐C12 | Philippines, Indonesia | |||||
| C13‐C16 | Indonesia | |||||
| D | D1 | Middle East, Central Asia |
Early HBeAg seroconversion D3 often associated with OBI |
Horizontal | High prevalence of precore mutations | |
| D2 | Europe, Lebanon | |||||
| D3 | Universal | |||||
| D4 | Pacific Islands, Papua New Guinea, Arctic Denes, India | |||||
| D5 | India | |||||
| D6 | Tunisia, Nigeria | |||||
| E | None | Africa—Western and Central | Horizontal | |||
| F | F1 | Argentina, Costa Rica, El Salvador, Alaska | Horizontal | |||
| F2 | Nicaragua, Venezuela, Brazil | |||||
| F3 | Venezuela, Colombia | |||||
| F4 | Argentina | |||||
| G | None | France, Germany, United States | Horizontal: sexual transmission in men who have sex with men | Two stop codons prevent HBeAg expression; requires another genotype, usually A, to establish CHB | ||
| H | None | Mexico, Japan, Nicaragua, United States | Often associated with OBI | |||
| I | I1 | Vietnam, Laos, China | Perinatal | Recombinant of A, C, G | ||
| I2 | Vietnam, Laos, India | |||||
| J (putative) | Japan/Borneo |
Biomarkers Associated With Risk for Progression
Natural History of CHB
CHB is a prolonged, dynamic infection influenced by host responses and risk factors, immune selection of emerging quasispecies and mutants, and viral genotype. The five recognized phases of infection, summarized in Fig. 2, are defined by HBsAg status, HBV DNA load, HBeAg status, and alanine aminotransferase (ALT) level.
Future Perspectives
Double‐Stranded Linear/Duplex‐Linear DNA
Double‐stranded linear/duplex‐linear DNA (dlDNA) is the genomic form of the DNA that is involved in integration. Assays for detecting dlDNA in serum may have the potential to reflect intrahepatic activity and in particular risk for HCC. Zhao et al.10 demonstrated increasing dlDNA levels in patients with cirrhosis, HCC, and while receiving interferon and/or NA therapy.
Spliced DNA/RNA
Splicing of DNA and RNA during replication may result in truncated variants which may be involved in liver disease including HCC and impaired response to interferon. PCR assays for detection of spliced DNA/RNA (spDNA/RNA) may reflect increased risk for HCC.11
Empty Particles
HBV excretes excessive quantities of genome‐free empty particles that consist of enveloped nucleocapsids, that is, HBsAg and HBcAg.12 These particles are not affected by NA therapy, and hence may prove a useful biomarker for cccDNA TA.
Conclusion
Several biomarkers are available to accurately diagnose this complex and evolving viral infection in clinical practice. However, as new therapies emerge, developmental and future biomarkers will be crucial to monitor clinical and virological outcomes.
Potential conflict of interest: Nothing to report.
References
- 1. Honer Zu Siederdissen C, Maasoumy B, Cornberg M. What is new on HBsAg and other diagnostic markers in HBV infection? Best Pract Res Clin Gastroenterol 2017;31:281–289. [DOI] [PubMed] [Google Scholar]
- 2. Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: from discovery to regulatory approval. Hepatology 2017;66:1296–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cornberg M, Wong VW, Locarnini S, Brunetto M, Janssen HLA, Chan HL. The role of quantitative hepatitis B surface antigen revisited. J Hepatol 2017;66:398–411. [DOI] [PubMed] [Google Scholar]
- 4. Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology 2010;51:1933–1944. [DOI] [PubMed] [Google Scholar]
- 5. Mak LY, Wong DK, Cheung KS, Seto WK, Lai CL, Yuen MF. Review article: hepatitis B core‐related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment Pharmacol Ther 2018;47:43–54. [DOI] [PubMed] [Google Scholar]
- 6. van Bommel F, Bartens A, Mysickova A, Hofmann J, Kruger DH, Berg T, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 2015;61:66–76. [DOI] [PubMed] [Google Scholar]
- 7. Wang J, Yu Y, Li G, Shen C, Meng Z, Zheng J, et al. Relationship between serum HBV‐RNA levels and intrahepatic viral as well as histologic activity markers in entecavir‐treated patients. J Hepatol; doi: 10.1016/j.jhep.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 8. Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014;57:141–150. [DOI] [PubMed] [Google Scholar]
- 9. Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol 2014;20:5427–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao XL, Yang JR, Lin SZ, Ma H, Guo F, Yang RF, et al. Serum viral duplex‐linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 2016;65:502–511. [DOI] [PubMed] [Google Scholar]
- 11. Bayliss J, Lim L, Thompson AJ, Desmond P, Angus P, Locarnini S, et al. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol 2013;59:1022–1028. [DOI] [PubMed] [Google Scholar]
- 12. Hu J, Liu K. Complete and incomplete hepatitis B virus particles: formation, function, and application. Viruses 2017;9:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peters MG, Locarnini S. New direct‐acting antiviral agents and immunomodulators for hepatitis B virus infection. Gastroenterol Hepatol (N Y) 2017;13:348–356. [PMC free article] [PubMed] [Google Scholar]


