Abstract
Gram-negative bacterial pathogens inject type III secreted effectors (T3SEs) directly into host cells to promote pathogen fitness by manipulating host cellular processes. Despite their crucial role in promoting virulence, relatively few T3SEs have well-characterized enzymatic activities or host targets. This is in part due to functional redundancy within pathogen T3SE repertoires as well as the promiscuity of individual T3SEs that can have multiple host targets. To overcome these challenges, we generated and characterized a collection of yeast strains stably expressing 75 T3SE constructs from the plant pathogen Pseudomonas syringae. This collection is devised to facilitate heterologous genetic screens in yeast, a non-host organism, to identify T3SEs that target conserved eukaryotic processes. Among 75 T3SEs tested, we identified 16 that inhibited yeast growth on rich media and eight that inhibited growth on stress-inducing media. We utilized Pathogenic Genetic Array (PGA) screens to identify potential host targets of P. syringae T3SEs. We focused on the acetyltransferase, HopZ1a, which interacts with plant tubulin and alters microtubule networks. To uncover putative HopZ1a host targets, we identified yeast genes with genetic interaction profiles most similar (i.e., congruent) to the PGA profile of HopZ1a and performed a functional enrichment analysis of these HopZ1a-congruent genes. We compared the congruence analyses above to previously described HopZ physical interaction datasets and identified kinesins as potential HopZ1a targets. Finally, we demonstrated that HopZ1a can target kinesins by acetylating the plant kinesins HINKEL and MKRP1, illustrating the utility of our T3SE-expressing yeast library to characterize T3SE functions.
Keywords: Pseudomonas syringae, Type III secreted effector, Pathogenic Genetic Array, Yeast screen, Pathogen-host interactions, HopZ1, Kinesin
Bacterial pathogens of both plants and animals subvert key host processes in order to suppress host immunity and manipulate nutrient supplies. Many Gram-negative bacterial pathogens achieve this goal by delivering type III secreted effectors (T3SEs) into the host cytosol where they manipulate the host in a variety of ways, including modulating signaling pathways, transcription, intracellular transport, cytoskeletal stability, and host defenses (Büttner and Bonas 2003; Jin et al. 2003; Cornelis 2006; Zhou and Chai 2008; Lewis et al. 2009). Although many bacterial T3SEs have been shown to generally suppress host immunity, we know relatively little about the specific virulence targets and mechanisms of action of most T3SEs. The difficulty in functional characterization of T3SE virulence mechanisms is due to a number of factors, including: (1) redundant targeting of a given host protein by multiple effectors which confounds analysis of individual T3SE deletion mutants; (2) promiscuous individual effectors which can target multiple host proteins, thereby making it difficult to ascribe a virulence function to any individual target (Lewis et al. 2011; Deslandes and Rivas 2012); (3) effectors often show no similarity to proteins or domains with characterized functions, limiting bioinformatic approaches to infer effector functions; and (4) effectors can trigger immune responses as a result of host recognition, which complicates virulence target identification.
In order to gain new insights into the biochemical functions and host targets of bacterial T3SEs, a number of research groups have utilized the model organism Saccharomyces cerevisiae (yeast) as a tool (Yoon et al. 2003; Jamir et al. 2004; Alto et al. 2006; Kramer et al. 2007; Slagowski et al. 2008; Alemán et al. 2009; Munkvold et al. 2009; Salomon and Sessa 2010). The rationale for using yeast to characterize bacterial effectors rests on the fact that many biological processes (for example central metabolism, the control of cytoskeleton dynamics, vesicle trafficking, signal transduction, DNA metabolism and cell cycle processes) are conserved among eukaryotes (Dolinski and Botstein 2007; Siggers and Lesser 2008; Curak et al. 2009; Botstein and Fink 2011). Therefore, effectors that target a conserved cellular process in a higher eukaryote may also act on the same cellular process in the simpler and genetically-tractable yeast system. This is particularly attractive if the original host is not readily amenable to high-throughput assays. Another advantage of studying bacterial T3SEs in the yeast system is that the expression of non-effector bacterial proteins does not generally affect yeast growth (Slagowski et al. 2008). This indicates that most fitness defects observed upon T3SE expression in yeast is specifically due to T3SE activity, and not simply due to the heterologous overexpression of bacterial proteins. Finally, the expression of translocated effector proteins from both plant and animal pathogens has been shown to inhibit yeast growth by targeting conserved eukaryotic proteins (Munkvold et al. 2008; Siggers and Lesser 2008; Curak et al. 2009; Salomon et al. 2011). For instance, the Yersinia T3SE YopJ has been shown to disrupt mammalian innate immunity by preventing the activation of MAPK kinase (MAPKK) and subsequently blocking the MAPK and NFκB signaling pathways (Orth et al. 1999; Orth et al. 2000). Even though yeast cells lack key components of the mammalian innate immune system, YopJ was shown to inhibit MAPK pathways in yeast by preventing the activation of MAPKK as previously observed in mammalian systems (Yoon et al. 2003).
A number of groups have developed yeast genomics tools to characterize bacterial effectors that target conserved eukaryotic cellular processes (Alto et al. 2006; Kramer et al. 2007). A very successful genetic approach is the Pathogenic Genetic Array (PGA), a variation of the well-established Synthetic Genetic Array (SGA) technology, which enables high-throughput genetic screens to identify conserved host targets (Alto et al. 2006; Kramer et al. 2007). The SGA technology involves a series of robotics-assisted cell matings to introduce any marked allele of interest into an array of mutants, allowing the systematic generation of double mutants and the interrogation of di-genic genetic interactions at a genome-wide scale (Tong et al. 2001; Tong et al. 2004; Costanzo et al. 2010). Genetic interactions between two mutations are inferred when the observed double mutant phenotype deviates from the expected phenotype of the combined single mutants. In extreme cases, a synthetic lethal interaction occurs when the combination of two non-lethal mutations causes cell death. Large scale, genome-wide SGA screens have provided global genetic interaction profiles in the yeast genome (Costanzo et al. 2010; Costanzo et al. 2016). Since genes within the same pathway or bioprocess tend to show very similar genetic interaction profiles, querying the genetic interactions of an unknown gene against the nearly complete SGA compendium of the yeast genome can be a powerful way to predict functions of uncharacterized genes (Costanzo et al. 2010).
Similar to SGA, PGA queries a pathogen effector against a collection of viable yeast deletion strains in a high-throughput array format to analyze effector functions. PGA identifies those yeast deletion mutants that interact genetically with T3SE, assessed by fitness of combined mutants showing greater or lower fitness than expected, and subsequently guides the inference of functional relationships between these yeast genes and the pathogen T3SEs (Alto et al. 2006; Kramer et al. 2007). This PGA strategy was first used to identify yeast deletion mutants that suppress Shigella T3SE IpgB2-induced toxicity (Alto et al. 2006). Consistent with the ability of IpgB2 to interfere with Rho1p signaling in mammalian cells, the genetic suppressors of IpgB2 in yeast are downstream of Rho1p, part of the cell wall integrity MAPK-signaling pathway (Alto et al. 2006). Overall this PGA screen revealed that IpgB2 functions as a G protein mimic, capable of activating the Rho1p pathway (Alto et al. 2006).
In this study, we hypothesized that T3SEs that target evolutionarily conserved plant processes can regulate the same processes in yeast. Furthermore, if this conserved process is important for optimal yeast growth, then the overexpression of T3SEs should decrease yeast fitness. We generated a library of 75 P. syringae T3SE-expressing yeast strains and identified 24 effectors that reduced yeast fitness in either standard rich media or under high osmotic stress. We performed PGA screens on five T3SEs and established genetic interaction profiles for three: HopF2PtoT1, HopX1PmaES4326 and HopZ1aPsyA2. We used HopZ1a as our proof-of-principle T3SE, and compared the genetic interaction profile of HopZ1a with previously generated SGA datasets (Costanzo et al. 2010) to identify yeast genes with interaction profiles similar (or congruent) to that of HopZ1a in order to identify potential HopZ1a targets. Among the yeast genes with interaction profiles congruent to HopZ1a were kinesins, which have been previously shown to physically interact with HopZ1a (Mukhtar et al. 2011; Lewis et al. 2012). These findings implicate kinesins as putative targets of HopZ1a. In support of this, we have demonstrated that HopZ1a can acetylate Arabidopsis thaliana (hereafter Arabidopsis) kinesin proteins. This study emphasizes the power of high-throughput heterologous screens for exploration of T3SE function and for identification of conserved eukaryotic processes that are targeted by diverse pathogens.
Materials and Methods
Cloning
Promoter-less coding sequences lacking stop codons of P. syringae T3SEs were PCR-amplified to include the addition of attB1 and attB2 linkers and cloned into the Gateway donor vector, pDONR207, using the Gateway BP reactions. T3SEs from PtoDC3000, PsyB728a and Pph1448a were generous gifts from J. Chang (Chang et al. 2005). The additional T3SEs from PmaES4326, as well as T3SEs from the HopZ and HopF families were cloned for this study. The pDONR207-T3SE collection was sequenced-confirmed via Sanger sequencing. These T3SEs were subcloned into the Gateway-compatible yeast integration vector, pBA2262 (Youn et al. 2017), using the Gateway LR reactions. To confirm the pBA2262-T3SE constructs, purified plasmids were digested with BsrGI or NotI and the restriction digest patterns were analyzed.
Promoter-less coding sequences of A. thaliana kinesins HINKEL (At1g18370) and MKRP1 (At1g21730) lacking stop codons were likewise PCR-amplified and cloned into pDONR207 and were subcloned by Gateway LR reactions into the autonomously-replicating, single-copy, Gateway-compatible yeast expression vector, pBA350V (Lewis et al. 2013; Youn et al. 2017).
Yeast strain construction, growth medium, immunoblot analyses
To integrate the PGAL1-T3SE-FLAG::NATR constructs into the yeast genome at the ho locus, the SGA query strain (Y7092, MATα, can1Δ::STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) was transformed with NotI-digested BA2262-T3SE plasmid DNA using the standard transformation method (Gietz and Woods 2002).
For immunoblot analyses, yeast strains expressing the FLAG-tagged T3SEs under control of the GAL1 promoter were grown overnight at 30° shaking (200 RPM) in 1 ml of YP broth with 2% raffinose (YPR) in deep-well plates with sterile glass beads in each well. The overnight cultures were subsequently diluted into deep-well plates containing 1 ml of YP broth with 2% galactose (YPG) at OD600 of 0.1. The cultures were induced for T3SE expression for 7 to 8 hr, or until the cultures reach OD600 of 1. The 1 ml-cultures were pelleted at 13,000 x g for 1 min, washed, and frozen at -20°. Whole cell extracts were prepared from trichloroacetic acid (TCA)-fixed cells as described (Kurat et al. 2009). The protein pellets were resuspended in 1X sample buffer and neutralized by addition of 2M Tris solution. The lysates were separated by 12% SDS-PAGE and immunoblot was performed with mouse anti-FLAG primary antibodies (Sigma, F3165, USA) via chemiluminescence (Amersham, USA).
Pathogenic genetic array
The pathogenic genetic array (PGA) analysis was based on a variation of the SGA method used for synthetic dosage lethality screens (Tong et al. 2001; Sopko et al. 2006). In brief, Y7092 (the SGA query strain) with integrated hoΔ::GAL1-T3SE-FLAG::NATR was mated into the 1536-density MATa deletion mutant array marked with KANR, which represents each single mutant colony four times on the array. Y7092 carrying hoΔ::NATR (SN851) was used as a negative control strain. The MATa/α diploids were selected on YPD supplemented with clonNAT (100 μg/ml) and G418 (200 μg/ml) at 30° for two days. Diploid cells were pinned onto enriched sporulation media (20 g/L agar, 10 g/L potassium acetate, 1 g/L yeast extract, 0.5 g/L glucose, 0.1 g/L amino acids-supplement) and allowed to sporulate at 22° for at least one week. The spores were pinned onto synthetic dextrose (SD) media (Tong et al. 2004) – His/Arg/Lys + clonNAT/canavanine/thialysine and incubated at 30° for two days to select for MATa haploid meiotic progeny. The drugs canavanine and thialysine were used at 50 μg/ml. The MATa haploid meiotic progeny were subsequently pinned onto SD – His/Arg/Lys + clonNAT/ canavanine/ thialysine/ G418 plates twice to select for the final MATa meiotic progeny carrying both the kanR (yeast deletion strains) and NATR (GAL1-T3SE-FLAG constructs) markers. To induce for T3SE expression, the MATa haploid meiotic progeny from final selection were pinned onto the synthetic galactose (SG) media – His/Arg/Lys + clonNAT/canavanine/thialysine/G418, and in the case of the HopZ1a screen the plates also contain 0.5M NaCl, followed by incubation of plates at 30° for two day.
In order to generate double mutants successfully using the SGA procedure, each array plate of haploid deletion strains contained a border of wild type yeast carrying the necessary selectable markers to correct for edge effects, where colonies toward the edge of the plate have greater access to nutrients and are therefore larger in size compared to colonies near the center of the plate (Baryshnikova et al. 2010; Wagih et al. 2013). Lastly, to ensure that the expression of effectors did not inhibit yeast mating or sporulation, all of the strain construction steps utilized glucose-containing media to repress effector expression.
After obtaining images of final plates, we quantified colony sizes and assessed fitness manually. In detail, we assessed the fitness of double mutants relative to the single mutants by comparing the colony size of each mutant on the experiment array (T3SE-expression combined with a yeast gene deletion; Figure 2C bottom panel) and the control array (no T3SE, fitness of yeast deletion mutant only; Figure 2C top panel), all on T3SE-expressing (galactose) media. We were able to indirectly assess T3SE-associated fitness by gauging the overall fitness of all the strains in the experimental plate.
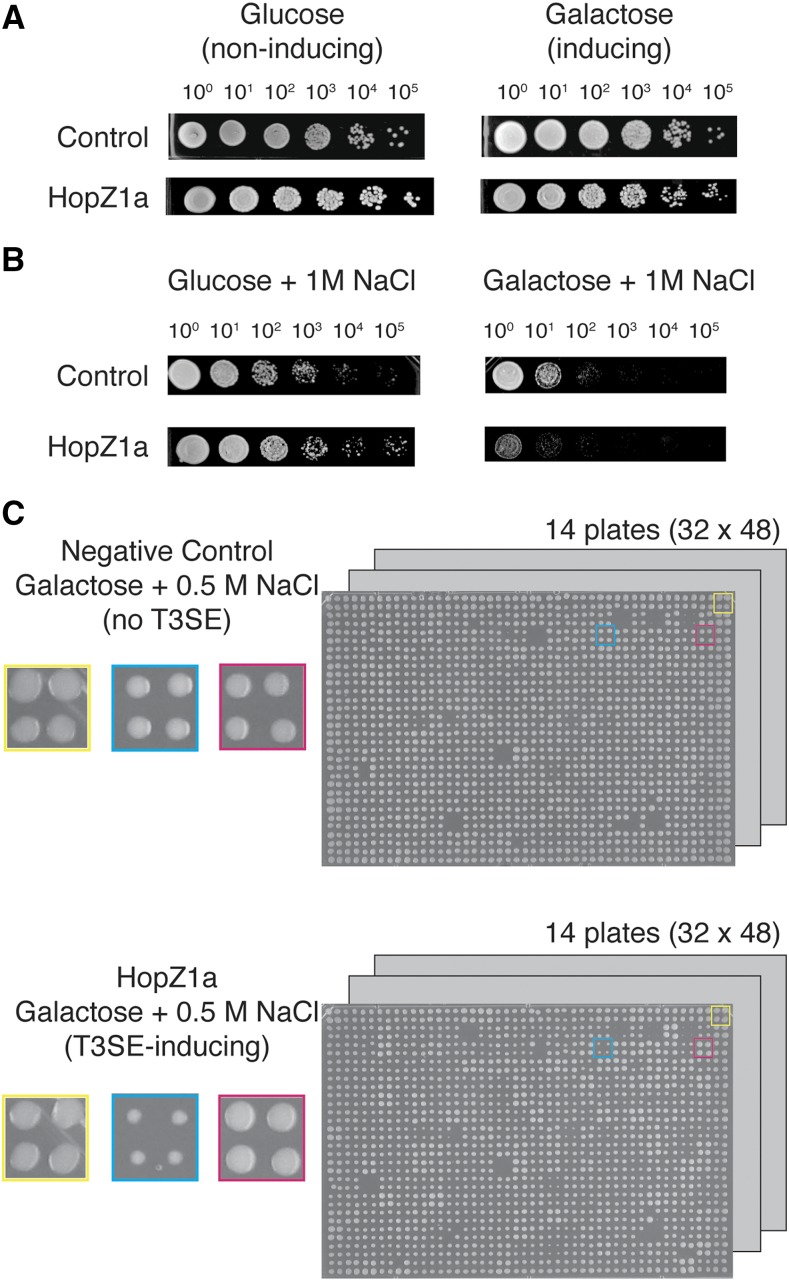
Figure 2.
Genome-wide phenotypic screens to identify yeast deletion strains that suppressed or were sensitized to P. syringae T3SE expression. (A) HopZ1a does not inhibit yeast growth on rich media as shown by spot dilution assay. The control strain has the NATR antibiotic cassette integrated at the ho locus. (B) The growth inhibition by HopZ1a compared to the negative control strain on rich media with 1M NaCl (galactose + 1M NaCl) by spot dilution assay is shown. (C) Yeast haploid deletion collection with the integrated GAL1-hopZ1a at the ho locus (hoΔ::GAL1-hopZ1a-FLAG::NATR) on galactose (HopZ1a-inducing) media with 0.5 M NaCl. The negative control array was also pinned on the galactose media with 0.5 M NaCl. Each deletion mutant was pinned in quadruplicate onto the appropriate solid media to generate four replicates in each screen. Colonies in the yellow square represent the border control strain, colonies in the blue square represent a yeast deletion strain that is sensitive to HopZ1a expression, while colonies in the red square represent a yeast deletion strain that suppresses fitness defects as a result of HopZ1a expression.
Confirmation of PGA interactors
Yeast deletion strains that were either putative suppressors or synthetic lethal interactors from the PGA screens were streaked out on YPD with 200 μg/ml of G418 (Invitrogen Life Technologies, USA) and incubated at 30° for 2 – 3 days. Single colonies of each deletion strain were patched onto YPD plates in 1 – 2 cm2 patches and incubated at 30° for 1 overnight to allow for actively growing yeast cultures. A single colony of wild type yeast from the deletion array border was also streaked out and patched onto YPD plates as control strains. Each yeast deletion strain was scraped off from the patches (∼108 – 109 cells) using sterile toothpicks and arrayed into a 96-well microtiter plate containing 200 μl of sterile water. Yeast cells were washed once with 200 μl of 0.1 M lithium acetate by centrifugation for 5 min at 1,500 x g at 20° in a centrifuge with a microtiter plate rotor. Each well of pelleted yeast cells was resuspended with 180 μl of transformation mix (120 μl of 50% w/v PEG-3350, 18 μl of 1 M lithium acetate, and 25 μl of boiled single-stranded carrier DNA). 60 μl each of resuspended cells were subsequently transferred to 96-well microtiter plates containing either 1 μl of purified plasmid DNA pBA350V (empty vector) (Lewis et al. 2013; Youn et al. 2017) or 1 μl of purified plasmid DNA (pBA350V-hopZ1a, pBA350V-hopF2 and pBA350V-hopX1). The remaining 60 μl of cells served as a mock transformation control. The 96-well microtiter plates were incubated at 30° for 30 min followed by heat shock at 42° for 30 min. Cells were harvested by centrifugation for 10 min at 1,500 x g at 20° and resuspended in 100 μl of SD. 50 μl of transformed or mock-transformed cells were plated on SD-Leu and incubated at 30° for 3 days. Transformants carrying either pBA350V or pBA350V-T3SE (pBA350V-hopZ1a, pBA350V-hopF2 and pBA350V-hopX1) were grown on SD-Leu plates and were subsequently used for confirmation by spot dilution assays. In order to confirm positive or negative genetic interactions, we used the number of spots to calculate the fitness of each single or double mutant in semi-quantitative manner, as described in (Youn et al. 2017).
Spot dilution assay
For spot dilution assay to determine growth inhibition of Y7092 expressing P. syringae T3SEs, 1 ml of cultures were grown at 30° and 200 RPM in YPR in deep-well plates that contain sterile glass beads in each well. Ten-fold dilution series of the overnight cultures were spotted onto YPD, YPG, YPD with 1 M sorbitol, YPG with 1 M sorbitol, YPD with 1 M NaCl, or YPG with 1 M NaCl.
For spot dilution assays to confirm the putative PGA hits as either suppressors or synthetic lethal interactors, the deletion strains carrying either the empty vector (pBA350V) or the effector of interest (pBA350V-T3SE) were grown in synthetic drop-out media lacking Leu with 2% raffinose (SR-Leu) for two overnights at 30° and 200 RPM. The overnight cultures were serially diluted 15-fold and spotted onto SD-Leu, SG-Leu, SD-Leu and 0.5 M NaCl, or SG-Leu and 0.5 M NaCl. Spot dilutions were grown for two to three days before being photographed. Spot assays were quantified using an unbiased visual toxicity score (between 1 to 5), where 1 represented the strongest toxicity (1 spot grew) and 5 represented the least toxicity (all 5 spots grew). A fitness defect score was subsequently calculated using the toxicity score to compare the expected fitness defect to the observed fitness defect of each mutant (Baryshnikova et al. 2010; Sharifpoor et al. 2012).
Gene Ontology (GO) Enrichment Analysis
GO enrichment analysis was performed by entering query genes (either HopZ1a PGA interactors or yeast mutants with congruent SGA interaction profiles as HopZ1a) into the GO Term Finder of the Saccharomyces Genome Database (https://www.yeastgenome.org/goTermFinder) using a gene universe (background gene set) consisting of the ∼4,400 deletion mutants tested. We analyzed the three different ontologies: GO Process, GO Function and GO Component, with default p-value (P < 0.01) and false discovery rate filter thresholds.
Yeast co-expression, immunoprecipitations and sample preparation
Yeast co-expression and immunoprecipitation was performed as described previously (Lewis et al. 2013). Briefly, overnight cultures of yeast strain Y7092 co-expressing FLAG-tagged HopZ1a (wild type or a catalytically-inactive mutant, C216A) with putative acetylation targets MKRP1 or HINKEL were diluted into fresh SD-Leu (2% raffinose) and allowed to grow at 30° for two doublings prior to inducing expression of effector and targets by addition of galactose to a final concentration of 2%. Following 15 h of induction, cultures were mechanically lysed and lysates were incubated with an anti-FLAG agarose resin (Sigma). The resin was washed in cell lysis buffer (50 mM Tris, pH = 8; 150 mM NaCl; 1.5 mM magnesium acetate; 5 mM EDTA; 0.15% NP-40) as described previously, with reduced NP-40 (0.015%) for the last of three washes (Lewis et al. 2013). After washing the resin to remove unbound proteins, FLAG-tagged proteins were eluted by incubating with 100 uL of FLAG peptide solution (150 ug/mL FLAG peptide in TBS) for one hour at 4°. Eluted material was dried to a pellet under vacuum and stored at -80° prior to subsequent mass spectrometry analysis. Dried protein samples were re-solubilized in 50 mM ammonium bicarbonate (pH 7.8) and then subjected to reduction with dithiothreitol at 56°, alkylation with iodoacetamide at room temperature, and overnight digestion with sequencing-grade trypsin (Promega, Madison, WI) at 37°. The enzymatic reactions were stopped with 3% formic acid, purified and concentrated with Pierce C18 Spin Columns (Thermo Scientific) and again dried to a pellet under vacuum. Peptide samples were then solubilized in 0.1% formic acid prior to LC-MS/MS analyses.
LC-MS/MS Analysis of Proteins, Chromatography and Mass Spectrometry
Subsequent analytical separation was performed on a homemade gravity-packed 75 µm internal diameter column (New Objective, Woburn, MA) packed with 10 cm of 100 Å, 5 µm Magic C18AQ particles (Michrom, Auburn, CA). Peptide samples were loaded onto the analytical column using a variable gradient with a flow rate of 300 nL/min. The gradient utilized two mobile phase solutions: A, water/0.1% formic acid; and B, 80% acetonitrile/0.1% formic acid. Samples were analyzed on a linear ion trap-Orbitrap hybrid analyzer outfitted with a nano spray source and EASY-nLC 1200 nano-LC system. The instrument method consisted of one MS full scan (400–1400 m/z) in the Orbitrap mass analyzer, an automatic gain control target of 500,000 with a maximum ion injection of 500 ms, one microscan, and a resolution of 60,000. Six data-dependent MS/MS scans were performed in the linear ion trap using the three most intense ions at 35% normalized collision energy. The MS and MS/MS scans were obtained in parallel fashion. In MS/MS mode automatic gain control targets were 10,000 with a maximum ion injection time of 100 ms. A minimum ion intensity of 1000 was required to trigger an MS/MS spectrum. The dynamic exclusion was applied using an exclusion duration of 145s.
Protein ID and Database Searching
Proteins were identified by searching all MS/MS spectra against a large database composed of the complete proteome of Saccharomyces cerevisiae strain S288C (ATCC 204508; UniProt proteome ID UP000002311) supplemented with sequences for P. syringae HopZ1a (WP_011152901.1), and the Arabidopsis kinesins HINKEL (NP_173273.2) and MKRP1 (NP_173592.3) (all retrieved from the NCBI database) using SEQUEST (Thermo Scientific Proteome Discoverer software). A fragment ion mass tolerance of 0.6 Da and a parent ion tolerance of 10 ppm were used. Up to two missed tryptic cleavages were allowed. Methionine oxidation (+15.99492 Da), cysteine carbamidomethylation (+57.02146 Da), and acetylation (+42.01057 Da) were set as variable modifications. The generated search results were imported into the Scaffold data analysis platform, an X!Tandem search (Beavis Informatics, Winnipeg, MA) was performed and the peptides were evaluated using a false discovery rate of 0.1% as determined using a reversed version of the database used in the original search. A mzident.xml file was generated from Scaffold and imported into Scaffold PTM (Proteome Software, Portland, OR) to evaluate and score the post translational modifications.
Yeast strains and Data availability
All yeast strains and plasmids described in this study are available upon request. Mass spectrometry data consisting of raw files and associated peak list and results files has been deposited in MassIVE as complete (Data Dependent Acquisition). Mass spectrometry data are available from MassIVE (https://massive.ucsd.edu) using Massive ID: MSV000083076. Table S1.xlsx: list and description of confirmed genetic interactions for HopZ1a. Table S2.xlsx: list and description of confirmed genetic interactions for HopF2. Table S3.xlsx: list and description of confirmed genetic interactions for HopX2. TableS4.xlsx: congruence scores for yeast genes with genetic interaction profiles similar to that of HopZ1a. TableS5.xlsx: congruence scores for yeast genes with genetic interaction profiles similar to that of HopF2. TableS6.xlsx: congruence scores for yeast genes with genetic interaction profiles similar to that of HopX1. FigureS1.tiff: immunoblot analysis of yeast strain Y7092 expressing P. syringae T3SEs. FigureS2.tiff: spot dilution assays to determine growth inhibition profiles of yeast expressing P. syringae T3SEs. FigureS3.pdf: extracted ion chromatograms, reversed phase chromatography and MS/MS spectra supporting identification of two distinct (singly) acetylated forms of the doubly charged HINKEL peptide, VFGPESLTENVYEDGVK. FigureS4.pdf: extracted ion chromatograms, reversed phase chromatography and MS/MS spectra supporting acetylation of the doubly and triply charged MKRP1 peptide, EISCLQEELTQLR. FigureS5.pdf: extracted ion chromatograms, reversed phase chromatography and MS/MS spectra supporting acetylation of the doubly and triply charged MKRP1 peptide, EIYNETALNSQALEIENLK. FigureS6.pdf: extracted ion chromatograms, reversed phase chromatography and MS/MS spectra supporting acetylation of the doubly and triply charged HopZ1a peptide, ELLDDETPSNTQFSASIDGFR. FigureS7.pdf: zoomed-in views of the extracted ion chromatograms presented in Figure S6. FigureS8.pdf: acetylated HINKEL residues are proximal to the kinesin ATP-binding site. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7318505.
Results
Generation and characterization of yeast strains carrying P. syringae T3SEs
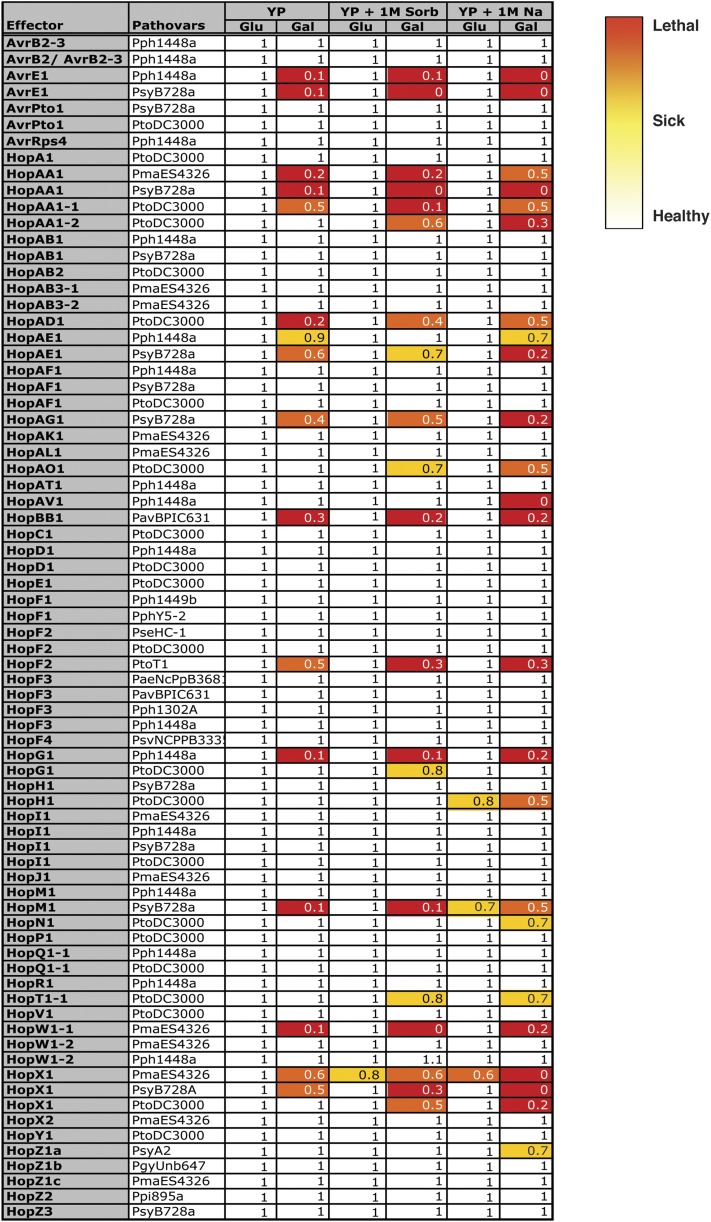
We generated a collection of 75 yeast strains each carrying an inducible P. syringae T3SE expression construct (Figure 1). The T3SEs included those from three widely studied P. syringae strains: 22 T3SEs from P. syringae pv. tomato DC3000 (PtoDC3000); 12 T3SEs from P. syringae pv. syringae B728a (PsyB728a); and 17 T3SEs from P. syringae pv. phaseolicola 1448A (Pph1448a). These three strains have finished genome sequences and represent three of the five major P. syringae phylogroups (phylogroups 1, 2, and 3, respectively) (Hwang et al. 2005). We also screened 12 T3SEs from P. syringae pv. maculicola ES4326 (PmaES4326), which belongs to phylogroup 4. Finally, we screened three additional T3SEs from the HopZ family and nine additional T3SEs from the HopF family, as these two effector families are of particular interest to our group (Figure 1) (Ma et al. 2006; Lewis et al. 2008; Wilton et al. 2010). Briefly, each T3SE construct was linked to a drug resistance cassette (NATR) and integrated at the ho locus – a neutral, dispensable locus not functionally required in stable haploid or diploid cells (Baganz et al. 1997; Youn et al. 2017). Each T3SE was tagged with a C-terminal FLAG epitope and expressed under the control of a galactose-inducible promoter. We confirmed galactose-dependent expression of the 75 T3SEs using western blot analysis (Figure S1).
Figure 1.
Growth inhibition profiles of yeast (Y7092) expressing 75 P. syringae T3SEs on rich media (Hong et al.), rich media with 1 M sorbitol (YP + 1M Sorb), and rich media with 1 M NaCl (YP + 1M Na). The growth inhibition by each T3SE in each condition is represented in numbers and heat map, with 1 (or white) corresponding to no growth inhibition to 0 (or red) corresponding to complete growth inhibition. The fitness numbers are calculated for every condition (Glu = glucose and T3SE-repressing, Gal = galactose and T3SE-expressing) by normalizing the fitness of yeast expressing T3SE to the negative control strain containing the integrated NATR antibiotic cassette at the ho locus.
Using the collection of T3SE-expressing yeast strains we performed a fitness-based screen to identify T3SEs that inhibit yeast growth. We examined the phenotypic consequence of T3SE expression in yeast using serial dilution spot assays on rich media with glucose (T3SE-repressing) or galactose (T3SE-expressing). As expected, we did not observe fitness defects on T3SE-repressing media (Figure 1 and S2) compared to the negative control strain (hoΔ::NATR), however, the expression of 16 out of 75 T3SEs inhibited yeast growth on T3SE-expressing rich media (Figures 1 and S2: AvrEPph1448a, AvrEPsyB728a, HopAA1PmaES4326, HopAA1PsyB728a, HopAA1-1PtoDC3000, HopAD1PtoDC3000, HopAE1Pph1448a, HopAE1PsyB728a, HopAG1PsyB728a, HopBB1PavBPIC631, HopF2PtoT1, HopG1Pph1448a, HopM1PsyB728a, HopW1-1PmaES4326, HopX1PsyB728a and HopX1PmaES4326).
To identify additional T3SEs that may target conserved cellular processes under stress conditions, we also performed fitness assays on media inducing hyperosmotic stress (containing 1 M sorbitol or 1 M NaCl). Nine additional T3SEs altered yeast fitness when expressed in the presence of high osmolytes, with yeast expressing HopW1-2Pph1448a showed a slightly increased fitness on 1 M sorbitol (Figures 1 and S2). Four of the PtoDC3000 T3SEs caused enhanced fitness defects in yeast both with 1 M sorbitol and with 1 M NaCl (HopAA1-2, HopAO1, HopT1-1, and HopX1). Although 1 M sorbitol and 1 M NaCl both activate the high osmolarity glycerol (Hohmann 2002) pathway by creating a high osmolarity environment, NaCl stress creates additional toxicity by altering the ion homeostasis in the cell (Giaever et al. 2002). We also identified a single T3SE that affected yeast fitness only in the presence of 1 M sorbitol (HopG1PtoDC3000) and three T3SEs that altered yeast fitness only in the presence of 1 M NaCl (HopAV1Pph1448a, HopN1PtoDC3000 and HopZ1aPsyA2).
Identifying genetic interactors of P. syringae T3SEs by PGA analysis
To further characterize P. syringae T3SE functions and their mechanisms of toxicity in yeast, we utilized the yeast PGA functional genomics approach on HopAA1, HopW1-1, HopZ1a, HopF2 and HopX1. To this end, we performed a PGA screen by crossing our integrated T3SE-expressing strain with ∼4400 haploid yeast non-essential gene deletion mutants (Giaever et al. 2002), looking for negative and positive genetic interactions in the context of T3SE expression (Figure 2C). We carried out a parallel control screen using a query strain (Youn et al. 2017) harboring a deletion in a benign locus (ho) instead of a T3SE (see Materials and Methods section for details) to obtain ‘control arrays’ that reflect the fitness of the yeast gene deletion mutants.
We manually scored positive or negative genetic interactions by observing changes in colony size (fitness) between the experimental and control plate (Figure 2C; see Materials and Methods for further details). We classified negative genetic interactors for those double mutants that grew more poorly than expected based on those of the single mutant fitness. In addition, if the double mutant grew much better than that of a single mutant (in this case, T3SE), we classified these interactions as suppression.
We observed potential genetic interactors in the PGA screens of HopF2 (132 suppressors and 73 synthetic lethal interactors) and HopX1 (88 synthetic lethal interactors), whereas HopAA1 and HopW1-1, with the most severe fitness defect, did not reveal any reproducible interactors (including suppressors) in our initial PGA analysis. As for HopZ1a, we observed no genetic interactions under standard PGA conditions, which prompted us to test genetic interactions in a condition that shows HopZ1a-induced fitness defect: high osmotic stress condition. Since 1 M NaCl drastically reduced the fitness of the HopZ1a-expressing yeast strain, we therefore assessed the fitness of ∼4400 double mutants on media containing a range of salt concentrations below 1 M NaCl. At 0.25 M and 0.5 M NaCl, we initially identified 137 deletion mutants with reduced HopZ1a toxicity (suppressors) and 53 deletion mutants with enhanced HopZ1a toxicity (negative genetic interactors; data not shown).
To confirm the genetic interaction phenotypes, we conducted a secondary screen by transforming the haploid yeast deletion strains that were identified in our primary PGA screen with single copy plasmid (pBA350V) carrying GAL-T3SE (HopZ1a, HopF2 or HopX1), and then used spot dilution assays to characterize fitness. We excluded any strains with deletions in dubious open reading frames (Winzeler et al. 1999; Giaever et al. 2002; Kramer et al. 2007) as well as galactose metabolism genes. For HopZ1a, 95 suppressors and 10 negative genetic interactors were confirmed by independent transformation and spot dilution assays on 0.5 M NaCl and galactose (Figure 3 and Table S1). For HopF2, 105 suppressors and 20 negative interactions were confirmed (Table S2), whereas 32 negative interactions (no suppressors) were confirmed for HopX1 (Table S3). Confirmed genetic interactors for HopZ1a, HopF2 and HopX1 can be found in Supplementary Tables S1-S3.
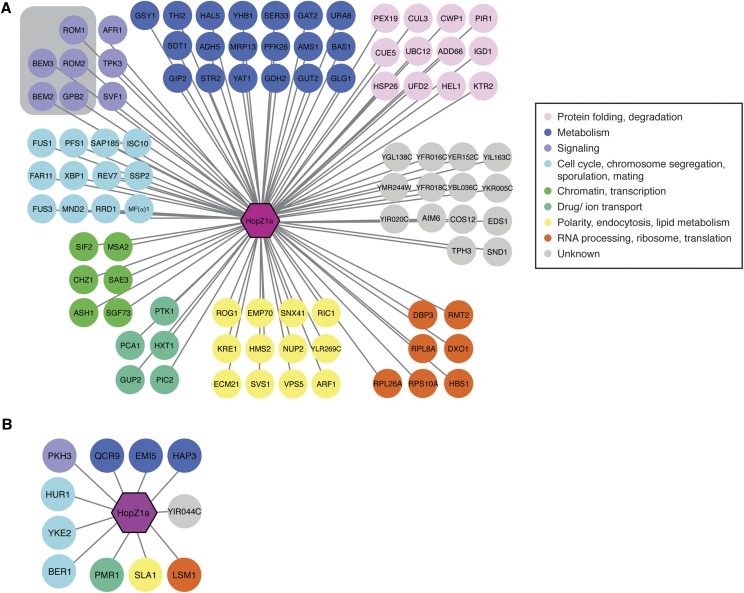
Figure 3.
Analysis of HopZ1a suppressors and negative genetic interactors in yeast. Diagram showing (A) suppressors and (B) negative genetic interactors of HopZ1a generated using Cytoscape. Nodes are color coded based on annotations of biological processes from Constanzo et al. (Costanzo et al. 2010). The HopZ1a suppressors showed an enrichment in GTPase-mediated signal transduction (gray-shaded box; P = 0.006).
Biological processes enriched in PGA profiles
We utilized the Gene Ontology (GO) vocabulary to identify biological processes associated with the confirmed HopF2, HopX1 and HopZ1a PGA interaction partners, since GO processes that are enriched within this genetic interaction data set may potentially illuminate functional processes that are influenced by effectors (Kramer et al. 2007; Baryshnikova et al. 2010). We analyzed positive and negative interactors separately.
Using the Saccharomyces Genome Database (SGD) GO Term Finder (Hong et al. 2008), we found a significant enrichment of genes involved in GTPase-mediated signal transduction and its regulation (Figure 3A; P = 0.006) in HopZ1a suppressors. Specifically, we identified two Rho GTPase activating proteins that are critical for cell polarity and cell division: BEM2 and BEM3; as well as two GDP/GTP exchange proteins: ROM1 and ROM2. In contrast, no significant GO enrichment was found for HopF2 suppressors (no HopX1 suppressors were identified).
Negative genetic interactors of HopF2 were enriched for various intracellular trafficking pathways (endosomal transport P < 0.004, vacuolar transport, P < 0.01, late endosome to vacuole transport via multi vesicular body sorting pathway, P < 0.06), whereas HopX1 negative interactors were enriched for protein complex assembly and biogenesis (P < 0.01), lipid tube assembly (P < 0.02), protein-lipid complex assembly (P < 0.02), mitochondrial respiratory chain complex IV assembly (P < 0.06). We did not identify significant enrichment of GO processes in the HopZ1a negative genetic interactors. However, two negative genetic interactors of HopZ1a, YKE2 and BER1, are involved in regulating tubulin folding and microtubule-related processes (Figure 3B). Additionally, we identified both suppressors (BEM2, BEM3 and RRD1) and negative genetic interactors (SLA1) that are involved in regulating the actin cytoskeleton (Figure 3A and B). Actin and microtubule cytoskeletons are both involved in fundamental processes such as cell division and intracellular trafficking, raising the possibility that our genetic interaction screen identified genes whose functions influence both of these two important cytoskeletal components.
Predicting HopZ1a targets by congruence analysis of genetic interactors
Previously we have shown that the T3SE HopZ1a can bind to tubulin and alter microtubule networks in planta (Lee et al. 2012). We were particularly interested in the role of HopZ1a in regulating microtubule dynamics (and potentially other processes) and we therefore focused our analysis on this effector as proof-of-principle that our genomic resource can be used to characterize P. syringae T3SE functions.
The analysis of the HopZ1a genetic interactors described above revealed several biological processes that may be disrupted by HopZ1a but provided limited insight regarding its direct targets. We therefore sought to predict direct targets by identifying yeast gene disruptions that show similar (i.e., congruent) genetic interaction profiles to HopZ1a. This approach is similar to one used previously to identify drug targets in yeast (Costanzo et al. 2010) and assumes that if HopZ1a activity disrupts a given target protein’s function in yeast, the HopZ1a PGA profile would be similar (or ‘congruent’) to the SGA profile of the corresponding gene knockout strain lacking this putative target (Figure 4A and B). We focused our congruency analyses on negative genetic interactors since previous work has indicated that these interactions are easier to interpret than suppressors (Ye et al. 2005).
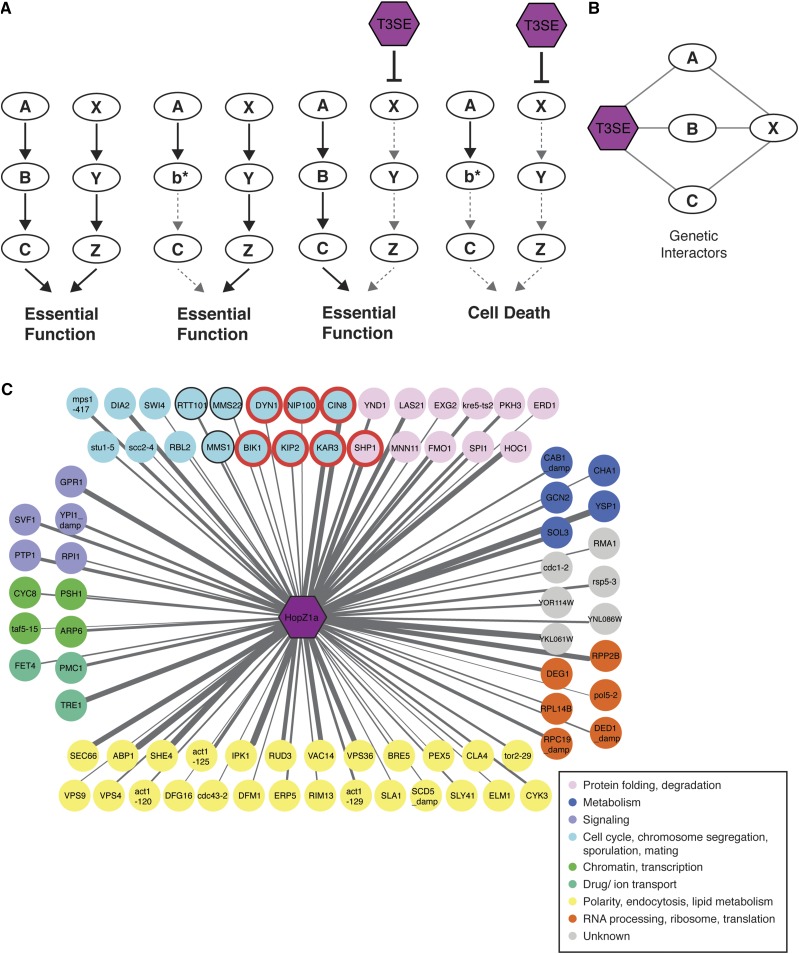
Figure 4.
Congruence gene analyses of HopZ1a negative genetic interactors identify microtubule motor proteins as potential targets. (A) A model for the molecular mechanism of enhancing T3SE toxicity by targeting redundant pathways. A mutation in either one of the parallel redundant pathways (b* or the inhibition of X by T3SE) does not alter cell viability. However, when both pathways are disrupted (b* and the inhibition of X by T3SE), the cells are not viable. (B) Congruence analysis predicts potential T3SE targets by identifying yeast genes (gene X) with similar genetic interaction profiles as the T3SE. (C) 81 congruent yeast genes with congruence score ≥ 2 are shown, with nodes color coded based on annotations of biological processes from Constanzo et al. (Costanzo et al. 2010). HopZ1a is congruent to yeast deletion strains that are enriched for replication fork processing (P < 0.0001) and microtubule-based processes (with a P < 0.0004) as analyzed by GOrilla tool (Eden et al. 2009). Genes enriched in microtubule-based processes are circled in red, and genes enriched for replication fork processing are circled in black. Edge thickness is proportional to congruence scores.
To identify yeast genes with HopZ1a-congruent genetic interaction profiles, we compared our HopZ1a genetic interaction profile with those of 1712 single yeast mutants (encompassing ∼170,000 interactions) and calculated the pairwise overlap of genetic interactions (Costanzo et al. 2010) using a previously established congruence score (Ye et al. 2005). In brief, congruence score is defined as the –log10 of the p-value for the number of shared genetic interaction profiles of two genes and provides a ranking of the degree of similarity in genetic interaction profiles. Therefore, for any particular congruent gene pair, the overlap in shared genetic interacting partners increases with increasing congruence score (Table S4 – S6). We identified 81 yeast genes with HopZ1a congruence scores ≥ 2, indicating similarity to the negative genetic interaction profile of HopZ1a (Figure 4C and Table S4). We performed GO biological process enrichment analysis on this set of yeast genes (those with interaction profiles congruent to HopZ1a), and we found significant GO enrichment for genes involved in replication fork processing (P < 0.0001) (Figure 4C; circled black) and for genes involved in microtubule-based processes (P < 0.0004) (Figure 4C; circled red). These GO enrichment categories were specific to HopZ1a since HopF2 congruent genes were enriched for vesicle-mediated transport (P < 0.08; Table S5) whereas those of HopX1 were enriched for cell cycle (P < 0.0005), mitotic cell cycle process (P < 0.02), double-strand break repair via homologous recombination (P < 0.07; Table S6; data not shown).
Given that HopZ1a has been shown to disrupt microtubule networks in Arabidopsis, we were particularly interested in the congruent genes involved in microtubule-based processes (P < 0.0004). These included several microtubule-directed motor proteins such as kinesins (i.e., CIN8, KIP2, VIK1, and KAR3), which have been shown to be HopZ-interacting proteins in two published yeast two-hybrid datasets between A. thaliana genes and P. syringae T3SEs (Mukhtar et al. 2011; Lewis et al. 2012). This overlap between the genetic and physical interactions observed for HopZ1a motivated further investigation into whether Arabidopsis kinesins represent direct targets of HopZ1a activity.
HopZ1a acetylates plant kinesins
Kinesins are microtubule-based motor proteins involved in many cellular processes, including intracellular transport, mitotic cell division, signaling, and microtubule organization (Zhu and Dixit 2012). There are 61 kinesins in Arabidopsis (Lee and Liu 2004) and nearly one quarter of these (15) are members of the kinesin 7 (Kin7) subfamily, which were shown to interact with the HopZ family (Richardson et al. 2006; Mukhtar et al. 2011; Lewis et al. 2012). Given the large number of potential targets, we focused on kinesins that 1) interact with the HopZ family and 2) related kinesins that have been demonstrated to regulate plant microtubule stability. The kinesins previously shown to interact with the HopZ family are the mitochondrially-localized MKRP1 (At1g21730) and MKRP2 (At4g39050) (Mukhtar et al. 2011; Lewis et al. 2012). Since mitochondrial localization of HopZ1a has not been observed (Lewis et al. 2008), we also investigated whether HopZ1a may target the related Kin7 kinesin HINKEL (also known as AtNACK1 or HIK), which is involved in regulating microtubule stability in plants (Strompen et al. 2002; Takahashi et al. 2010; Komis et al. 2011).
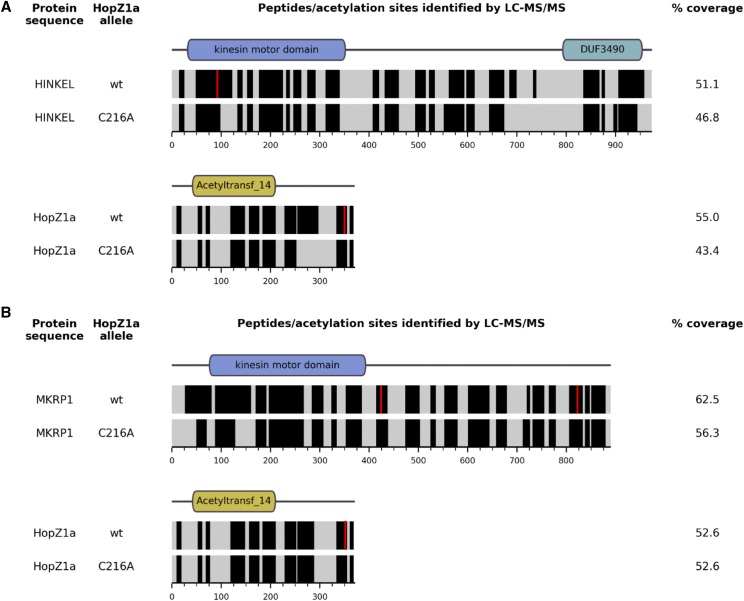
HopZ1a is an acetyltransferase with multiple eukaryotic targets, including tubulin and the A. thaliana pseudokinase ZED1 (Lee et al. 2012; Lewis et al. 2013). To test whether HopZ1a acetylates kinesins in vivo in a heterologous yeast system, we co-expressed HopZ1a in yeast with each candidate kinesin, all as FLAG-tagged recombinant proteins, as previously described for Arabidopsis pseudokinase, ZED1 (Lewis et al. 2013). We used liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify acetylated peptides of both HINKEL and MKRP1. LC-MS/MS analysis of anti-FLAG immunoprecipitates identified acetylated peptides from both kinesins (mass increases in multiples of 42 Daltons) present when co-expressed with wild type HopZ1a but not with the catalytically inactive mutant, HopZ1aC216A (Figure 5). Candidate acetylation sites were confirmed by manual inspection of extracted ion chromatograms and MS/MS spectra (Figures S3-S7 and data not shown). In this way we identified two distinct acetylated species of the same HINKEL peptide (VFGPESLTENVYEDGVK; residues 83-99) - ‘peptide A’ (VFGPE[S-Ac]LTENVYEDGVK, acetylated at S88) and ‘peptide B’ (VFGPESL[T-Ac]ENVYEDGVK, acetylated at T90) (Figures 5A, S3). We also identified acetylated peptides from two distinct sites in MKRP1 - ‘peptide C’ (EISCLQEEL[T-Ac]QLR; residues 416-428; acetylated at T425) and ‘peptide D’ (EIYNE[T-Ac]ALNSQALEIENLK; residues 815-33; acetylated at T820) (Figures 5B, S4 and S5). Similar analysis of the HopZ1a-derived peptides from those cells co-expressing MKRP1 or HINKEL indicates auto-acetylation of HopZ1a at three sites in close proximity (T342, S344, T346) (Figures 5, S6, S7), consistent with a recent report that also described auto-acetylation of T346 (Ma et al. 2015).
Figure 5.
HopZ1a acetylates A. thaliana kinesins HINKEL and MKRP1. Predicted domain architectures (as annotated by the NCBI Conserved Domain Database; Marchler-Bauer et al. 2015) for HINKEL (A), MKRP1 (B) and HopZ1a (A and B) are indicated above horizontal bands representing the mass spectrometry sequence coverage for each protein. Black bands indicate sequences identified with high confidence, while gray bands indicate sequences that were not reliably detected. Red vertical stripes indicate the position of acetylated residues.
Discussion
In this study, we report the generation of a yeast strain collection stably expressing T3SEs from the plant pathogen P. syringae and demonstrate its utility for functional characterization of T3SEs. Out of 75 P. syringae T3SEs in yeast, we identified 24 effectors that altered yeast fitness on rich media or under high osmolarity conditions, including HopZ1a. Using HopZ1a as proof-of-principle, we took advantage of the genetic tractability of yeast to perform a high-throughput PGA screen to look for conserved biological processes that are targeted by HopZ1a. Exploiting a large genetic interaction dataset that covers ∼75% of all yeast genes, we performed congruency analysis to delineate conserved targets of HopZ1a in yeast and combined this with previously-described physical interaction datasets that include HopZ family members to implicate kinesins as potential targets of the T3SE, HopZ1a.
Previous studies have identified bacterial phytopathogen T3SEs that altered yeast fitness (Jamir et al. 2004; Munkvold et al. 2008; Salomon et al. 2011). Of the 27 PtoDC3000 effectors tested by Munkvold et al. 7 inhibited yeast growth (Munkvold et al. 2008; Munkvold et al. 2009). We tested 20 of these same 27 PtoDC3000 effectors and observed fitness phenotypes consistent with these previous data in all cases except for HopAO1, HopD1 and HopN1 (Munkvold et al. 2008; Munkvold et al. 2009). While we integrated T3SEs into the yeast genome and expressed them as single copy genes, Munkvold et al. expressed T3SEs on a high-copy plasmid. Differences in gene dosage may have contributed to these differences.
Our initial screen provides numerous interesting leads for further study. Notably, P. syringae T3SEs encoded in the conserved effector locus (CEL) caused severe fitness defects in yeast (Figure 1). T3SEs of the CEL are conserved across most P. syringae strains and typically include the evolutionarily unrelated T3SEs AvrE, HopM1, and HopAA1 (Alfano et al. 2000). Pph1448a has nonfunctional alleles of HopM1 and HopAA1 (Joardar et al. 2005), while PtoDC3000 contains an additional effector in its CEL, HopN1 (O’Brien et al. 2011). The CEL has been shown to play an important role in bacterial virulence (Alfano et al. 2000; Badel et al. 2003; Munkvold et al. 2009) and in the suppression of salicylic acid (SA)-mediated basal immunity (DebRoy et al. 2004). However, with the exception of HopM1 (Nomura et al. 2006; Nomura et al. 2011), the host targets and the mechanisms by which T3SEs in the CEL promote virulence are not well characterized. Our results suggest that most CEL T3SEs may have evolved to target conserved components of eukaryotic processes. The yeast fitness defects induced by expression of CEL T3SEs observed in this study will provide an important tool to help identify virulence targets of this ubiquitous class of phytopathogen T3SEs.
The PGA approach can be used to infer the function of T3SEs by identifying those yeast genes whose deletions either suppress or enhance T3SE lethality. Intuitively, deletion strains that suppress T3SE lethality (known as suppressors) can reveal genes involved in the same pathways as putative T3SE targets. This can be particularly informative when the T3SE activates a pathway resulting in toxicity, as was observed with the Shigella T3SE IpgB2 which activates the Rho1p GTPase signaling pathway in yeast (Alto et al. 2006). However, one caveat of the suppressor screen is that we may identify mutants that suppress T3SE lethality by a general mechanism (i.e., by induction of a general stress response); such genes are unlikely to be informative for the inference of T3SE function.
Deletion mutants that exacerbate the fitness cost of T3SE activity can be explained by either of two alternate mechanisms resulting in ‘negative genetic interactions’. In one case, the T3SE acts in the same pathway as the ‘negative genetic interactor’, resulting in cumulative insults to an essential pathway or complex (Boone et al. 2007; Dixon et al. 2009). Alternatively, the T3SE and ‘negative genetic interactor’ may act on parallel pathways, which redundantly contribute to an essential function (Figure 3A) (Boone et al. 2007; Dixon et al. 2009). Our analysis of both suppressors and negative genetic interactors revealed enrichment of signal transduction pathways involving small-GTPases and may reflect an ability of HopZ1a to influence these cellular processes. Similarly, HopF2 and HopX1 may influence cellular trafficking and lipid metabolism, respectively. In order to gain further insight into the direct targets of HopZ1a we applied a congruence analysis to compare SGA interaction profiles of 1,712 yeast genes, including 334 conditional alleles of essential genes (Costanzo et al. 2010) with the HopZ1a PGA interaction profile described in this study. This approach is conceptually similar to the integration of chemical-genetic and SGA datasets for identification of drug targets (Costanzo et al. 2010); functional inhibition of a target protein by drug or by T3SE is expected to mimic the consequences of the corresponding target gene’s deletion, resulting in similar/congruent genetic interaction profiles.
Applying these principles, we identified SGA profiles that were most similar to the HopZ1a PGA profile and analyzed them for functional enrichment. Genes involved in replication fork processing (P < 0.0001) and microtubule-based processes (P < 0.0004) were enriched in the subset with HopZ1a-congruent interaction profiles. We were particularly interested in microtubule-associated processes since HopZ1a can disrupt microtubules in plants and interacts with tubulin in both plant and animal cells (Lee et al. 2012). Indeed, kinesins (known microtubule-guided motor proteins) were identified not only through our congruence analysis, but also by two independent yeast two-hybrid screens for Arabidopsis proteins that bind to related HopZ family members. The fact that kinesins are found at the intersection of these three independent datasets indicates that members of this family may indeed represent bona fide, direct targets of HopZ1a. In support of this possibility, HopZ1a can acetylate both of the Arabidopsis kinesins HINKEL and MKRP1 (Figure 5).
The acetylated sites (S88, T90) of HINKEL are found within its kinesin motor domain (Figure 5A), and mapping these to the corresponding positions in the structure of human kinesin CENP-E (Garcia-Saez et al. 2004) reveals a close proximity to the nucleotide-binding pocket (Figure S8). In A. thaliana, HINKEL activates the ANP1/ANQ1/MPK4 MAPK pathway that ultimately regulates microtubule-bundling proteins (e.g., MAP65) via phosphorylation (Komis et al. 2011). Our data suggest a possible mechanism for HopZ1a-mediated antagonism of this pathway whereby nucleotide binding and/or hydrolysis activity is altered following acetylation of sites proximal to the nucleotide-binding pocket of HINKEL.
Although HopZ1a has not been detected in mitochondria, we cannot rule out the possibility that the mitochondrial kinesins identified by yeast two-hybrid assays are also targeted by HopZ1a, especially considering that they are targeted by the P. syringae T3SE HopG1 and are involved in plant immunity (Shimono et al. 2016). HopZ1a acetylates MKRP1 at two distinct sites: T425 is just ‘downstream’ of the kinesin motor domain while T820 is near its C-terminus (Figure 5B). In Nicotiana, the HINKEL ortholog NACK1 is phosphorylated near the C-terminus at residues T675, T690 and T836 by cyclin-dependent kinases to regulate microtubule dynamics during cytokinesis (Sasabe et al. 2011). Although reasonable speculation might suggest that C-terminal acetylation could disrupt hypothetical phosphorylation sites of MKRP1 and other kinesins, MKRP1 however lacks the C-terminal DUF3490 domain common to HINKEL and NACK1 (Figure 5) and we did not detect HopZ1a acetylation at the C-terminus of HINKEL.
Additional acetylation sites may exist on HINKEL and MKRP1 (and HopZ1a) since LC-MS/MS analysis is unable to detect all peptides generated from trypsin digests of the proteins of interest; we only acquired 47–51% coverage of HINKEL, 56–63% coverage of MKRP1, and 43–55% coverage of HopZ1a (Figure 5). Thus, our acetylation analysis is conservative and it remains possible that HopZ1a acetylates additional residues of HINKEL and/or MKRP1 that we were unable to observe. Although HINKEL is acetylated within its kinesin motor domain at positions S88 and T90, the corresponding residues were not acetylated in MKRP1. The acetylation sites of MKRP1 are not present in HINKEL (not shown) and HINKEL has a C-terminal DUF3490 domain that is absent from MKRP1 (Figure 5). Thus, if acetylation of these two kinesins is an important function of HopZ1a in planta, they are likely to be regulated by contrasting mechanisms. Nevertheless, these data indicate that HopZ1a can target A. thaliana Kinesin 7 family members.
Overall, we believe that the library of T3SE-expressing yeast strains developed in this study represents a powerful resource to functionally characterize T3SE from P. syringae.
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada awards to D.S.G. and D.D; a Canada Research Chair in Plant-Microbe Systems Biology (D.D.) or Comparative Genomics (D.S.G.); the Centre for the Analysis of Genome Evolution and Function (D.D. and D.S.G.). In memory of Dr. Jianfeng Zhang.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7318505.
Communicating editor: D. Baltrus
Literature Cited
- Alemán A., Fernández-Piñar P., Pérez-Núñez D., Rotger R., Martín H., et al. , 2009. A yeast-based genetic screen for identification of pathogenic Salmonella proteins. FEMS Microbiol. Lett. 296: 167–177. 10.1111/j.1574-6968.2009.01630.x [DOI] [PubMed] [Google Scholar]
- Alfano J. R., Charkowski A. O., Deng W. L., Badel J. L., Petnicki-Ocwieja T., et al. , 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97: 4856–4861. 10.1073/pnas.97.9.4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., et al. , 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124: 133–145. 10.1016/j.cell.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Badel J. L., Nomura K., Bandyopadhyay S., Shimizu R., Collmer A., et al. , 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49: 1239–1251. 10.1046/j.1365-2958.2003.03647.x [DOI] [PubMed] [Google Scholar]
- Baganz F., Hayes A., Marren D., Gardner D. C., Oliver S. G., 1997. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast 13: 1563–1573. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., et al. , 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 7: 1017–1024. 10.1038/nmeth.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B. J., 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8: 437–449. 10.1038/nrg2085 [DOI] [PubMed] [Google Scholar]
- Botstein D., Fink G. R., 2011. Yeast: an experimental organism for 21st Century biology. Genetics 189: 695–704. 10.1534/genetics.111.130765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D., Bonas U., 2003. Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Plant Biol. 6: 312–319. 10.1016/S1369-5266(03)00064-5 [DOI] [PubMed] [Google Scholar]
- Chang J. H., Urbach J. M., Law T. F., Arnold L. W., Hu A., et al. , 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102: 2549–2554. 10.1073/pnas.0409660102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R., 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4: 811–825. 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., VanderSluis B., Koch E. N., Baryshnikova A., Pons C., et al. , 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353: aaf1420 10.1126/science.aaf1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curak J., Rohde J., Stagljar I., 2009. Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 12: 18–23. 10.1016/j.mib.2008.11.004 [DOI] [PubMed] [Google Scholar]
- DebRoy S., Thilmony R., Kwack Y. B., Nomura K., He S. Y., 2004. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA 101: 9927–9932. 10.1073/pnas.0401601101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Rivas S., 2012. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17: 644–655. 10.1016/j.tplants.2012.06.011 [DOI] [PubMed] [Google Scholar]
- Dixon S. J., Costanzo M., Baryshnikova A., Andrews B., Boone C., 2009. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 43: 601–625. 10.1146/annurev.genet.39.073003.114751 [DOI] [PubMed] [Google Scholar]
- Dolinski K., Botstein D., 2007. Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 41: 465–507. 10.1146/annurev.genet.40.110405.090439 [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez I., Yen T., Wade R. H., Kozielski F., 2004. Crystal structure of the motor domain of the human kinetochore protein CENP-E. J. Mol. Biol. 340: 1107–1116. 10.1016/j.jmb.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A., 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- Hohmann S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. 10.1128/MMBR.66.2.300-372.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. L., Balakrishnan R., Dong Q., Christie K. R., Park J., et al. , 2008. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 36: D577–D581. 10.1093/nar/gkm909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang M. S., Morgan R. L., Sarkar S. F., Wang P. W., Guttman D. S., 2005. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71: 5182–5191. 10.1128/AEM.71.9.5182-5191.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir Y., Guo M., Oh H. S., Petnicki-Ocwieja T., Chen S., et al. , 2004. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37: 554–565. 10.1046/j.1365-313X.2003.01982.x [DOI] [PubMed] [Google Scholar]
- Jin Q., Thilmony R., Zwiesler-Vollick J., He S. Y., 2003. Type III protein secretion in Pseudomonas syringae. Microbes Infect. 5: 301–310. 10.1016/S1286-4579(03)00032-7 [DOI] [PubMed] [Google Scholar]
- Joardar V., Lindeberg M., Jackson R. W., Selengut J., Dodson R., et al. , 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187: 6488–6498. 10.1128/JB.187.18.6488-6498.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G., Illes P., Beck M., Samaj J., 2011. Microtubules and mitogen-activated protein kinase signalling. Curr. Opin. Plant Biol. 14: 650–657. 10.1016/j.pbi.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Kramer R. W., Slagowski N. L., Eze N. A., Giddings K. S., Morrison M. F., et al. , 2007. Yeast functional genomic screens lead to identification of a role for a bacterial effector in innate immunity regulation. PLoS Pathog. 3: e21 10.1371/journal.ppat.0030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., et al. , 2009. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33: 53–63. 10.1016/j.molcel.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Lee A. H., Hurley B., Felsensteiner C., Yea C., Ckurshumova W., et al. , 2012. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 8: e1002523 10.1371/journal.ppat.1002523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. R., Liu B., 2004. Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiol. 136: 3877–3883. 10.1104/pp.104.052621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Abada W., Ma W., Guttman D. S., Desveaux D., 2008. The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J. Bacteriol. 190: 2880–2891. 10.1128/JB.01702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Guttman D. S., Desveaux D., 2009. The targeting of plant cellular systems by injected type III effector proteins. Semin. Cell Dev. Biol. 20: 1055–1063. 10.1016/j.semcdb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Lee A., Ma W., Zhou H., Guttman D. S., et al. , 2011. The YopJ superfamily in plant-associated bacteria. Mol. Plant Pathol. 12: 928–937. 10.1111/j.1364-3703.2011.00719.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Lee A. H., Hassan J. A., Wan J., Hurley B., et al. , 2013. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. USA 110: 18722–18727. 10.1073/pnas.1315520110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Wan J., Ford R., Gong Y., Fung P., et al. , 2012. Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics 13: 8 10.1186/1471-2164-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K. W., Jiang S., Hawara E., Lee D., Pan S., et al. , 2015. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 208: 1157–1168. 10.1111/nph.13528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Dong F. F., Stavrinides J., Guttman D. S., 2006. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2: e209 10.1371/journal.pgen.0020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., et al. , 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., et al. , 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold K. R., Martin M. E., Bronstein P. A., Collmer A., 2008. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol. Plant Microbe Interact. 21: 490–502. 10.1094/MPMI-21-4-0490 [DOI] [PubMed] [Google Scholar]
- Munkvold K. R., Russell A. B., Kvitko B. H., Collmer A., 2009. Pseudomonas syringae pv. tomato DC3000 type III effector HopAA1–1 functions redundantly with chlorosis-promoting factor PSPTO4723 to produce bacterial speck lesions in host tomato. Mol. Plant Microbe Interact. 22: 1341–1355. 10.1094/MPMI-22-11-1341 [DOI] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y. H., Pumplin N., Jones J., et al. , 2006. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. 10.1126/science.1129523 [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y. N., Imboden L. A., Chang J. H., et al. , 2011. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. 10.1073/pnas.1103338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien H. E., Desveaux D., Guttman D. S., 2011. Next-generation genomics of Pseudomonas syringae. Curr. Opin. Microbiol. 14: 24–30. 10.1016/j.mib.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Orth K., Palmer L. E., Bao Z. Q., Stewart S., Rudolph A. E., et al. , 1999. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science 285: 1920–1923. 10.1126/science.285.5435.1920 [DOI] [PubMed] [Google Scholar]
- Orth K., Xu Z., Mudgett M. B., Bao Z. Q., Palmer L. E., et al. , 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290: 1594–1597. 10.1126/science.290.5496.1594 [DOI] [PubMed] [Google Scholar]
- Richardson D. N., Simmons M. P., Reddy A. S., 2006. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7: 18 10.1186/1471-2164-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D., Dar D., Sreeramulu S., Sessa G., 2011. Expression of Xanthomonas campestris pv. vesicatoria type III effectors in yeast affects cell growth and viability. Mol. Plant Microbe Interact. 24: 305–314. 10.1094/MPMI-09-10-0196 [DOI] [PubMed] [Google Scholar]
- Salomon D., Sessa G., 2010. Identification of growth inhibition phenotypes induced by expression of bacterial type III effectors in yeast. J. Vis. Exp. 37: e1865 10.3791/1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Boudolf V., De Veylder L., Inze D., Genschik P., et al. , 2011. Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc. Natl. Acad. Sci. USA 108: 17844–17849. 10.1073/pnas.1110174108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifpoor S., van Dyk D., Costanzo M., Baryshnikova A., Friesen H., et al. , 2012. Functional wiring of the yeast kinome revealed by global analysis of genetic network motifs. Genome Res. 22: 791–801. 10.1101/gr.129213.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M., Lu Y. J., Porter K., Kvitko B. H., Henty-Ridilla J., et al. , 2016. The Pseudomonas syringae type III effector HopG1 induces actin remodeling to promote symptom development and susceptibility during infection. Plant Physiol. 171: 2239–2255. 10.1104/pp.16.01593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers K. A., Lesser C. F., 2008. The Yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4: 8–15. 10.1016/j.chom.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagowski N. L., Kramer R. W., Morrison M. F., LaBaer J., Lesser C. F., 2008. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 4: e9 10.1371/journal.ppat.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21: 319–330. 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Strompen G., El Kasmi F., Richter S., Lukowitz W., Assaad F. F., et al. , 2002. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12: 153–158. 10.1016/S0960-9822(01)00655-8 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y., 2010. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51: 1766–1776. 10.1093/pcp/pcq135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Wagih O., Usaj M., Baryshnikova A., VanderSluis B., Kuzmin E., et al. , 2013. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 41: W591–W596. 10.1093/nar/gkt400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., et al. , 2010. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107: 2349–2354. 10.1073/pnas.0904739107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Ye P., Peyser B. D., Pan X., Boeke J. D., Spencer F. A., et al. , 2005. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol Syst Biol 1: 2005 0026. 10.1038/msb4100034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Liu Z., Eyobo Y., Orth K., 2003. Yersinia effector YopJ inhibits yeast MAPK signaling pathways by an evolutionarily conserved mechanism. J. Biol. Chem. 278: 2131–2135. 10.1074/jbc.M209905200 [DOI] [PubMed] [Google Scholar]
- Youn J. Y., Friesen H., Nguyen Ba A. N., Liang W., Messier V., et al. , 2017. Functional Analysis of Kinases and Transcription Factors in Saccharomyces cerevisiae Using an Integrated Overexpression Library. G3 (Bethesda) 7: 911–921. 10.1534/g3.116.038471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. M., Chai J., 2008. Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11: 179–185. 10.1016/j.mib.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Zhu C., Dixit R., 2012. Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 249: 887–899. 10.1007/s00709-011-0343-9 [DOI] [PubMed] [Google Scholar]