Abstract
Small Tail Han Sheep is a highly valued local breed in China because of their precocity, perennial estrus, and high fecundity. The average annual lambing rate of ewes is as high as 180–270%, the semen of ram has characteristics of high yield, high density, and good motility. To reveal the key miRNAs and miRNA-targets underlying testis development and spermatogenesis in male Small Tail Han Sheep, integrated analysis of miRNA and mRNA expression profiles in 2-, 6-, and 12-month-old testes was performed by RNA-seq technology and bioinformatics methods. The results showed that total of 153 known sheep miRNAs and 2712 novel miRNAs were obtained in 2-,6 - and 12-month-old Small Tail Han Sheep testes; 5, 1, and 4 differentially expressed (DE) known sheep miRNAs, and 132, 105, and 24 DE novel miRNAs were identified in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, respectively. We combined miRNA results of this study and the mRNA results obtained in our previous study to predict the target mRNAs of DE known sheep miRNAs; 131, 10, and 15 target mRNAs of DE known sheep miRNAs and 76, 1, and 11 DE miRNA–targets were identified in the three groups, respectively. GO and KEGG analyses showed that: in 2- vs. 6-month-olds, the target genes of DE known sheep miRNAs were involved in 100 biological processes and 11 signaling pathways; in 6- vs. 12-month-olds, the target genes of DE known sheep miRNAs were involved in 4 biological processes; and in 2- vs. 12-month-olds, the target genes of DE known sheep miRNAs were involved in 17 biological processes and 4 signaling pathways. Three miR–target regulatory networks were constructed based on these DE miRNA–targets. The key miRNA-Targets involved in testis development and spermatogenesis were screened. 6 known sheep miRNAs and 6 novel miRNAs were selected to validate the accuracy of miRNA sequencing data by qRT-PCR. The binding sites of oar-miR-379-5p with WNT8A was validated by a dual luciferase reporter gene detection system.
Keywords: Small Tail Han Sheep, testis development, spermatogenesis, miRNAs, mRNAs
In recent years, the sheep industry has developed rapidly. Reproductive efficiency has a major impact on profitability. At present, improving the reproductive efficiency of sheep is an issue of great concern for farmers and herdsmen. Reproductive traits typically have low to medium heritability, the speed of genetic progress obtained by traditional breeding method is slow and the effect is not stable (Abdoli et al. 2016). This can be improved by molecular genetic marker and marker-assisted selection (Abdoli et al. 2013). Sheep reproductive traits are influenced by many factors, such as genetic background, age, season, environment, and nutrition (Cahill 1981; Makawi et al. 2007; Sezenler et al. 2011; Pickering et al. 2012; Canché et al. 2016). Reproductive traits are quantitative traits, which could be controlled by lots of minor genes or by some major genes (Drouilhet et al. 2010). Focusing on development of reproductive traits will have a long term effect on profitability in the sheep production (Pramod et al. 2013). A lot of researches have demonstrated that high fecundity of ewes is mainly related to some major fecundity genes, such as BMP15, BMPR1B, GDF9, B4GALNT2, ESR1, FSHR, POU1F1, OLKUSKA and BELLE-ILE (Liu et al. 2014; Abdoli et al. 2016). There is little research to select the molecular markers in ram. Small Tail Han Sheep is a highly valued local breed in China that has excellent breeding characteristics, such as large size, early sexual maturity, perennial estrus, and multiple births (Chu et al. 2011). The average annual lambing rate of ewes is as high as 180–270%, and the semen of ram has characteristics of high yield, high sperm density, and good sperm motility. Male Small Tail Han Sheep can be used as a good research subject. Sperm quality is closely related to testis tissue structure, testis development and spermatogenesis are obvious factors that affect male reproductive efficiency (Gong et al. 2013; Zhu et al. 2016). Therefore, understanding the molecular regulatory mechanisms and finding out the molecular marker underlying testis development and spermatogenesis in Small Tail Han Sheep will provide theoretical basis for improving male sheep reproductive efficiency.
With the wide application of molecular biology technology in animal genetics and breeding, it is possible to systematically clarify the molecular regulatory mechanisms underlying gene functions (Miao and Qin 2015). Gene regulation is at the core of all biological behaviors and phenotypes, which is a multi-mode spatial and temporal dynamic process and mainly regulated at transcriptional and post-transcriptional levels (Wang et al. 2016). Generally, transcriptome refers to the sum of all RNAs transcribed by an organism in a particular environment or physiological state, including mRNA and noncoding RNA (such as rRNA, tRNA, microRNA, siRNA, piRNA, circle RNA, and lncRNA). miRNAs are important noncoding RNAs that regulate gene expression and participate in many biological and metabolic processes, such as growth, development, reproduction, and disease, by specifically binding to 3′untranslated regions (UTRs) of mRNA to degrade a target gene or inhibit target gene translation (Ambros 2004; Ran et al. 2014). Increasing evidence has shown that miRNAs play key roles in testis development and spermatogenesis (Kotaja 2014; Pratt and Calcatera 2016). Testis development and spermatogenesis have obvious stage specificity, miRNA expression changes with testis development and spermatogenic cell development (Zhang et al. 2010; Lian et al. 2012; Wu et al. 2014; Kasimanickam and Kasimanickam 2015; Kasimanickam and Kasimanickam 2015; Tariq et al. 2016). To date, many miRNAs have been identified and studied in different species, such as humans, mice, pig, cattle and chicken (Yan et al. 2007; Zhang et al. 2010; Huang et al. 2011; Luo et al. 2015; Wang and Xu 2015; Wu et al. 2017b). However, miRNAs identification and molecular mechanism exploration has not been investigated in sheep testis as in other livestock species. Therefore, it is necessary to investigate miRNA and mRNA expression profiles to understand the molecular regulatory mechanism and identify the key miRNA-targets underlying testes development and spermatogenesis in Small Tail Han Sheep.
In our previous publication, histological structure and mRNA expression profiles of 2-, 6-, and 12-month-old Small Tail Han Sheep testes were investigated using hematoxylin–eosin staining and RNA-seq technology, respectively (Bai et al. 2017). In this study, we analyzed miRNA expression profiles in 2-, 6-, and 12-month-old Small Tail Han Sheep testes, integrated the miRNA expression profile in this study and the mRNA expression profile obtained in our previous study to construct miRNA–target regulatory networks, selected the key miRNAs and their target mRNAs involved in testis development and spermatogenesis in Small Tail Han Sheep. These results of our study can help to identify the molecular markers affecting reproductive efficiency of male Small Tail Han Sheep and provide theory evidence for further finding out the molecular markers affecting male sheep reproductive ability.
MATERIALS AND METHODS
Sample preparation
Two-, 6-, and 12-month-old male Small Tail Han Sheep from the nucleus herd bred by Songyuan Sheng Hua Animal Husbandry Co. Ltd (Jilin, China) were used for this experiment. Three different rams of each age were selected for sample collection. Left testis of each ram was collected by surgical operation and immediately frozen in liquid nitrogen. Intratesticular tissues were used for total RNA extraction. Samples were the same as those used in our previous study (Bai et al. 2017).
miRNA library construction and sequencing
Total RNA was extracted from testis tissues using Trizol Reagent (Invitrogen, USA). RNA integrity number (RIN) and RNA quality were determined using Agilent Tapestation 2200 (Agilent, USA) and NanoDrop 2000 (Thermo Scientific, USA) instruments. RNA samples with RIN ≥6.0 were considered acceptable for miRNA library construction. The miRNA library of each sample was prepared using an Ion Total RNA-seq Kit v2 (Life Technologies, USA) following the manufacturer’s instructions. Briefly, 13-34nt RNAs were captured and enriched using magnetic beads with oligo (dt); subsequently, 13-34nt RNAs were fractionated into short fragments using RNase III and an Ion adaptor. The short RNA fragments were ligated with 3′ and 5′ adaptors. Then, the short mRNA fragments were reverse-transcribed and amplified to synthetize double-stranded cDNA. Emulsion PCR was performed using the miRNA cDNA library as the template. RNA-seq was conducted on an ABI Ion Proton instrument by NovelBio Bio-Pharm Technology Co. Ltd (Shanghai, China). Raw sequencing data were assessed by Fast-QC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) (Ramayo-Caldas et al. 2012).
DE miRNA analysis
The raw sequencing reads were filtered, and the clean reads were mapped to miRBase 21.0 (http://www.mirbase.org/) to identify known sheep miRNAs (oar-miRNAs). Then, the remaining clean reads that were not mapped to sheep miRBase were mapped to sheep genome Oar3.1 to predict sheep novel miRNAs by miRDeep 2.0.0.8. The remaining clean reads that did not map to sheep genome Oar3.1 were mapped to human, rat, and mouse miRBase using the BWA algorithm to predict novel miRNAs. Only the mapped reads were used for miRNA expression analysis. miRNA expression levels were quantified by the reads per kilobase of transcript per million mapped reads (RPKM) method. The RPKM value was used to calculate expression, and the upper-quartile algorithm was used to correct expression to help produce accurate results for some miRNAs that occurred in low abundance. The DE miRNAs were identified by the EB-Seq algorithm (criteria:|log2FoldChange|>0.585; false discovery rate, FDR < 0.05); FDR was calculated to correct the P-value (Wright and Simon 2003).
Venn, Series-cluster and hierarchical clustering analysis of DE miRNAs
Venn analysis was performed to visualize the DE mRNA and miRNA changes by stage using Microsoft Excel 2016. Series-cluster analysis was performed to identify the expression profiles of miRNAs using the STEM program (http://www.cs.cmu.edu/∼jernst/st/). The DE miRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes were categorized into 8 expression profiles. Hierarchical cluster analysis was performed with Cluster 3.0 and Tree View 1.6 programs (http://rana.lbl.gov/eisen).
Target prediction of DE miRNAs and integrative analysis
The TargetScan and miRanda algorithms were used to predict putative target mRNAs of DE miRNAs. The putative target mRNAs of DE miRNAs were crossed with DE mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, respectively, which were obtained in our previous study (Bai et al. 2017). Negative correlation analysis was performed to determine the relation of DE miRNAs and mRNAs. Pearson correlation coefficients were used to determine whether the expression levels of each particular miRNA and its target mRNAs were negatively correlated (correlation < 0, P < 0.05, FDR < 0.05). If a miRNA and its target mRNA were negatively correlated, the mRNA was identified as a candidate target of miRNA.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
The GO database (http://geneontology.org/) was used to analyze the main functions of candidate target mRNAs. Biological process (BP) terms were annotated for candidate target mRNAs. Similarly, the KEGG database (http://www.genome.jp/kegg/) was used to analyze the pathways in which the candidate target mRNAs are involved. Fisher’s exact and χ2 tests were used to classify the significant GO terms and pathways of DE mRNAs (P < 0.05), and FDR was calculated to correct the P-value (Wang et al. 2014). All of the candidate target mRNAs were analyzed using the GO and KEGG pathway databases.
miRNA–target regulatory network construction
To identify all possible miRNA–mRNA interactions, miRNA–target regulatory networks were built with Cytoscape 2.8.3. The expression values for DE miRNAs and their candidate target mRNAs were imported into Cytoscape to generate and visualize functional networks.
RNA-seq data validation
Six known sheep miRNAs and 6 novel mRNAs were randomly selected and analyzed to validate RNA-seq data using qRT-PCR. Synthesized cDNA was used as the template for qRT-PCR. qRT-PCR amplification for miRNAs was performed using the miRcute Plus miRNA qPCR Detection Kit (FP411, TIANGEN BIOTECH, China). The 20-µl reaction system contained 10µl 2×miRcute Plus miRNA Premix, 0.4 µl universal reverse primer (200nM) provided by the kit, 0.4 µl miRNA sequence-specific forward primer (200nM), 2µl cDNA, and 7.2 µl ddH2O. The reaction conditions were 95° for 15 min, and 40 cycles of 94° for 20s, 60° for 34s; the reaction was completed with a melting curve analysis according to the manufacturer’s instructions. qRT-PCR was performed on a Bio-Rad CFX96 system (Bio-Rad, USA). All qRT-PCRs were completed with a melting curve analysis. The 2−ΔΔCt method and internal control gene U6 was used to calculate miRNA expression levels. Specific primers were designed and synthesized by Sangon Biotech (Shanghai, China); primer information is shown in Table 1.
Table 1. qRT-PCR primer information.
| Name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| hsa-miR-3529-5p | ACGAGGTAGACTGGGATTTGTTGT T | Universal reverse* |
| hsa-miR-6820-3p | TGTGACTTCTCCCCTGCCACAG | Universal reverse* |
| hsa-miR-3615 | CTCTCGGCTCCTCGCGGC | Universal reverse* |
| mmu-miR-1957b | CGCGTGGAATGTAACTCAGTGGTA T | Universal reverse* |
| mmu-miR-182-3p | CGGTGGTTCTA ACTTGCCAA CT | Universal reverse* |
| mmu-miR-1929-3p | CAGCTCATGGAGACCTAGGTGG | Universal reverse* |
| oar-miR-379-5p | CTGGTAGACTATGGAACGTAGGC | Universal reverse* |
| oar-miR-665-3p | CCAGTAGGCCGAGGCCCC | Universal reverse* |
| oar-miR-200b | ACGCTAATACTGCCTGGTAATGATG | Universal reverse* |
| oar-miR-409-5p | CGAATGTTGCTCGGTGAACCCC | Universal reverse* |
| oar-miR-495-3p | AGCGAAACAAACATGGTGCACTTCTT | Universal reverse* |
| oar-miR-200c | CGTAATACTGCCGGGTAATGATGG | Universal reverse* |
| U6 | CGAGGATGTGAAGACACCAAGAC | Universal reverse* |
Universal reverse was provided by manufacture (miRcute Plus miRNA qPCR Detection Kit, TIANGEN BIOTECH, China).
Cell culture
HEK-293T cells were resuscitated and cultured in DMEM containing 10% fetal bovine serum, 1.5 mM L-glutamine, 100U/ml penicillin, and 100μg/ml streptomycin in a 5% CO2 saturated humidity incubator at 37°. Adherent cells were passaged daily with 0.05% trypsin–EDTA.
Plasmid vector construction and transfection
The sequence fragment of WNT8A-3′UTR that binds to oar-miR-379-5p (wild-type), WNT8A-3′UTR with an oar-miR-379-5p binding site mutation (mutant-type), and positive control (miRNA inhibitor) were designed by Target Scan 7.1 and RNA22 v2.0, and synthesized by total gene synthesis. Sequence information of wild- and mutant-type target gene fragments and the miRNA inhibitor fragment are shown in Table 2. The pmirGLO Dual-Luciferase miRNA Target Expression Vector was used to confirm the putative binding site of WNT8A-3′UTR with oar-miR-379-5p. pmirGLO was digested by restriction endonuclease SacI and XhoI. pmirGLO and target gene fragments were ligated by T4 DNA ligase. 293T cells were washed and incubated with 4ml serum-free medium for 24h. The miRNA (or miRNA inhibitor) was mixed with 250µl serum-free medium and 1.6µg recombinant vector, and Lipofectamine 2000 transfection reagent (Life Technologies, USA) was mixed with 250µl serum-free medium. Then, the 2 mixtures were combined and incubated for 25min at room temperature. The Lipofectamine 2000–miRNA mixture was added to the cells and incubated at 37° for 5h. Subsequently, 500µL DMEM containing 10% fetal bovine serum was added to the cells and incubated in 5% CO2 at 37°. The cells were collected after culturing for 24h and 48h. Then, 7 experimental groups were set up: WNT8A-WT+mimics NC, WNT8A-WT+oar-miR-379-5p, WNT8A-MUT+mimics NC, WNT8A-MUT+oar-miR-379-5p, PC+mimics NC, PC+ oar-miR-379-5p, and a blank group. There were 3 replicates of each group.
Table 2. Sequence information of wild- and mutant-type target gene fragments and positive control fragments.
| Gene name | Sequence information (5′-3′) |
|---|---|
| WNT8A-WT | Sense: CGGAAGTTGGCATCTCAAGAAAAACCATAAGCAGGTTCTTTGCAAGTCTACCCTTATCTCTGTTTTGC |
| Antisense: TCGAGCAAAACAGAGATAAGGGTAGACTTGCAAAGAACCTGCTTATGGTTTTTCTTGAGATGCCAACTTCCGAGCT | |
| WNT8A-MUT | Sense: CGGAAGTTGGCATCTCAAGAAAAACCATAAGCAGGTTGTAAGCATCAGATGGCTTATCTCTGTTTTGC |
| Antisense: TCGAGCAAAACAGAGATAAGCCATCTGATGCTTACAACCTGCTTATGGTTTTTCTTGAGATGCCAACTTCCGAGCT | |
| PC (oar-miR-379-5p inhibitor) | Sense: CGCCTACGTTCCATAGTCTACCAACCGGTGCCTACGTTCCATAGTCTACCAC |
| Antisense: TCGAGTGGTAGACTATGGAACGTAGGCACCGGTTGGTAGACTATGGAACGTAGGCGAGCT |
Dual luciferase reporter detection
Luciferase and Renilla activities were measured using a Dual-Luciferase Reporter Assay Kit (Promega Corporation, USA) after transfection for 24h and 48h, respectively.
Statistical analysis
The data were analyzed using SPSS 18.0, and all results were presented as means ± SD. The significance of differences between groups were assessed by ANOVA. Differences were considered statistically significant when P < 0.05.
Data availability
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number: GSE107803. All data supporting the results of this article are contained within the article and its Supplemental Materials. Table S1, miRNA Sequence Quality Control. Table S2, read mapping summary of known sheep miRNA. Table S3, the DE novel miRNAs and DE sheep known miRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S4, the union of DE miRNA in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S5, predicted target mRNAs of DE known sheep miRNAs. Table S6, DE miRNA-Targets. Table S7, GO-analysis of candidate target mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S8, pathway analysis of candidate target mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S9, GO and pathway terms that WNT8A involved in related to testis development and spermatogenesis. Supplemental figure 1, Hierarchical clustering of DE mRNAs and DE miRNAs in testis samples. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7473047.
RESULTS
RNA-seq data summary
The miRNA sequence quality control results are shown in Supplementary Table S1. A total of 11.3, 9.6, 16.4, 9.2, 5.7, 6.7, 8.7, 9.3, and 23.8 million high-quality clean reads of miRNAs were obtained from 2- (A1, A2, A3), 6- (B1, B2, B3), and 12-month-old (C1, C2, C3) testes, respectively. For each sample, 1.8–11.5% of miRNA reads were mapped to sheep miRbase (Table S2). Because of the limited information contained in sheep miRBase, the miRNA mapping rate was low.
DE miRNAs and hierarchical clustering analysis
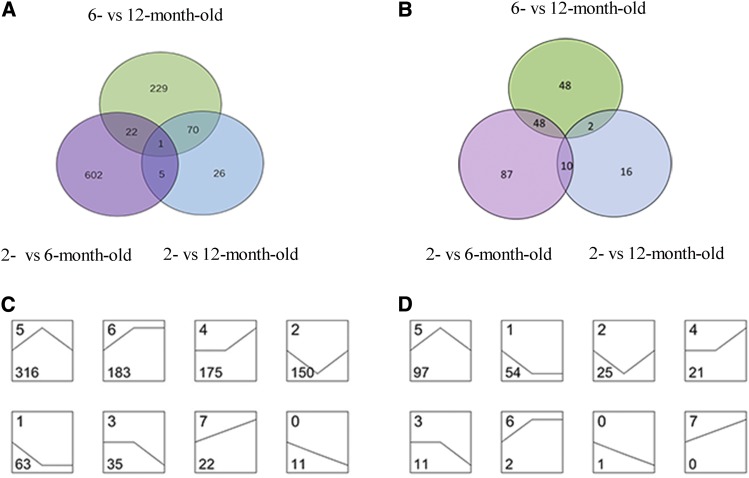
In our previous publication, we obtained 630, 322, and 102 DE mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, respectively; in total, there were 955 DE mRNAs (Bai et al. 2017). In this study, we obtained 5, 1, and 4 DE known sheep miRNAs, and 132,105, and 24 DE novel miRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, respectively (Table S3). In total, 211 DE miRNAs were identified in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, as shown in Supplementary Table S4.
In Venn analysis, DE mRNA and miRNA changes can be visualized by stage (Figure 1 a b). In series-cluster analysis, we categorized the 955 DE mRNAs and 211 DE miRNAs into 8 possible expression profiles to observe the overall expression tendencies (Figure 1c d). Subsequently, based on these DE mRNAs and DE miRNAs, heat maps were generated by hierarchical clustering analysis (Supplemental figure 1 a b).
Figure 1.
Venn and series-cluster analyses of DE mRNAs and DE miRNAs in 2-, 6-, and 12-month-old testes. a. Venn diagram of DE mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. b. Venn diagram of DE miRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. c. Eight profiles of DE mRNAs in 2-, 6-, and 12-month-old testes. d. Eight profiles of DE miRNAs in 2-, 6-, and 12-month-old testes. Profile number is shown at the top left corner of each square. Number of DE mRNAs or miRNAs grouped in each profile is shown at the bottom left corner of each square. Profiles are ordered based on the number of mRNAs or miRNAs assigned.
Target prediction of DE miRNAs and integrative analysis
In total, 131, 10, and 15 predicted target mRNAs of DE known sheep miRNAs were obtained from 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, as shown in Table S5. Combined with DE mRNAs, the predicted target mRNAs and DE mRNAs were crossed, and significantly DE target mRNAs were retained. In total, 76, 1, and 11 candidate target mRNAs of DE known sheep miRNAs were identified in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes, respectively (Table S6). For some DE miRNAs, only one target mRNA was identified, however, most of DE miRNAs can target several mRNAs. In addition, several miRNAs can target one mRNA. For example, oar-miR-379-5p targeted 16 different mRNAs; however, GRIK5 is targeted by 2 miRNAs (oar-miR-409-3p and oar-miR-665-3p).
Candidate target mRNA GO analysis
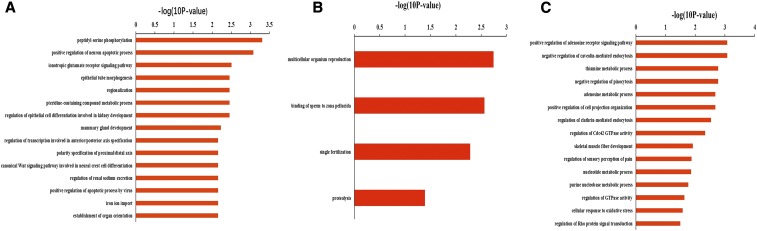
The GO analysis results are shown in Table S7. In 2- vs. 6-month-old testes, the candidate target mRNAs were significantly enriched in 100 BP terms, such as apoptotic process, meiosis I, regulation of acrosome reaction, protein phosphorylation, and epithelial cell maturation; the top 15 BP terms are shown in Figure 2 a: peptidyl-serine phosphorylation, positive regulation of neuron apoptotic process, ionotropic glutamate receptor signaling pathway, epithelial tube morphogenesis, regionalization, pteridine-containing compound metabolic process, regulation of epithelial cell differentiation involved in kidney development, mammary gland development, regulation of transcription involved in anterior/posterior axis specification, polarity specification of proximal/distal axis, canonical WNT signaling pathway involved in neural crest cell differentiation, regulation of renal sodium excretion, positive regulation of apoptotic process by virus, iron ion import, and establishment of organ orientation. In 6- vs. 12-month-old testes, the candidate target mRNAs were significantly enriched in 4 BP terms, as shown in Figure 2 b: multicellular organism reproduction, binding of sperm to zona pellucida, single fertilization, and proteolysis. In 2- vs. 12-month-old sheep testes, the candidate target mRNAs were significantly enriched in 17 BP terms, such as adenosine metabolic process, nucleotide metabolic process, purine nucleobase metabolic process, cellular response to oxidative stress, and RNA processing; the top 15 BP terms are shown in Figure 2 c: positive regulation of adenosine receptor signaling pathway, negative regulation of caveolin-mediated endocytosis, thiamine metabolic process, negative regulation of pinocytosis, adenosine metabolic process, positive regulation of cell projection organization, regulation of clathrin-mediated endocytosis, regulation of Cdc42 GTPase activity, skeletal muscle fiber development, regulation of sensory perception of pain, nucleotide metabolic process, purine nucleobase metabolic process, regulation of GTPase activity, cellular response to oxidative stress, and regulation of Rho protein signal transduction.
Figure 2.
Top 15 BP terms of candidate target mRNAs. a. Two- vs. 6-month-old testes; b. 6- vs. 12-month-old testes; c. 2- vs. 12-month-old testes; P < 0.05.
Candidate target mRNA pathway analysis
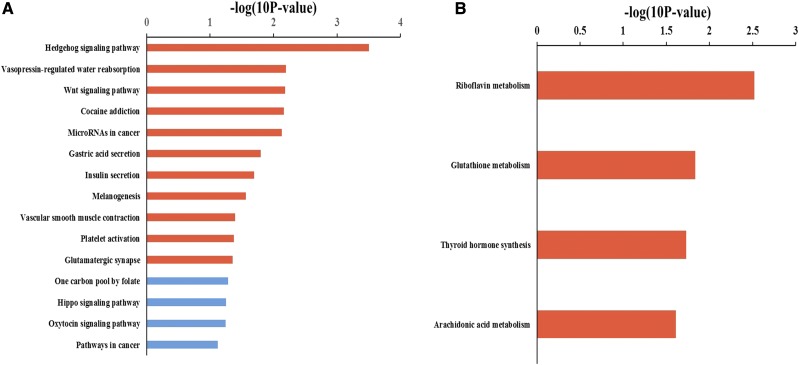
The results of pathway analysis are shown in Table S8 and Figure 4. In 2- vs. 6-month-old testes, the candidate target mRNAs were significantly involved in 11 pathways, which are shown in Figure 3 a: Hedgehog signaling pathway, vasopressin-regulated water reabsorption, WNT signaling pathway, cocaine addiction, microRNAs in cancer, gastric acid secretion, insulin secretion, melanogenesis, smooth muscle contraction, platelet activation, and glutamatergic synapse. In 6- vs. 12-month-old testes, there was only one candidate target mRNA of DE known sheep miRNAs that was not enriched in any signaling pathway. In 2- vs. 12-month-old testes, the candidate target mRNAs were significantly involved in 4 pathways, as shown in Figure 3 b: riboflavin metabolism, glutathione metabolism, thyroid hormone synthesis, and arachidonic acid metabolism.
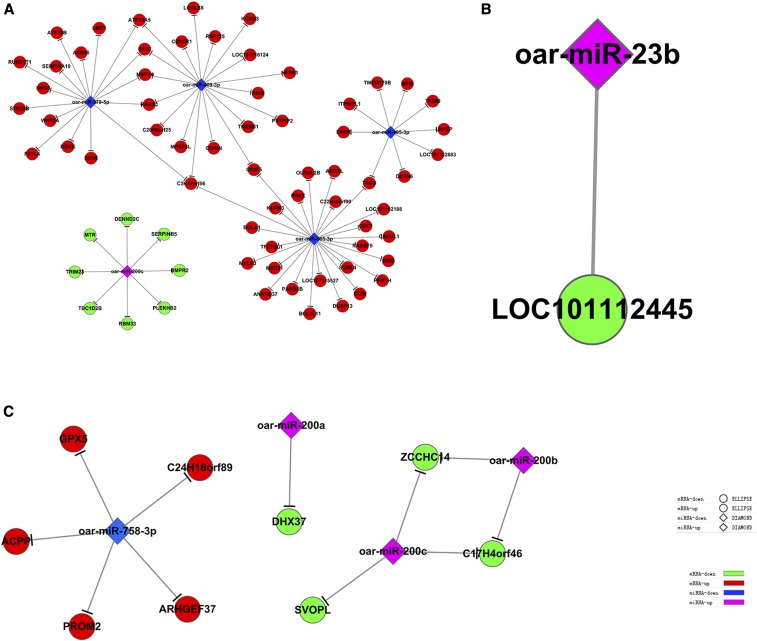
Figure 4.
miRNA-Target regulatory networks in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. a. Two- vs. 6-month-old testes; b. 6- vs. 12-month-old testes; c. 2- vs. 12-month-old testes.
Figure 3.
Top 15 pathways of candidate target mRNAs. a. Two- vs. 6-month-old testes; b. 2- vs. 12-month-old testes; red terms represent P < 0.05.
miRNA–target regulatory networks
miRNA–target regulatory networks in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes were generated as shown in Figure 4. miRNA–target regulatory networks contained networks of downregulated miRNAs with upregulated mRNAs and networks of upregulated miRNAs with downregulated mRNAs in different testis development stages. In 2- vs. 6-month-old testes, 4 downregulated miRNAs with 60 upregulated mRNAs and 1 upregulated miRNA with 8 downregulated mRNAs were identified (Figure 4 a); in 6- vs. 12-month-old testes, 1 upregulated miRNA with 1 downregulated mRNA was identified (Figure 4 b); and in 2- vs. 12-month-old testes, 3 upregulated miRNAs with 3 downregulated mRNAs and 1 downregulated miRNA with 5 upregulated mRNAs were identified (Figure 4 c).
RNA-seq data validation
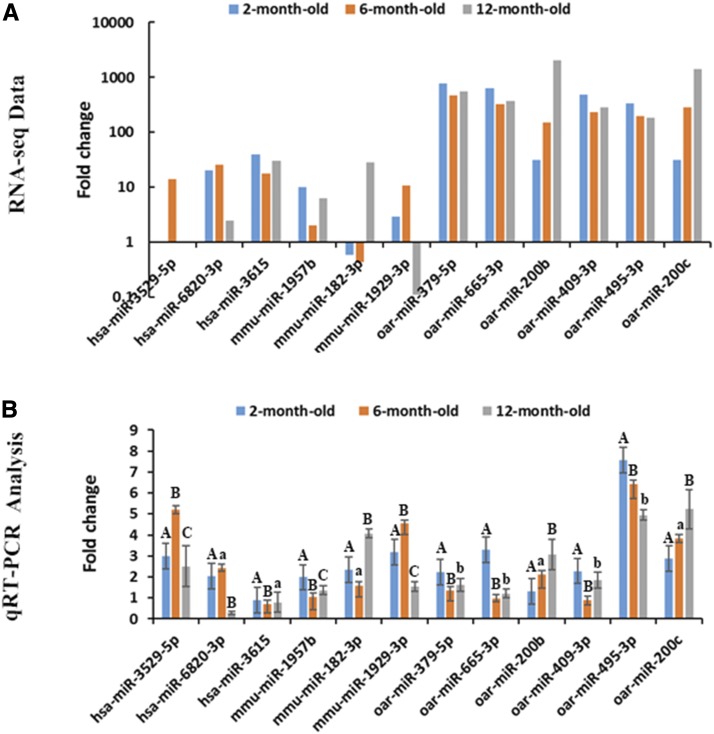
Six known sheep miRNAs (oar-miR-379-5p, oar-miR-665-3p, oar-miR-409-5p, oar-miR-495-3p, oar-miR-200b, and oar-miR-200c) and six novel miRNAs (hsa-miR-3529-5p, hsa-miR-6820-3p, hsa-miR-3615, mmu-miR-1957b, mmu-miR-182-3p, and mmu-miR-1929-3p) were randomly selected to validate RNA-seq data. qRT-PCR was performed to detect DE miRNA expression. The qRT-PCR results consistent with RNA-seq data (Figure 5 a b).
Figure 5.
qRT-PCR validation of RNA-seq data for miRNAs. a. RNA-seq data. Values were calculated and normalized by EB-Seq algorithm if the fold changes were >1.5 and FDR < 0.05. b. RT-PCR analysis of 6 novel miRNAs and 6 known miRNAs. The fold change in each time point was relative to 2 months. The expression values were calculated by the 2−ΔΔCt method. Different capital letters represent extremely significant differences (P < 0.01); different letters indicate significant differences (P < 0.05), whereas the same letters indicate no significant differences (P > 0.05).
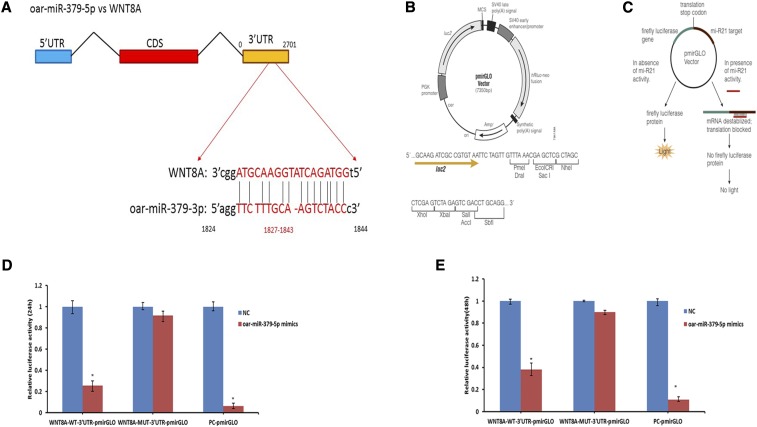
oar-miR-379-5p targets WNT8A
According to GO and pathway analysis, 78 target mRNAs of 10 DE known sheep miRNAs in 3 groups were further studied to identify the testis development- and spermatogenesis-related biological processes and signaling pathways in which these target mRNAs were involved. It was found that WNT8A was closely related to tissue development processes, such as multicellular organismal development, cell morphogenesis, and cell fate commitment, and signaling pathways such as WNT, melanogenesis, and Hedgehog signaling pathways; the GO and pathway terms associated with WNT8A are shown in Table S9. Thus, oar-miR-379-5p and WNT8A were selected to verify their targeted regulatory relationship. The binding site of WNT8A-3′UTR with oar-miR-379-5p is shown in Figure 6 a. pmirGLO plasmid vector and its working principle are shown in Figure 6 b c. Luciferase assays were performed to confirm the potential binding between oar-miR-379-5p and WNT8A to further validate the interaction of miRNA–mRNA. The luciferase activity of the WNT8A receptor decreased 74.2% and 61.8% upon co-transfection with oar-miR-379-5p mimics (P < 0.05) compared with co-transfection with negative control mimics at 24h and 48h, respectively (Figure 6 d e), which indicated that oar-miR-379-5p can directly target WNT8A-3′UTR.
Figure 6.
Detection of interactions between oar-miR-379-5p and WNT8A by dual luciferase reporter system. a. Binding site sequence of miRNA and target gene WNT8A. b. Structure of pmirGLO plasmid vector. c. Working principle of pmirGLO plasmid vector. The dual luciferase reporter gene was inserted the 3′UTR sequence of the target gene, which can bind with miRNA into the downstream of the luciferase gene in pmirGLO plasmid vector, the translation of luciferase from firefly will be inhibited and the luciferase activity will eventually decrease when endogenous miRNA or introduced exogenous miRNA are combined with the inserted 3′UTR sequence. d. Relative luciferase activity after transfecting oar-mir-3799-5p and WNT8A recombinant plasmid for 24h. e. Relative luciferase activity after transfecting oar-mir-3799-5p and WNT8A recombinant plasmid for 48h.
DISCUSSION
Sheep reproductive efficiency is important for livestock husbandry; testis development and spermatogenesis are key factors influencing male sheep reproductive efficiency, many functional genes regulate testis development and spermatogenesis (Gong et al. 2013; Ding et al. 2016). The aim of this study was to identify the key miRNAs and functional miRNA-targets in developing testis of Small Tail Han Sheep using integrated analysis of miRNA and mRNA expression profiles. The analysis will help us understanding the molecular regulating mechanism in testis development and reveal the molecular markers related to reproductive efficiency of male Small Tail Han Sheep.
In this study, 2-, 6-, and 12-month-old corresponding to infant, adolescent and adult stages in male Small Tail Han Sheep presenting the dynamic process of testis development. Only 5, 1, and 4 DE known sheep miRNAs were identified in 2- vs. 6-, 6-vs 12-, and 2-vs 12-month-old testes, respectively, which is because of the limited information in sheep miRBase. Novel miRNAs were predicted from sheep genome, human, rat, and mouse miRBase; therefore, more DE novel miRNAs were identified than DE known sheep miRNAs. DE novel miRNAs can help to enrich our knowledge about the key miRNAs involved in sheep testis development and provide new references for the sheep miRBase. The DE known sheep miRNAs were reported in previous studies, miR-409-3p, miR-665-3p, miR-379-5p, miR-495-3p, miR-23b, and miR-758-3p are all involved in cell proliferation, differentiation, migration, and apoptosis (Weng et al. 2012; Dong et al. 2016; Chen et al. 2017a; Chen et al. 2017b; Jiang et al. 2017; Xie et al. 2017). Based on our results, we can infer that oar-miR-409-3p, oar-miR-665-3p, oar-miR-379-5p, oar-miR-495-3p, oar-miR-23b, and oar-miR-758-3p may play key roles in growth, proliferation, and apoptosis of testis cells. oar-miR-200a, oar-miR-200b, and oar-miR-200c belong to the miR-200 family, which plays a key role in epithelial–mesenchymal transition (Mongroo and Rustgi 2010; Pan et al. 2017). It can be conferred that oar-miR-200a, miR-200b, and miR-200c may be associated with seminiferous epithelium development. Currently, these miRNAs are mainly investigated in the context of invasion and migration of cancer, the specific functions of these miRNAs in sheep testes need to be further explored and validated.
GO and KEGG pathway analyses can help elucidate the functions and pathways that candidate target genes involved in. As our results shown: in 2- vs. 6-month-old testes, most of the GO terms were related to spermatogenesis (such as apoptotic process, meiosis I, and acrosome reaction), and most of the pathways were related to tissue development (such as Hedgehog and WNT signaling pathways); in 6- vs. 12-month-old testes, GO terms were related to male reproductive processes (multicellular organism reproduction, binding of sperm to zona pellucida, single fertilization, and proteolysis). In 2- vs. 12-month-old sheep testes, most GO terms and pathways were related to testis metabolic and biosynthesis processes.
Integrative analysis of miRNA and mRNA expression profiles can help elucidate molecular regulatory mechanisms (Miao and Qin 2015). This study is the first attempt to identify key miRNA–targets by integrated analysis of miRNA and mRNA expression profiles in Small Tail Han Sheep testes. Regulatory networks of DE known sheep miRNAs and their target mRNAs can help us more precisely identify the functions of these miRNAs in testis development and spermatogenesis in sheep. In this study, 3 miRNA–target networks were constructed and several miRNA–targets were identified. According to GO and pathway analysis, WNT8A (the target of oar-miR-379-5p) was determined to be involved in biological processes such as multicellular organismal development, endoderm development, and palate development, and signaling pathways such as the WNT, Hedgehog, and Hippo signaling pathways. It was previously reported that miR-379 is a key miRNA that inhibits proliferation, invasion, and migration of tumor cells, and plays an important role in tumor occurrence and development; miR-379 can also inhibit proliferation, invasion, and migration of vascular smooth muscle cells by targeting IGF-1 and regulate breast and liver cancer processes by inhibiting the expression of Cyclin B1 and IGF-1R (Khan et al. 2013; Li et al. 2017a). Studies have shown that miR-379-5p expression was significantly downregulated in cancer tissues, such as liver, bladder, and breast cancer tissues, and closely related to tumor stage; moreover, miR-379-5p can also inhibit the transformation and metastasis of endothelium to mesenchymal cells in liver cancer (Chen et al. 2016; Wu et al. 2017a). To date, miR-379 has been widely studied in human cancer, and its research on reproductive organ development has not been reported yet. WNT8A is a member of the WNT ligand family, which participates in the WNT, Hedgehog, and Hippo signaling pathway. Most studies reported that WNT8A gene plays a vital role on embryonic development and early organ formation (Narayanan 2012; Vendrell et al. 2013; Wylie et al. 2014; Hino et al. 2018). It is reported that the WNT pathway is involved in the major events during testis development, including primordial germ cell specification, proliferation and migration, testis determination, spermatogenesis, and somatic cell regulation (Dong et al. 2015). It was also reported that the WNT signaling pathway is involved in male reproductive organ development (Sun and Wang 2003), the Hedgehog signaling pathway is involved in testicular cord and spermatogonial stem cell development (Sahin et al. 2014; Li et al. 2017b), the Hippo signaling pathway is involved in regulating growth, proliferation, and apoptosis of cells, and regulating proliferation and maintenance of stem cells to control organ size (Pan 2007; Zhang et al. 2008). Therefore, it can be inferred that oar-miR-379-5p may be involved in Small Tail Han Sheep testis development and spermatogenesis by downregulated WNT8A, which involved in these pathways.
Currently, the dual luciferase reporter gene method is the most convenient method for verifying the relationship between miRNAs and their target mRNAs. The results demonstrated that oar-miR-379-5p can decrease WNT8A expression by targeting WNT8A-3′UTR. It is the first time to verify that WNT8A is the target gene of oar-miR-379-5p. However, the hypothesis that oar-miR-379-5p is involved in testicular development and spermatogenesis by targeting WNT8A needs to be further studied and verified. The results of this study will provide theory evidence for identifying the molecular markers in Small Tail Han Sheep testis development and further finding out the molecular markers affecting male sheep reproductive ability.
CONCLUSION
This study is the first integrative analysis of miRNA and mRNA expression profiles in Small Tail Han Sheep testis development. We identified the key miRNAs and miRNA–targets in 2- vs. 6-, 6-vs 12-, and 2-vs 12-month-old testes, which will provide theoretical basis for elucidating the molecular regulatory mechanisms underlying sheep testis development and identifying the biomarkers that affect reproductive efficiency of male Small Tail Han Sheep. Our study is the first to reveal that WNT8A was targeted by oar-miR-379-5p. We present a hypothesis that oar-miR-379-5p is involved in sheep testis development and spermatogenesis by targeting WNT8A and we will further investigate and verify this hypothesis.
ACKNOWLEDGMENTS
This work was supported by Jilin Province Science and Technology Development Project (20160204018NY), Jilin Province modern sheep industry technology system project, Jilin Province Department of Education“13th Five-Year” Science and Technology Development Project (JJKH20180691). We appreciate the Jilin Sheng Hua Animal Husbandry Co. Ltd for providing the experimental animals and Shanghai Novel Bio-Pharm Technology Co. Ltd for help with the RNA-sequencing.
Ethics approval and consent to participate
Experimental animals were provided Songyuan Sheng Hua Animal Husbandry Co. Ltd (Jilin, China). The methods were carried out in accordance with relevant guidelines from the Ministry of Agriculture of the People’s Republic of China. All experimental protocols were approved by the Jilin Laboratory Animal Specialized Committee.
Authors’ contributions
MB and HZJ conceived and designed the experiments. MB, LMS, CJ, JRL, YH, and HL performed the experiments. MB analyzed the data, and wrote the paper. YC gave some pertinent suggestions on the experiment and writing. All authors read and approved the final manuscript.
Competing interests
The authors declare that there are no competing interests.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7473047.
Communicating editor: D. J. de Koning
Literature Cited
- Abdoli R., Zamani P., Deljou A., Rezvan H., 2013. Association of BMPR-1B and GDF9 genes polymorphisms and secondary protein structure changes with reproduction traits in Mehraban ewes. Gene 524: 296–303. 10.1016/j.gene.2013.03.133 [DOI] [PubMed] [Google Scholar]
- Abdoli R., Zamani P., Mirhoseini S. Z., Ghavi H. N., Nadri S., 2016. A review on prolificacy genes in sheep. Reprod. Domest. Anim. 51: 631–637. 10.1111/rda.12733 [DOI] [PubMed] [Google Scholar]
- Ambros V., 2004. The functions of animal microRNAs. Nature 431: 350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Bai M., Sun L., Zhao J., Xiang L., Cheng X., et al. , 2017. Histological analysis and identification of spermatogenesis-related genes in 2-, 6-, and 12-month-old sheep testes. Naturwissenschaften 104: 84 10.1007/s00114-017-1505-1 [DOI] [PubMed] [Google Scholar]
- Cahill L. P., 1981. Folliculogenesis in the sheep as influenced by breed, season and oestrous cycle. J. Reprod. Fertil. Suppl. 30: 135–142. [PubMed] [Google Scholar]
- Canché J. E. T., Monforte J. G. M., Correa J. C. S., 2016. Environmental effects on productive and reproductive performance of Pelibuey ewes in Southeastern México. J. Appl. Anim. Res. 44: 508–512. 10.1080/09712119.2015.1102730 [DOI] [Google Scholar]
- Chen J. S., Li H. S., Huang J. Q., Dong S. H., Huang Z. J., et al. , 2016. MicroRNA-379–5p inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in hepatocellular carcinoma. Cancer Lett. 375: 73–83. 10.1016/j.canlet.2016.02.043 [DOI] [PubMed] [Google Scholar]
- Chen M., Shi J., Zhang W., Huang L., Lin X., et al. , 2017a. MiR-23b controls TGF-β1 induced airway smooth muscle cell proliferation via direct targeting of Smad3. Pulm. Pharmacol. Ther. 42: 33–42. 10.1016/j.pupt.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Chen Y., Luo D., Tian W., Li Z., Zhang X., 2017b. Demethylation of miR-495 inhibits cell proliferation, migration and promotes apoptosis by targeting STAT-3 in breast cancer. Oncol. Rep. 37: 3581–3589. 10.3892/or.2017.5621 [DOI] [PubMed] [Google Scholar]
- Chu M., Jia L., Zhang Y., Jin M., Chen H., et al. , 2011. Polymorphisms of coding region of BMPR - IB gene and their relationship with litter size in sheep. Mol. Biol. Rep. 38: 4071–4076. 10.1007/s11033-010-0526-z [DOI] [PubMed] [Google Scholar]
- Ding H., Luo Y., Liu M., Huang J., Xu D., 2016. Histological and transcriptome analyses of testes from Duroc and Meishan boars. Sci. Rep. 6: 20758 10.1038/srep20758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Du Q., Wang Z., Wang Y., Wu S., et al. , 2016. MicroRNA-665 suppressed the invasion and metastasis of osteosarcoma by directly inhibiting RAB23. Am. J. Transl. Res. 8: 4975. [PMC free article] [PubMed] [Google Scholar]
- Dong W. L., Tan F. Q., Yang W. X., 2015. Wnt signaling in testis development: Unnecessary or essential? Gene 565: 155–165. 10.1016/j.gene.2015.04.066 [DOI] [PubMed] [Google Scholar]
- Drouilhet L., Lecerf F., Bodin L., Fabre S., Mulsant P., 2010. Fine mapping of the FecL locus influencing prolificacy in Lacaune sheep. Anim. Genet. 40: 804–812. 10.1111/j.1365-2052.2009.01919.x [DOI] [PubMed] [Google Scholar]
- Gong W., Pan L., Lin Q., Zhou Y., Xin C., et al. , 2013. Transcriptome profiling of the developing postnatal mouse testis using next-generation sequencing. Science China 56: 1–12. 10.1007/s11427-012-4411-y [DOI] [PubMed] [Google Scholar]
- Hino H., Nakanishi A., Seki R., Aoki T., Yamaha E., et al. , 2018. Roles of maternal wnt8a transcripts in axis formation in zebrafish. Dev. Biol. 434: 96–107. 10.1016/j.ydbio.2017.11.016 [DOI] [PubMed] [Google Scholar]
- Huang J., Ju Z., Li Q., Hou Q., Wang C., et al. , 2011. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 7: 1016–1026. 10.7150/ijbs.7.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Cho W., Li Z., Xu X., Qu Y., et al. , 2017. MiR-758–3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 96: 535. 10.1016/j.biopha.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Kasimanickam V. R., Kasimanickam R. K., 2015. Differential expression of microRNAs in sexually immature and mature canine testes. Theriogenology 83: 394–398.e1. 10.1016/j.theriogenology.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Khan S., Brougham C. L., Ryan J., Sahrudin A., O’Neill G., et al. , 2013. miR-379 regulates Cyclin B1 expression and is decreased in breast cancer. PLoS One 8: e68753 10.1371/journal.pone.0068753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N., 2014. MicroRNAs and spermatogenesis. Fertil. Steril. 101: 1552–1562. 10.1016/j.fertnstert.2014.04.025 [DOI] [PubMed] [Google Scholar]
- Li K., Wang Y., Zhang A., Liu B., Jia L., 2017a. miR-379 Inhibits Cell Proliferation, Invasion, and Migration of Vascular Smooth Muscle Cells by Targeting Insulin-Like Factor-1. Yonsei Med. J. 58: 234–240. 10.3349/ymj.2017.58.1.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., M. Wang, Y. Chen, W. Wang, J. Wu et al., 2017b Role of the Hedgehog Signaling Pathway in Regulating the Behavior of Germline Stem Cells. Stem Cells International, 2017, (2017–8-13) 2017: 1–9. [DOI] [PMC free article] [PubMed]
- Lian C., Sun B., Niu S., Yang R., Liu B., et al. , 2012. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. 279: 964–975. 10.1111/j.1742-4658.2012.08480.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Pan Z., Wang X., Wenping H. U., Ran D. I., et al. , 2014. Progress on major genes for high fecundity in ewes. Frontiers of Agricultural Science & Engineering 1: 282 10.15302/J-FASE-2014042 [DOI] [Google Scholar]
- Luo Z., Liu Y., Chen L., Ellis M., Li M., et al. , 2015. microRNA profiling in three main stages during porcine spermatogenesis. J. Assist. Reprod. Genet. 32: 451–460. 10.1007/s10815-014-0406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makawi S. A., Elsharif B. A., Babiker E. A., 2007. Effect of Season on Freezability of Semen from Two Breed-Types of Desert Sheep in the Sudan. 6: 846–849. [Google Scholar]
- Miao X., Qin Q. L. X., 2015. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Mol. Cell. Endocrinol. 402: 32–42. 10.1016/j.mce.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Mongroo P. S., Rustgi A. K., 2010. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 10: 219–222. 10.4161/cbt.10.3.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, A., 2012 Molecular Mechanisms of Wnt8a Regulation: Insights Into Vertebrate Mesoderm Development and Patterning. Doctoral Dissertation, Texas A&M University.
- Pan D., 2007. Hippo signaling in organ size control. Genes Dev. 21: 886–897. 10.1101/gad.1536007 [DOI] [PubMed] [Google Scholar]
- Pan Q., Meng L., Ye J., Wei X., Shang Y., et al. , 2017. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT). Cancer Lett. 392: 26–38. 10.1016/j.canlet.2017.01.039 [DOI] [PubMed] [Google Scholar]
- Pickering N. K., Dodds K. G., Blair H. T., Hickson R. E., Johnson P. L., et al. , 2012. Genetic parameters for production traits in New Zealand dual-purpose sheep, with an emphasis on dagginess. J. Anim. Sci. 90: 1411–1420. 10.2527/jas.2011-4163 [DOI] [PubMed] [Google Scholar]
- Pramod R. K., Sharma S. K., Kumar R., Rajan A., 2013. Genetics of ovulation rate in farm animals. Vet. World 6: 833–838. 10.14202/vetworld.2013.833-838 [DOI] [Google Scholar]
- Pratt S. L., Calcatera S. M., 2016. Expression of microRNA in male reproductive tissues and their role in male fertility. Reprod. Fertil. Dev. 29: 24 10.1071/RD16293 [DOI] [PubMed] [Google Scholar]
- Ramayo-Caldas Y., Mach N., Esteve-Codina A., Corominas J., Castelló A., et al. , 2012. Liver transcriptome profile in pigs with extreme phenotypes of intramuscular fatty acid composition. BMC Genomics 13: 547 10.1186/1471-2164-13-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran M., Chen B., Yin J., Yang A., Jiang M., 2014. Advances in miRNA research related to testis development and spermatogenesis. Yi Chuan 36: 646–654. 10.3724/SP.J.1005.2014.0646 [DOI] [PubMed] [Google Scholar]
- Sahin Z., Szczepny A., Mclaughlin E. A., Meistrich M. L., Zhou W., et al. , 2014. Dynamic Hedgehog signalling pathway activity in germline stem cells. Andrology 2: 267–274. 10.1111/j.2047-2927.2014.00187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezenler T., Yildirir M., Ceyhan A., Yüksel M. A., Önal A. R., et al. , 2011. The Effects of Body Condition Score and Age of Ewes on the Reproductive Performance in Kivircik, Sakiz and Gokceada Sheep. J. Anim. Sci. Adv. 1: 94–99 [Google Scholar]
- Sun X., Wang Y., 2003. Wnt signaling pathways in mammalian reproduction. Progress in Biochemistry & Biophysics 30: 180–184. [Google Scholar]
- Tariq K., Peng W., Saccone G., Zhang H., 2016. Identification, characterization and target gene analysis of testicular microRNAs in the oriental fruit fly Bactrocera dorsalis. Insect Mol. Biol. 25: 32–43. 10.1111/imb.12196 [DOI] [PubMed] [Google Scholar]
- Vendrell V., Vázquezecheverría C., Lópezhernández I., Alonso B. D., Martinez S., et al. , 2013. Roles of Wnt8a during formation and patterning of the mouse inner ear. Mech. Dev. 130: 160–168. 10.1016/j.mod.2012.09.009 [DOI] [PubMed] [Google Scholar]
- Wang J., Lv Q., Li X., Liu Y., Liu K., et al. , 2016. Post-transcriptional and translational regulation modulates gene co-expression behavior in more synchronized pace to carry out molecular function in the cell. Gene 587: 163–168. 10.1016/j.gene.2016.04.055 [DOI] [PubMed] [Google Scholar]
- Wang L., Xu C., 2015. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction 149: R127–R137. 10.1530/REP-14-0239 [DOI] [PubMed] [Google Scholar]
- Wang, W., M. Meng, Y. Zhang, C. Wei, Y. Xie et al., 2014 Global transcriptome-wide analysis of CIK cells identify distinct roles of IL-2 and IL-15 in acquisition of cytotoxic capacity against tumor. BMC Medical Genomics, 7,1(2014–08–09) 7: 49. [DOI] [PMC free article] [PubMed]
- Weng C., Dong H., Chen G., Zhai Y., Bai R., et al. , 2012. miR-409–3p inhibits HT1080 cell proliferation, vascularization and metastasis by targeting angiogenin. Cancer Lett. 323: 171–179. 10.1016/j.canlet.2012.04.010 [DOI] [PubMed] [Google Scholar]
- Wright G. W., Simon R. M., 2003. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19: 2448–2455. 10.1093/bioinformatics/btg345 [DOI] [PubMed] [Google Scholar]
- Wu D., Niu X., Tao J., Li P., Lu Q., et al. , 2017a. MicroRNA-379–5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol. Rep. 37: 3502–3508. 10.3892/or.2017.5607 [DOI] [PubMed] [Google Scholar]
- Wu J., Zhu H., Song W., Li M., Liu C., et al. , 2014. Identification of conservative microRNAs in Saanen dairy goat testis through deep sequencing. Reprod. Domest. Anim. 49: 32–40. 10.1111/rda.12217 [DOI] [PubMed] [Google Scholar]
- Wu N., Gaur U., Zhu Q., Chen B., Xu Z., et al. , 2017b. Expressed microRNA associated with high rate of egg production in chicken ovarian follicles. Anim. Genet. 48: 205–216. 10.1111/age.12516 [DOI] [PubMed] [Google Scholar]
- Wylie A. D., Fleming J. A., Whitener A. E., Lekven A. C., 2014. Post-transcriptional regulation of wnt8a is essential to zebrafish axis development. Dev. Biol. 386: 53–63. 10.1016/j.ydbio.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Xie X., Li Y. S., Xiao W. F., Deng Z. H., He H. B., et al. , 2017. MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targetting EIF4G2. Biosci. Rep. 37: BSR20160542 10.1042/BSR20160542 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yan N., Lu Y., Sun H., Tao D., Zhang S., et al. , 2007. A microarray for microRNA profiling in mouse testis tissues. Reproduction 134: 73–79. 10.1530/REP-07-0056 [DOI] [PubMed] [Google Scholar]
- Zhang J., Qiang L., Wei Z., Li J., Zheng L., et al. , 2010. Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim. Biophys. Sin. (Shanghai) 42: 145–153. 10.1093/abbs/gmp116 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B., et al. , 2008. The TEAD/TEF Family of Transcription Factor Scalloped Mediates Hippo Signaling in Organ Size Control. Dev. Cell 14: 377–387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Li C., Yang S., Tian R., Wang J., et al. , 2016. Dynamics of the Transcriptome during Human Spermatogenesis: Predicting the Potential Key Genes Regulating Male Gametes Generation. Sci. Rep. 6: 19069 10.1038/srep19069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number: GSE107803. All data supporting the results of this article are contained within the article and its Supplemental Materials. Table S1, miRNA Sequence Quality Control. Table S2, read mapping summary of known sheep miRNA. Table S3, the DE novel miRNAs and DE sheep known miRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S4, the union of DE miRNA in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S5, predicted target mRNAs of DE known sheep miRNAs. Table S6, DE miRNA-Targets. Table S7, GO-analysis of candidate target mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S8, pathway analysis of candidate target mRNAs in 2- vs. 6-, 6- vs. 12-, and 2- vs. 12-month-old testes. Table S9, GO and pathway terms that WNT8A involved in related to testis development and spermatogenesis. Supplemental figure 1, Hierarchical clustering of DE mRNAs and DE miRNAs in testis samples. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7473047.