Abstract
During exon ligation, the S. cerevisiae spliceosome recognizes the 3´-splice site (3´SS) of precursor mRNA through non-Watson-Crick pairing with the 5´SS and the branch adenosine, in a conformation stabilised by Prp18 and Prp8. Here we present the 3.3 Å cryoEM structure of a human post-catalytic spliceosome just after exon ligation. The 3´SS docks at the active site through conserved RNA interactions in the absence of Prp18. Unexpectedly, the metazoan-specific FAM32A directly bridges the 5´-exon and intron 3´SS of pre-mRNA and promotes exon-ligation, as shown by functional assays. CACTIN, SDE2, and NKAP – factors implicated in alternative splicing – further stabilize the catalytic conformation of the spliceosome during exon ligation. Together these four proteins act as exon ligation factors. Our study reveals how the human spliceosome has co-opted additional proteins to modulate a conserved RNA-based mechanism for 3´-splice site selection and to potentially fine-tune alternative splicing at the exon ligation stage.
The spliceosome excises introns from pre-messenger RNAs (pre-mRNAs) to produce mature mRNA in two sequential transesterifications – branching and exon ligation – catalyzed at a single active site (1–3). The spliceosome assembles de novo on each pre-mRNA from component snRNPs and undergoes numerous conformational changes mediated by trans-acting proteins and DEAx/H-box ATPases (4). A series of cryo-EM structures of Saccharomyces cerevisiae (yeast hereafter) spliceosomes at different stages of assembly, catalysis and disassembly have rationalized decades of biochemical and genetic data and have provided considerable mechanistic insights into how the spliceosome achieves these two transesterification reactions (1, 5–9). During initial assembly, the U1 snRNP base-pairs with the 5´-splice site (5´SS) while the U2 snRNP forms the branch helix through pairing around the branch point (BP) adenosine. Prespliceosome formation, involving minimal interaction between the U1 and U2 snRNPs in yeast, brings the 5´SS and the BP sequence into one assembly. In mammals, formation of the prespliceosome is promoted and regulated by many alternative splicing factors (10,11). The prespliceosome then associates with the U4/U6-U5 tri-snRNP to form the pre-B complex, which is converted via B to Bact when U1 and U4 snRNPs dissociate by the activities of Prp28 and Brr2 followed by binding of the multi-subunit Prp19-associated (NTC) and Prp19-related (NTR) complexes. The 5´SS is handed off to the U6 snRNA and the catalytic core is formed during this conversion. The catalytic core of the spliceosome comprises U6 and U2 snRNAs folded into a compact structure that binds two catalytic divalent ions (12–14). The 5´SS is positioned precisely at the catalytic metal ions by pairing between the conserved 5´-intron sequence, GUAUGU, and the ACAGAGA sequence of U6 snRNA and between the 5´-exon and U5 snRNA loop I (15,16). During Prp2-induced remodeling to B*, the branch helix is docked into the active site by the branching factors, Cwc25 and Yju2, which allows the 2´-hydroxyl group of the BP adenosine to attack the 5´SS, producing the free 5´-exon and a lariat intron-3´exon intermediate (1). Prp16-induced dissociation of the branching factors from the resulting C complex promotes rotation of the branch helix out of the active site (17). Exon ligation factors lock the branch helix into its new position in the resulting C* complex (5,6). The 3´SS is positioned at the catalytic metal ions by non-Watson-Crick basepairing between the last intron nucleotide G and the first intron nucleotide G as well as between the penultimate intron nucleotide A and the BP adenosine. This configuration allows the 3´-hydroxyl group of the 5´-exon to attack the 3´SS, ligating the 5´- and 3´-exons into mRNA (7–9). The DEAH-box ATPase Prp22 then releases the resulting mRNA from the post-catalytic P complex (18,19) and finally the ATPase Prp43 disassembles the spliceosome for new rounds of splicing (1–3).
Human spliceosomes are larger than their yeast counterparts and contain many additional proteins (3,20,21). CryoEM structures of the human spliceosomes captured at near atomic resolution in different states confirm that the general architecture of the spliceosome is largely conserved between yeast and human, and reveal how some additional human proteins are integrated into the conserved architecture of the spliceosome (22–27). However, the functions of these proteins have not been determined experimentally. It is also not known if these proteins are constitutive components of the human spliceosome or some of them regulate alternative splicing of subsets of pre-mRNAs in a tissue-specific manner. Here we report the cryoEM structure of the human post-catalytic spliceosome, which shows that the 3´SS is recognized through RNA-RNA interactions conserved between humans and yeast. Our high-resolution structure reveals that four proteins, never observed in human spliceosome structures before, stabilize the branch helix and the docked 3´SS to facilitate exon ligation.
Purification and overall structure of the human P complex
The P complex spliceosome was assembled on MINX pre-mRNA in HeLa nuclear extract supplemented with recombinant hPrp22 (DHX8) mutant (K594A; see Supplementary Note 1, fig. S1) to prevent release of ligated exons. Oligonucleotide-directed RNaseH digestion was targeted to the region of the 3´-exon protected only when the 3´SS is docked into the active site. The resulting P complex was affinity-purified on amylose-resin using three MS2 aptamers attached to the 3´-exon to eliminate contaminating C* complex (Supplementary methods; figs. S1-2) (7).
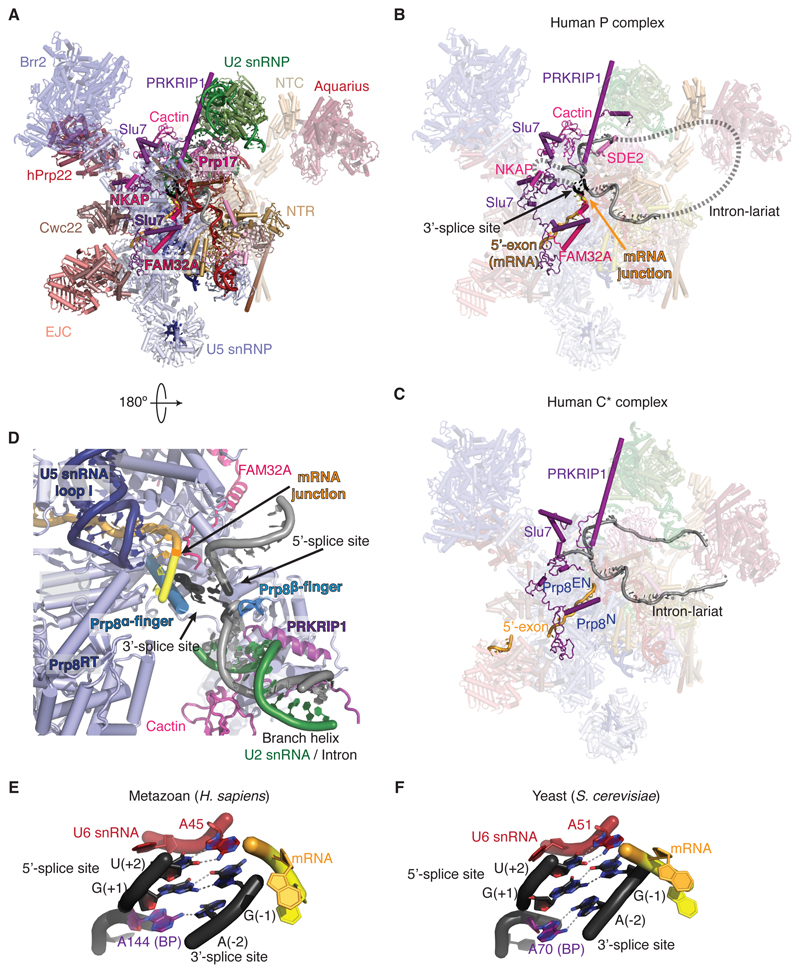
The overall architecture of the human P complex obtained by cryoEM reconstruction at 3.3 Å resolution (Supplementary Material PyMOL session, figs. S2-5) is similar to that of the human C* complex determined at an average resolution of 3.76 Å (22) and 5.9 Å (26) (Fig. 1). The higher resolution of our cryoEM density map of the human P complex allowed us to build more complete models of proteins in the peripheral region (table S2) and parts of four additional proteins (Cactin, FAM32A, SDE2, and NKAP) not present in S. cerevisiae (Fig. 1B,C; figs. S5-6). The remaining parts of these proteins are predicted to be largely disordered. The densities for Cactin and FAM32A were partially visible in the map of the C* complex (22) but were not of sufficient quality for model building. The higher resolution map of our P complex allowed us to build the C-terminal half of FAM32A based on density alone but the highly charged N-terminal half is disordered (fig. S6).
Figure 1. Structure of a human P complex reveals unexpected exon ligation factors.
(A) Overview of the human P complex spliceosome complex. EJC, Exon junction complex; NTC, Prp19-associated complex; NTR, Prp19-related complex. (B and C) Comparison of the P (present work) and C* (22) complexes reveals novel factors. Note the presence of mRNA and the docked 3´-splice site in our P-complex structure. Dashed lines indicate possible path of the intron not visible in the density. The intron is shown in gray, the 5´-exon in orange, and the 3’-exon in yellow. Prp8EN, Prp8 Endonucelase domain; Prp8N, Prp8 N-terminal domain. (D) Binding of the substrate in the active site cavity of P complex. Prp8RT, Prp8 reverse-transcriptase domain. (E) The 3´SS is recognized by the 5´SS and the BP adenosine in the human P complex. (F) 3´SS recognition in the yeast P complex (7).
A conserved 3´SS recognition mechanism
The RNA-based active site of the human P complex is essentially unchanged compared to C*, with the U2 and U6 snRNAs forming a triple helix that binds two catalytic Mg2+ ions (fig. S7). In the human P complex the newly formed mRNA remains bound at the active site through its 5´-exon pairing to U5 snRNA (fig. S7A). The new phosphodiester bond connecting the 5´-exon to the first two nucleotides of the 3´-exon is clearly visible confirming that our sample represents the genuine P complex (fig. S5B). Clear density extending from the intron G(+1) and the BP adenosine could be modeled as the last 3 nucleotides of the 3´SS (Fig. 1E, fig. S5B). As in yeast, the Hoogsteen edge of the 3′SS G(–1) forms a base pair with the Watson-Crick edge of the 5′SS G(+1). Additionally, N7 of the 3´SS A(-2) forms a H-bond with N6 of the BP adenosine. Thus the 3´SS is recognized, as in yeast (Fig. 1E,F), through pairing with the 5´SS and the BP adenosine. The 5´SS U(+2) pairs with the U6 snRNA A51, which stacks on the 3´SS G(-1), an interaction that was not modeled in the human C* complex (22) and which allows the 3´-hydroxyl of 3´SS G(-1) to project into the active site. Docking of the 3´SS onto the 5´SS is stabilized by the Prp8 alpha-finger and beta-finger – another feature similar to yeast (Fig. 1D). However, Prp18 – which in yeast projects into the active site and stabilizes the intron upstream of the 3´SS at positions -3 to -5 – was not observed in our map and was not detected by mass spectrometry either in our sample or in previous mass-spectrometric studies of C* and P complexes (20,26,27). Indeed, beyond the 3´SS C(-3), the intron becomes disordered in our map. The remaining nucleotides between the 3´SS and the branch helix loop out of the spliceosome and their path is likely guided by mammalian-specific proteins, as described below.
FAM32A is a metazoan-specific exon ligation factor
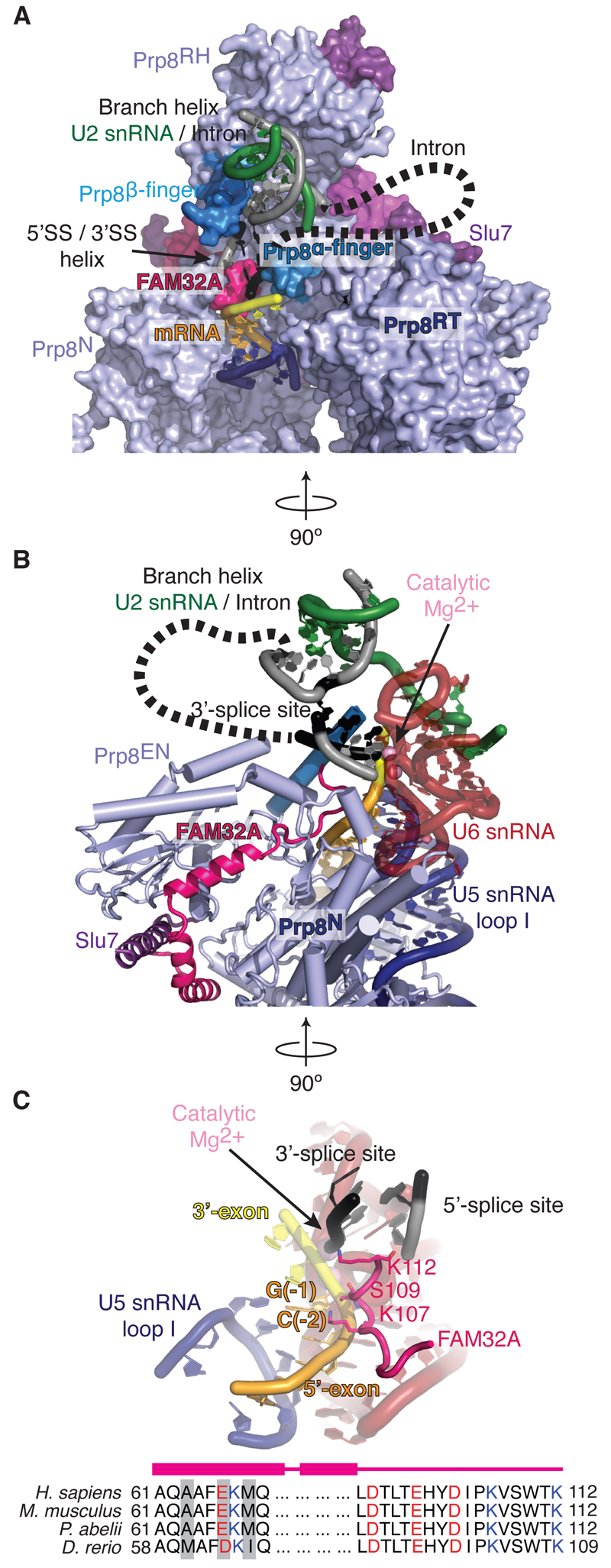
The most striking finding in our structure is that FAM32A (figs. S5C, S6), a poorly characterized protein of 13 kDa, binds between the endonuclease (EN) and N-terminal (N) domains of Prp8 and projects its C-terminus deep into the active site (Fig. 2A,B). Here FAM32A stabilizes the pairing between the 5´SS, the 3´SS, and the BP adenosine together with the alpha-finger and beta-finger of Prp8 (Fig. 2A). Importantly, the C-terminus of FAM32A binds along the 5´-exon through direct contacts between K107 and S109 and the phosphates of C(-2) and G(-1), respectively and stabilizes its base-pairing to loop I of U5 snRNA (Fig. 2C). The positively charged side chain of its C-terminal K112 extends into the space where 5´SS, 3´SS and BP come together to promote docking of the 3´SS (Fig. 2C). FAM32A is also known as ovarian tumor associated gene-12 (OTAG-12) and is down-regulated in a mouse model of ovarian tumor development (28). The OTAG-12 gene is expressed as three splice isoforms, OTAG-12a, OTAG-12b and OTAG-12c, in mice (figs. S5C, S6, Supplementary Note 2) and expression of the full-length OTAG-12b in ovarian cancer and HEK293 cells suppressed cell growth whereas OTAG-12c with N-terminal deletion or OTAG-12a with altered C-terminal sequence had no such effect. FAM32A (OTAG-12b, fig. S6B) bound in the P complex promotes mRNA formation for pro-apoptotic genes, acting as a tumor-suppressor. Indeed, the entire C-terminus of FAM32A is essentially invariant in metazoans from zebrafish to humans (Fig. 2C), consistent with a role in regulating splicing.
Figure 2. FAM32A is a component of the P-complex active site.
FAM32A is a component of the P-complex active site. (A and B) FAM32A binds Prp8 and projects its C-terminus into the RNA catalytic core. Prp8RH, Prp8 RNaseH domain, Prp8N, N-terminal domain of Prp8. (C) FAM32A stabilizes the 5´-exon onto U5 snRNA loop I, in proximity to the docked 3’SS. Note striking conservation of the FAM32A C-terminus across metazoans; variable residues are shaded gray. Dashed lines indicate possible path of the intron not visible in the density.
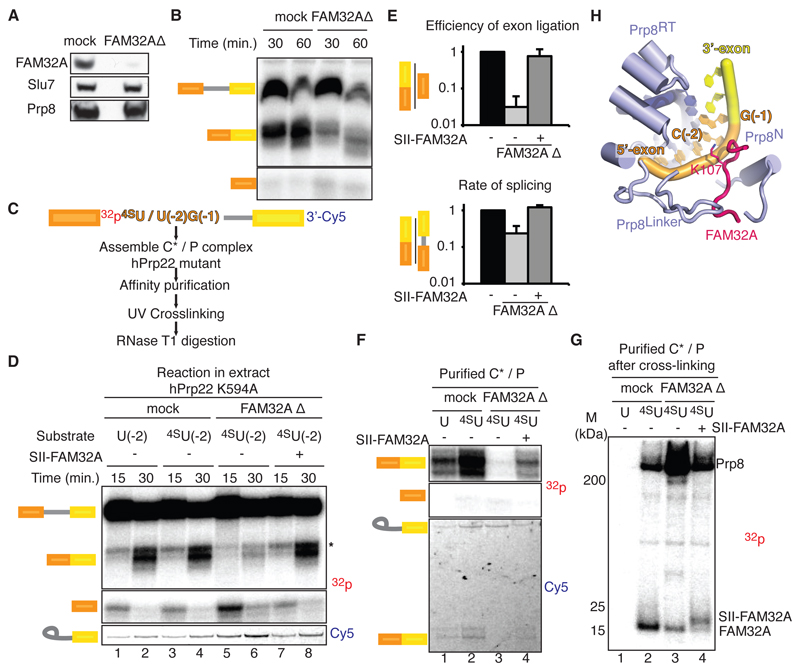
Depletion of FAM32A from HeLa nuclear extracts impaired exon ligation (Fig. 3A-E, fig. S8A-C), causing accumulation of cleaved 5´-exon at the C* stage (fig. S8D,E). Recombinant FAM32A restored efficient mRNA formation (Fig. 3D, fig. S8B,C), demonstrating that FAM32A promotes splicing by facilitating exon ligation, in agreement with our structure. UV cross-linking using pre-mRNA containing a single 4-thioU substitution at position -2 of the 5´-exon (Fig. 3C,D,F,G) produced two major cross-links (Fig. 3F,G). The one above 200 kDa represents Prp8, while the crosslink between 15 and 25 kDa was confirmed to be FAM32A by depletion and addition of slightly larger, tagged FAM32A (Fig. 3G,H). P complexes assembled in FAM32A-depleted extracts contained mostly lariat-intermediate and cleaved 5´-exon, which cross-linked to residual FAM32A, demonstrating that FAM32A also binds the 5´-exon in the pre-catalytic C* complex (Fig. 3F,G). Thus, FAM32A is a bona fide exon ligation factor that stabilizes docking of the 3´SS into the active site and promotes splicing in mammals.
Figure 3. FAM32A promotes exon ligation by binding the 5’-exon.
FAM32A promotes exon ligation by binding the 5´-exon. (A and B) Depletion of FAM32A impairs exon ligation. (C) Overview of the UV cross-linking experiment. Note that C(-2) was changed to U(-2) for these experiments. (D) FAM32A promotes exon ligation. (E) Effect of FAM32A depletion on exon ligation efficiency. Experiments were performed using a substrate with a single 32P at U(-2) of the 5´-exon. Error bars represent standard deviation (n=3). (F) C* complexes accumulate in FAM32A-depleted extracts. Shown is RNA extracted from affinity-purified P complexes (see also fig. S8). (G) FAM32A cross-links to the 5´-exon. SDS-PAGE of proteins labeled through cross-linking. SII-FAM32A, Strep-tactin-tagged FAM32A; 32p, 32P radioactive phosphate; 4SU, 4-thio-uridine. (H) Positioning of FAM32A and Prp8 around C(-2) of the 5´-exon rationalizes the cross-linking results.
NKAP and FAM32A stabilize Slu7 binding
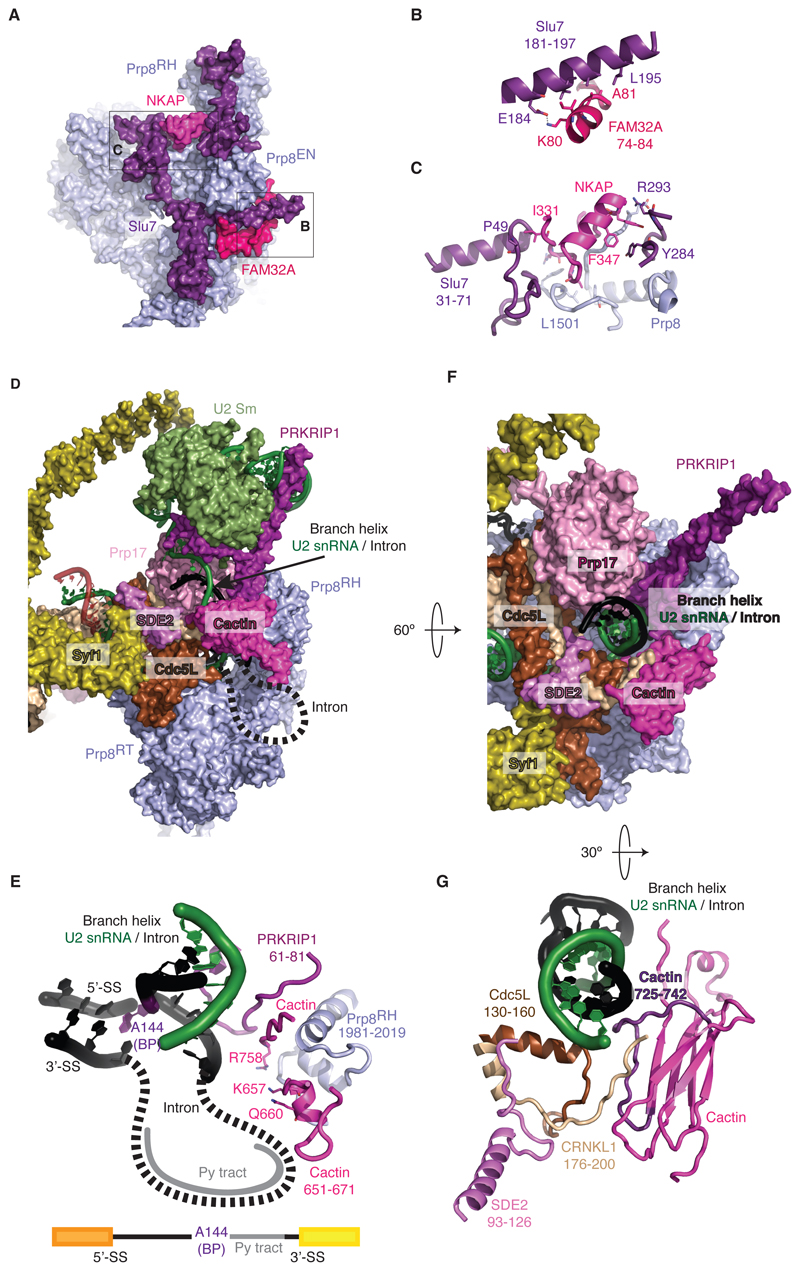
As in yeast, Slu7 rigidifies the C*/P conformation by binding across the Prp8 EN and N domains (Fig. 1C, 4A-C). Binding of the central region of Slu7 to the Prp8 EN domain is stabilized by FAM32A (Fig. 2B), while NF-κB-activating protein (NKAP) – a factor identified for the first time in our map – promotes binding of the Slu7 N-terminus onto Prp8 (Fig. 4C, fig. S5D). NKAP is a 415 residue protein implicated in T-cell development; it consists of highly charged repetitive sequences like Ser-Arg and poly-Lys and is expected to be intrinsically disordered through almost its entire length. However, residues 329-358 form a short helix that bridges the N- and C-terminal fragments of Slu7 bound to Prp8 and stabilizes the P complex. Indeed, NKAP binds exon sequences genome-wide, associates with mRNA in vitro, and depletion of NKAP in vivo reduces splicing efficiency, consistent with a role in promoting mRNA formation (29).
Figure 4. Unexpected factors stabilize the P-complex conformation.
(A to C) FAM32A and NKAP promote binding of Slu7 to the P complex. Prp8EN, Prp8 endonuclease domain. (D and E) Cactin stabilizes the position of the branch helix for exon ligation. Dashed lines indicate possible path of the intron not visible in the density. Py tract, polypyrimidine tract. (F) Novel factors position the branch helix. (G) SDE2 promotes Cactin binding near the branch helix. The loop of Cactin that projects a positive surface onto the branch helix is highlighted in magenta.
Cactin, SDE2 and PRKRIP1 stabilize the branch helix
The branch helix is locked into position by the WD40 domain of Prp17, CDC5L (Cef1) and CRNKL1 (Clf1) as in yeast C* and P complexes (5,7–9) (Fig. 4D-G). Unexpectedly, the human P complex structure revealed that the branch helix is further secured in its exon ligation conformation by Cactin, SDE2 and PRKRIP1. Our cryoEM map enabled us to build residues between 637 and 756 of Cactin, which folds into a β-sandwich domain. Its N-terminal region has long stretches of charged and polar amino acids suggesting these regions are intrinsically disordered. Its C-terminus and a short α-helix protruding from the β-sandwich domain interact with the Prp8 RNaseH domain (Fig. 4D,E), allowing Cactin to project a series of charged residues towards the branch helix, near the predicted path of the intron between the BP and the docked 3´SS (Fig. 4E) stabilizing 3´SS docking. Finally, a loop just before the C-terminal β strand of Cactin forms an extensive positively charged surface with the N-terminal region of CRNKL1 and with Cdc5L that surrounds the branch helix (Fig. 4F,G). This surface is stabilized by an α-helix of SDE2 that interacts with CRNKL1 and Cdc5L (Figs. 4G, S9A). Our map also enabled us to build 30 additional residues of PRKRIP1 (22) which reveals its striking structure (fig. S9B,C). The N-terminal residues 28-39 form an α-helix which bridges between the U2 Sm ring and U2 snRNA at the tip of the branch helix. Together with the long C-terminal α-helix bound to stem IV of U2 snRNA it locks the branch helix into the exon-ligation orientation (fig. S9B). The intervening loop inserts into the active site and interacts with the Prp8 RNaseH domain and the C-terminus of Cactin to stabilize the branch helix and promote exon-ligation.
SDE2 is first synthesized as an inactive precursor containing an N-terminal ubiquitin-fold-domain, which is cleaved to produce activated Sde2-C. Our structure shows that the N-terminal ubiquitin domain of unprocessed SDE2 would clash with the branch helix (Fig. 4F,G), explaining why the full-length protein cannot be incorporated into the spliceosome as shown in Schizosacchoromyces pombe (S. pombe). S. pombe cells that cannot produce Sde2-C show defects in splicing of the same specific introns as cells lacking Cactin (30), suggesting that Cactin and Sde2 binding to the spliceosome is highly cooperative. Indeed cells lacking Sde2-C show reduced Cactin binding to the spliceosome (30). The P-complex structure shows how SDE2 guides CRNKL1 to bind Cactin (Fig. 4G) thus rationalising the functional observations in S. pombe.
Discussion
The human P-complex structure shows that the 3´SS is recognised and docked into the active site of the spliceosome based on the same base-pairing interactions seen in the yeast P complex (7–9). Similarly to yeast, a set of conserved factors including the RNaseH domain of Prp8, Prp17, and Slu7 rigidify the position of the branch helix in the P-complex and promote 3´SS docking (7). Unexpectedly, our high resolution map of human P complex enabled us to build four additional proteins – FAM32A, Cactin, SDE2 and NKAP, which have been identified by mass-spectrometry but never found in any of the cryoEM structures of human spliceosomes (22–26). Three of these factors – NKAP, Cactin, and Sde2 – cooperate with PRKRIP1 to lock the branch helix in place in P complex (Fig. 5). Indeed Cactin and Sde2 promote splicing of the same specific subset of introns in S. pombe (31), highlighting their role in exon ligation, although the basis of their specificity is not understood. Together these factors partially compensate for the absence of Yju2, which stabilizes the branch helix in the yeast P complex (fig. S10B) but which dissociates during the C to C* transition in humans (Fig. 5) (7).
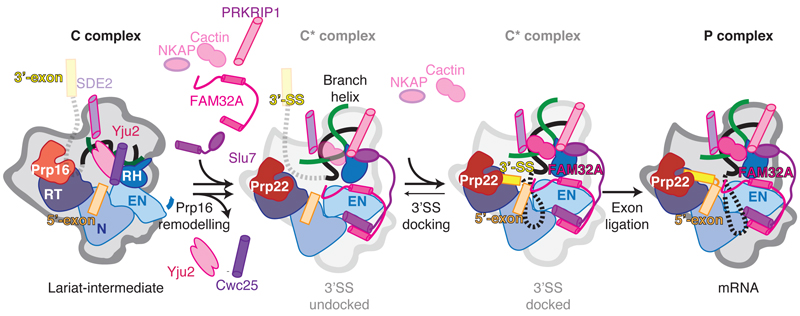
Figure 5. Model for the action of exon ligation factors in metazoans.
After Prp16 dissociates Cwc25 and Yju2 from C complex, Slu7, PRKRIP1, and FAM32A can bind the remodeled C* conformation. Cactin may bind before, or concomitantly with, docking of the 3´-exon at the catalytic core and associates more strongly upon 3´SS docking. SDE2 is likely present already in the C complex, as it interacts with the NTC, remains bound throughout the catalytic stage and promotes Cactin binding after remodeling by Prp16. FAM32A binds the 5´-exon and likely stabilizes docking of the 3´SS onto the 5´SS and BP adenosine.
In yeast, docking of the 3´SS is stabilized by Prp18, which abuts the 3´SS and guides Slu7 binding to Prp8. Indeed, in a subset of the yeast P complex particles lacking Prp18 and Slu7 (7), the 3´SS is not stably docked in the active site and the branch helix shows weaker density, suggesting that the branch helix is mobile (7). In contrast Prp18 was not detected by mass-spectrometric analysis of the human C* and P complex spliceosome assembled on MINX pre-mRNAs (20,26,27) and Prp18 is absent in the cryoEM structure of the human C* complex (22). The human P complex structure presented here also lacks Prp18 (fig. S10A,B, table S3).
Strikingly, FAM32A penetrates into the active site of the P-complex spliceosome assembled on MINX pre-mRNA and promotes 3´SS docking, thus partly substituting for Prp18. In contrast, depletion of Prp18 from HeLa extracts abolishes exon ligation of β-globin pre-mRNA (32) raising the intriguing possibility that Prp18 promotes splicing of a subset of human transcripts, acting as in yeast. Indeed, in S. pombe genetic depletion of Prp18 abolishes splicing in an intron-specific manner (33). Docking of the yeast Prp18 structure onto our human P complex indicates that Prp18 binding can be accommodated while FAM32A is bound in the active site of the human P complex (fig. S10C,D). Hence both Prp18 and FAM32A could influence alternative splicing of specific pre-mRNAs at the exon ligation stage. Consistent with this idea, Slu7 has been shown to influence selection of competing 3´SS by regulating docking of the 3´SS at the P complex stage (34). Intriguingly, Slu7 does not closely approach the active site in our human P complex but binds FAM32A, which enters the active site. Thus FAM32A could be responsible, at least in part, for the effects of Slu7 on 3´SS selection. Therefore, several exon ligation factors could modulate 3´SS choice during the catalytic stage.
Our P-complex structure highlights how in mammal specific proteins regulate a conserved mechanism for 3´SS recognition and provides a framework to expand mechanistic studies of the human spliceosome to different cell types and different metabolic or developmental states.
Supplementary Material
One Sentence Summary.
The human spliceosome adopts a more complex solution for exon ligation compared to the yeast spliceosome.
Acknowledgments
We thank G. Cannone, S. Chen, G. McMullan, R. Brown, J. Grimmett, and T. Darling for smooth running of the EM and computing facilities; A. Murzin and T. Anderee for discussion; R. Thompson and Y. Chaban for assistance with data collection at Leeds and eBIC; the mass spectrometry facility for help with protein identification; J. Richardson for advice; and the members of the spliceosome group for help and advice throughout the project. We thank J. Löwe, D. Barford, S. Scheres and R. Henderson for their continuing support.
Funding: The project was supported by the Medical Research Council (MC_U105184330) and ERC Advanced Grant (AdG-693087-SPLICE3D). S.M.F. was supported by EMBO and Marie Sklodowska-Curie fellowships and the ERC grant. M.E.W was supported by a Cambridge-Rutherford Memorial PhD Scholarship.
Footnotes
Author contributions: S.M.F. designed the strategy to purify human P complex, purified proteins, prepared the sample, made EM grids, collected and processed EM data, and carried out all functional assays. S.M.F. performed initial docking and rebuilding of previously assigned complex components. M.E.W. identified Cactin, C.O. identified FAM32A, and S.M.F. identified NKAP and SDE2. S.M.F., M.E.W. and C.O. completed model building and refinement. S.M.F. and A.J.N. designed and carried out UV cross-linking. S.M.F., M.E.W., C.O. and K.N. analyzed the structure and S.M.F. and K.N. drafted a manuscript and finalized with input from all authors. K.N. coordinated the spliceosome project.
Competing interests: Authors declare no competing interests.
Data and materials availability: Cryo-EM maps are deposited in the Electron Microscopy Data Bank under accession numbers EMD-4525 (stalled with DHX8 K594A mutant, overall map), EMD-4526 (stalled with DHX8 S717A mutant, overall map), EMD-4527 (stalled with DHX8 K594A mutant, focused refinement of core), EMD-4528 (stalled with DHX8 S717A mutant, focused refinement of core), EMD-4529 (focused refinement of Aquarius and Syf1), EMD-4530 (focused refinement of Brr2), EMD-4532 (focused refinement of DHX8), EMD-4533 (focused refinement of Prp19), EMD-4534 (focused refinement of U2 snRNP), EMD-4535 (focused refinement of U5 Sm); the atomic model is deposited in the Protein Data Bank under accession 6QDV.
References
- 1.Plaschka C, Newman AJ, Nagai K. Structural basis of Nuclear pre-mRNA splicing: lessons from yeast. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan C, Wan R, Shi Y. Molecular mechanisms of pre-mRNA splicing through structural biology. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastner CL, Will H, Stark R, Lührmann R. Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb Perspect Biol. doi: 10.1101/cshperspect.a.032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol. 2013;10:83–95. doi: 10.4161/rna.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355:149–155. doi: 10.1126/science.aak9979. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson ME, Fica SM, Galej WP, Norman CM, Newman AJ, Nagai K. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science. 2017;358:1283–1288. doi: 10.1126/science.aar3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Li X, Zhang L, Jiang J, Hill RC, Cui Y, Hansen KC, Zhou ZH, Zhao R. Structure of the yeast spliceosomal postcatalytic P complex. Science. 2017;358:1278–1283. doi: 10.1126/science.aar3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai R, Yan C, Wan R, Lei J, Shi Y. Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae. Cell. 2017;171:1589–1598. doi: 10.1016/j.cell.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 10.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe JB. Deep surveying of alternative splicing complexity in the human transcriptome by high throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 11.Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 12.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol. 2014;21:464–471. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 16.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 17.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 19.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–39. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y. An atomic structure of the human spliceosome. Cell. 2017;169:918–929. doi: 10.1016/j.cell.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Lührmann R, Stark H. Structure and conformational dynamics of the human spliceosomal Bact Complex. Cell. 2018;172:454–464. doi: 10.1016/j.cell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Yan C, Zhan X, Li L, Lei J, Shi Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018;28:307–322. doi: 10.1038/cr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan X, Yan C, Zhang X, Lei J, Shi Y. Structure of a human catalytic step I spliceosome. Science. 2018;359:537–545. doi: 10.1126/science.aar6401. [DOI] [PubMed] [Google Scholar]
- 26.Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Luhrmann R. CryoEM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 27.Ilagan J, Chalkley RJ, Burlingame AL, Jurica MS. Rearrangements within human spliceosomes captured after exon ligation. RNA. 2013;19:400–412. doi: 10.1261/rna.034223.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Zhang H, Aravindakshan JP, Gotlieb WH, Sairam MR. Anti-proliferative and pro-apoptotic actions of a novel human and mouse ovarian tumor-associated gene OTAG-12: downregulation, alternative splicing and drug sensitization. Oncogene. 2011;30:2874–2887. doi: 10.1038/onc.2011.11. [DOI] [PubMed] [Google Scholar]
- 29.Burgute BD, Peche VS, S A-L, Glöckner G, Gaßen B, Gehring NH, Noegel AA. NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins. Nucleic Acids Res. 2009;42:3177–3193. doi: 10.1093/nar/gkt1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakran P, Pandit PA, Datta S, Kolathur KK, Pleiss JA, Mishra SK. Sde2 is an intron-specific pre-mRNA splicing regulator activated by ubiquitin-like processing. EMBO J. 2018;37:89–101. doi: 10.15252/embj.201796751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzi LE, Bah A, Wischnewski H, Shchepachev V, Soneson C, Santagostino M, Azzalin CM. Fission yeast Cactin restricts telomere transcription and elongation by controlling Rap1 levels. EMBO J. 2015;34:115–29. doi: 10.15252/embj.201489559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horowitz DS, Krainer AR. A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]
- 33.Vijaykrishna N, Melangath G, Kumar R, Khandelia P, Bawa P, Varadarajan R, Vijayraghavan U. The fission yeast pre-mRNA processing factor 18 (prp18+) has intron-specific splicing functions with links to G1-S cell cycle progression. J Biol Chem. 2016;291:27387–27402. doi: 10.1074/jbc.M116.751289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3’ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 35.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Nafría J, Watson JF, Greger IH. IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. Sci Rep. 2016;6 doi: 10.1038/srep27459. 27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayeda A, Krainer AR. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 38.Mayeda A, Krainer AR. Mammalian in vitro splicing assays. Methods Mol Biol. 1999;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci USA. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pikielny CW, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 41.Zivanov J, Nakane T, Forsberg B, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. RELION-3: new tools for automated high-resolution cryo-EM structure determination. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–7065. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 51.Shinoda K, Tomita M, Ishihama Y. emPAI Calc--for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics. 2010;26:576–7. doi: 10.1093/bioinformatics/btp700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.