Abstract
The spliceosome excises introns from pre-messenger RNAs using an RNA-based active site cradled by a dynamic protein scaffold. A recent revolution in cryo-electron microscopy has led to near-atomic structures of key spliceosome complexes, which provide insight into the mechanism of activation, splice site positioning, catalysis, protein rearrangements, and ATPase-mediated dynamics of the active site. The structures rationalize decades of genetics and biochemistry and provide a molecular framework for future functional studies.
Introduction
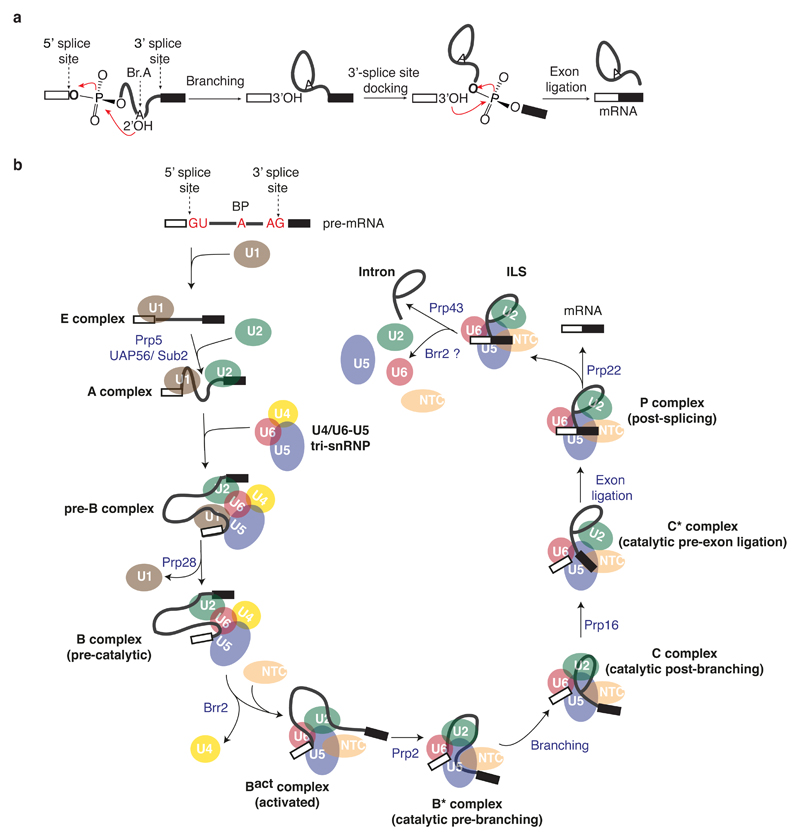
During eukaryotic gene expression, genes are first transcribed into pre-messenger RNAs (pre-mRNAs), in which the coding information, represented by exons, is interrupted by introns. To produce mature messenger RNAs (mRNAs) with an uninterrupted protein coding sequence, introns are excised from pre-mRNAs by two sequential phosphoryl transfer reactions: branching and exon ligation1–4 (Fig. 1a). During branching the 2’ hydroxyl of a conserved adenosine called the branch point (BP) attacks a phosphate at the 5’-splice site (5’-SS), producing a free 5’-exon and a lariat intron—3’-exon intermediate, while during exon ligation the new 3’ hydroxyl group of the 5’-exon attacks a phosphate at the 3’-splice site, ligating the exons and releasing the lariat intron. These reactions are chemically simple but catalysed by a dynamic ribonucleoprotein enzyme, the spliceosome, comprising five small nuclear RNAs (snRNAs) and over 70 proteins in yeast. The spliceosome is not a pre-formed enzyme and the active site is created only after the spliceosome, assembled on pre-mRNA from many components, has undergone extensive conformational and compositional changes5,6 (Figs 1b and 2). Because of its dynamic nature, understanding the molecular mechanism of splicing has been an enormous challenge for structural biologists.
Figure 1. A functional view of the splicing cycle.
a, Two step mechanism of pre-mRNA splicing. b, Assembly and catalytic cycle of the spliceosome.
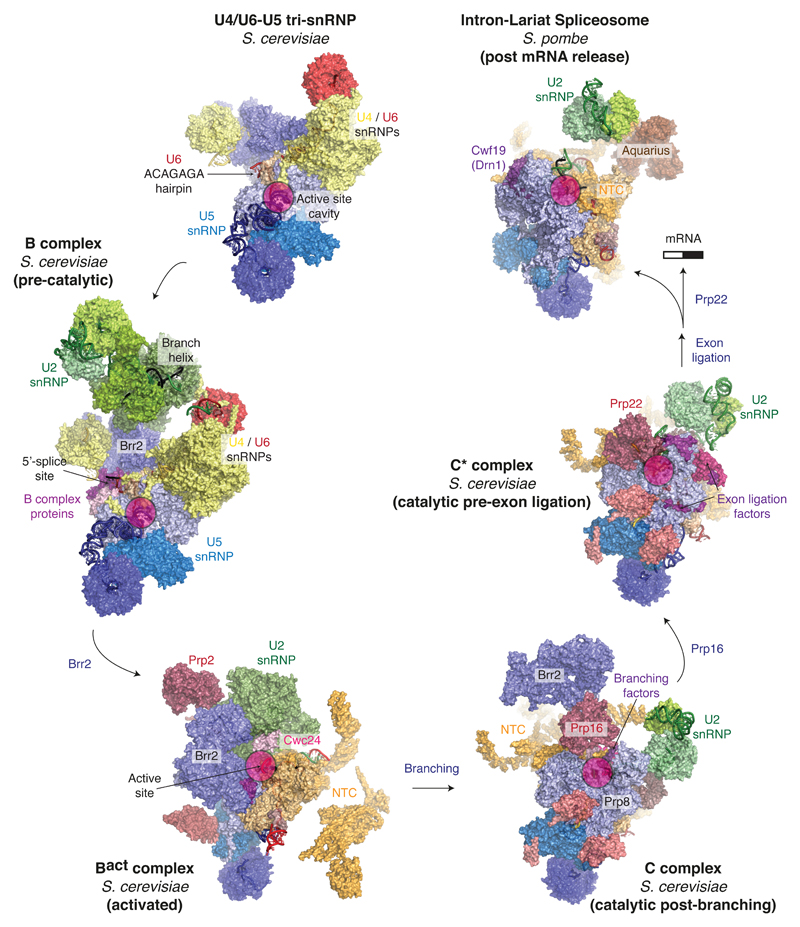
Figure 2. A structural view of the splicing cycle.
Complexes for which high-resolution structures were solved by cryo-EM are shown in surface representation. Key features are indicated for each complex (e.g. the location of the active site). Major sub-complexes are coloured as follows: U5 snRNP, blue; U6 snRNP, red; U4 snRNP and U4/U6 proteins, yellow; U2 snRNP, green; NTC and NTC-associated factors, orange; trans-acting protein factors, magenta. The following PDB entries were used: 5GAN (U4/U6•U5 tri-snRNP); 5NRL (B complex); 5GM6 (Bact complex); 5LJ5 (C complex); 5MQ0 (C* complex); 3JB9 (ILS).
The spliceosome assembles de novo around each intron of pre-mRNA in a stepwise manner from individual small nuclear ribonucleoprotein particles – snRNPs – composed of snRNA and associated proteins5,6. Five snRNPs (U1, U2, U4, U5 and U6 snRNPs), named after their snRNA component, associate with a pre-mRNA (Fig.1b). Several key RNA recognition and remodelling events occur during activation, in addition to numerous changes in protein composition, to create the active site5–6. Four decades of biochemical and genetic studies have established a series of assembly and remodelling steps that underlie the precise functioning of this intricate molecular machine. The U1 and U2 snRNPs first pair with the 5’-SS and the BP of pre-mRNA respectively7–10, leading to formation of the A complex11 (Fig. 1b). Assembly then proceeds with association of the U4/U6•U5 tri-snRNP (ref. 11. Figs 1b and 2), in which U6 snRNA is extensively base-paired with U4 snRNA (ref. 12). In the resulting pre-B complex the U1 snRNP remains bound to the 5’-SS, but the ATPase Prp28 promotes dissociation of U1 snRNP (ref. 13), leading to a stable B complex14 (Figs 1b,2 and 5a). To enable catalytic activation, the ATPase Brr2 dissociates U4 snRNA15,16, allowing U6 snRNA to adopt a catalytically active structure together with U2 snRNA17 and to pair with the 5’-end of the intron18,19 (Figs 4d and 5b). Elucidation of base-pairing between U6, U2 snRNAs and pre-mRNA revealed RNA elements structurally similar to those present in group II self-splicing intron20,21 suggesting that their active sites might be structurally similar (Fig. 4). The Bact complex containing U2, U5 and U6 snRNAs formed after Brr2-dependent remodelling is stabilised by numerous proteins termed the NTC, together with NTC-related factors22–24 (Figs 2 and 3). The ATPase Prp2 then promotes binding of branching factors and juxtaposition of the 5’-SS and BP for branching25–27 (Figs 4 and 5). The resulting B* complex catalyses branching, forming the C complex (Figs 2, 4e and 5c). Following the first catalytic reaction, the ATPase Prp16 remodels the spliceosome to allow dissociation of branching factors28,29 and enables docking of 3’-splice site into the active site27,30 with the help of exon ligation factors25,31. The 5’- and 3’-exons are aligned by the U5 snRNA32,33 and the resulting spliceosomal C* complex performs the second catalytic step (Figs 1, 2 and 4). The ATPase Prp22 then releases the mRNA34,35 and the ATPases Prp43 and Brr2 disassemble the spliceosome36,37 (Figs 1 and 2).
Figure 5. Movement of the Prp8 RNaseH-like domain and its interaction with active site elements.
a-e, Surface representation of the position and key interacting partners of the Prp8 RNaseH-like domain (Prp8RH) in specific spliceosomal complexes, relative to the Prp8 Large domain (Prp8L). The insets show the relative movement of the domain; RH, RNaseH domain; L, Large domain. The RNaseH-like domain of Prp8 rotates during the catalytic phase of splicing and mediates conformational changes in the active site.
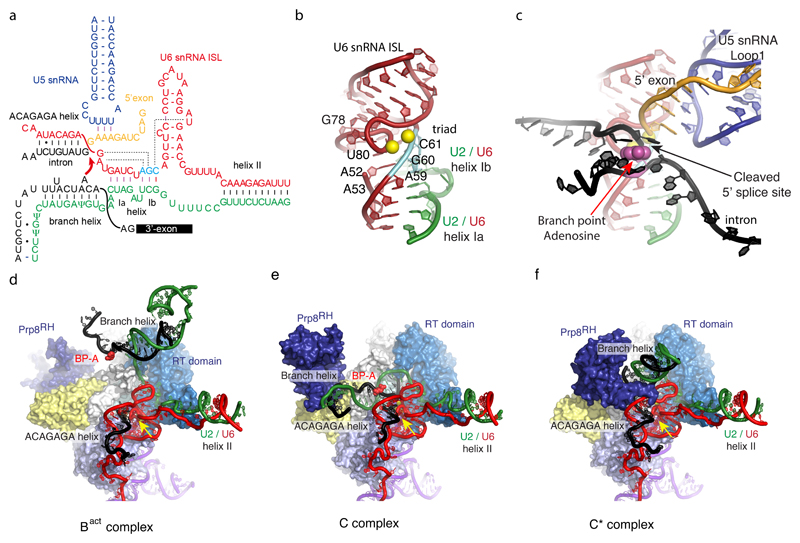
Figure 4. The active site of the spliceosome and its interaction with substrate.
a, The RNA interaction network prior to the first trans-esterification reaction is shown. U6 snRNA forms the intra-molecular stem-loop (ISL) and helices Ia and Ib with U2 snRNA. The three nucleotides in cyan (catalytic triad) form three consecutive triple base-pairs with U80, U52 and U53 and U2 snRNA nucleotides (catalytic triplex). The first six nucleotides of the intron, GUAUGU, are base-paired with the ACAGAGA box in U6 snRNA. The conserved UACUAAC (where A represents the branch point adenosine)_sequence in the intron basepairs with U2 snRNA to form the branch helix from which the branch point adenosine bulges out. The hydroxyl group of this adenosine functions as a nucleophile that attacks the 5’ splice site. RNAs are colour coded: red, U6 snRNA; cyan, catalytic triad of U6 snRNA; green, U2 snRNA; blue, U5 snRNA; orange, 5’exon; black, intron. b, three-dimensional structure of the active site RNA in C complex. Magnesium ions are represented by two yellow spheres located between the backbone of the catalytic triad and the highly twisted backbone at the bulge in ISL. c, the 5’-exon and branched intron bound to the active site (overlaid on b). The base of the branch point adenosine is shown in magenta and sphere representation. The 5’ phosphate of the first intron nucleotide (G+1) forms a 2’-5’ phosphodiester bond with the 2’hydroxyl of the branch-point adenosine. The branch helix is not depicted, for clarity. d-f, Interaction of the catalytic core of the spliceosome and movement of the branch helix in Bact, C and C* complexes. RNAs are colour-coded as above. Domains of Prp8 are colour-coded42 as follows: light blue, the N-terminal domain; marine, the Reverse transcriptase domain; white, Linker domain; light yellow, endonuclease domain; deep blue, RNaseH domain. The branch-point adenosine is coloured in magenta. Note that the active site RNAs remain unchanged but the branch helix shows large movements between Bact, C and C* complexes. The branch helix is stablised by SF3b (Bact), step 1 factors (C) and step 2 factors (C*), respectively (See Figure 5). Yellow arrow indicates the active site metals.
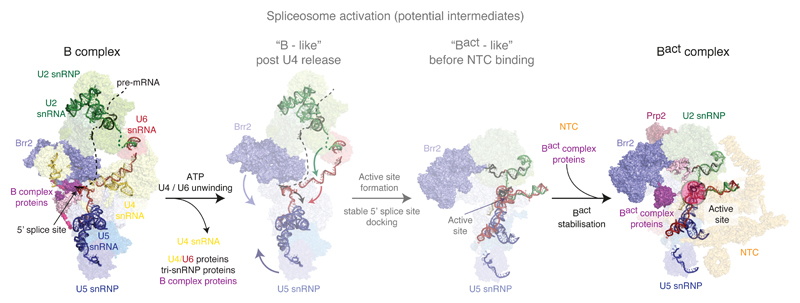
Figure 3. Activation of the spliceosome.
A fully assembled spliceosome, pre-B complex, is converted to stable B complex upon release of U1 snRNP by the action of the ATPase Prp28. Within the B complex, the single-stranded region of U4 snRNA is already bound to the active site of the Brr2 helicase, ready to translocate along U4 snRNA and free U6 snRNA from U4 snRNA and U4/U6 di-snRNP proteins. U6 snRNA folds, pairs with U2 snRNA and interacts with NTC and NTR proteins. During this process, U2, U5 and U6 snRNAs, together with the pre-mRNA substrate, forms the catalytic RNA core, reminiscent of the group II intron active site (see Fig. 4).
A cryo-EM revolution
Crystal structures of key protein components such as Prp8 (ref. 38–42) and Brr2 (ref. 43–46), the Sm47 and LSm48 protein complexes, the Snu13/Prp31/U4 snRNA complex49, SF3a (ref. 50) and SF3b (ref. 51) complexes and the functional core of U1 snRNP52–54f have provided structural insights and complemented the functional studies. However, an in-depth mechanistic understanding of spliceosome catalysis and dynamics requires high-resolution structures of fully-assembled spliceosomes.
Previous negative stain and cryoEM studies of spliceosomes and spliceosomal snRNPs had revealed their overall shapes at low resolution and the location of some specific tagged components55–61. Recent advances in cryo-EM data acquisition and processing have ushered in a so-called “resolution revolution” that has allowed structures of heterogeneous macromolecular assemblies to be determined to near-atomic resolution62. In the past two years three laboratories have applied the new EM methods to the structural study of the spliceosome. The outcome has been a series of near atomic-resolution snapshots of fully-assembled spliceosomes captured at key steps along the splicing pathway which are now allowing an unprecedented molecular view of the splicing cycle (Fig. 2).
Assembly and Activation of the spliceosome
U1 and U2 snRNPs recognise the 5’ splice site52,53 and BP of pre-mRNA leading to formation of the A complex, which associates with U4/U6•U5 tri-snRNP to form the fully assembled pre-B complex14 (Fig. 1b). Following the Prp28-dependent U1 snRNP release and association of the B complex proteins the pre-B complex is converted to the stable B complex14 (Figs 1b, 2 and 5a), which subsequently undergoes an extensive rearrangement of the RNA components as well as changes in protein composition63 to become the catalytically activated Bact complex (Figs 2, 3 and 4d). The structures of B (ref. 64) and Bact (ref. 65,66) complexes provide a first glimpse into the activation mechanism (Fig. 3). In B complex U2/U6 helix II, formed between the 5’-end of U2 snRNA and the 3’-end of U6 snRNA, holds U2 snRNP and tri-snRNP firmly together in addition to more flexible interactions between the protein components of U4/U6•U5 tri-snRNP and the U2 snRNP-bound pre-mRNA (ref. 64) (Figs 2 and 3). The conserved UACUAAC sequence around the BP of pre-mRNA pairs with U2 snRNA forming the branch helix within the context of U2 snRNP (Fig. 4c, 6a). The HEAT repeat of Hsh155, a component of SF3b complex, binds the branch helix (Fig. 5a). The active site of the N-terminal helicase cassette of Brr2 is bound to the single-stranded region of U4 snRNA ready to translocate along U4 snRNA and unwind the U4/U6 snRNA duplex (Fig. 3). A U6 snRNA hairpin containing the ACAGAGA 5’-SS binding region, which was stabilised by Dib1 in the structure of free tri-snRNP (ref. 67–69), is further stabilised by B complex proteins and now weakly interacts with the 5’-SS (Figs 2 and 3). This suggests that, in the absence of NTC proteins, Prp28 activity is not sufficient for stable U6 exchange at the 5’-SS, consistent with functional studies13. Nonetheless B complex proteins stabilise this initial 5’-exon tethering and, through interactions with Brr2, may couple initial weak recognition of the 5’-SS to Brr2 activation and U4/U6 unwinding. Although still base-paired with U4, U6 snRNA already forms U2/U6 helix II in B complex (Fig. 3), a structure that likely primes U6 and U2 snRNAs to fold into the catalytically active RNA structure when U6 snRNA is freed from U4 snRNA by Brr2 action64. During this process at least 24 proteins, including the U4/U6•U5 tri-snRNP and B complex proteins, dissociate and numerous NTC and NTR complex proteins join the spliceosome63 (Fig. 3).
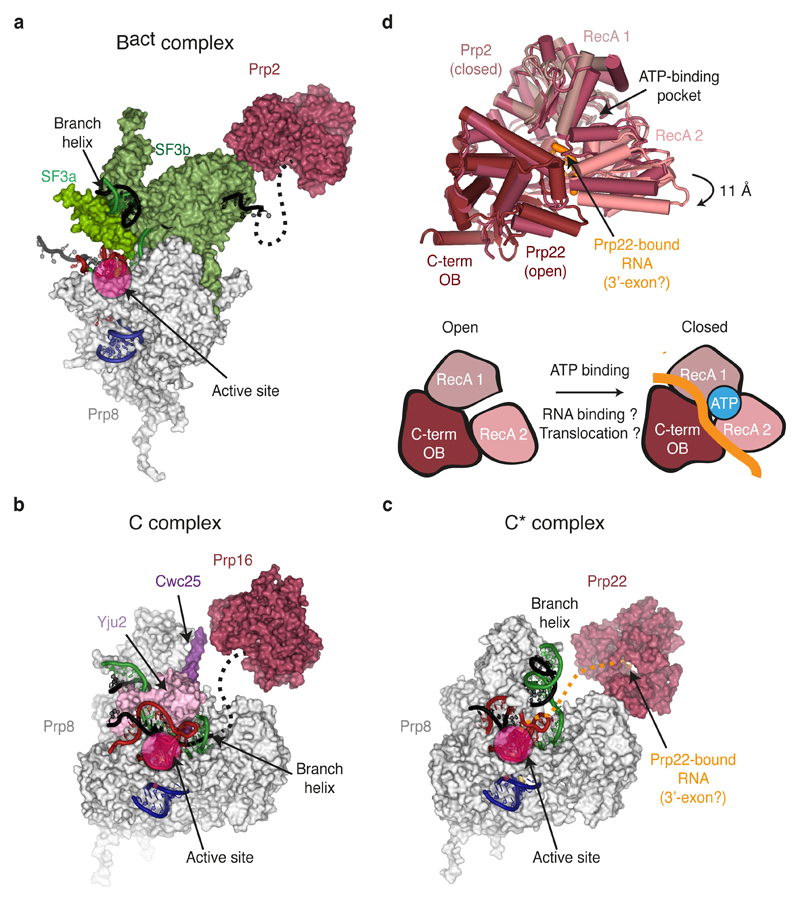
Figure 6. Binding of DEAH-box ATPases to specific spliceosomal complexes.
a-c, Surface representation of the positions and key interacting partners of DEAH-box ATPases in specific spliceosomal complexes, relative to Prp8. Key RNA components are shown in cartoon representation. The likely paths of the intron 3’ of the BP in Bact and C, and of the 3’-exon in C* are indicated as dashed lines. d, Different conformations of Prp2 and Prp22 observed in the cryo-EM maps of Bact and C* complexes. Note that for Prp2 no bound RNA was modelled and the RecA cassettes are present in a closed conformation, while for Prp22, bound RNA could be observed and the RecA cassettes are open as a result of a downward movement of RecA2 relative to the RecA1 domain. The open and closed conformations are shown schematically in the lower diagram, where bound RNA is coloured.
The active site of the spliceosome, reminiscent of the group II intron active site20,21, (discussed below) is fully formed during the transition from B to Bact complex and remains unchanged during the two phosphoryl transfer reactions (Fig. 4d-f). The 5’-SS is correctly positioned in the active site by pairing between the 5’-end of the intron and the U6 snRNA ACAGAGA sequence (ref. 18,19) as well as tethering of the 5’-exon to the U5 snRNA loop 1 (ref. 32,33)(Fig. 4a). The 5’-exon is further clamped together with Cwc21 between the N-terminal and Linker domains of Prp8 when the U5 snRNP foot domain undergoes a rotation that may be induced by dissociation of B complex proteins and initial unwinding of the U6 ACAGAGA stem. The Bact complex is kept inactive by the U2 snRNP SF3b subcomplex, which encircles the BP and sequesters the branch helix more than 50Å from the catalytic Mg2+ ions65,66 (Figs 3, 4d and 5b). The overall architecture of the Bact complex is held together by the NTC (Figs 2 and 3), whose components act as a multipronged clamp that restrains the many intricate RNA interactions crucial for catalysis. Indeed, specific NTC-associated factors such as Cwc24 replace B complex proteins (Fig. 2) and appear to regulate recognition of the 5’-SS and its docking at the catalytic Mg2+ site; functional studies have further confirmed the importance of such factors in stabilising Bact before Prp2 activity70. Together the B and Bact structures reveal key transitions of the snRNAs during creation of the active site and they provide a molecular basis for understanding the role of NTC and NTR proteins in spliceosome activation (Fig. 3).
The active site
Cryo-EM structures of Saccharomyces cerevisiae C complex71,72 have revealed the structure of the active site in a catalytically active spliceosome bound by the products of the first phosphoryl-transfer reaction (branching): a free 5’-exon and a lariat—3’-exon intermediate (Figs 2 and 4). U6 snRNA forms an intramolecular stem-loop (ISL) and helices Ia and Ib with U2 snRNA as first demonstrated by elegant genetic experiments17. This produces a highly twisted backbone of the bulged nucleotides of the ISL, which together with the backbone of the U6 catalytic triad (AGC), forms binding sites for two catalytic magnesium ions, consistent with the two-metal ion mechanism73 (Figs 4a and 4b). Indeed, the catalytic Mg2+-coordinating phosphate oxygens identified by metal rescue experiments are in perfect agreement with the structure74,75. The bases of A53, G52 and U80 form three consecutive triple base-pairs76 with U6/U2 helix Ib involving the catalytic triad (A59, G60 and C61) and the stacking of these triple base-pairs stabilises the folded RNA structure as in the active site of the group II intron20,21 (Fig. 4b).
As a result of the branching reaction the substrate pre-mRNA is cleaved at the 5’-SS and the 5’-phosphate of the first intron nucleotide (G+1) forms a new 2’-5’ phospho-diester bond with the 2’-hydroxyl group of the BP adenosine, producing a lariat intron structure (Figs 1a and 4c). In C complex both the 3’OH group of the 5’-exon and the 5’-phosphate of the first intron nucleotide remain close to the catalytic Mg2+ ions suggesting that the configuration of the B* complex active site (prior to branching) could be restored readily with minimal structural changes. The 5’ exon is tethered by the conserved loop 1 of U5 snRNA as first demonstrated by genetics and cross-linking32,33. The first six intron nucleotides (GUAUGU) are stringently conserved in yeast and the Watson-Crick and non-Watson-Crick basepairs between this hexanucleotide and the U6 snRNA ACAGAGA box position the 5’-SS in the active site70,71 (Fig. 4a). In yeast the pre-mRNA sequence around the BP adenosine is conserved to be UACUAAC (A denotes the BP adenosine) and this sequence pairs with U2 snRNA to form the branch helix with the bulged BP adenosine10. The base of the BP adenosine (A70 in UBC4 pre-mRNA) is flipped out and interacts with surrounding protein residues in B and Bact complexes63–65 whereas in C complex it forms hydrogen bonds with the U68 base, creating an unusual backbone structure that projects the 2’OH group towards the 5’-SS. The branch helix in C complex is significantly distorted from canonical A form and docked into the active site by the branching-specific proteins, Cwc25, Yju2, and Isy1 (Figs. 2, 4e and 5c) to insert the 2’-OH group of the BP adenosine into the active centre.
The group II intron active site is stabilised by a network of interactions provided by the RNA scaffold20,21. In the absence of such an RNA scaffold, the active site RNA of the spliceosome is stabilised by surrounding proteins (Fig. 4d-f). Prp8 is the largest and most conserved protein in the spliceosome, folded into four major domains connected by flexible linkers42,68: the N-terminal domain (N); the Large domain comprising the reverse-transcriptase (RT), Linker (L), and Endonuclease domains (EN); the RNaseH-like domain (RH); and the Jab1/MPN domain (Jab). The RT domain is expanded by the helix bundle domain attached to its N-terminus42,68. The catalytic RNA core is accommodated in the active site cavity formed by the RT, L, EN, and N domains42 (Fig. 4d-f) and clamped onto Prp8 by NTC and NTR proteins such as Cwc2, Bud31, Ecm2, Cef1 and Clf1 and Syf2 (ref. 71,72) (Fig. 5c). These proteins act as a cradle for the active site RNA and remain bound throughout the catalytic phase of the spliceosome, and hence the structure of the RNA active site changes little between Bact and C* complexes65,66,71,72,77,78. In contrast in C complex the branch helix in docked into the active site by the branching specific factors (Cwc25, Yju2 and Isy1)71,72 (Figs 4e and 5c). In the absence of these proteins the branch helix is free to move and change its orientation. The U5 snRNA stems I and II are firmly bound to the N-terminal domain of Prp8 and secured by a Prp8 polypeptide fitted into the minor groove65–69,71,72,77,78. The exon binding loop 1 attached to stem I projects into the active site.
Remodelling for exon ligation
Cross-linking studies showed that the 5’- and 3’-exons are aligned by U5 snRNA loop 1 during exon ligation to allow the 3’OH group of the 5’-exon to attack the 3’-splice site32,33. The structure of the stalled C complex provides the first structural insights into remodelling of the active site induced by the action of spliceosomal DEAH-box ATPases71 (Fig 6). Prp16 is poised to bind and translocate the intron downstream of the branch helix to destabilise branching-specific factors71 (Figs 2 and 6b). In C complex the 5’-exon is tethered by loop 1 of U5 snRNA and its 3’OH group is already positioned near the catalytic Mg2+ ions. However, the branch helix docked into the active site by branch-specific factors prevents access of an incoming 3’-exon to the active site (Figs 4c, 4e and 5c). This shows that the active site of the spliceosome has to be remodelled to create a space for 3’-exon binding. The structure of a C* spliceosome stalled right after Prp16 action but prior to exon ligation further elucidated the consequences of Prp16 action77,78 (Figs 5c and 6b). As in C complex, the catalytic RNA core is fastened onto Prp8 in C* (Figs. 1c, 2, 3d) by proteins common to both steps, while the branch helix has rotated by approximately 75° compared to complex C and is held in a new position by the Prp8 RNaseH-like (Prp8RH) domain and by Slu7 and Prp18 together with the repositioned Prp17 WD40 domain (Figs 4f, 5d and 6c). A β-hairpin protruding from the Prp8RH domain is likely to play an important functional role as several mutations that affect the first and second steps of splicing are mapped to it. The Prp8RH domain has rotated by approximately 80° and extends its β-hairpin through the minor groove of the branch helix towards Cef1 (Figs 4f, 5d and 6c). The Prp17 WD40 domain binds across the β-finger of the Prp8RH domain and Cef1 stabilising the rotated Prp8RH domain (Figs 4f and 5d). Slu7 is essential for exon ligation but is dispensable when the distance between the BP and 3’-splice site is shorter than 9 nucleotides77,79; the precise function of Slu7 remains unclear. The rotation of the branch helix moves the BP adenosine out of the active site together with the attached 5’-end of the intron linked to the BP adenosine, creating a space for 3’-exon docking and reorganising the pairing between the 5’-SS and the U6 ACAGAGA region77,78 (Fig. 4a and 4f).
Structural basis for remodelling by DEAH-box ATPases
It has been proposed that, during the catalytic stage, the spliceosome exists in a dynamic equilibrium between several conformations80, which is modulated both by the action of trans-acting factors such as Prp2, Prp16, and Prp22 and by co-acting step-specific factors (ref. 80–83) (Figs 5 and 6). The cryoEM structures of B, Bact, C, and C* complexes have visualised these conformational changes, including the dramatic movement of the Prp8RH domain, Prp17, Syf1 and Clf1 and a more subtle movement of the Prp8 endonuclease (Prp8EN) domain (Figs 2, 4f and 5d). To modulate such transitions throughout the catalytic stage, DEAH-box ATPases bind the intron at a similar position 3’ of the BP (or the ligated exon junction for Prp22) and induce movement of the branch helix as well as these proteins (Fig. 6), underscoring a common remodelling mechanism, as discussed below. These trans-acting ATPases have also been implicated in proofreading correct transitions through the pathway84–86. Biochemical studies suggested that DEAH-box ATPases such as Prp16 and Prp22 act at a distance through a “winching” mechanism involving translocation towards, but not necessarily through, their remodelling targets27.
In Bact, Prp2 binds the intron downstream of the branch helix which is held by Hsh155 within SF3b complex65,66 (Fig. 5b and 6a). Thus translocation of Prp2 toward the BP could dissociate SF3a and SF3b from the intron, allowing the branch helix to dock into the active site87. In C complex Prp16 binds on Prp8 in proximity to Cwc25 and is poised to bind the intron 3’ of the BP71 (Fig. 5c and 6b). Translocation towards the BP would destabilise Cwc25 from the Prp8RH domain, thus allowing binding of Slu7 and Prp18 to the Prp8RH domain77. Importantly, this model for action at a distance implies that transient destabilization of the branch helix and thus both Prp16- and Prp2-mediated remodelling would depend on the stability of the branch helix, consistent with both Prp2 and Prp16 proofreading usage of the correct BP83,85. More broadly, the large movement of the branch helix from Bact to C and C* (Fig. 4d-f) promoted by Prp2 and Prp16, respectively, could affect the stability of the RNA active site76,88, enabling Prp2 and Prp16 to proofread the integrity of the active site. Indeed Prp16 proofreads catalytic interactions between U6 snRNA and the catalytic Mg2+ (ref.77,89).
Interestingly, the cryo-EM structures revealed both “open” and “closed” conformations for the DEAH-box ATPases (Fig. 6d). As the “open” conformation observed for Prp22 allows RNA binding, toggling between “open” and “closed” states could underlie the mechanism of RNA translocation upon ATP hydrolysis (Fig. 6d).
Dynamics of the Prp8 RNaseH-like domain
The structure of the Prp8RH domain was one of the first domain structures of Prp8 to be determined and attracted much attention39–41. Based on cross-linking and genetic experiments it was proposed that the Prp8RH domain might be part of the spliceosome active site90–92. The cryoEM structures of Bact, C and C* now show that this is not the case. In the crystal structure of Prp8 the linker between the preceding Large domain and the Prp8RH domain was disordered and suggested that the Prp8RH domain could change its position with respect to the Large domain during the splicing cycle42. Cryo-EM structures have now revealed that the Prp8RH domain moves significantly during the splicing cycle (Fig 5). Dissociation of SF3b induced by Prp2 allows rotation of the branch helix and docking at the active site65,66,71,72,87 (Figs 2, 4f and 5b), while dissociation of Cwc24 and Cwc27 permits binding of Cwc25, Yju2, and Isy1 to clamp the branch helix in the conformation necessary for branching71,72 (Figs 4e, 5c and 6a). In this C conformation the Prp8RH domain moves into the body of the complex, while the β-hairpin binds along the extended branch helix and stabilises its position in cooperation with Yju2 (Figs 4e and 5c). The N-terminus of Cwc25 interacts with the Prp8RH domain and together they triangulate interactions that lock the branch helix in its first step conformation. Importantly the Prp8RH domain together with the NTC factor Syf1 holds the 3’ domain of U2 snRNP. Thus, the Prp8RH domain has now moved closer to the active site and could impact the conformation of the branch helix. Indeed, following Prp16 action, the branch helix in C* complex has undergone a dramatic conformational change forming an extended helix rotated approximately 75° from its position in C complex77 (Figs 4f and 5d). This is accompanied by an 80° inward rotation of the Prp8RH domain. The β-hairpin now extends in the minor groove of the branch helix, contacts Cef1 and straddles the interface between the branch helix and the ACAGAGA helix, placing it proximal to the active site (Fig. 3h), though not directly part of the active site. Here the β-hairpin stabilises the reorganised interaction between the 5’-SS and the U6 snRNA ACAGAGA sequence, particularly the new base-pair between U(+2) of the intron and A51 in U6 snRNA, which forms in C*. Indeed the β-hairpin interacts genetically with mutations at both U(+2) and A51 in U6 (ref. 91,92). The exon ligation factors Slu7 and Prp18 bind on the surface of the Prp8RH domain and lock it in the exon ligation conformation, thus contributing to stabilization of the rotated branch helix.
Finally, in the post-splicing intron-lariat spliceosome (ILS) from S. pombe93, the Prp8RH domain has rotated outward from the branch helix (Fig. 5e). The disassembly factor Cwf19 (Drn1 in yeast), which is important for recruitment of the debranching enzyme Dbr1 to the spliceosome94, wedges between the Prp8RH domain and the branch helix. These conformational changes are likely the result of mRNA release following Prp22 activity. Overall the structures of specific spliceosomal complexes reveal that while remodelling the complex, the ATPases Prp2, Prp16, and Prp22 each cause dramatic movements of the Prp8RH domain. The Prp8RH domain stabilises and modulates the conformation of the branch helix during the catalytic stage, with its β-hairpin likely affecting the Prp16-mediated transition between C and C* complexes (Figs 5 and 6).
Human spliceosome
The mechanism of splicing is likely to be universal but some details between yeast and human may differ, particularly with regard to splice site selection. The sequences of the 5’-SS and BP are stringently conserved in yeast but in humans they are much more degenerate. The 3’-SS is preceded by a poly-pyrimidine tract in human introns. These sequences are interrogated more than once during the splicing cycle and the human spliceosome tolerates more sequence variability (ref. 6). The structures of the human C* complex95,96 provided a first glimpse of the human spliceosome revealing association of the Exon Junction Complex (EJC) through binding of Cwc22, which is deposited by the spliceosome approximately between -20 and -25 nucleotides upstream of the exon-exon junction and removed by the ribosome during translation. The structure is in good agreement with modelling based on the yeast C complex71 and consistent with biochemistry95. It is interesting to note that human RBM22 protein shares homology with both yeast Cwc2 and Ecm2 even though the human spliceosome lacks individual homologues of yeast Cwc2 and Ecm2 (ref. 71). Additionally, mammalian-specific factors such as PRKRIP1 stabilize C* and may impact the active site, while NTC-related factor RBM22 likely cooperates with the ATPase Aquarius to guide the intron.
Structurally, free human U4/U6•U5 tri-snRNP differs significantly from its yeast counterpart. Sad1 keeps the Brr2 helicase away from its substrate U4 snRNA and Prp28 is found in the open conformation near the N-terminal and Large domains of Prp8 (ref. 98). The pre-B complex that forms when U4/U6•U5 tri-snRNP associates with A complex is remodeled by Prp28, which dissociates U1 snRNP from the 5’-SS to form stable B complex. A previous low-resolution EM study showed that the structure of U4/U6•U5 tri-snRNP in the human B complex is similar to its yeast counterpart99. Now cryo-EM revealed that the structure of human B complex is remarkably similar to that of yeast B complex. Indeed Brr2 helicase has moved to a position found in yeast tri-snRNP where the substrate U4 snRNA is bound in the active site of the Brr2 N-terminal helicase cassette100, suggesting a conserved mechanism of pre-catalytic spliceosome assembly and activation.
Conclusions
The recent cryo-EM snapshots of the spliceosome allow a nearly complete structural view of key intermediates in the splicing pathway and provide an atomic framework to rationalise genetic and biochemical research from the last four decades. The structures reveal how this intricate molecular machine uses a single RNA-based active site to catalyse the branching and exon ligation reactions that excise introns from pre-mRNA. The structural snapshots visualise for the first time how the substrates and products of these two reactions are progressively docked and undocked at the active site using the ATP-powered actions of RNA helicases and how movement of specific domains in Prp8 promotes conformational toggling of the spliceosome.
Our challenge now is to use structural information to design further experiments to uncover the detailed inner workings and energetics of this dynamic machine, which will keep us busy for many years to come.
Acknowledgements
This work was funded by the Medical Research Council (MC_U105184330) and European Research Council Advanced Grant (693087 - SPLICE3D). S.M.F. has been supported by EMBO and Marie Skłodowska-Curie fellowships. We thank Andy Newman, Clemens Plaschka, Clément Charenton, Lisa Strittmatter and Wojtek Galej for discussion and critical reading of the manuscript, and Clemens Plaschka for drawing Figure 3.
References
- 1.Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 2.Domdey H, Apostol B, Lin RJ, Newman A, Brody E, Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984;39:611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- 3.Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–31. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez JR, Pikielny CW, Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984;39:603–10. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- 5.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 6.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 9.Siliciano PG, Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 10.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 11.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 12.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 13.Staley JP, Guthrie C. An RNA switch at the 5' splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 14.Boesler C, et al. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nature Commun. 2016;7 doi: 10.1038/ncomms11997. 11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 17.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 18.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 19.Kandels-Lewis S, Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 20.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–197. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 23.Chan S-P, Cheng S-C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 24.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu Y-F, et al. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol. 2009;29:5671–5678. doi: 10.1128/MCB.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan R, et al. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol. 2013;20:1450–1457. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [This biochemical study proposed that DEAH-box ATPases bind away from their spliceosomal target and pull on the intron at a distance to remodel the spliceosome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng C-K, Liu H-L, Cheng S-C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warkocki Z, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 30.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James S-A, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5‘ and 3’ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 33.Sontheimer EJ, Steitz JA. The U5 and U6 Small Nuclear RNAs as Active Site Components of the Spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 34.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 35.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fourmann J-B, et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013;27:413–428. doi: 10.1101/gad.207779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie DB, et al. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol. 2008;15:1199–1205. doi: 10.1038/nsmb.1505. [DOI] [PubMed] [Google Scholar]
- 40.Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J. 2008;27:2929–2940. doi: 10.1038/emboj.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K, Zhang L, Xu T, Heroux A, Zhao R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos KF, et al. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci USA. 2012;109:17418–17423. doi: 10.1073/pnas.1208098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mozaffari-Jovin S, et al. Inhibition of RNA Helicase Brr2 by the C-Terminal Tail of the Spliceosomal Protein Prp8. Science. 2013;341:80–84. doi: 10.1126/science.1237515. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen THD, et al. Structural Basis of Brr2-Prp8 Interactions and Implications for U5 snRNP Biogenesis and the Spliceosome Active Site. Structure. 2013;21:910–919. doi: 10.1016/j.str.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pena V, et al. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Leung AKW, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–539. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Hang J, Zhou Y, Wan R, Lu G, Yin P, Yan C, Shi Y. Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA. Nature. 2014;506:116–120. doi: 10.1038/nature12803. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 50.Lin P-C, Xu R-M. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J. 2012;31:1579–1590. doi: 10.1038/emboj.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cretu C, et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell. 2016;64:307–319. doi: 10.1016/j.molcel.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 52.Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kondo Y, Oubridge C, van Roon A-MM, Nagai K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition. Elife. 2015;4:e04986. doi: 10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–4184. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark H, Lührmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 56.Sander B, et al. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 57.Boehringer D, et al. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol. 2004;11:463–468. doi: 10.1038/nsmb761. [DOI] [PubMed] [Google Scholar]
- 58.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- 59.Golas MM, et al. 3D cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Mol Cell. 2010;40:927–938. doi: 10.1016/j.molcel.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 60.Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc Natl Acad Sci USA. 2007;104:3195–200. doi: 10.1073/pnas.0611591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen-Krausz S, Sperling R, Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J Mol Biol. 2007;368:319–27. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 62.Kühlbrandt W. Cryo-EM enters a new era. Elife. 2014:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabrizio P, et al. The Evolutionarily Conserved Core Design of the Catalytic Activation Step of the Yeast Spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 64.Plaschka C, Lin P-C, Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [This paper determined the cryo-EM structure of the spliceosomal B complex, providing insight into Brr2-mediated activation of the spliceosome.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. [The structure of yeast spliceosome after Brr2-mediated activation] [DOI] [PubMed] [Google Scholar]
- 66.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen TH, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [The cryoEM structure of the yeast U4/U6.U5 tri-snRNP revealed the organization of protein and RNA components in a large spliceosomal complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen THD, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [This paper described a complete architecture of the 1.5 MDa U4/U6.U5 tri-snRNP consisting of 3 snRNAs and 30 proteins and the mechanism of U4/U6 unwinding by the Brr2 helicase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan R, et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [This paper also describes the structure of essentially the same yeast U4/U6.U5 tri-snRNP.] [DOI] [PubMed] [Google Scholar]
- 70.Wu N-Y, Chung C-S, Cheng S-C. Role of Cwc24 in the First Catalytic Step of Splicing and Fidelity of 5' Splice Site Selection. Mol Cell Biol. 2017;37:e00580–16. doi: 10.1128/MCB.00580-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [This study revealed the cryo-EM structure of a catalytic spliceosome with the products of the first catalytic reaction bound to the active site, uncovered novel RNA interactions that position. the branch adenosine for catalysis, and provided support for action at a distance by the Prp16 ATPase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan R, et al. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science. 2016;353:895–904. doi: 10.1126/science.aag2235. [This paper also presented the cryo-EM structure of the catalytic spliceosome after the first step of splicing.] [DOI] [PubMed] [Google Scholar]
- 73.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yean SL, Wuenschell G, Termini J, Lin R-J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [This study identified U6 snRNA phosphate oxygens that coordinate catalytic metal ions for branching and exon-ligations and shows that both reactions are catalyzed by a single RNA-based active site.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol. 2014;21:464–471. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [This paper presented the cryo-EM structure of a spliceosome just before the second catalytic step showed that Prp16-induced structural change undocks the branch helix from the active site and create a space for 3’-exon docking for exon ligation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355:149–155. doi: 10.1126/science.aak9979. [This paper also describes the cryoEM structure of the yeast C* spliceosome.] [DOI] [PubMed] [Google Scholar]
- 79.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3' splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 80.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 81.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 82.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 83.Query CC, Konarska MM. CEF1/CDC5 alleles modulate transitions between catalytic conformations of the spliceosome. RNA. 2012;18:1001–1013. doi: 10.1261/rna.029421.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 85.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu Y-Z, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lardelli RM, Thompson JX, Yates JR, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wlodaver AM, Staley JP. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA. 2014;20:282–294. doi: 10.1261/rna.042598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5' splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5' splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [The cryoEM structure of S. pombe intron lariat spliceomse was the first high-resolution structure of a spliceosome and provided a first view of the Prp19-associated complex.] [DOI] [PubMed] [Google Scholar]
- 94.Garrey SM, et al. A homolog of lariat-debranching enzyme modulates turnover of branched RNA. RNA. 2014;20:1337–1348. doi: 10.1261/rna.044602.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [The structure of a human spliceosome is remarkably similar to its yeast counterpart but interestingly two yeast proteins,Cwc2 and Ecm2, are fused to one polypeptide.] [DOI] [PubMed] [Google Scholar]
- 96.Zhang X, et al. An Atomic Structure of the Human Spliceosome. Cell. 2017;169:918–929.e14. doi: 10.1016/j.cell.2017.04.033. [This paper presented a high-resolution structure of a human spliceosome and uncovered mammalian-specific protein factors that stabilize the human C* conformation.] [DOI] [PubMed] [Google Scholar]
- 97.De I, et al. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat Struct Mol Biol. 2015;22:138–144. doi: 10.1038/nsmb.2951. [DOI] [PubMed] [Google Scholar]
- 98.Agafonov DE, et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351:1416–1420. doi: 10.1126/science.aad2085. [The cryoEM structure of the human tri-snRNP shows that Sad1 tethers the Brr2 helicase at pre-activation position and keeps it away from the substrate U4 snRNA.] [DOI] [PubMed] [Google Scholar]
- 99.Wolf E, et al. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 2009;28:2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bertram K, et al. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell. 2017;170:1–13. doi: 10.1016/j.cell.2017.07.011. [DOI] [PubMed] [Google Scholar]