Abstract
Little is known about the potential for ion channels to regulate cellular behaviors during tissue regeneration. Here, we utilized an amphibian tail regeneration assay coupled with a chemical genetic screen to identify ion channel antagonists that altered critical cellular processes during regeneration. Inhibition of multiple ion channels either partially (anoctamin1/Tmem16a, anoctamin2/Tmem16b, KV2.1, KV2.2, L-type CaV channels and H/K ATPases) or completely (GlyR, GABAAR, KV1.5 and SERCA pumps) inhibited tail regeneration. Partial inhibition of tail regeneration by blocking the calcium activated chloride channels, anoctamin1&2, was associated with a reduction of cellular proliferation in tail muscle and mesenchymal regions. Inhibition of anoctamin 1/2 also altered the post-amputation transcriptional response of p44/42 MAPK signaling pathway genes, including decreased expression of erk1/erk2. We also found that complete inhibition via voltage gated K+ channel blockade was associated with diminished phagocyte recruitment to the amputation site. The identification of H+ pumps as required for axolotl tail regeneration supports findings in Xenopus and Planaria models, and more generally, the conservation of ion channels as regulators of tissue regeneration. This study provides a preliminary framework for an in-depth investigation of the mechanistic role of ion channels and their potential involvement in regulating cellular proliferation and other processes essential to wound healing, appendage regeneration, and tissue repair.
1. Introduction
Ion channels are known for traditional physiological functions, including muscle contraction, nerve conduction, and maintenance of ionic homeostasis. However, ion channels modulate membrane ion conductance across all cells and tissues, establishing electrical fields (EF) that affect cellular behaviors under normal conditions, during critical periods of development, and in response to tissue injury. Understanding how ion channels function within different biological contexts is central to identifying the molecular basis of channelopathies and for exploiting ion channels for wound healing and tissue repair. Bioelectric signaling via ion channels and control of cellular membrane potentials in planarian and Xenopus regeneration have significantly contributed to our understanding of ionic influences on regenerative processes (Levin, 2007; Levin, 2009; Tseng et al., 2010; Levin, 2014). Despite these advances, little is known about the individual channels that are important during regeneration and the specific cellular functions that they influence.
Amphibians and fish provide powerful models to investigate the role of EF and ion channels on cellular processes that are activated during appendage regeneration. Typically in these vertebrates, amputated body parts are flawlessly repaired via the activation, proliferation, and patterning of progenitor cells (McCusker and Gardiner, 2014; Tanaka, 2016). Wound-induced EFs likely affect the behavior of immune and progenitor cells because they are enacted during the early wound-healing response, and interruption or reversal of an EF is detrimental to regeneration (Borgens et al., 1979; Borgens et al., 1977; Jenkins et al., 1996). However, exactly how cells detect and translate EF information to elicit specific cell behaviors is not well understood. One possibility is that information from an EF is modulated and transduced by ion channels. Consistent with this idea, Özkucur et al. (2010)) showed that ion concentrations are modulated in cells near the amputation site during axolotl (Ambystoma mexicanum) tail regeneration. This observation suggests that ion channels affect cellular behaviors by regulating ion concentration-dependent signaling cascades during regeneration (Wei et al., 2011; Mao et al., 2009; Wondergem et al., 2001; Wang et al., 2002; Tao et al., 2008).
In this study, we performed a chemical genetic screen to identify ion channels whose functions are required for normal axolotl tail regeneration. We administered tail amputations to axolotl embryos and treated groups with ion channel antagonists. This type of hierarchical drug screen has been previously described, and chemical genetics approaches have proven to be robust for identifying bioelectric mechanisms that affect regeneration (Adams and Levin, 2006; Sengupta et al., 2015). Multiple channels were identified from several ion channel families that either delayed the regenerative process or inhibited the initiation of regeneration. Two of these channels, anoctamin 1 and voltage-gated K+ channels, were investigated further to determine their effects on cellular proliferation and the activity of phagocytic cells.
2. Methods
2.1. Anesthesia
Experiments were conducted using pre-feeding, hatchling stage axolotl embryos (RRID:AGSC_100H, 9–11 mm in length) obtained from the Ambystoma Genetic Stock Center (RRID:SCR_006372), Department of Biology, University of Kentucky. These nearly mature embryos were removed from chorions immediately before the onset of the experimental protocol. Ethical animal procedures performed in this study were approved by the University of Kentucky IACUC committee (protocol 00907L2005). The skeletal muscle-specific myosin inhibitor N-benzylp-toluene sulfonamide (BTS) was used to anesthetize embryos before all amputations and imaging procedures. This was chosen over benzocaine, a voltage gated Na+ channel inhibitor, as to not confound the results of the ion channel antagonist being investigated. Embryos were placed in modified Holtfreter’s solution (59 mM NaCl, 0.67 mM KCl, 0.76 mM CaCl2, 2.4 mM NaHCO3) with 10 μM BTS at least 15 min before conducting any procedures.
2.2. Regeneration assay
Axolotl tails were imaged under a dissecting microscope and amputated approximately halfway between the tail tip and cloaca (~2 mm). Embryos were then placed in individual wells of a 24-well tissue culture plate with 3 mL of Holtfreter’s solution containing various ion channel antagonists or a modified Cl− free Holtfreter’s solution (15 mM NaC6H11O7, 0.2 mM KC6H11O7, 0.2 mM Ca(C6H11O7)2, 0.2 mM MgSO4·7H20, 0.6 mM NaHCO3). The drug screen consisted of three tiers of ion channel antagonist ranging from broad scale general ion conductance inhibitors (tier 1), to antagonists of ion channel sub families (tier 2), and finally, specific ion channel blockers (tier 3, Table 1). All drugs were initially evaluated at four concentrations (10−4, 10−5, 10−6 and 10−7 M) with 6 biological replicates for each concentration; some concentrations were lethal and this resulted in n < 6 for some drug treatments. Subsequently, ranges of drug concentrations were evaluated on a case-by-case basis and conclusions regarding a drug’s impact on regeneration were only drawn from nontoxic concentrations of drugs. Drugs that exhibited systemic toxicity (identified by general atrophy, lethargy and/or tissue degeneration) or lethality at all concentrations were excluded from the study. Lethal or toxic drug concentrations are emphasized in bold in Table 1. At 7 days post-amputation (dpa), embryos were anesthetized, imaged, and tail length was measured. To assess the extent of regeneration, tail length at day 0 (Fig. S1A) was subtracted from tail length at day 7 (Fig. S1B). Following a positive result, drugs were assayed for their impact on normal development in unamputated axolotl embryos and drugs found to adversely affect normal developmental growth were excluded from the study. Results for each pharmacological agent were analyzed in Sigma Plot Statistical Software (SPSS) using a one-way ANOVA with Dunnett’s test for post hoc analysis. Significant results were qualitatively assigned as either delayed/reduced regeneration, inhibited regeneration, or toxic/lethal (Fig. S1B–E).
Table 1.
This table summarizes all compounds and the concentrations of each compound screened for an effect on regeneration. Every concentration of each compound was screened using 6 or more animals. Concentrations were deemed to be lethal and/or systemically toxic (listed in bold) if 60% or greater of the animals in the screen either died or exhibited characteristics of toxicity (anatomical abnormalities in the gills, reduced blood flow in gills, general atrophy and degeneration).
| Tier 1 | Tier 2 | Tier 3 | ||||
|---|---|---|---|---|---|---|
| Drug | Concentrations (μM) | Drug | Concentrations (μM) | Drug | Concentrations (μM) | |
| Sodium | Amiloride | 0.1, 1, 10 & 100 | ||||
| Lidocaine | 0.1, 1, 10 & 100 | |||||
| Tetrodotoxin | 0.01, 0.1, 1, 10 & 100 | |||||
| Chloride | DIDS | 0.1, 1, 10 & 100 | CaCCinh-A01 | 0.1, 1, 2, 4, 6, 8, 10 & 100 | T16Ainh-A01 | 0.1, 1, 10 & 100 |
| Benzbromarone | 0.01, 0.05, 0.1, 0.33, 0.66, 1, 10 & 100 | |||||
| ClC channels | GaTx2 | 0.001, 0.01, 0.1, 1 & 10 | ||||
| DNDS | 0.1, 1, 10 & 100 | Ligand gated Cl channels | Picrotoxin | 0.1, 1, 10 & 100 | ||
| Bicuculine Methiodide | 0.1, 1, 10 & 100 | |||||
| Niflumic acid | 0.1, 1, 10 & 100 | Volume-sensitive anion channel | DCPIB | 0.1, 1, 10 & 100 | ||
| Cystic fibrosis transmembrane receptor | CFTRinh-172 | 0.1, 1, 10 & 100 | ||||
| Potassium | Tetraethylammonium | 0.1, 1, 10 & 100 | 4-Aminopyridine | 0.1, 1, 2, 4, 6, 8, 10 & 100 | Cytochalasin B | 0.001, 0.01, 0.1, 1 & 10 |
| Dapoxetine | 0.1, 1, 10 & 100 | |||||
| Phrixotoxin-1 | 0.0001, 0.001, 0.01, 0.1, 1 & 10 | |||||
| Guangxitoxin-1E | 0.001, 0.01, 0.1, 1 & 10 | |||||
| Terfenadine | 0.1, 1, 2, 4, 6, 8, 10 & 100 | |||||
| Apamin | 0.01, 0.1, 1 & 10 | |||||
| Glibenclamide | 0.1, 1, 10 & 100 | |||||
| Calcium | Bepridil HCl | 0.1, 1, 10 & 100 | Benidipine HCl | 0.1, 1, 10 & 100 | Amlodipine | 0.1, 1, 10 & 100 |
| Diltiazem | 0.1, 1, 10 & 100 | |||||
| Cinnarizine | 0.1, 1, 10 & 100 | |||||
| ω-Conotoxin MVIIC | 0.001, 0.01, 0.1, 1 & 10 | ω-Agatoxin | 0.01, 0.05, 0.1, 1 & 10 | |||
| Pumps/transporters | Esomeprazole | 0.1, 1, 10 & 100 | ||||
| Pantoprazole | 0.1, 1, 10 & 100 | |||||
| Furosemide | 0.1, 1, 10 & 100 | |||||
| Ouabain | 0.1, 1, 10, 50 & 100 | |||||
| tBuBHQ | 0.1, 0.5, 1, 2, 4, 6, 8, 10 & 100 | |||||
2.3. Phagocyte imaging
Live imaging of phagocytes was accomplished by staining with the non-toxic dye, neutral red. Neutral red stains lysosomes which are present in many cell types but are especially rich in phagocytes such as macrophages. Also, neutral red has been confirmed as an effective and specific stain for macrophages in zebrafish and axolotl using the protocol described below (Herbomel et al., 2001; Davis and Ramakrishnan, 2009; Godwin et al., 2013). Prior to imaging, embryos were placed in Holtfreter’s solution containing 5 μg/mL of neutral red dye for 6 h and then de-stained in normal Holtfreter’s solution for 24 h. Following this staining/de-staining procedure, embryos were anesthetized and imaged under bright-field (Herbomel et al., 2001; Godwin et al., 2013; Carradice and Lieschke, 2008). Images were quantified by counting all labeled cells within 500 μm of the amputation plane and these values were normalized by tissue section area. Results were analyzed in SPSS using a one-way ANOVA with Dunnett’s test for post hoc analysis (control was t = 0).
2.4. Proliferation assay
Axolotl embryos were amputated and placed in 24 well plates as outlined above. At 3 dpa, embryos were incubated for 16 h in 10 μM 5-ethynyl-2′-deoxyuridine (EdU), a BrdU analog. After this incubation period, embryos were euthanized and tails were amputated at the cloaca, immediately fixed in 4% paraformaldehyde, and stored in 70% EtOH. Tissues were then embedded in paraffin wax and cut into 5-micron sagittal sections. The Click-iT® EdU Alexa Fluor® 488 Imaging Kit (C10337, invitrogen) was used to visualize EdU incorporation into the DNA of proliferating cells per the prescribed protocol. Hoechst was used to counter stain the sections. All cells staining positive for EdU incorporation within 500 μm of the amputation plane were counted as well as all cells staining positive for Hoechst within the same anatomical area. Proliferative index was determined by dividing the number of EdU+ cells by the number of Hoechst + cells in each tissue section. Results were analyzed in SPSS using either a one-way ANOVA with Dunnett’s test for post hoc analysis or, in the case of tissue specificity analysis, a two-way ANOVA (factors were tissue type and drug treatment) with Student-Newman-Keuls (SNK) test used for post hoc analysis.
2.5. Apoptosis assay
Axolotl embryos were administered tail amputations and placed in 24 well plates and allowed to regenerate for 3 days. Embryos were euthanized and tails were amputated at the cloaca, immediately fixed in 4% paraformaldehyde, and stored in 70% EtOH. Tissues were then embedded in paraffin wax and cut into 5-micron transverse sections. The Click-iT® TUNEL Alexa Fluor® 594 Imaging Kit (C10246, invitrogen) was used to assay for fragmented DNA, a measure of apoptosis (Gavrieli et al., 1992). Tissue sections were grouped as either 0–100 μm or 150–250 μm from the tail tip. TUNEL positive cells in each group were counted and divided by the total number of cells in each group (Hoechst) to give a measure of apoptotic index. Results were analyzed in SPSS using one-way ANOVA and SNK test for post hoc analysis.
2.6. RNA extraction and quantitative real-time PCR
Embryos were administered amputations and placed in 24-well plates as before and allowed to regenerate for 3 days in either modified Holtfreter’s or modified Holtfreter’s supplemented with the anoctamin 1 inhibitor, T16A(inh)-A01 (10 μM). The distal 1 mm of the tail was re-amputated and collected in ice-cold RNAlater (25 mM C6H7NaO7, 10 mM EDTA, 5.3 M (NH4)2SO4 and pH 5.2) and stored at −20°C. To isolate mRNA, 2 tails were placed in 100 μL of Trizol, homogenized via sonication, and purified via Zymo Direct-zol™ RNA MiniPrep (R2050) kit and treated with DNase 1. RNA concentration and purity were determined via Nanodrop ND-1000 spectrophotometry (Nanodrop Technologies). cDNA was generated using qScript™ cDNA Supermix (Quanta Biosciences 95,048) and used for PCR. PCR primers were designed using sequence data available on Sal-Site and are listed in Table 2 (Baddar et al., 2015). Quantitative real-time PCR was performed using the StepOnePlus™ Real-Time PCR system (Applied Biosystems®). Samples were loaded into 96-well plates in triplicate with probes and Power SYBR® Green PCR Master Mix (4367659) to detect PCR products. Relative gene expression was calculated using the comparative CT method (2−ΔΔCT) in StepOne software v2.3 per the manufacturer’s instructions (Schmittgen and Livak, 2008). Relative quantifications were compared in SPSS using a one-way ANOVA and Student–Newman–Keuls (SNK) method for post hoc analysis.
Table 2.
This table summarizes the primers used to measure gene transcription by quantitative real-time PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | ACGTCTCTGTGGTTGACTTG | TTCCCTTCATTGGTCCATCAG |
| PKC-γ | GGCAGTCGTGAGATGAGTTT | ACCGATACAAGCTGAGTGAAG |
| PKC-α | CGTAGAATGCACGATGGTAGAA | TCCTCAGTTTGGAAGCAAGAA |
| PKC-β | CGCATGAAGCTATCCGACTT | CATAGAGCTCGTCAGTACCTTTC |
| RRAS | GTCCACATTAAGCCGGATCT | CCTACATAGACAGGTGCCAAA |
| RAF1 | AGGAGACCAAGTTTCAGATGTT | GTCCCTGTGGATGATGTTCTT |
| MAPKK1 | AGCTCCTGTGAAGCGTATTC | CCTAGATCTGCCCTGCATTT |
| MAPKK2 | GAGGAAGGGAAACCGAACATAA | CTTAGCTCGTCTACAGCCAATC |
| MAPK1/ERK2 | CGGGCACCAGAGATAATGTT | GGAAGATGGGTCTGTTAGATAGC |
| MAPK3/ERK1 | CGCATTGGATCTGCTGGATAA | GGTCGTAGTACTGTTCCAGGTA |
3. Results
3.1. Pharmacological screen of ion channel groups
3.1.1. Chloride
Nonspecific chloride conductance was examined with the broad-scale Cl− channel blockers 4,4′-Diisothiocyanatostilbene-2,2′-disulfonic acid (DIDS), 4,4′-dinitrostilbene-2,2′-disulfonic acid (DNDS), and niflumic acid (NFA). Embryos treated with any of these broad scale Cl− channel antagonists exhibited decreased tail regeneration at all concentrations tested, with the exception of DNDS at 1 μM. For example, embryos exposed to 10 μM DIDS, DNDS, or NFA regenerated 0.71 ± 0.31 mm, 1.11 ± 0.32 mm, and 0.63 ± 0.24 mm respectively, significantly less than untreated embryos (Fig. 1A). Calcium-activated Cl− channels (CaCC) were evaluated with the family-specific inhibitor CaCCinh-A01. CaCCinh-A01 was lethal at concentrations N2 μM but reduced regeneration at 0.1 μM (1.01 ± 0.24 mm) and 1 μM (1.09 ± 0.17 mm, Fig. 1B) without observable toxic effects. Anoctamins 1&2 (aka Tmem16A/B, Ano1, Ano2) were inhibited with benzbromarone (BBM) or T16a(inh)-A01 (A01). Embryos treated with either BBM or A01 had decreased regenerative capacity at 100 nM (0.56 ± 0.18 mm) and 10 μM (0.88 ± 0.21 mm, Fig. 1C&D). The ligand gated Cl− channels, GABAA receptors (GABAAR) and glycine receptors (GlyR), were inhibited with picrotoxin (PTx) and bicuculline methiodide (BCU, GABAAR only) and both compounds caused reduced regenerative capacity at all concentrations tested. Both PTx (0.645 ± 0.27 mm) and BCU (0.618 ± 0.32 mm) elicited complete inhibition of regeneration at a concentration of 100 μM (Figs. 1E&F). All other Cl− channel blockers were either toxic or did not affect regeneration (Fig. S2).
Fig. 1.
(A) All broad scale inhibitors of chloride channels (DNDS, DIDS and NFA) robustly reduced tail regeneration at concentrations of 100 nM and above. (B–D) Inhibitors of calcium activated chloride currents with CaCCinh-A01, T16a(inh)-A01 and Benzbromarone all reduced regenerative growth in a concentration dependent manner with maximal responses at 2, 100 and 0.33 μM respectively. (E & F) PTx and BCU were used to investigate the role of ligand-gated chloride channels and both exhibited robust inhibition of regeneration at all concentrations tested (* indicates p < 0.05 compare to control or concentration = 0; error bars are standard deviations, n values depicted on bar graphs represent biological replicates).
3.1.2. Potassium
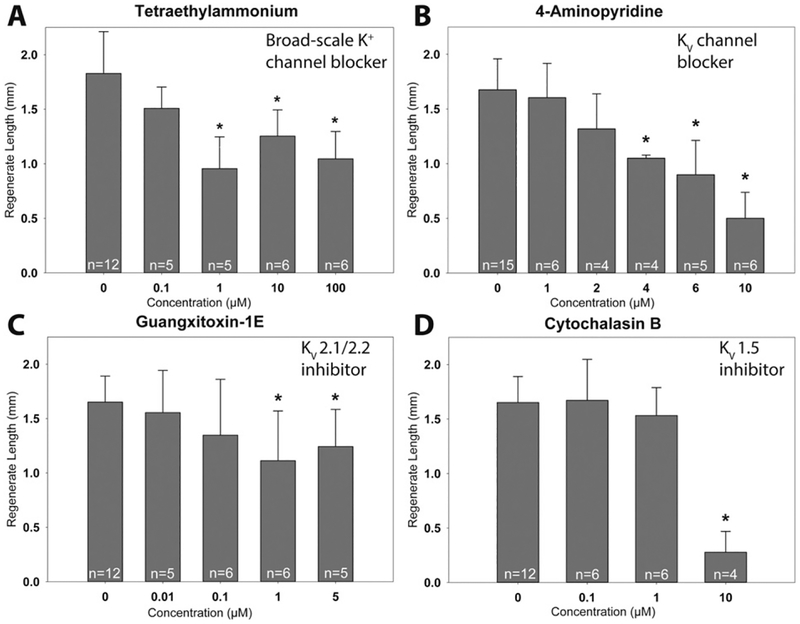
General potassium conductance was initially examined with the broad-scale K+ channel blocker tetraethylammonium (TEA). Compared to control embryos (1.83 ± 0.38 mm), TEA treated embryos at a concentration of 1 μM or higher decreased regenerative growth (0.96 ± 0.29 mm, Fig. 2A). The voltage-gated K+ channel (KV) blocker 4-aminopyridine (4-AP) reduced regenerative outgrowth at 4 μM (1.05 ± 0.03 mm) and completely inhibited regeneration at 10 μM (0.50 ± 0.24 mm) and 100 μM (0.59 ± 0.27 mm, Fig. 2B). KV2.1/2.2 blockade with Guangxitoxin-1E (GTx-1E) slightly reduced regeneration at 1 μM (1.11 ± 0.46, Fig. 4C). KV1.5 blockade with cytochalasin B (CytB) had no effect on regeneration when embryos were incubated at 0.1 or 1 μM, but tail outgrowth was inhibited at 10 μM (0.16 ± 0.11 mm, Fig. 2D). All other K+ channel blockers used were either toxic or did not affect regeneration (Fig. S3).
Fig. 2.
(A) While the broad scale potassium channel inhibitor TEA only reduced regenerative growth (B) the voltage-gated K+ channel blocker 4-AP exhibited concentration dependent inhibition. (C) KV2.1/2.2 channels that were inhibited with GTx-1E exhibited a slight reduction of regenerative growth at 1 and 5 μM but was lethal at all concentrations above 5 μM and (D) CB exhibited complete inhibition at 10 μM (* indicates p < 0.05 compare to control or concentration = 0; error bars are standard deviations, n values depicted on bar graphs represent biological replicates).
Fig. 4.
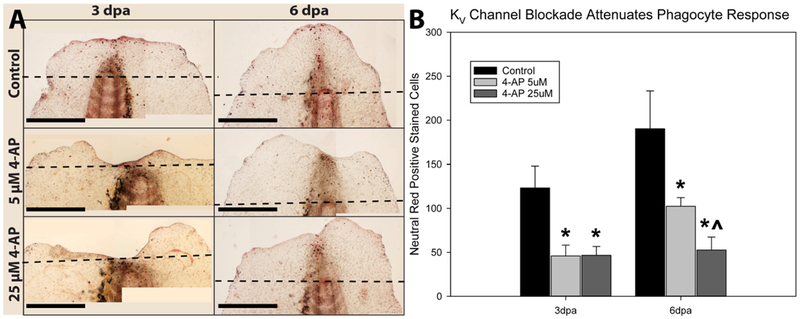
Phagocytes were stained in live animals with neutral red at 3 and 6 dpa. Panel (A) depicts representative micrographs of all time points and treatments (amputation plane indicated by black dashed line). (B) 4AP inhibits phagocyte activation and recruitment to the wound site at concentrations of either 5 or 25 μM. This response is concentration dependent at 6dpa but not 3dpa (* indicates p < 0.05 compared to control and ^ indicates p < 0.05 compared animals treated with 5 μM 4AP; scale bars represent 1 mm; error bars are standard deviations).
3.1.3. Sodium
Sodium was assessed using only tier 2 antagonists as no tier 1 drugs were available (Table 1). The epithelial sodium channel (ENaC) blocker amiloride did not affect regeneration at any concentration tested. The voltage-gated sodium channel (NaV) antagonists, tetrodotoxin (TTx) and lidocaine, also did not affect regeneration at any concentration tested, although the highest concentration of TTx (100 μM) was lethal in 100% of embryos (Fig. S4).
3.1.4. Calcium
Calcium conductance was examined with the broad-scale Ca2+ channel blocker bepridil (BPD). BPD was 100% lethal at every concentration tested. The L, N & T-type calcium channel inhibitor, benidipine HCl (BNP), completely inhibited regeneration at 100 nM (0.60 ± 0.35 mm) and all other concentrations were lethal. L-type channel blockers amlodipine (CaV1.3) and diltiazem (CaV1.1/1.2/1.4) diminished the regenerative response, with a maximal effect observed 10 μM (1.03 ± 0.38 mm) and 1 μM (1.04 ± 0.50 mm), respectively. The P & Q-type blocker, ω-conotoxin MVIIC, completely inhibited regeneration with a maximal effect at 10 nM (0.57 ± 0.19 mm). P-type specific inhibitor, ω-agatoxin IVA, had no effect on the regenerative response at any concentration, thus eliminating Q-type channels as affecting regeneration (Fig. S5).
3.2. Pumps/transporters
The sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) was examined with 2,5-di-(tert-butyl)-1,4-benzohydroquinone (tBuBHQ), which completely inhibited regeneration with maximal inhibition at 2 μM (0.41 ± 0.15 mm). Incubation with the H+/K+-ATPase inhibitor, pantoprazole, resulted in a slight reduction of regenerative growth at 100 μM (1.14 ± 0.05 mm). All other transporter inhibitors used were either toxic or did not affect regeneration (Fig. S6).
3.3. Phagocyte activation is dependent on KV channel signaling
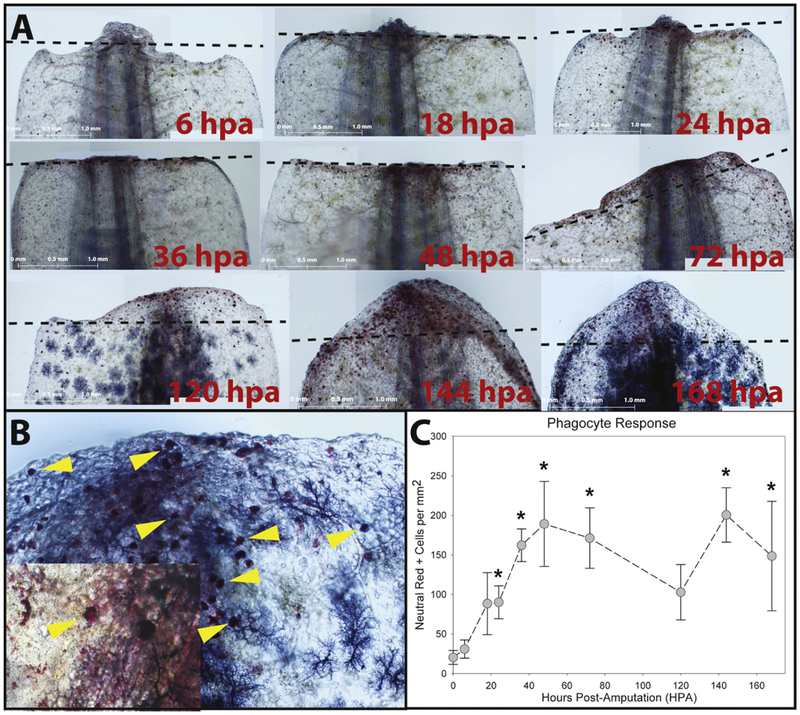
Macrophage activation is required for regeneration to proceed normally and KV channels are known to influence many functions of macrophages in vitro (Godwin et al., 2013; Kotecha and Schlichter, 1999; Moreno et al., 2013; Qiu et al., 2002; Vicente et al., 2005; Li et al., 2016). This suggests that KV’s may influence tail regeneration through modulation of the macrophage response. To test this hypothesis, it was first necessary to characterize the normal response of phagocytic cells during tail regeneration. Using neutral red (NR) to label phagocytic cells, a significant accumulation of phagocytes was observed at the wound site beginning 24 hours post amputation (hpa, 90.1 ± 20.8 NR+ cells/mm2 tissue) and initially peaking at 48 hpa (189.2 ± 53.7 NR+ cells/mm2 tissue). This was followed by a decline concurrent with tissue outgrowth and then a second peak at 144 hpa (200.6 ± 34.4 NR+ cells/mm2 tissue, Fig. 3). NR stained cells were confirmed to be phagocytic by repeating the NR staining procedure on embryos that had been previously injected with liposome encapsulated clodronate. In comparison to control embryos, clodronate treated embryos exhibited a significant reduction of NR+ cells (Fig. S7).
Fig. 3.
Phagocytes were stained in live regenerating axolotl embryos with neutral red at 6, 18, 24, 36, 48 (2 days), 72 (3 days), 120 (5 days), 144 (6 days) & 168 (7 days) hpa. Panel (A) depicts representative micrographs of each time point (amputation plane indicated by black dashed line) with enlarged photos in panels (B & B′) showing individual phagocytes. Representative (but not all) NR+ cells are indicated by yellow arrowheads. (C) Phagocytes were counted from 500 μM proximal of the amputation plane to the tip of the tail and normalized by tissue cross-sectional area. Phagocyte density steadily increased over the first few days following amputation peaking between days 2 and 3. This was followed by a trough in phagocyte density between days 3 and 5 that corresponded to increased tissue outgrowth, and then a second phagocyte density peak at day 6 (* indicates p < 0.05 compared to phagocyte density immediately following amputation; scale bars represent 1 mm; error bars are standard deviations). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
While CytB is a potent inhibitor of KV1.5 channel activity, it has off target effects, such as disruption of actin polymerization that could also influence regeneration. To further assess the role of KV channels on phagocyte activity we used an alternate KV channel blocker, 4-AP. Embryos were incubated at 2 concentrations of 4-AP (5 μM and 25 μM) beginning at 12 hpa and harvested at either 3 or 6 days post amputation (dpa). At 3 dpa, embryos incubated in either 5 μM (45.88 ± 12.35 NR+ cells) or 25 μM (46.63 ± 10.07 NR+ cells) exhibited a marked reduction in phagocyte recruitment to the wound site compared to control embryos (123.33 ± 15.67 NR+ cells) but there was no difference between the two concentrations tested. At 6 dpa, the reduction of phagocyte recruitment persisted compared to control (190.29 ± 43.02 NR+ cells) and there was a concentration dependent response between the 5 μM group (102.38 ± 9.89 NR+ cells) and 25 μM group (52.71 ± 14.74 NR+ cells, Fig. 4). To confirm this result, embryos were subjected to one of three treatments: (1) Encapsome®/Fluorosome®-DiI 50:50 mixture IP injection 24 h before amputation (24hba), (2) Clodrosome®/Fluorosome®-DiI 50:50 mixture IP injection 24hba or (3) Encapsome®/Fluorosome®-DiI 50:50 mixture IP injection 24hba and incubation in 25 μM 4-AP beginning at 12 hpa. Embryos were then harvested at 3 dpa and whole mount imaged under fluorescent and bright field conditions. Embryos subjected to both the phagocyte depletion protocol (17.045 ± 17.30 DiI+ cells/mm2) and KV channel inhibition (31.80 ± 19.35 DiI+ cells/mm2) exhibited reduced phagocyte recruitment following amputation compared to control conditions (145.20 ± 77.17 DiI+ cells/mm2, Fig. 5). Notably, the number of phagocytes at the amputation site at 3 dpa under control conditions identified by Fluorosome®-DiI incorporation was consistent with the number identified by NR staining at the same time point.
Fig. 5.
Phagocytes were labeled by IP injection of Fluorosome®-DiI 24 h prior to amputation and co-injected with either Encapsome® as a vehicle control (B-B″; n = 10), Clodrosome® for macrophage depletion (C-C″, n = 15) and a final group co-injected with Encapsome® and then incubated with 25 μM 4-AP following amputation for KV channel blockade (D-D”; n = 14). (* indicates p < 0.05 compared to control; error bars are standard deviations) Both macrophage depletion prior to amputation (C-C″) and KV channel blockade following amputation (D-D″) reduced the number of DiI+ cells at the wound site.
3.4. Anoctamin 1 blockade delays regeneration via inhibition of proliferation
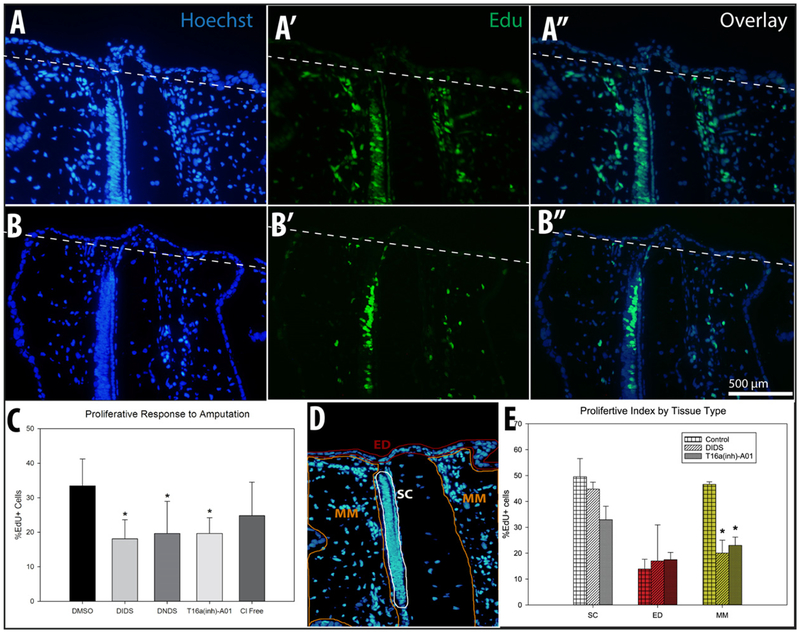
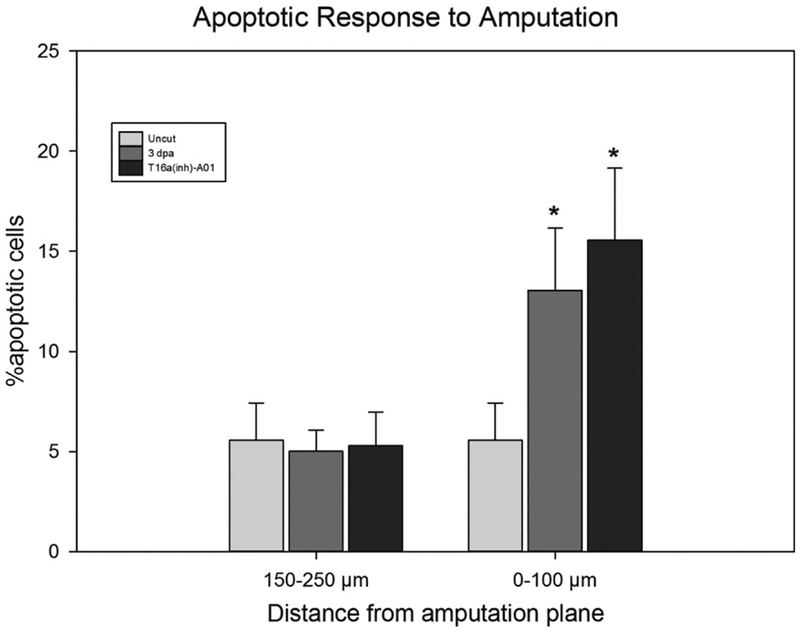
Cell proliferative responses to amputation were assessed by measuring EdU incorporation at 3 dpa under several experimental interventions. EdU incorporation was observed in 34.00 ± 6.44% of cells within 500 μm of the amputation plane under control conditions (n = 4, Fig. 6 A–A″ & C). In contrast, broad-scale Cl− channel blockade (DIDS: n = 4, 18.13 ± 5.52%& DNDS: n = 5, 19.65 ± 9.29%, Fig. 6C) or specific inhibition of anoctamin 1 (A01: n = 5, 19.65 ± 4.54%, Fig. 6 B–B″ & C) significantly reduced the percentage of cells proliferating within this same anatomical area. Embryos incubated in Cl− free Holtfreter’s exhibited a trend towards reduced proliferation (24.80 ± 9.74%, n = 5, Fig. 6C) but the difference was not statistically significant relative to controls. We note that apoptosis was assessed using a TUNEL assay; no differences were observed between embryos subjected to anoctamin 1 blockade and controls (Fig. 7).
Fig. 6.
Compared to controls (n = 4) (A-A″), the proliferative response to amputation was reduced by broad scale chloride channel blockade with either DIDS (n = 4) or DNDS (n = 5) (C) as well as anoctamin-1 channel blockade with T16a(inh)-A01 (n = 5) (B-B″)) but not when animals were incubated in chloride free medium (n = 5) (C) (amputation plane indicated by white dashed line). Taken together these data suggest that chloride channel signaling is a critical step in the mechanism(s) driving proliferation in response to amputation. (* indicates p < 0.05 compared to control; error bars are standard deviations) (D&E) Neither DIDS nor T16a(inh)-A01 had an effect on proliferation in spinal cord (SC) or epidermal (ED) tissues. In mesenchymal tissues (MM) directly underlying the wound epidermis and within 500 μm of the amputation plane, proliferation was reduced from 46.6 ± 0.7% in control animals to either 20.0 ± 3.5% (DIDS) or 23.0 ± 2.9% (T16a(inh)-A01) in treated animals (* indicates p < 0.05 compared to control of same tissue type; error bars are standard deviations).
Fig. 7.
Apoptosis was assessed following amputation in 3 dpa animals and compared to apoptotic rates in tissue from un-amputated axolotl tails. Tissues were cut into transverse sections and then divided into groups consisting of sections of either from the tail tip to 100 μM distal of the amputation plane and a second group consisting of tissue from 150 to 250 μM distal of the amputation plane. No differenced were observed in the latter group but the former exhibited similar increases in apoptotic cells in both the control 3 dpa animals and 3 dpa animals treated with the Ano1 inhibitor (* indicates p < 0.05 compared to un-amputated tail tissue).
To more finely examine proliferation spatially, cell counts were obtained for epidermal (ED), spinal cord (SC), and mesenchymal & muscle (MM) regions of the tail. Neither broad-scale Cl− channel blockade (DIDS & DNDS) nor anoctamin 1 inhibition (A01) treatments affected proliferation rates within the ED and SC tissues; the reduced proliferation response was only observed in MM tissues. These results suggest that functional chloride flux mediated by Ano1 is critical for regulating the proliferative response in the mesenchyme but not in other tissues (Fig. 6 D&E).
3.5. Anoctamin 1 blockade diminishes activation of p44/42 MAPK signaling pathway
To investigate gene expression changes associated with anoctamin 1 inhibition of tail regeneration, quantitative real-time PCR was used to measure mRNA expression of p44/42 MAPK signaling pathway genes, a pathway activated during cellular proliferation. After amputation, pkcγ, mek1, mek2, erk1 and erk2 showed a higher percent increase in transcription in control embryos vs anoctamin 1 treated embryos at 3 dpa. In contrast, expression was downregulated more strongly in control embryos than embryos subjected to anoctamin 1 inhibition by A01 at 3 dpa for early p44/42 pathway genes (pkcα, pkcβ, rras and raf1) (Fig. 8). These results show that anoctamin 1 affects the transcription of p44/42 MAPK pathway genes that regulate cellular proliferation.
Fig. 8.
Quantitative Real-Time PCR was used to assess the mRNA expression of genes associated with the p44/42 MAPK (Erk 1/2) signaling pathway at 72 h post amputation (hpa) in regenerating axolotl tails and at the same time point in animals treated with 20 μM T16Ainh-A01. Expression in tail tissues from un-amputated animals was used as control/baseline. All genes other than PKCγ exhibit modulated expression at 3 dpa, which was attenuated by Ano1 blockade (* indicates p < 0.05 compared to control and ^ indicates p < 0.05 compared to normal 72 hpa animals; error bars are standard deviations).
4. Discussion
In this study, we identified ion channel antagonists that partially (anoctamin1/Tmem16a, anoctamin2/Tmem16b, KV2.1, KV2.2, L-type CaV channels and H/K ATPases) or completely (GlyR, GABAAR, KV1.5 and SERCA pumps) inhibited axolotl tail regeneration. Below we separately discuss the affected ion channels from each of these categories (Fig. 9).
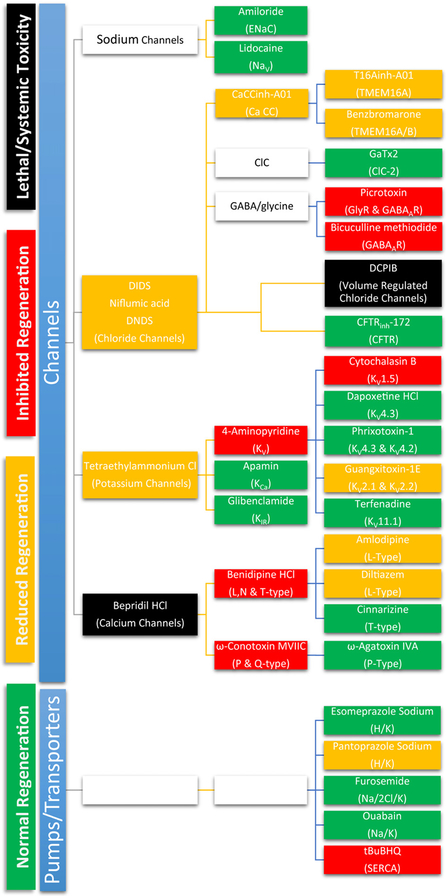
Fig. 9.
A comprehensive screen of ion channel blockers was used to identify specific channels involved in critical cellular processes of growth and regeneration such as proliferation, migration and differentiation. Drugs that did not affect regeneration are depicted in green, drugs that resulted in partial regeneration are depicted in yellow, drugs that completely inhibited regeneration (for any of the concentrations tested) are depicted in red and drugs that were lethal at all concentrations or caused systemic toxicity (identified by general atrophy, lethargy and/or tissue degeneration) are depicted in black. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.1. Partially inhibiting channels & transporters
4.1.1. Anoctamins 1/2 (Tmem16A/B)
Our results suggest a role for CaCCs in the tail regeneration process. Regeneration was delayed when embryos were subjected to either broad-scale Cl− channel blockade (DIDS, DNDS & niflumic acid) or CaCC inhibition (CaCCinh-A01, T16a(inh)-A01 & Benzbromarone). CaCC currents were described over 25 years ago but their molecular basis was only recently discovered (Hartzell et al., 2005). In 2008, three independent researchers identified anoctamin 1 and anoctamin 2 channels as mediators of CaCC currents (Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008). Anoctamin 1 was first discovered as an overexpression marker for gastrointestinal stromal tumors (then known as DOG1) and has since been shown to promote other forms of cancer (Britschgi et al., 2013; West et al., 2004). Blockade of anoctamin 1 suppresses tumor growth and invasion in multiple human cancer lines (Jia et al., 2015; Liu et al., 2012). There is evidence that anoctamin 1 manifested these effects through regulation of both cell migration and proliferation (Qu et al., 2014; Ruiz et al., 2012). Our results support this idea because inhibition of anoctamin 1 significantly decreased the number of proliferating cells in the regenerating tail. This suggests that anoctamin 1 channel function is directly or indirectly required to sustain cell proliferation at a level that is typical of normal tail regeneration.
Our results further suggest that anoctamin 1 affected tail regeneration by modulating the p44/42 MAPK pathway, which is a well-known regulator of cell proliferation (Zhang and Liu, 2002). Following tail amputation, Erk1/2 signaling pathway genes were transcriptionally upregulated to a higher degree in control embryos than embryos treated with anoctamin 1 antagonist. Thus, the magnitude of erk1/erk2 transcription correlated on one hand with anoctamin 1 function, and conversely, with the magnitude of cell proliferation. We propose the following model: anoctamin 1, activated by an initial calcium surge, acts as a countercurrent ion channel to amplify calcium signaling. This amplified surge subsequently activates the Erk1/2 pathway leading to increased cellular proliferation. This model assumes there is a burst in intracellular [Ca2+] following tail amputation and a dependence of intracellular Ca2+ flux on chloride conductance. Özkucur et al. (2010) downplayed the role of Ca2+ in their model of axolotl tail regeneration. However, they reported significant increases in Ca2+ fluorescence at 48 hpa, preceding the time when the tail shows measureable regenerative outgrowth. This Ca2+ flux at 48 hpa supports our proposed model (Özkucur et al., 2010; Barro-Soria et al., 2010; Wang and van Breemen, 1999).
4.1.2. KV2.1/2.2
KV2 channel blockade with TEA or GTx-1E also reduced regeneration. KV2 channels traditionally act as the primary delayed rectifier involved in regulating the excitability of neurons and facilitating exocytosis in neurons and neuroendocrine cells (Feinshreiber et al., 2009; Misonou et al., 2005). These functions of KV2 channels could be associated with the release of neurotrophic factors that are required for regeneration, the so-called neurotrophic requirement for regeneration (Kumar and Brockes, 2012; Singer, 1974). However, it is also important to consider the direct influence of KV2 channels on the cellular dynamics of non-excitable cells (Kumar and Brockes, 2012). KV2 acts as a promoter of cell migration in cultured HEK293, CHO, and bone derived mesenchymal stem cells (MSC) via phosphorylation of focal adhesion kinase (Hu et al., 2011; Wei et al., 2008). MSCs also require functioning KV2 channels for proper progression through the cell cycle by facilitating plasma membrane-endoplasmic reticulum contact sites (Cobb et al., 2015; Deng et al., 2007). These MSC specific characteristics of KV2 channels are particularly interesting considering their resemblance to progenitor cells of the blastema, but more research is required to determine the specific role of KV2 channels within the context of appendage regeneration.
4.2. L-type Ca2+ channels (CaV1.1–1.4)
The L-type channel blockers, amlodipine and diltiazem, partially inhibited tail regeneration. These channels are mostly known for their roles in excitable cells, where they couple plasma membrane depolarization with increases in Ca2+ conductance across the membrane. Recent evidence also suggests that they are important physiological components of many non-excitable cell types (Davenport et al., 2015; Wen et al., 2012). Ca2+ is a ubiquitous second messenger in all cell types and can influence many different cellular processes (Berridge, 1995; Berridge et al., 2000; Clapham, 2007; Zayzafoon, 2006; Zheng and Poo, 2007). Because of the widespread nature of L-type Ca2+ channel expression and the complicated features of intracellular Ca2+ signaling, these channels may not present the best targets for future investigations. But, if the model we proposed above is correct, the anoctamin 1/2 Cl− channels may provide an indirect path to modulate intracellular calcium flux in a specific cell population.
4.2.1. H+/K+ ATPases
Our study found that H+ pump inhibition with pantoprazole sodium reduced regenerative outgrowth. Proton pumps have been classically associated with their roles in gastric and renal function. However, over the past several decades they have been identified as critical mediators of wound healing and regeneration (Adams et al., 2007; Walan et al., 1989; Balestrini et al., 2017). Most of this influence has been attributed to the proton pump’s involvement in establishing and maintaining trans-epithelial electrical potentials that drive wound-induced electrical fields (Nuccitelli, 2003). A recent chemical genetic screen performed using planarian head regeneration also identified H+/K+ ATPase as important for initiating the regenerative process through control of cellular membrane potentials (Vmem) (Beane et al., 2011). H+ pump driven changes in Vmem are required for regeneration to proceed normally in the Xenopus model of regeneration (Adams et al., 2007). Our findings further validate the requirement of H+ pumps in organisms capable of appendage regeneration.
4.3. Inhibiting channels & transporters
4.3.1. KV1.5
Blockade of KV1.5 with 4-AP or CytB resulted in a robust and concentration dependent total inhibition of regeneration. These channels are widely expressed in a large number of tissues and are involved in regulating Vmem and electrophysiological properties of a variety of cell types (Archer et al., 1998; Olson et al., 2006; Tabarean, 2014). In addition to these electrophysiological functions, KV1.5 channels have been implicated in regulating cell cycle progression and migration (Kotecha and Schlichter, 1999; Vallejo-Gracia et al., 2013; Villalonga et al., 2008). Of particular interest is their role in regulating immune cell dynamics. Macrophages are required for amphibian regeneration and the invertebrate homologue of macrophages (hemocytes) respond to exogenously applied electrical fields in vivo in a KV dependent manner (Godwin et al., 2013; Franklin et al., 2016). Consistent with this line of reasoning, phagocyte activation was severely diminished during tail regeneration after KV channel blockade with 4-AP. This suggests that KV channels may be critical regulators of macrophage/monocyte populations during regeneration. However, further studies are necessary to further clarify the role for Kv channels in macrophage activation during tissue regeneration and whether or not this mechanism is a cell autonomous.
4.3.2. GlyR/GABAAR
GlyR and GABAAR blockade with picrotoxin or bicuculline resulted in strong inhibition of regeneration at relatively low concentrations. These ligand-gated chloride channels act primarily as receptors for inhibitory neurotransmitters in the CNS, with GABAAR acting mostly in the brain and GlyR functioning primarily in the brainstem and spinal cord (Lynch, 2004; Sigel and Steinmann, 2012). This immediately implicates nerves and their required neurotrophic factors for appendage regeneration since modulation of either of these receptors would disrupt normal neuronal activity during regeneration. There is evidence that these receptors influence neural progenitor cells during early development and regulate many critical cellular processes in multiple cell types, mostly via modulation of intracellular Ca2+ concentrations (Nguyen et al., 2001; Van Den Eynden et al., 2009). Importantly, misexpression of GlyR has recently been shown to disrupt patterning and development in neural, muscle and vascular tissues via physiological modulation of membrane potentials (Lobikin et al., 2012; Lobikin et al., 2014; Pai et al., 2015). It will be interesting to examine the expression patterns of these receptors in cells of regenerating axolotl tissues to see if they are altered following tissue injury. More research will be necessary to assess the exact role of GlyR/GABAAR during axolotl regeneration.
4.3.3. SERCA pump
SERCA inhibition with tBuBHQ completely inhibited regeneration at low concentrations, although higher concentrations of this drug were lethal. The sacroplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA) is expressed in virtually all cell types and distributes Ca2+ ions against their concentration gradient into sarcoplasmic and endoplasmic reticula. This allows for quick sequestration of cytoplasmic free Ca2+ following a Ca2+ signaling event and for the generation of intracellular calcium stores that allow for a more rapid and robust Ca2+ surge upon cellular stimulation (Wuytack et al., 2002). As discussed above, Ca2+ signaling is associated with many cytosolic signaling pathways and physiological functions, all of which require precise control of intracellular Ca2+ concentration pulses in terms of both magnitude and duration, and these Ca2+ dependent pathways are operative in models of epimorphic regeneration (Berridge et al., 2000; Rao et al., 2014). The ubiquitous nature of SERCA expression may explain why inhibition of this ion channel completely inhibited regeneration, while incomplete regeneration was observed for L-type Cav channel inhibition. Based on the extensive network of pathways influenced by Ca2+ signaling and the pervasive nature of SERCA pumps, SERCA pumps will present challenging targets to investigate mechanisms of tissue regeneration.
5. Summary
A chemical genetic screen was performed to identify ion channel antagonists that inhibit axolotl tail regeneration. Then, experiments were performed to determine how select antagonists affected cellular behaviors (cell proliferation and phagocyte activation) that are required for successful tail regeneration. The antagonists that were identified from the screen targeted the following ion channels: anoctamins 1/2 (Tmem16a/b), GlyR, GABAAR, KV1.5, KV2.1, KV2.2, L-type CaV channels, H/K ATPases and SERCA pumps. An association was established between KV channel blockade and phagocyte activation, and thus a possible mechanism for KV channel mediated inhibition of axolotl tail regeneration. Also, blockade of the anoctamins reduced cellular proliferation, and this was associated with modulated transcription of Erk1/2 signaling pathway genes. It has been shown previously that the transcriptional response to bioelectric signaling is conserved among axolotl regeneration, Xenopus development and human progenitor cells (Pai et al., 2015). This speaks to the broad relevance of the data presented in this report and lends credence to the idea that the antagonists and ion channels prioritized from our study will provide useful tools and targets for investigating mechanisms of tissue regeneration.
Supplementary Material
Acknowledgements
This work was funded by the Biomedical Sciences Research Group, LLC and the NIH (R24OD021479). Axolotl embryos used in this study were obtained from the Ambystoma Genetic Stock Center (AGSC) at the University of Kentucky. The AGSC is funded by the NIH (P40OD019794). The Authors would like to thank Deirdre Mcdonnell, for her expertise in quantitative PCR and other general lab work that contributed to this project.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mod.2017.06.001.
References
- Adams DS, Levin M, 2006. Inverse drug screens: a rapid and inexpensive method for implicating molecular targets. Genesis 44 (11), 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Masi A, Levin M, 2007. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development 134 (7), 1323–1335. [DOI] [PubMed] [Google Scholar]
- Archer SL, et al. , 1998. Molecular identification of the role of voltage-gated K+ channels, Kv1. 5 and Kv2. 1, in hypoxic pulmonary vasoconstriction and control of resting membrane potential in rat pulmonary artery myocytes. J. Clin. Inv 101 (11), 2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddar NWAH, et al. , 2015. Sal-site: research resources for the Mexican Axolotl In: Kumar A, Simon A (Eds.), Salamanders in Regeneration Research: Methods and Protocols. Springer New York, New York, NY, pp. 321–336. [Google Scholar]
- Balestrini L, et al. , 2017. The natural compound sanguinarine perturbs the regenerative capabilities of planarians. Int. J. Dev. Biol 61 (1–2), 43–52. [DOI] [PubMed] [Google Scholar]
- Barro-Soria R, et al. , 2010. ER-localized bestrophin 1 activates Ca2+-dependent ion channels TMEM16A and SK4 possibly by acting as a counterion channel. Pflugers Arch. 459 (3), 485–497. [DOI] [PubMed] [Google Scholar]
- Beane WS, et al. , 2011. A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem. Biol 18 (1), 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, 1995. Calcium signalling and cell proliferation. BioEssays 17 (6), 491–500. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD, 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell. Biol 1 (1), 11–21. [DOI] [PubMed] [Google Scholar]
- Borgens RB, Vanable JW, Jaffe LF, 1977. Bioelectricity and regeneration: large currents leave the stumps of regenerating newt limbs. Proc. Natl. Acad. Sci 74 (10), 4528–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgens RB, Vanable JW Jr., Jaffe LF, 1979. Role of subdermal current shunts in the failure of frogs to regenerate. J. Exp. Zool 209 (1), 49–56. [DOI] [PubMed] [Google Scholar]
- Britschgi A, et al. , 2013. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. U. S. A 110 (11), E1026–E1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo A, et al. , 2008. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322 (5901), 590–594. [DOI] [PubMed] [Google Scholar]
- Carradice D, Lieschke GJ, 2008. Zebrafish in hematology: sushi or science? Blood 111 (7), 3331–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE, 2007. Calcium Signaling. Cell 131 (6), 1047–1058. [DOI] [PubMed] [Google Scholar]
- Cobb MM, et al. , 2015. Cell cycle-dependent changes in localization and phosphorylation of the plasma membrane Kv2.1 K+ channel impact endoplasmic reticulum membrane contact sites in COS-1 cells. J. Biol. Chem 290 (49), 29189–29201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport B, et al. , 2015. Signature channels of excitability no more: L-type channels in immune cells. Front. Immunol 6, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L, 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136 (1), 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XL, et al. , 2007. Cell cycle-dependent expression of potassium channels and cell proliferation in rat mesenchymal stem cells from bone marrow. Cell Prolif. 40 (5), 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinshreiber L, et al. , 2009. Voltage-gated potassium channel as a facilitator of exocytosis. Ann. N. Y. Acad. Sci 1152, 87–92. [DOI] [PubMed] [Google Scholar]
- Franklin BM, Maroudas E, Osborn JL, 2016. Sine‐wave electrical stimulation initiates a voltage‐gated potassium channel‐dependent soft tissue response characterized by induction of hemocyte recruitment and collagen deposition. Physiol. Rep 4 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA, 1992. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol 119 (3), 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA, 2013. Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci 110 (23), 9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell C, Putzier I, Arreola J, 2005. Calcium-activated chloride channels. Annu. Rev. Physiol 67 (1), 719–758. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C, 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol 238 (2), 274–288. [DOI] [PubMed] [Google Scholar]
- Hu X, et al. , 2011. Hypoxic preconditioning enhances bone marrow mesenchymal stem cell migration via Kv2.1 channel and FAK activation. Am. J. Physiol. Cell Physiol 301 (2), C362–C372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins LS, Duerstock BS, Borgens RB, 1996. Reduction of the current of injury leaving the amputation inhibits limb regeneration in the red spotted newt. Dev. Biol 178 (2), 251–262. [DOI] [PubMed] [Google Scholar]
- Jia L, et al. , 2015. Inhibition of calcium-activated chloride channel ANO1/TMEM16A suppresses tumor growth and invasion in human lung cancer. PLoS One 10 (8), e0136584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha SA, Schlichter LC, 1999. A Kv1.5 to Kv1.3 switch in endogenous hippocampal microglia and a role in proliferation. J. Neurosci 19 (24), 10680–10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Brockes JP, 2012. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 35 (11), 691–699. [DOI] [PubMed] [Google Scholar]
- Levin M, 2007. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 17 (6), 261–270. [DOI] [PubMed] [Google Scholar]
- Levin M, 2009. Bioelectric mechanisms in regeneration: unique aspects and future perspectives Seminars in Cell & Developmental Biology. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, 2014. Endogenous bioelectrical networks store non‐genetic patterning information during development and regeneration. J. Physiol 592 (11), 2295–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Levin M, Kaplan DL, 2016. Bioelectric modulation of macrophage polarization. Sci Rep 6, 21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. , 2012. Inhibition of Ca2+-activated Cl− channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett. 326 (1), 41–51. [DOI] [PubMed] [Google Scholar]
- Lobikin M, et al. , 2012. Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo. Phys. Biol 9 (6), 065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobikin M, et al. , 2014. Selective depolarization of transmembrane potential alters muscle patterning and muscle cell localization in Xenopus laevis embryos. Int. J. Dev. Biol 59 (7–9), 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JW, 2004. Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev 84 (4), 1051–1095. [DOI] [PubMed] [Google Scholar]
- Mao J, et al. , 2009. Volume-activated chloride channels contribute to cell-cycle-dependent regulation of HeLa cell migration. Biochem. Pharmacol 77 (2), 159–168. [DOI] [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM, 2014. Understanding positional cues in salamander limb regeneration: implications for optimizing cell-based regenerative therapies. Dis. Model. Mech 7 (6), 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Trimmer JS, 2005. Kv2.1: a voltage-gated k+ channel critical to dynamic control of neuronal excitability. Neurotoxicology 26 (5), 743–752. [DOI] [PubMed] [Google Scholar]
- Moreno C, et al. , 2013. Modulation of voltage-dependent and inward rectifier potassium channels by 15-epi-lipoxin-A4 in activated murine macrophages: implications in innate immunity. J. Immunol 191 (12), 6136–6146. [DOI] [PubMed] [Google Scholar]
- Nguyen L, et al. , 2001. Neurotransmitters as early signals for central nervous system development. Cell Tissue Res. 305 (2), 187–202. [DOI] [PubMed] [Google Scholar]
- Nuccitelli R, 2003. Endogenous electric fields in embryos during development, regeneration and wound healing. Radiat. Prot. Dosim 106 (4), 375–383. [DOI] [PubMed] [Google Scholar]
- Olson TM, et al. , 2006. Kv1. 5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum. Mol. Genet 15 (14), 2185–2191. [DOI] [PubMed] [Google Scholar]
- Özkucur N, Epperlein H-H, Funk RHW, 2010. Ion imaging during axolotl tail regeneration in vivo. Dev. Dyn 239 (7), 2048–2057. [DOI] [PubMed] [Google Scholar]
- Pai VP, et al. , 2015. Endogenous gradients of resting potential instructively pattern embryonic neural tissue via notch signaling and regulation of proliferation. J. Neurosci 35 (10), 4366–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu MR, Campbell TJ, Breit SN, 2002. A potassium ion channel is involved in cytokine production by activated human macrophages. Clin. Exp. Immunol 130 (1), 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, et al. , 2014. The Ca2+-activated Cl− channel, ANO1 (TMEM16A), is a double-edged sword in cell proliferation and tumorigenesis. Cancer Med. 3 (3), 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, et al. , 2014. Proteomic analysis of fibroblastema formation in regenerating hind limbs of Xenopus laevis froglets and comparison to axolotl. BMC Dev. Biol 14 (1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, et al. , 2012. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One 7 (8), e43265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ, 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protocol 3 (6), 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schroeder BC, et al. , 2008. Expression cloning of TMEM16A as a calcium-activated chlo-ride channel subunit. Cell 134 (6), 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhang L, Mumm JS, 2015. Chemical genetics and regeneration. Future Med. Chem 7 (16), 2263–2283. [DOI] [PubMed] [Google Scholar]
- Sigel E, Steinmann ME, 2012. Structure, function, and modulation of GABAA receptors. J. Biol. Chem 287 (48), 40224–40231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, 1974. Neurotrophic control of limb regeneration in the NEWT*. Ann. N. Y. Acad. Sci 228 (1), 308–321. [DOI] [PubMed] [Google Scholar]
- Tabarean IV, 2014. Electrical remodeling of preoptic GABAergic neurons involves the Kv1.5 subunit. PLoS One 9 (5), e96643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Elly M., 2016. The molecular and cellular choreography of appendage regeneration. Cell 165 (7), 1598–1608. [DOI] [PubMed] [Google Scholar]
- Tao R, et al. , 2008. Regulation of Cell Proliferation by Intermediate-Conductance Ca2+-Activated Potassium and Volume-Sensitive Chloride Channels in Mouse Mesenchymal Stem Cells. 295 pp. C1409–C1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng A-S, et al. , 2010. Induction of vertebrate regeneration by a transient sodium current. J. Neurosci 30 (39), 13192–13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Gracia A, et al. , 2013. Emerging role for the voltage-dependent K+ channel Kv1.5 in B-lymphocyte physiology: expression associated with human lymphoma malignancy. J. Leukoc. Biol 94 (4), 779–789. [DOI] [PubMed] [Google Scholar]
- Van Den Eynden J, et al. , 2009. Glycine and glycine receptor signalling in non-neuronal cells. Front. Mol. Neurosci 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente R, et al. , 2005. Pattern of Kv beta subunit expression in macrophages depends upon proliferation and the mode of activation. J. Immunol 174 (8), 4736–4744. [DOI] [PubMed] [Google Scholar]
- Villalonga N, et al. , 2008. Cell cycle-dependent expression of Kv1. 5 is involved in myoblast proliferation. Biochim. Biophys. Acta 1783 (5), 728–736. [DOI] [PubMed] [Google Scholar]
- Walan A, et al. , 1989. Effect of omeprazole and ranitidine on ulcer healing and relapse rates in patients with benign gastric ulcer. N. Engl. J. Med 320 (2), 69–75. [DOI] [PubMed] [Google Scholar]
- Wang X, van Breemen C, 1999. Depolarization-mediated inhibition of Ca(2+) entry in endothelial cells. Am. J. Phys 277 (4 Pt 2), H1498–H1504. [DOI] [PubMed] [Google Scholar]
- Wang G-L, et al. , 2002. Deficiency in ClC-3 chloride channels prevents rat aortic smooth muscle cell proliferation. Circ. Res 91 (10), e28–e32. [DOI] [PubMed] [Google Scholar]
- Wei J-F, et al. , 2008. Formation of Kv2.1-FAK complex as a mechanism of FAK activation, cell polarization and enhanced motility. J. Cell. Physiol 217 (2), 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W-C, et al. , 2011. The potassium–chloride cotransporter 2 promotes cervical cancer cell migration and invasion by an ion transport-independent mechanism. J. Physiol 589 (22), 5349–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, et al. , 2012. L-type calcium channels play a crucial role in the proliferation and osteogenic differentiation of bone marrow mesenchymal stem cells. Biochem. Biophys. Res. Commun 424 (3), 439–445. [DOI] [PubMed] [Google Scholar]
- West RB, et al. , 2004. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am. J. Pathol 165 (1), 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondergem R, et al. , 2001. Blocking swelling-activated chloride current inhibits mouse liver cell proliferation. J. Physiol 532 (3), 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuytack F, Raeymaekers L, Missiaen L, 2002. Molecular physiology of the SERCA and SPCA pumps. Cell Calcium 32 (5), 279–305. [DOI] [PubMed] [Google Scholar]
- Yang YD, et al. , 2008. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455 (7217), 1210–1215. [DOI] [PubMed] [Google Scholar]
- Zayzafoon M, 2006. Calcium/calmodulin signaling controls osteoblast growth and differentiation. J. Cell. Biochem 97 (1), 56–70. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu HT, 2002. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12 (1), 9–18. [DOI] [PubMed] [Google Scholar]
- Zheng JQ, Poo M. m., 2007. Calcium signaling in neuronal motility. Annu. Rev. Cell Dev. Biol 23 (1), 375–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.