Abstract
Putrescine belongs to the large group of polyamines, an essential class of metabolites that exists throughout all kingdoms of life. The Salmonella speF gene encodes an inducible ornithine decarboxylase that produces putrescine from ornithine. Putrescine can be also synthesized from arginine in a parallel metabolic pathway. Here, we show that speF expression is controlled at multiple levels through regulatory elements contained in a long leader sequence. At the heart of this regulation is a short open reading frame, orf34, which is required for speF production. Translation of orf34 interferes with Rho-dependent transcription termination and helps to unfold an inhibitory RNA structure sequestering speF ribosome-binding site. Two consecutive arginine codons in the conserved domain of orf34 provide a third level of speF regulation. Uninterrupted translation of orf34 under conditions of high arginine allows the formation of a speF mRNA structure that is degraded by RNase G, whereas ribosome pausing at the consecutive arginine codons in the absence of arginine enables the formation of an alternative structure that is resistant to RNase G. Thus, the rate of ribosome progression during translation of the upstream ORF influences the dynamics of speF mRNA folding and putrescine production. The identification of orf34 and its regulatory functions provides evidence for the evolutionary conservation of ornithine decarboxylase regulatory elements and putrescine production.
Author summary
Polyamines are widely distributed in nature, they bind nucleic acids and proteins and although their exact mechanism of action is not clear, their effect on fundamental cellular functions is well documented. The canonical biosynthesis pathway of polyamines is conserved and begins with speF encoding ornithine decarboxylase, an inducible enzyme that produces putrescine from ornithine. Putrescine can also be produced from arginine in an alternative metabolic pathway. Here, we show that the rate of ribosome progression during translation of a short ORF (ORF34) upstream of speF influences the dynamics of speF mRNA folding and thus putrescine production. Uninterrupted translation of orf34 carrying two consecutive arginine codons, under conditions of high arginine, results in the formation of a speF mRNA structure that is degraded by RNase G, whereas ribosomes slow-down at the consecutive arginine codons in the absence of arginine enables the formation of an alternative structure that is unsusceptible to RNase G and thus results in putrescine production. The study of Salmonella speF regulation provides evidence that, despite variations in the mechanistic details, RNA-based regulation of putrescine biosynthesis and ornithine decarboxylase is conserved from bacteria to mammals.
Introduction
Regulatory RNAs are now recognized as important players in many adaptive and physiological responses in bacteria. RNA regulators allow bacterial cells to fine-tune their metabolism during growth [1], sense population density [2], modulate and modify cell-surface properties [3], and regulate virulence gene expression [4]. With respect to the molecular mechanisms involved, there are four main classes of regulatory RNAs in bacteria to date [5]. One extensively studied class are mRNA leaders, attenuators and riboswitches. This class of riboregulators control expression in cis by adopting altered structural conformations in response to cellular and/or environmental signals. For example, mRNA leaders of RNA thermosensors undergo significant changes in response to elevated temperatures [6] and RNA structural changes caused by stalled ribosomes affect transcription elongation through formation of terminator or antiterminator structures [7]. In contrast, riboswitches were defined as leader sequences that change their structure upon binding to small molecule ligands such as amino acids, carbohydrates, coenzymes and nucleobases [8, 9]. Riboswitches modulate gene expression at the level of transcription termination/elongation, translation initiation, or splicing [10, 11]. In many cases, this regulation involves the highly conserved transcription termination factor Rho [12]. For the majority of riboswitches, impaired translation promotes transcript termination by Rho. For example, translational repression of thiMD genes caused by the binding of TPP to thiM riboswitch promotes Rho-dependent premature termination in a region located between codons 20 and 34 of thiM [13]. A similar Rho-dependent transcriptional polarity was described for ChiX sRNA regulation of chiPQ operon. Inhibition of chiP translation by ChiX causes the nascent mRNA to become susceptible to Rho action, thereby preventing RNA polymerase from reaching chiQ [14]. The binding of Mg+2 and the FMN precursor to mgtA and rib, respectively, promotes structural changes in the RNA leader that facilitate interaction with Rho [15]. In case of the mgtA riboswitch, the leader mgtL also encodes a small, proline-rich peptide. Contrary to the canonical mechanism in which impaired translation favors Rho binding, complete translation of the mgtL peptide promotes association of Rho and premature termination [16, 17].
Searching for new riboswitches and other regulatory RNA motifs in α-proteobacteria, Corbino and colleagues [18] discovered a putative novel RNA element in the leader sequence of speF. The speF gene encodes an inducible ornithine decarboxylase catalyzing the initial step in putrescine production [19]. Putrescine can be produced from the precursor ornithine, via ornithine decarboxylase or from arginine through agmatine. Consequently, arginine and ornithine compete over the production of putrescine by being precursors of alternative pathways [20, 21]. Putrescine belongs to the large group of polyamines, an essential class of metabolites that exists throughout all kingdoms of life. In the absence of polyamines, a plethora of cellular processes are impaired, including gene expression, cell growth and/or proliferation and stress resistance [22]. The intracellular levels of polyamines are carefully controlled and so is the expression of ornithine decarboxylase, the limiting factor for putrescine biosynthesis. The expression of ornithine decarboxylase homologs are monitored at multiple levels, including transcription, translation and protein stability by highly conserved regulaory mechanisms involving feedback loops [22]. In mammals, translation of ornithine decarboxylase is repressed by high levels of polyamines via a small upstream ORF (uORF) encoded in a highly structured 5’UTR. The uORF and the long structured 5’UTR are conserved, regulating the expression of ornithine decarboxylase homologs in yeast, plants and mammals [22, 23].
Here we show that Salmonella speF-potE operon carries a long mRNA leader, which encodes an ORF of 34 amino acids (here denoted orf34). Regulatory elements encoded within the mRNA leader including two consecutive arginine codons facilitate speF expression control at multiple levels including premature transcription termination, translation initiation and mRNA decay.
Results
Translation of orf34 prevents premature transcription termination by Rho
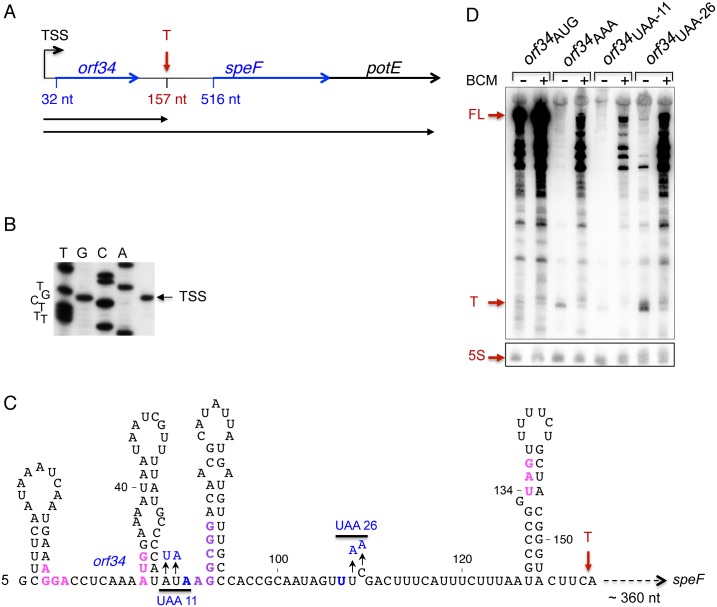
To investigate a possible role of the 5’UTR in post-transcriptional regulation of speF in Salmonella, we first mapped the transcriptional start site (TSS) using primer extension. We discovered only a single initiation site located 516 nucleotides upstream of the speF ORF (Fig 1A and 1B), which is in line with previous reports [24]. A speF-lacZ transcriptional reporter revealed that the gene is transcribed in rich media and that deletion of the speF promoter strongly inhibited expression (Table 1). The long 5’UTR of speF prompted us to explore its sequence. The search revealed a short putative ORF of 34 amino acids (orf34) encoded close to the transcription start site, at position +32 of the mRNA (Fig 1A). Transcription and translation reporter fusions demonstrated that ORF34 is efficiently translated. Replacing the initiation codon of orf34 with AAA abolished translation of orf34-lacZ fusion (Table 1; rows 4,5). We used this mutant to examine the influence of orf34 translation on speF expression. Lack of orf34 translation strongly reduced speF-lacZ expression; both the transcription and the translation of speF-lacZ decreased dramatically (Table 2; rows 1,2). Furthermore, premature translation termination caused by replacing amino acids 11 and 26 of orf34 with stop codons (orf34UAA11 and orf34UAA26 respectively) weakened expression of speF-lacZ to a similar extent, indicating that expression of orf34 is required for transcription of the speF gene (Fig 1C and Table 2).
Fig 1. Characteristics of the speF locus.
(A) Schematic representation of the speF locus including orf34, speF and potE. The transcription start site (TSS) is indicated. The initiation codons of orf34 and speF are located at nucleotides 32 and 516 of the mRNA, respectively. The red vertical arrow indicates the site of rho-dependent premature transcription termination (T). Horizontal arrows indicate the short transcript obtained due to Rho (157 nt) and the full-length polycistronic mRNA. (B) Start site determination. The transcription start site of speF operon was determined by primer extension using primer 1614. RNA was extracted form cultures grown in LB to OD600 of 0.5. (C) The sequence of the transcript obtained by premature termination. The Shine-Dalgarno (AGGA), the initiation codon (AUG) and the stop codon (UGA) of orf34 are in magenta. The consecutive arginine codons AGG;CGG are in purple. UAA11 and UAA26 indicate mutated codons leading to premature translation termination and thus to premature transcription termination by Rho. (D) Translation of the entire orf34 prevents premature transcription termination by Rho. Cultures carrying Ptac-orf34-speF plasmids wild type and translational mutants as indicated (orf34AAA; orf34UAA11 and orf34UAA26), were grown in LB to OD600 of 0.2 and then treated with 20 μg/ml BCM for 60 minutes (+) or were left untreated (-). Northern blot of RNA samples (10 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (1614) and 5S rRNA (459) specific primers. 5S RNA serves as a loading control. The Full-length (FL) and the short transcript obtained by premature termination (T) are indicated in red.
Table 1. Transcription and translation at the orf34-speF locus.
| Genotypea | Transcription fusion | Translation fusion |
|---|---|---|
| 1. P-orf34-speF-lacZ | 5263±820 | 7081±1491 |
| 2. ΔP-orf34-speF-lacZ | 65 ± 7 | 5 ± 2 |
| 3. pRS551/pRS552 | 10 ± 2 | 3 ± 2 |
| 4. P-orf34-lacZ* | 145 ± 6 | 4977 ± 130 |
| 5. P-orf34AAA-lacZ* | 115 ± 29 | 3 ± 2 |
aThe orf34-speF fusions harbor the promoter region (P), orf34 and speF fused to lacZ in PRS551 (transcription) and PRS552 (translation) fusion vectors. In ΔP-orf34-speF-lacZ, the promoter has been deleted. In P-orf34-lacZ, the orf34 is directly fused to lacZ. Where indicated, the initiation codon of orf34 (AUG) has been replaced by AAA (orf34AAA).
*Because of the high LacZ activity of orf34-lacZ translation fusion, all orf34 fusions were inserted in the chromosome as a single copy. The cultures were grown in LB to OD600 of 0.9. Results are displayed as mean of 2 colonies ± standard deviation.
Table 2. Translation of orf34 prevents premature termination caused by Rho.
| Genotypea | Transcription fusion | Translation fusion | ||
|---|---|---|---|---|
| -BCM | +BCM | -BCM | +BCM | |
| 1. P-orf34AUG-speF-lacZ | 5263±820 | 5413 ± 171 | 7081±1491 | 5116 ± 125 |
| 2. P-orf34AAA-speF-lacZ | 53 ± 4 | 1762 ± 176 | 1 ± 6 | 82 ± 15 |
| 3. P-orf34UAA-11-speF-lacZ | 61 ± 21 | 3216 ± 1407 | 4 ± 4 | 169 ± 56 |
| 4. P-orf34UAA-26-speF-lacZ | 72 ± 3 | 3635 ± 1139 | 5 ± 1 | 135 ± 43 |
aThe orf34-speF fusions harbor the promoter region (P), orf34 and speF fused to lacZ. In orf34AAA, the initiation codon of orf34 was replaced by AAA. In orf34UAA-11 and orf34UAA-26, premature stop codons (UAA) were inserted in orf34 at amino acid positions 11 and 26, respectively. Cultures grown in 7 ml LB medium to OD600 of 0.2 were treated with 20μg/ml of Bicyclomycin for 60 min. Results are displayed as mean of 2 colonies ± standard deviation.
The ~100-fold reduction in activity of the transcriptional reporter (5263±820 to 53±4, Table 2) prompted us to examine the pattern of RNA species in wild type and orf34 translational mutants. To this end, plasmid-borne expression of the orf34-speF locus (Ptac-orf34-speF) carrying wild type and orf34 translational mutants was monitored using Northern Blots. The data in Fig 1D show that the translational mutants (orf34AAA; orf34UAA11 and orf34UAA26) produced mainly a short RNA species of ~157 nucleotides (indicated by T) as determined by 3’-end RACE. Therefore, in the absence of orf34 translation or upon premature translation termination, the transcription terminates before it reaches the speF coding-region. Given that sequence analysis of the speF 5’UTR did not reveal a rho-independent transcription terminator, we speculated that in the absence of orf34 translation, the transcription termination factor Rho terminated elongation. We tested this hypothesis by using bicyclomycin (BCM), which inhibits the ATPase activity of Rho [25]. Northern Blot analysis demonstrated that upon addition of BCM, transcription of orf34-speF (Ptac-orf34-speF) carrying orf34 translational mutants (AAA, UAA11 and UAA26) extended into the speF ORF (Fig 1D). Likewise, the levels of the orf34-speF-lacZ transcriptional fusions carrying orf34 mutations increased with BCM, indicating that complete orf34 translation is required to prevent premature transcription termination by Rho (Table 2; transcription fusion +BCM). Moreover, as UAA-26 includes RNA species that result from escaped transcription elongation, the closer the site of the stop codon is to the end of the peptide, the lower the activity of Rho.
Translation of orf34 facilitates downstream translation of speF
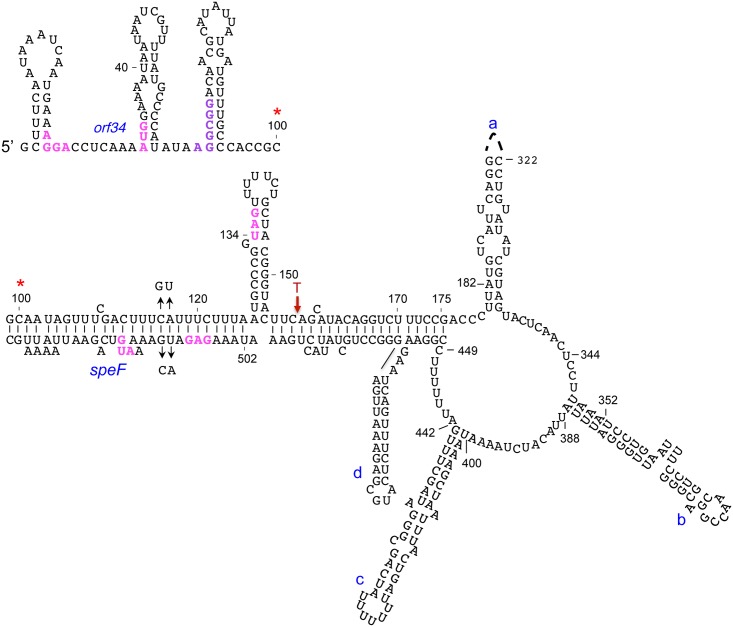
The data presented in Table 2 showing that the activity of the orf34AAA-speF-lacZ transcriptional reporter increased with BCM, whereas the levels of the corresponding translation fusion remain low, indicated that unlike the ribosome-binding site (RBS) of lacZ, the RBS of speF mRNA is translationally inactive under these conditions. These results suggest that translation of orf34 has a positive effect on speF translation initiating approximately 500 nts downstream and motivated a search for potential long-range RNA interactions. The RNA-fold program [26] predicted such a potential long-range interaction covering the 5’-end of the transcript and the sequence surrounding the RBS of speF (Fig 2). The predicted structure contains a ring-like organization encompassing nucleotides 175 to 449 followed by a long helix in which nucleotides 99–125 of orf34 base-pair with nucleotides 502–535 overlapping the ribosome binding site of speF and nucleotides 155–175 upstream of orf34 base-pair with nucleotides 450 to 454 and 482 to 500 upstream of the ribosome binding site of speF (Fig 2). To validate basepairing around the RBS of speF, we exchanged nucleotides cytidine 116 and adenine 117 to guanine and uracil, respectively in orf34 and uracil 510 and guanine 511 with adenine and cytidine on the opposite strand. These mutations when combined together are predicted to restore helix formation. To examine basepairing per se, we prevented upstream translation using orf34AAA, while allowing transcription elongation into speF using BCM. The translation fusions of these mutants demonstrated that disruption of hybrid formation increased speF translation (Table 3). Furthermore, using the corresponding complementary mutations restoring basepairing resulted in decreased speF translation (Table 3). Together, our data demonstrate that upstream translation of orf34 prevents premature termination upstream of speF and facilitates translation of the speF mRNA by disrupting an inhibitory RNA structure sequestering the RBS of speF.
Fig 2. Putative long-range interaction at the speF locus as predicted by RNAfold.
The sequence starts at the upper left side of the slide and the red asterisk indicates its continuation. The Shine-Dalgarno sequences of orf34 and speF, both AUGs and orf34 stop codon are indicated in magenta. The structure predicted harbors a ring-like structure encompassing nucleotides 175 to 449 followed by a long helix in which nucleotides 99–125 of orf34 basepair with nucleotides 502–535 overlapping the ribosome binding site of speF and nucleotides 155–175 upstream of orf34 basepair with nucleotides 450 to 454 and 482 to 500 upstream of ribosome binding site of speF. Mutations that disrupt the helix and compensatory mutations that restore helix-formation are indicated by arrows (GU at position 116,117 and CA at position 510,511). Premature transcription termination site by Rho is indicated (T).
Table 3. Translation of orf34 facilitates translation of speF.
| Genotypea | Translation fusion +BCM |
|---|---|
| P-orf34AAA-speF-lacZ | 95 ± 33 |
| ΔP-orf34AAA C116G;A117U-speF-lacZ | 1037 ± 202 |
| ΔP-orf34AAA U510A;G511C-speF-lacZ | 309 ± 74 |
| ΔP-orf34AAA C116G;A117U, U510A;G511C -speF-lacZ | 66 ± 11 |
aThe orf34-speF fusions harbor the promoter region (P), orf34 and speF fused to lacZ. All fusions carry orf34AAA mutation in addition to the mutations that prevent formation of the hybrid as mentioned. BCM was added at OD600 of 0.2 for 1 hour. Results are displayed as mean of 4 colonies ± standard deviation
Positive and negative regulation of speF by ornithine and arginine, respectively
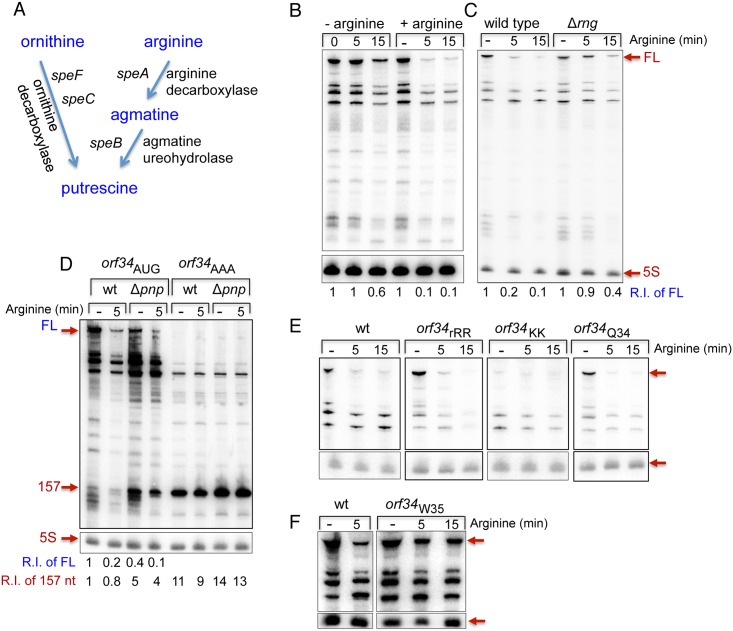
Putrescine can be produced from its precursor ornithine by speF encoding ornithine decarboxylase, an inducible enzyme, or from arginine in a metabolic pathway that involves both arginine decarboxylase (speA) and agmatine ureohydrolase (speB) (see diagram in Fig 3A). Whereas, ornithine is converted directly to putrescine, arginine is first converted to agmatine and subsequently to putrescine [20, 21]. We tested the effect of ornithine and arginine on speF expression using orf34-speF-lacZ fusions. The activity of orf34-speF-lacZ increased in minimal medium supplemented with ornithine (Table 4 row 1) and decreased in medium supplemented with arginine (Table 4 row 4). To determine which of the genetic elements mediated the effect of the precursors, we examined activity of the speF promoter (P-speF-lacZ) and orf34 (P-orf34-lacZ) in the presence and absence of ornithine and arginine. Our data revealed that neither the speF promoter, nor orf34 were influenced by ornithine or arginine (Table 4 rows 2,3 and 5,6). In summary, our results show that ornithine induces expression of ornithine decarboxylase, however in presence of the alternative precursor arginine, the inducible pathway producing putrescine via speF is turned off.
Fig 3. Arginine-dependent mRNA destabilization.
(A) A partial diagram of polyamine biosynthesis. Putrescine can be produced from ornithine via speC and speF encoding a constitutive and inducible ornithine decarboxylase, respectively or from arginine in two steps via speA and speB encoding arginine decarboxylase agmatine ureohydrolase respectively. (B) speF mRNA levels decrease in the presence of arginine. Cultures of wild type carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg/ml) or left untreated for the indicated time. Northern blots as in Fig 1D. Relative intensity of the full-length RNA measured by ImageQuant TL 1D v8.1 is shown (R.I of FL). (C) RNase G destabilizes the full-length mRNA in the presence of arginine. Cultures of wild type and RNase G mutant (Δrng) carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg/ml) for the indicated time. Relative intensity of the full-length RNA vs. 5S is shown. RNA samples taken from cells at OD600 of 0.2 before exposure to arginine are indicated as (-). (D) The 3’-end exonuclease PNPase degrades the short transcript produced by premature termination. Cultures of wild type and PNPase mutant (Δpnp) carrying Ptac-orf34-speF and Ptac-orf34AAA-speF speF were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg/ml) for the indicated time as described in C. Relative intensity of the 157 nt long transcript measured by ImageQuant TL 1D v8.1 is shown (R.I of 157 nt). The short transcript accumulates in Δpnp mutant (5 fold). Unlike the 157 nt transcript, the full-length transcript (R.I of FL) decreases by 2 fold in Δpnp compared to wild type. (E) The regulation by arginine depends on the arginine codons at positions 12 and 13 but not on the arginine codon at position 34. Cultures with Ptac-orf34-speF carrying mutations in the arginine codons as indicated were grown and treated as described above. The Northern blot was probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. (F) mRNA destabilization by arginine depends on the position of the stop codon. Cultures carrying wild type Ptac-orf34-speF and Ptac-orf34W35-speF in which the stop codon of orf34 was replaced with Trp codon were grown and treated as described above. The Northern blot was probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers.
Table 4. Regulation of orf34 and speF by ornithine and arginine (β-galactosidase activity).
| Genotypea | Transcription fusion | Translation fusion | |||
|---|---|---|---|---|---|
| -Ornithine | + Ornithine | Control | - Ornithine | + Ornithine | |
| 1. P-orf34AUG-speF-lacZ | 908 ± 35 | 5624 ± 760 | + | 900 ± 351 | 6777 ± 1666 |
| 2. P-lacZ* | 3047 ± 51 | 2721 ± 175 | - | ||
| 3. P-orf34-lacZ* | 482 ± 70 | 414 ± 22 | - | 17466 ± 3347 | 16121 ± 2883 |
| -Arginine | +Arginine | -Arginine | +Arginine | ||
| 4. P-orf34AUG-speF-lacZ | 908 ± 35 | 281 ± 92 | + | 900 ± 351 | 33 ± 30 |
| 5. P-lacZ* | 2182 ± 112 | 2050 ± 92 | - | ||
| 6. P-orf34-lacZ* | 385 ± 60 | 433 ± 33 | - | 15052 ± 4641 | 14612 ± 5508 |
| 7. P-orf34rRR -speF-lacZ | 465 ± 52 | 251 ± 47 | + | 213 ± 104 | 20 ± 20 |
| 8. P-orf34fRR-speF-lacZ | 1015 ± 123 | 410 ± 91 | + | 860 ± 290 | 99 ± 40 |
| 9. P-orf34KK-speF-lacZ | 243 ± 53 | 239 ± 31 | - | 18 ± 2 | 9 ± 2 |
| 10. P-orf34Q34-speF-lacZ | 857 ± 137 | 180 ± 121 | + | 582 ± 267 | 133 ± 124 |
| 11. P-orf34W35-speF-lacZ | 2577 ± 423 | 1917 ± 210 | - | 2277 ± 326 | 1809 ± 422 |
aThe orf34-speF fusions harbor the promoter region (P), orf34 and speF fused to lacZ. In P-orf34-lacZ, the orf34 is directly fused to lacZ. *Because of the high LacZ activity of the translation fusion orf34-lacZ, all orf34 fusions were inserted in the chromosome as a single copy. In P-lacZ the promoter of speF was directly fused to lacZ and the fusion was inserted in the chromosome as a single copy. In orf34rRR, orf34fRR and orf34KK, the two rare arginine codons were replaced by other identical rare arginine, frequent arginine and Lys codons, respectively. In orf34Q34 the rare arginine codon was replaced by Gln codon. In orf34W35 the stop codon was replaced by Trp codon. The orf was extended by 23 amino acids till it reached a new stop codon. Fusions regulated (+) and unregulated (-) by the precursors. Results are displayed as mean of 3 (P-lacZ*), 5–26 (all others) colonies ± standard deviation. Cultures were grown for 17 hours from a single colony in 5 ml (50 ml tubes) of E Minimal supplemented with ornithine or arginine (100 μg /ml).
Two consecutive arginine codons in orf34 mediate speF regulation by arginine
Exploring the orf34 sequence, we found that ORF34 homologues are conserved among γ-proteobacteria carrying a highly conserved domain of unknown function 2618 (DUF2618; pfam: PF10940) (S1 Fig). The core of DUF2618 is conserved carrying two consecutive arginine residues (S1 Fig: HIRRT*HIMM). In Salmonella, the coding sequence of DUF2618 core harbors two consecutive, rare arginine codons (AGG and CGG) in the 5’-end of orf34 and an additional one (CGG) adjacent to its stop codon (Fig 1C). The conservation of the arginine codons as well as of the position of ORF34 homologues i.e. in front of speF prompted us to hypothesize that ribosome stalling at these codons alters the structure of the speF mRNA and that this structural change is required for down-regulation of speF. To investigate if the arginine codons are involved in arginine sensing, we changed the consecutive rare arginine codons with either two other rare arginine codons (CGACGA; rRR), with frequent arginine codons (CGCCGC; fRR), or with lysine codons (AAGAAG; KK). orf34-speF-lacZ carrying consecutive other rare (rRR) or frequent (fRR) arginine codons was regulated by arginine similar to wild type, indicating that arginine codons either rare or frequent mediate the regulation by arginine (Table 4). Furthermore, changing these codons with lysine codons abolished speF regulation by arginine. Changing the rare arginine codon positioned adjacent to the stop codon, with Gln (Q34) had no effect on speF expression regulation by arginine (Table 4; rows 4, 7–10). The conserved core amino acids of ORF34 also prompted us to examine whether translation of ORF34 in trans affects expression of speF. The activity of transcription and translation fusions of P-Δ(orf34)-speF-lacZ and P-orf34AAA-speF-lacZ was unaffected by arginine when ORF34 was expressed in trans (S1 Table). Together, the data indicate that two consecutive arginine codons at positions 12 and 13 independent of whether rare or frequent sense and transduce the signal of arginine availability.
speF mRNA destabilization by Arginine
To characterize the effect of arginine on speF expression, we analyzed speF mRNA levels in the absence and upon addition of arginine. The northern shows that exposure of cells to arginine led to about 10-fold decrease in speF mRNA within the first 5 minutes of treatment while speF mRNA of untreated cell was stable (Fig 3B). Given that arginine had no effect on promoter activity of speF (Table 4 row 5) and did not induce premature transcription termination (Fig 3B), we concluded that arginine turns off speF pathway by affecting speF mRNA stability. To determine which ribonucleases are involved in speF mRNA decay, we examined the effects of rne, rng and pnp encoding endoribonuclease E, endoribonuclease G and 3’-end exoribonuclease PNPase, respectively. Analysis of the RNA levels upon exposure to arginine in wild type and in rng mutant revealed a significant decrease in speF mRNA in wild type while the decrease in speF mRNA in Δrng cells was minimal, indicating that endoribonuclease G is involved in speF mRNA decay (Fig 3C). Kinetics studies to monitor the decrease in speF mRNA levels upon exposure of arginine showed that the levels of this mRNA decreased rapidly in wild type cells reaching ~10% of its initial level by 10 min of exposure, whereas in rng mutant speF RNA decay was significantly slow reaching ~ 40% of its initial levels by 10 min of exposure (S2 Fig). Mapping of RNase G cleavage sites by primer extension of RNA samples extracted from untreated wild type and Δrng cells and samples of cells exposed to arginine for 2 and 10 min revealed a number of cleavage sites at 10 min of exposure at which mRNA decay was more prominent. Notably, whereas a few of the sites may have resulted from secondary cleavage, the cleavage sites detected within the ring structure were specific to adenines reminiscent of the RNase E consensus site (G/A N↓A/U U/C U/A) [27] (S3 & S4 Figs). Unlike rng, the full length of speF mRNA was not detected in a temperature sensitive mutant of RNase E (S5 Fig). It is possible that in the absence of RNAse E other ribonucleases took over this function.
RNA analysis of speF mRNA (orf34AUG) in wild type and Δpnp demonstrates that the 3’ exoribonuclease PNPase affects the stability of the 157 nt long RNA species as well as of RNA species obtained by degradation upon exposure to arginine (Fig 3D). Quantification of the levels of the 157 nt transcripts in wild type vs. Δpnp shows that the transcript produced by orf34AUG is ~5 fold higher in Δpnp compared to wild type (Fig 3D). These results indicate that a significant portion of the speF transcript produced under conditions of low and high arginine terminates prematurely and that the terminated transcript is quickly degraded and thus its abundance is reduced in wild type cells (Fig 3D). The truncated transcript produced by orf34AAA is only slightly higher (1.35 fold) in Δpnp compared to wild type. However, the basal level of the transcript produced by orf34AAA in wild type cells is ~10 fold higher than that of the transcript produced by orf34AUG. Thus, we suspect that its degradation by PNPase could be masked by other 3’-end exoribonucleases such as RNase II or RNase R.
In addition, we examined the effect of arginine on speF mRNA carrying rare arginine codon mutants (orf34rRR and orf34Q34). The northern showed that similarly to wild type, and as indicated by the lacZ fusions, orf34rRR-speF and orf34Q34-speF mRNAs produced by Ptac-orf34-speF were destabilized by arginine (Fig 3E). Intriguingly, only low levels of full-length mRNA were detected in orf34KK-speF in which the consecutive arginine codons were changed with lysine, indicating that translation of orf34 in the absence of the arginine codons at positions 12 and 13 results in transcript degradation (Fig 3E). Likewise, wild type orf34 translation through the consecutive arginine codons under conditions of high arginine results in mRNA degradation.
The position of the stop codon of orf34 affected regulation by arginine
Our data suggested that complete translation of the orf34 results in transcript destabilization and prompted us to examine whether the position of the stop codon of orf34 affected regulation by arginine. By replacing the stop codon at the end of orf34 with Trp codon (W35), orf34 was extended by 23 amino acids when reaching a new stop codon. Transcription and translation fusion levels of orf34W35-speF-lacZ demonstrate that moving the position of the stop codon further downstream abolishes regulation by arginine (Table 4 compare lanes 1 and 11). Northern Blot analysis showed that the mutant is no longer subjected to arginine regulation, as the transcript remains stable in the presence of arginine (Fig 3F). Therefore, the position of the stop codon together with full translation of orf34 to its native stop codon determines transcript stability.
Formation of an alternative structure
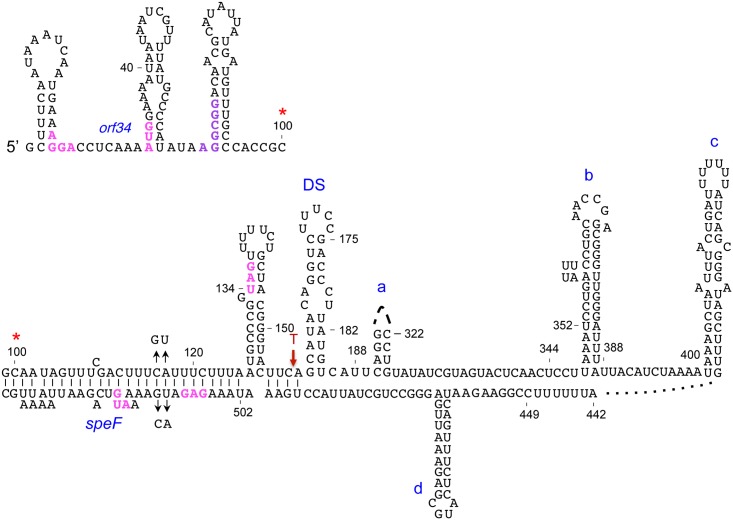
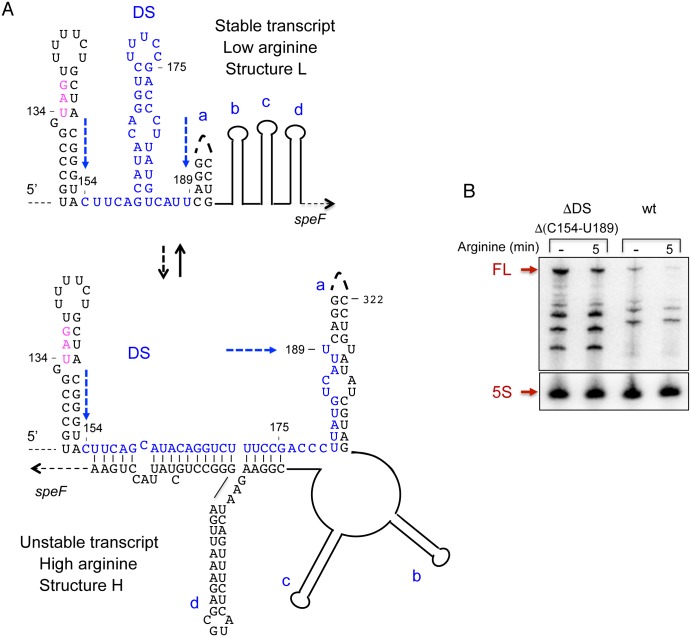
The observations that orf34-speF locus can produce both stable and unstable transcripts and that ribosome pausing due to amino acid deficiency affect speF mRNA decay indicated that the speF mRNA could form alternative RNA structures. As the final result of RNA folding algorithms is typically different from the RNA structures occurring during the continuous process of transcript elongation during transcription, we searched for alternative structures using shorter sequences. The RNAfold program consistently predicted formation of a new hairpin right downstream of the hairpin encompassing orf34 stop codon (Fig 4). We hypothesized that formation of a hairpin structure downstream of the stop codon (DS) prevented annealing of the proximal strand of this hairpin with a sequence downstream of the ring-like structure, while the distal strand could no longer be part of the helix a. Thus, formation of hairpin DS would interfere with the formation of the alternative structure (Structure H, Fig 5A). To test this hypothesis, we deleted the DS sequence and investigated RNA stability. Specifically, deleting this sequence is unlikely to affect the structure formed under low arginine conditions (Structure L, Fig 5B), but would prevent formation of the structure predicted to form under high arginine conditions (Structure H) (Fig 5A). Northern Blot analysis of ΔDS (ΔC154-U189) with and without arginine revealed significantly increased RNA levels when compared to the wild type construct, indicating that hairpin DS plays a role in the formation of the unstable structure H and that the stable L structure forms under low arginine conditions (Fig 5B).
Fig 4. Putative alternative structure predicted to form under low arginine.
The sequence starts at the upper left side of the slide and the red asterisk indicates its continuation at the lower part of the slide. In this structure the hairpin downstream of stop (DS) is formed, preventing formation of the ring-like structure and of the helix engaging nucleotides 155–175 and 450–500 (see also Fig 2). The Shine-Dalgarno sequence of orf34, the AUG and orf34 stop codon are indicated in magenta. The consecutive arginine codons AGG CGG are in purple.
Fig 5. Deleting hairpin DS prevents formation of the ring-like structure and the RNA remains stable.
(A) Structures predicted to form under low and high arginine conditions (L and H, respectively) are shown. The sequence of hairpin DS is marked in blue. (B) RNA analysis of Ptac-orf34-speF wild type and deletion mutant missing nucleotides C154 to U189 with and without arginine as indicated.
The results showing that RNAs produced by the mutants; orf34W35-speF and ΔDS are stable and unaffected by arginine suggest that ribosomes pausing at the natural position of orf34 stop codon lead to the formation of the metabolically unstable alternative structure via destabilization of the DS hairpin. In vivo structure probing of the stop codon hairpin and the neighboring downstream DS hairpin of wild type and orf34W35-speF showed that wild type RNA was significantly more accessible to DMS modification, while the mutant RNA was highly inaccessible to DMS modification and the region preceding the stop codon remained unmodified (S6 Fig). These results indicate that ribosome pausing at the natural stop codon leads to destabilization of the downstream DS hairpin.
The ring-like structure in the speF 5’ UTR affects RNA destabilization by arginine
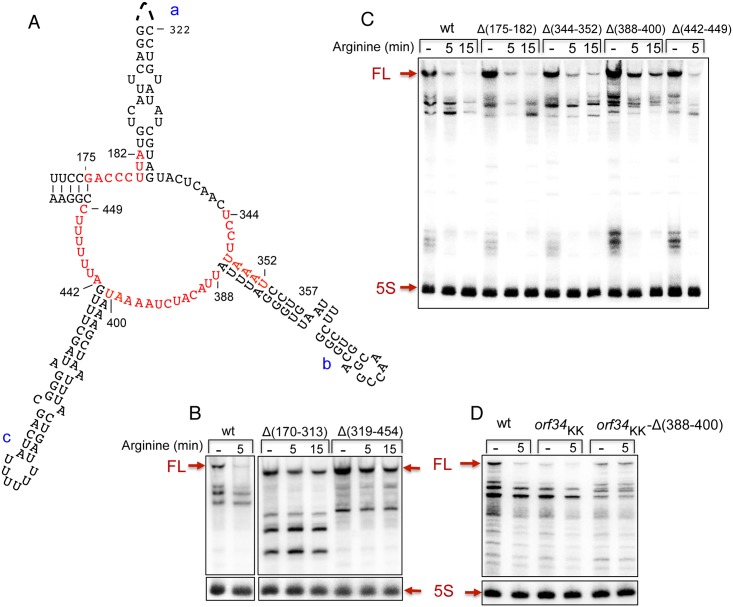
To examine the involvement of the putative ring-like structure and its sequence in RNA destabilization by arginine, we deleted two sequence elements of 143 (Δ170–313) and 135 (Δ319–454) nucleotides that comprise the 5‘ (proximal strand of hairpin a) and the 3’ (distal strand of hairpin a, and hairpins b and c) ends of the ring, respectively. RNA analysis of the two deletion mutants demonstrated that these regions play a role in mRNA destabilization in response to arginine (Fig 6A and 6B). To refine our deletion mapping, we also deleted the sequences constituting parts of the ring. RNA produced from the orf34-Δ388-400-speF construct in the presence of arginine displayed increased stability when compared to the wild type construct, indicating that deleting 12 nucleotides from position 388 to 400 affected the response to arginine and transcript stability (Fig 6C). Moreover, combing the Δ388–400 mutant that reduced RNA destabilization by arginine with the orf34KK mutant that exhibits no full-length transcript (Fig 3C) resulted in increased basal levels of the orf34KK-speF mRNA that was unaffected by arginine (Fig 6D). These results indicate that formation of the alternative ring-like structure facilitates speF mRNA decay.
Fig 6. The ring-like structure affects RNA destabilization by arginine.
(A) The ring structure as predicted by RNAfold is presented (see also Fig 2). The nucleotides deleted in C are marked in red. Long (B) and Short (C) deletions of ring sequences. Cultures with Ptac-orf34-speF carrying deletions as indicated were grown and treated by arginine as described in Fig 3. The Northern blot was probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. (C) Deleting nucleotides 388–400 negates RNA destabilization by arginine. (D) Deleting nucleotides 388–400 negates the effect of orf34KK mutation. RNA analysis of wild type, Ptac-orf34KK-speF and a double mutant Ptac-orf34KK-Δ388-400-speF as indicated.
Discussion
In this study we showed that expression of speF in Salmonella is controlled at multiple levels through regulatory elements contained in the long leader sequence of the mRNA. A short open reading frame of 34 amino acids harboring a conserved domain functions at the heart of this regulation, required for speF production.
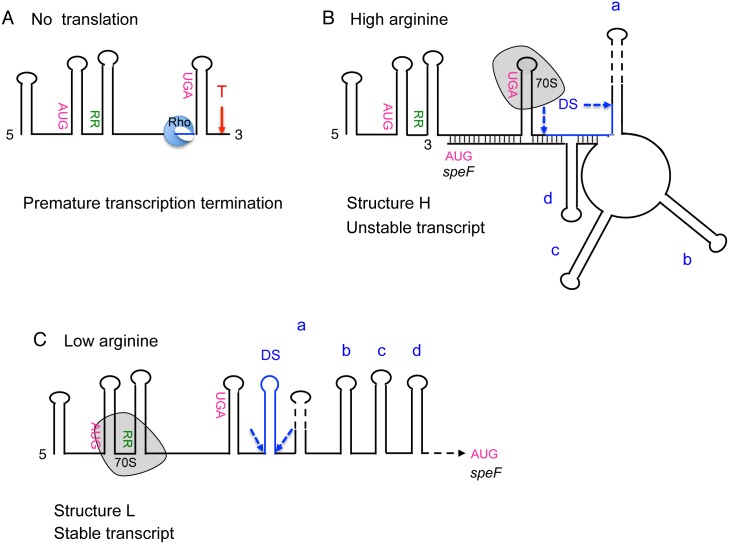
A key factor for speF regulation is the transcription termination factor Rho. Translation of orf34 prevents Rho from transcription termination at the 5’ proximal region of the operon and unfolds an inhibitory structure that sequesters speF ribosome binding site. Rho contacts the RNA at Rho utilization (rut) sites, characterized by high pyrimidine residues with a preference for cytidine and relatively little secondary structure [28, 29]. The sequence of speF harbors a 12 nt long CU-rich domain opposite of the RBS of speF positioned at nucleotides 112 to 124. This sequence is similar to the CU-rich octamer identified in Salmonella chiPQ locus as the main rut site [14]. We propose that in the absence of translation or upon premature translational stop (UAA11 and UAA26), Rho binds the newly synthesized as yet unpaired CU-rich sequence opposite of the RBS, leading to premature transcription termination (Fig 7). Conversely, ribosomes translating orf34 under conditions of high arginine, by reaching its stop codon sequester the putative Rho/anti-SD binding site preventing premature termination.
Fig 7. Post-transcriptional regulation of speF expression.
(A) In the absence of orf34 translation, or upon premature translation termination, the transcription termination factor Rho (in blue) binds orf34 RNA, possibly at the CU-rich sequence that precedes the stop codon (blue line) leading to premature transcription termination of orf34-speF operon (T in red). (B) Under conditions of high arginine ribosomes translate orf34 to its stop codon. Pausing at the stop codon prevents the formation of the ‘downstream of stop’ hairpin (DS; blue line) leading to the formation of the alternative structure H in which a ring-like structure is susceptible to degradation and the RBS of speF is inaccessible (C) Under low arginine ribosome attenuation at the consecutive arginine codons enables formation of hairpin DS (in blue), which in turn interferes with the formation of the ring-like structure. Consequently, the long transcript formed under low arginine (structure L) is stable and translationally active.
The consecutive arginine codons in the conserved domain of orf34 provide an additional level of speF regulation. Under conditions of high arginine, uninterrupted translation of orf34 results in the formation of a structure that is degraded by RNase G. Specifically, we propose that ribosomes pausing at the stop codon of orf34 prevent formation of the ‘downstream of stop’ hairpin (DS), thus enabling the formation of an alternative conformation in which a ring-like RNA structure is formed. In this RNA structure the RBS of speF is inaccessible to the ribosome and becomes susceptible to degradation (Fig 7). Moving the orf34 stop codon further downstream (orf34W35-speF) abolished regulation by arginine and the transcript produced remained stable in the presence of arginine (Fig 3D). Given that ribosomes translating beyond the natural position of orf34 stop codon prevented regulation by arginine further confirms that ribosome pausing at this position is linked to the formation of an mRNA structure that is susceptible to decay. Furthermore, the structure probing data showing that the stop codon and the DS hairpins of wild type RNA were significantly more accessible to DMS modification than the RNA of orf34W35-speF mutant indicate that ribosomes pausing at the stop codon affect formation of the secondary structure downstream not by direct sequestration of the sequence required to form the DS hairpin but indirectly by affecting the rate of RNA polymerase progression and thus the dynamic of speF RNA folding. Numerous studies have shown that cooperation between the translating ribosomes and RNA polymerase influences the rate of transcription elongation and that direct binding between RNA polymerase and the first ribosome trailing the transcribing enzyme allows the polymerase to monitor translation rate [30–32]. We hypothesize that pausing of the translational machinery at the stop codon influences the dynamics of speF mRNA folding by affecting the rate of transcription elongation, hence leading to the formation of the alternative ring-like structure.
Unlike the speF transcript formed under high arginine conditions, the transcript formed under low arginine conditions is stable. Ribosome slow-down at the consecutive arginine codons in the absence of arginine enables the formation of hairpin DS, thus preventing formation of the ring-like RNA structure (Fig 7). Consequently, the transcript formed under low arginine is stable and translationally active. Deleting hairpin DS prevents formation of the unstable transcript, i.e. structure H, shifting the equilibrium towards structure L and the RNA remains stable in the presence of arginine. Overall, these results indicate that the rate of ribosome progression influences the dynamics of speF mRNA folding and thus the susceptibility of the transcript to ribonucleolytic degradation.
In addition to the long stable transcript produced under low arginine conditions, ribosome attenuation due to low arginine may also enable rho-dependent premature termination. Our results showing that compared to the full-length mRNA, the levels of the truncated transcripts increase by 12–13 fold in Δpnp vs. wild type, indicate that indeed upon ribosome attenuation a large portion of the transcripts terminate. These transcripts undergo decay in wild type cells by 3’-end exoribonucleases and thus are undetectable.
For many riboswitches, e.g. the thiM riboswitch of E. coli, mRNA decay is triggered as a consequence of translation inhibition [33]. In contrast, in the lysC riboswitch, these two regulatory activities, translation initiation and mRNA decay, are independently controlled using the same conformational switch [33]. When bound to lysine the lysC riboswitch adopts a conformation that inhibits translation and promotes RNase E-mediated cleavage. In the absence of lysine, the transcript adopts an alternative conformation that allows translation initiation and sequesters the RNase E cleavage sites. Whereas for thiM, lysC and other riboswitches impaired translation is accompanied by mRNA decay, in speF regulation, uninterrupted translation of orf34 results in mRNA decay. Translation of orf34 under conditions of high arginine results in formation of a ring-like alternative RNA structure and our data indicate that this conformation is susceptible to degradation by RNase G. Deleting 12 nucleotides from position 388 to 400 of the ring affected the response to arginine and transcript stability. RNA produced from this mutant was more stable compared to wild type and basal RNA levels produced by the double mutant combing Δ388–400 with orf34KK were higher than those of orf34KK. Furthermore, mapping of the RNase G cleavage sites revealed a number of weak and strong sites at 10 min of exposure to arginine at which mRNA decay was more prominent. Notably, one strong site A392 resides within the sequence 388–400, further indicating that RNase G, by cleaving the ring-like sequence mediates mRNA decay of speF in the presence of arginine.
The speF gene encodes an inducible ornithine decarboxylase to produce putrescine, which can also be produced from arginine in an alternative pathway. The production of putrescine from arginine requires two steps, whereas ornithine produces putrescine in one-step and yet, arginine down-regulates speF expression. As the Km value of arginine decarboxylase is two-fold lower than that of ornithine decarboxylase [34, 35], it is reasonable that in the presence of arginine the alternative "shorter" pathway is turned off.
Unlike speF regulation by arginine in which a dramatic drop in speF mRNA levels was observed within a few minutes of exposure at exponential phase, induction by ornithine was detected upon a long exposure into stationary phase. Treating cultures at exponential phase with ornithine showed no increase in the levels of speF mRNA (S7A Fig). In contrast, a dramatic increase in the levels of this mRNA was detected in cultures grown to stationary phase in the presence of ornithine compared to untreated cultures. As speF promoter is unaffected by ornithine, we propose that the mRNA undergoes decay in untreated cultures growing to stationary phase, whereas in the presence of ornithine speF mRNA accumulates. Deleting the 5’-end region of the ring-like structure rendered the RNA more stable increasing its basal levels, further indicating that the ring-like structure is involved in speF decay and that the unstable mRNA of stationary phase wild type cells is stabilized in the presence of ornithine (S7B Fig). As induction by ornithine of Δ388–400 construct that is unresponsive to arginine and for the same reason to RNase G cleavage is similar to that of wild type, speF mRNA decay during growth to stationary phase is unrelated to RNase G mediated regulation in response to arginine. In addition, since northern analysis of stationary phase speF mRNA (Ptac-orf34-speF) exhibits very low levels, whereas the levels of speF-lacZ fusions (PspeF-orf34-speF-lacZ) are relatively high, it is conceivable that the β- galactosidase value of ~800 Miller units detected at 17 hours of growth is over represented due to the extended stability of the lacZ fusion transcript.
Based on our data we propose that consecutive arginine codons at positions 12 and 13, either rare or frequent, sense and transduce the signal of arginine availability. The 2-fold decrease in the basal levels of the orf34rRR-speF-lacZ transcription fusion carrying the consecutive identical CGACGA low usage arginine codons could be due to the phenomenon called 5’ ‘translational blockage’. Gao and collogues previously reported that upon ribosome attenuation due to consecutive identical low usage arginine codons positioned near the 5’ end of the message (at codon 13, as in speF) the ratio of free to bound mRNA was increased by 2-fold indicating that some mRNA was released from the ribosomes, undergoing decay [36]. Accordingly we compared the RNA levels of wild type and orf34rRR-speF at 17 hr. of growth, conditions under which the β- galactosidase activity assays were performed. The northern showed that the mRNA levels of the two strains were low and yet the levels of orf34rRR-speF were lower than that of wild type, which could explain the lower levels of LacZ activity (S8 Fig). Since the RNA levels of wild type and orf34rRR-speF mutant at exponential phase were comparable (Fig 3E and S8 Fig), we suspect that orf34rRR-speF transcript becomes more susceptible to decay as the activity of ribosome attenuation at the consecutive identical rare arginine codons overlaps with stationary phase.
The elements for the arginine-dependent control reside within the conserved core of ORF34 that is widespread among γ-proteobacteria, many of which are pathogenic, and the homologues of ORF34 typically precede the inducible version of ornithine decarboxylase. We find that addition of ornithine or arginine supports the growth of wild type Salmonella as compared to Δorf34-speF mutant (S9 Fig) indicating that this regulatory cis-element and the availability of arginine and/or ornithine play a role in bacterial metabolism under normal growth conditions. Whether this conserved regulatory cis-element plays a role in bacterial virulence during infection or the rapid switch between the two alternative biosynthetic pathways is used as a checkpoint affecting both the host and its assailant is an intriguing thought for future studies.
Polyamines are widely distributed in nature, they bind nucleic acids and proteins and although their exact mechanism of action is not clear, their effect on fundamental cellular functions is well documented [22]. Accordingly, the intracellular concentrations of polyamines are tightly controlled and so are the enzymes involved in polyamine production. The canonical biosynthesis pathway of polyamines is conserved and begins with ornithine decarboxylase that forms putrescine. Not surprisingly, the intracellular levels of ornithine decarboxylase are strictly regulated at multiple levels by various mechanisms. Our results show that despite the variations in the mechanistic details, the regulatory elements of ornithine decarboxylase i.e., the upstream ORF and the structured 5’UTR are evolutionary conserved from bacteria to mammals.
Materials and methods
Bacterial growth conditions
Salmonella Typhimurium SL1344 cells were grown at 37°C (200 rpm) in Luria-Bertani (LB) broth (pH 6.8) or in E-minimal medium (Vogel-Bonner medium). The salt composition of E-min is MgSO4(H2O) (0.2 mg/ml), citric acid monohydrate (2 mg/ml), K2HPO4 (10 mg/ml), NaNH4PO4 (3.5 mg/ml). Histidine (100 μg/ml), glucose (0.4%) and vitamin B1 (2 μg/ml) were added after autoclave. Where indicated, L-ornithine (25, 50 or 100 μg/ml) or arginine (100 μg/ml) was added. Ampicillin (100 μg/ml) and kanamycin (40 μg/ml) were added where appropriate.
Plasmid construction
To construct Ptac-orf34-speF, a fragment of 616 nt of orf34-speF, spanning from the transcription start site to nucleotide +101 with respect to the speF AUG was PCR amplified from S. typhimurium SL1344 chromosomal DNA using primers 1788 and 1790 and cloned into the EcoRI and HindIII restriction sites of pRI. To construct PspeF-orf34-speF-lacZ transcriptional and translational fusions, a fragment of 817 nt (from nucleotide 201 upstream of speF transcription start site to nucleotide +101 with respect to the speF AUG) carrying the promoter of speF, orf34 and part of speF was PCR amplified from S. typhimurium SL1344 chromosomal DNA using primers 1636 and 1637 and cloned into the EcoRI and BamHI restriction sites of pRS551 and pRS552, respectively. To construct PspeF-orf34-lacZ transcriptional and translational fusions, a fragment of 334 nucleotides (from nucleotide 201 upstream of speF transcription start site to nucleotide +102 with respect to the orf34 AUG) carrying the promoter of speF and orf34 was PCR amplified from S. typhimurium SL1344 chromosomal DNA using primers 1636 and 1639 and cloned into the EcoRI and BamHI restriction sites of pRS551, pRS552 respectively. To fuse the promoter of speF operon to lacZ (PspeF-lacZ) a fragment of 215 nucleotides (from nucleotide 201 upstream of speF transcription start site to nucleotide 14 down stream of it) was PCR amplified from S. typhimurium SL1344 chromosomal DNA using primers 1636 and 1898 and cloned into the EcoRI and BamHI restriction sites of pRS551. To construct PLtetO-1-orf34 (p15A origin), the orf34 sequence was PCR amplified from +5 of the transcription start site up to 9 nt downstream of the stop codon using primers 2368 (KpnI) and 2369 (phosphorylated). The two PCR fragments (orf34 and pZA31) were then ligated.
Strain construction
speF deletion mutant in S. typhimurium SL1344 strain was constructed based on the gene disruption method [37]. Chloramphenicol cassette was amplified by PCR using primers 1831 and 1832 containing sequences homologous to orf34-speF. The DNA generated was transformed into LB5010 cells carrying pKD46 plasmid and chloramphenicol resistant colonies were selected. speF gene disruption was examined by PCR using flanking primers, 1833 and 1834. To insert the lacZ fusions as single copies in the hisG gene sequence in the chromosome, DNA fragments including kanamycin and speF-lacZ fusions were PCR amplified using 1514 and 1515 primers. Ampicillin sensitive and kanamycin resistant cells were selected. The insertions into hisG gene were examined by PCR using flanking primers, 1517 and 1518. The chromosomal fusions and the deletion mutant were moved into S. typhimurium SL1344 cells by P22 transduction.
Site directed mutagenesis
Mutants PspeF-orf34AAA-speF, PspeF-orf34UAA16-speF, PspeF-orf34UAA26-speF, PspeF-orf34rRR-speF, PspeF-orf34fRR-speF, PspeF-orf34KK-speF, PspeF-orf34CA116,117GT-speF, PspeF-orf34TG510,511AC-speF were generated by whole plasmid PCR using pGEM4 carrying PspeF-orf34AUG-speF and two tail-to-tail divergent primers of which, one or both, carried the desired mutation. The PCR product was subjected to blunt end ligation and transformed into MC4100. DNA fragments carrying the mutations were digested from pGEM4 using EcoRI and BamHI and cloned into pRS551 and pRS552. The deletion mutants were generated using divergent primers spaced by the desired deletion.
β-galactosidase assays
Overnight cultures were diluted 1/100 in 7 ml LB medium supplemented with ampicillin and kanamycin in non-aerated 50 ml tubes and grown to OD600 of 0.9. Where indicated, bicyclomycin (BCM 20 μg /ml) was added at OD600 of 0.2 for 1 hour. To examine regulation by arginine or ornithine, colonies were inoculated in 5ml E-minimal medium in non-aerated 50 ml tubes for 17 hours. Each colony was inoculated evenly in arginine plus and minus tubes. Arginine or ornithine (100 μg /ml) was added where indicated from the beginning of growth.
RNA extraction
Overnight cultures of S. typhimurium SL1344 carrying Ptac-orf34-speF plasmids were diluted 1/100 in 20 ml LB medium supplemented with ampicillin in 125ml flasks and grown to OD600 of 0.2. Where indicated, BCM (20 μg/ml) was added at OD600 of 0.2 for 1 hour. To examine regulation by arginine or ornithine, overnight cultures were diluted 1/50 in 20 ml E-minimal medium in 125 ml flasks and grown to OD600 of 0.2 at which arginine or ornithine (100 μg/ml) was added for 5 or 15 minutes. For ornithine induction at stationary phase, cultures were grown in E-minimal medium in the presence of ornithine (25 or 50 μg/ml). Total RNA was purified using TRI reagent (Sigma) according to the manufacturer’s protocol.
Northern analysis
RNA samples (10 μg) were denatured for 10 min at 70°C in 98% formamide loading buffer, separated on 8M urea-6% polyacrylamide gels and transferred to Zeta Probe GT membranes (Bio-Rad Laboratories) by electroblotting. To detect orf34-speF mRNA, the membranes were hybridized in modified CHURCH buffer using [32P]-end labeled 1614 primer ([38]. Since 1614 primer overlaps the orf34 consecutive arginine codons, orf34 arginine codon mutants were detected using 2411 primer. 5S rRNA was used as a loading control using primer 459. After 2 hours at 45°C, the membranes were washed for 20 min. in 3XSSC at 45°C.
3' RACE
The site of premature termination in orf34AAA mutant was determined by 3'-RACE as described before [39] using 10 μg of RNA extracted from S. typhimurium SL1344 culture carrying Ptac-orf34AAA-speF plasmid. RNA adapter (1579) and primer 2211 were used for reverse transcription reaction. 1 μl of the cDNA product was PCR amplified using primers 2212 and 2213 and cloned into the AatII and HindIII restriction sites of pBR plasmid. The ligation product of pBR with speF-E1 insert was electro-transformed into MC4100 cells. Plasmid was purified and sequenced to detect the exact position of premature termination of speF.
Supporting information
(A) Sequence studied in this work is marked in purple. Representative proteins were chosen from NCBI collection of DUF2618 containing proteins. Additional protein sequences were obtained from full genomes found by tBlastn using orf34 coding sequence as query. Multiple sequence alignments were created using ClustalX v.2.1 [1] Maximum-likelihood phylogenetic trees were constructed using the phylogeny.fr pipeline [2], including the PhyML v.3.0 [3] and the WAG substitution model for amino acids [4]. One hundred bootstrap replicates were performed for each analysis. Branches having branch support value smaller than 85% confidence were collapsed. (B) Multiple sequence alignment of DUF2618 containing representatives by ClustalX v 2.1 (C) Protein logo of DUF2618 containing proteins. Proteins containing DUF2618 (PF10940) alignment was generated from NCBI (43 non-redundant proteins) using Pfam (http://pfam.xfam.org/) [5]. Alignment was used to generate a consensus protein logo (https://rth.dk/resources/plogo/) [6],[7]. Amino acids are marked by one letter, the polarity of the amino acids side chain marked by color; positive (blue), negative (red), uncharged (green) or hydrophobic (black). The numbers represent the corresponding position in ORF34.
(PDF)
(A) Cultures of wild type and rng mutant carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg /ml) for the indicated time. Northern blot of RNA samples (10 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. 5S RNA serves as a loading control. Full-length (FL) is indicated in red. Relative intensity of the full-length RNA as quantified using ImageQuant TL 1D v8.1 is shown (R.I of FL). (B) Graphs of relative band intensity of the data presented in A.
(PDF)
(PDF)
Arrowheads and arrows indicate weak and strong cleavage sites respectively. Not all site are shown.
(PDF)
prior to the arginine treatment (100 μg /ml). The membranes were probed with end-labeled orf34 (1614) and 5S rRNA (459) specific primers.
(PDF)
In vivo structure probing. Cultures of RNase G mutant (Δrng) carrying Ptac-orf34-speF and Ptac-orf34W35-speF in which the stop codon was changed to a Trp codon were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg /ml) for 5 min and DMS (1/700) for 2.5 min (overlapping). Primer extension of 2 μg (no DMS) and 8 μg (plus DMS) of total RNA using 1789 primer. The data probing obtained with wild type RNA are displayed on the structure on the left. Red and blue asterisks indicate strong and weak modification sites. Data obtained with orf34W35 mutant are displayed on the structure on the right. Purple and grey asterisks indicate weak and faint modification sites. Note that wild type RNA is much more accessible to DMS modification including the region upstream of the stop codon, while the mutant RNA is much less accessible to DMS modification and the region upstream of the stop codon remains unmodified.
(PDF)
(A) Cultures carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then exposed to ornithine (100 μg /ml) for the indicated time or left untreated (-). (B) Cultures carrying Ptac-orf34-speF wild type, Δ(170–313) and Δ(388–400) were grown in E-minimal media to OD600 of 2.0 in the presence of ornithine (25 or 50 μg /ml) or left untreated (-). Northern blot of RNA samples (10 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (1614) and 5S rRNA (459) specific primers. 5S RNA serves as a loading control. Full-length (FL) and 5S are indicated in red.
(PDF)
Cultures carrying wild type Ptac-orf34-speF and Ptac-orf34rRR-speF were grown in E-minimal media to OD600 of 0.2 (Log) or for 17 hours to stationary phase (Stationary). Northern blot of RNA samples (left panel 10 μg and right panel 8 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. Left panel was exposed for 2 hours while the right panel was exposed for 8.5 hours. 5S RNA serves as a loading control. Full-length (FL) and 5S are indicated in red.
(PDF)
Cultures of wild type and ΔspeF were grown in E-minimal medium supplemented with ornithine (100 μg /ml) or arginine (100 μg /ml) as indicated or were left untreated. Note that in the absence of ornithine or arginine the growth rate of the mutant is similar to that of the wild type, whereas upon addition of ornithine or arginine wild type cells grow better than ΔspeF. Average of two biological experiments ± standard deviation.
(PDF)
Cultures were grown for 17 hours from a single colony in 5 ml (50 ml tubes) of E-Minimal supplemented with arginine (100 μg /ml). PLtetO and PLtetO-orf34 are P15A origin.
(DOCX)
(DOCX)
aPrimers used for fragment amplification. bPrimers used to generate mutations.
(DOCX)
(A) aPlus (+) and minus (-) strands are indicated. (B) aPlus (+) and minus (-) strands are indicated. (C) aPlus (+) and minus (-) strands are indicated. bAll mutant constructs are based on 616 nt fragment (1636–1637) consisting of PspeF-orf34-speF'-lacZ. cPositions of the mutations or deletions are relative to the transcription start site of speF operon transcript.
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Max Gottesman for generously providing bicyclomycin (BCM).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
S.A. acknowledges funding by the German-Israeli Foundation (G-1311-416.13/2015); the Israel Science Foundation founded by The Israel Academy of Sciences and Humanities (711/13), the Israel Centers of Research Excellence (ICORE), Chromatin and RNA (1796/12) and by Deutsch- lsraelische Projektkooperation (AM 441/1-1 SO 568/1-1). K.P acknowledges funding by the German Research Foundation (DFG – SPP2002 and Exc114-2), the Human Frontier Science Program (CDA00024/2016-C), German-Israeli Foundation (G-2411-416.13/2016), and the European Research Council (StG-758212). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bobrovskyy M, Vanderpool CK. Regulation of Bacterial Metabolism by Small RNAs Using Diverse Mechanisms. Annu Rev Genet. 2013;47:209–32. Epub 2013/09/11. 10.1146/annurev-genet-111212-133445 . [DOI] [PubMed] [Google Scholar]
- 2.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Micro. 2016;14(9):576–88. 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein G, Raina S. Regulated Control of the Assembly and Diversity of LPS by Noncoding sRNAs. Biomed Res Int. 2015;2015:153561 10.1155/2015/153561 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8(1):116–27. 10.1016/j.chom.2010.06.008 . [DOI] [PubMed] [Google Scholar]
- 5.Wagner EG, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. 10.1016/bs.adgen.2015.05.001 . [DOI] [PubMed] [Google Scholar]
- 6.Klinkert B, Narberhaus F. Microbial thermosensors. Cell Mol Life Sci. 2009;66(16):2661–76. Epub 2009/05/12. 10.1007/s00018-009-0041-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naville M, Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief Funct Genomic Proteomic. 2009;8(6):482–92. Epub 2009/08/03. 10.1093/bfgp/elp025 . [DOI] [PubMed] [Google Scholar]
- 8.Henkin TM. RNA-dependent RNA switches in bacteria. Methods Mol Biol. 2009;540:207–14. 10.1007/978-1-59745-558-9_15 . [DOI] [PubMed] [Google Scholar]
- 9.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–34. 10.1146/annurev.biochem.78.070507.135656 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastet L, Dubé A, Massé E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol Microbiol. 2011;80(5):1148–54. Epub 2011/04/20. 10.1111/j.1365-2958.2011.07654.x . [DOI] [PubMed] [Google Scholar]
- 11.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2). Epub 2012/02/01. 10.1101/cshperspect.a003566 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriner MA, Sevostyanova A, Groisman EA. Learning from the Leaders: Gene Regulation by the Transcription Termination Factor Rho. Trends Biochem Sci. 2016;41(8):690–9. Epub 2016/06/17. 10.1016/j.tibs.2016.05.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastet L, Chauvier A, Singh N, Lussier A, Lamontagne AM, Prévost K, et al. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 2017;45(12):7474–86. 10.1093/nar/gkx434 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 2012;26(16):1864–73. 10.1101/gad.195412.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109(14):5376–81. Epub 2012/03/19. 10.1073/pnas.1112211109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142(5):737–48. Epub 2010/09/04. 10.1016/j.cell.2010.07.046 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 2011;30(8):1485–96. Epub 2011/03/11. 10.1038/emboj.2011.66 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6(8):R70 Epub 2005/08/01. 10.1186/gb-2005-6-8-r70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266(31):20922–7. . [PubMed] [Google Scholar]
- 20.Boyle SM, Markham GD, Hafner EW, Wright JM, Tabor H, Tabor CW. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (speA, speB, speC and metK). Gene. 1984;30(1–3):129–36. . [DOI] [PubMed] [Google Scholar]
- 21.Nakada Y, Itoh Y. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology. 2003;149(Pt 3):707–14. 10.1099/mic.0.26009-0 . [DOI] [PubMed] [Google Scholar]
- 22.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J Mol Biol. 2015;427(21):3389–406. Epub 2015/07/05. 10.1016/j.jmb.2015.06.020 . [DOI] [PubMed] [Google Scholar]
- 23.Shantz LM, Pegg AE. Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int J Biochem Cell Biol. 1999;31(1):107–22. . [DOI] [PubMed] [Google Scholar]
- 24.Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A. 2012;109(20):E1277–86. Epub 2012/04/25. 10.1073/pnas.1201061109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magyar A, Zhang X, Kohn H, Widger WR. The antibiotic bicyclomycin affects the secondary RNA binding site of Escherichia coli transcription termination factor Rho. J Biol Chem. 1996;271(41):25369–74. . [DOI] [PubMed] [Google Scholar]
- 26.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26 Epub 2011/11/24. 10.1186/1748-7188-6-26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, et al. In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-coding RNA Pathways. Mol Cell. 2017;65(1):39–51. 10.1016/j.molcel.2016.11.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152(Pt 9):2515–28. 10.1099/mic.0.28982-0 . [DOI] [PubMed] [Google Scholar]
- 29.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3'-end chronicles. J Mol Biol. 2011;412(5):793–813. Epub 2011/03/23. 10.1016/j.jmb.2011.03.036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–8. 10.1126/science.1184939 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356(6334):194–7. 10.1126/science.aal3059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan H, Conn AB, Williams PB, Diggs S, Hahm J, Gamper HB, et al. Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 2017;45(19):11043–55. 10.1093/nar/gkx719 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Massé E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci U S A. 2012;109(50):E3444–53. Epub 2012/11/19. 10.1073/pnas.1214024109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flamigni F, Guarnieri C, Caldarera CM. Characterization of highly purified ornithine decarboxylase from rat heart. Biochim Biophys Acta. 1984;802(2):245–52. . [DOI] [PubMed] [Google Scholar]
- 35.Nam KH, Lee SH, Lee J. Purification and characterization of arginine decarboxylase from soybean (Glycine max) hypocotyls. Plant Cell Physiol. 1997;38(10):1150–5. . [DOI] [PubMed] [Google Scholar]
- 36.Gao W, Tyagi S, Kramer FR, Goldman E. Messenger RNA release from ribosomes during 5'-translational blockage by consecutive low-usage arginine but not leucine codons in Escherichia coli. Mol Microbiol. 1997;25(4):707–16. . [DOI] [PubMed] [Google Scholar]
- 37.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97(11):5978–83. 10.1073/pnas.100127597 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36(6):1913–27. Epub 2008/02/13 10.1093/nar/gkn050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fröhlich KS, Papenfort K, Berger AA, Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40(8):3623–40. Epub 2011/12/17. 10.1093/nar/gkr1156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Sequence studied in this work is marked in purple. Representative proteins were chosen from NCBI collection of DUF2618 containing proteins. Additional protein sequences were obtained from full genomes found by tBlastn using orf34 coding sequence as query. Multiple sequence alignments were created using ClustalX v.2.1 [1] Maximum-likelihood phylogenetic trees were constructed using the phylogeny.fr pipeline [2], including the PhyML v.3.0 [3] and the WAG substitution model for amino acids [4]. One hundred bootstrap replicates were performed for each analysis. Branches having branch support value smaller than 85% confidence were collapsed. (B) Multiple sequence alignment of DUF2618 containing representatives by ClustalX v 2.1 (C) Protein logo of DUF2618 containing proteins. Proteins containing DUF2618 (PF10940) alignment was generated from NCBI (43 non-redundant proteins) using Pfam (http://pfam.xfam.org/) [5]. Alignment was used to generate a consensus protein logo (https://rth.dk/resources/plogo/) [6],[7]. Amino acids are marked by one letter, the polarity of the amino acids side chain marked by color; positive (blue), negative (red), uncharged (green) or hydrophobic (black). The numbers represent the corresponding position in ORF34.
(PDF)
(A) Cultures of wild type and rng mutant carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg /ml) for the indicated time. Northern blot of RNA samples (10 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. 5S RNA serves as a loading control. Full-length (FL) is indicated in red. Relative intensity of the full-length RNA as quantified using ImageQuant TL 1D v8.1 is shown (R.I of FL). (B) Graphs of relative band intensity of the data presented in A.
(PDF)
(PDF)
Arrowheads and arrows indicate weak and strong cleavage sites respectively. Not all site are shown.
(PDF)
prior to the arginine treatment (100 μg /ml). The membranes were probed with end-labeled orf34 (1614) and 5S rRNA (459) specific primers.
(PDF)
In vivo structure probing. Cultures of RNase G mutant (Δrng) carrying Ptac-orf34-speF and Ptac-orf34W35-speF in which the stop codon was changed to a Trp codon were grown in E-minimal media to OD600 of 0.2 and then treated with arginine (100 μg /ml) for 5 min and DMS (1/700) for 2.5 min (overlapping). Primer extension of 2 μg (no DMS) and 8 μg (plus DMS) of total RNA using 1789 primer. The data probing obtained with wild type RNA are displayed on the structure on the left. Red and blue asterisks indicate strong and weak modification sites. Data obtained with orf34W35 mutant are displayed on the structure on the right. Purple and grey asterisks indicate weak and faint modification sites. Note that wild type RNA is much more accessible to DMS modification including the region upstream of the stop codon, while the mutant RNA is much less accessible to DMS modification and the region upstream of the stop codon remains unmodified.
(PDF)
(A) Cultures carrying Ptac-orf34-speF were grown in E-minimal media to OD600 of 0.2 and then exposed to ornithine (100 μg /ml) for the indicated time or left untreated (-). (B) Cultures carrying Ptac-orf34-speF wild type, Δ(170–313) and Δ(388–400) were grown in E-minimal media to OD600 of 2.0 in the presence of ornithine (25 or 50 μg /ml) or left untreated (-). Northern blot of RNA samples (10 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (1614) and 5S rRNA (459) specific primers. 5S RNA serves as a loading control. Full-length (FL) and 5S are indicated in red.
(PDF)
Cultures carrying wild type Ptac-orf34-speF and Ptac-orf34rRR-speF were grown in E-minimal media to OD600 of 0.2 (Log) or for 17 hours to stationary phase (Stationary). Northern blot of RNA samples (left panel 10 μg and right panel 8 μg) separated using 6% urea-polyacrylamide gels. The membranes were probed with end-labeled orf34 (2411) and 5S rRNA (459) specific primers. Left panel was exposed for 2 hours while the right panel was exposed for 8.5 hours. 5S RNA serves as a loading control. Full-length (FL) and 5S are indicated in red.
(PDF)
Cultures of wild type and ΔspeF were grown in E-minimal medium supplemented with ornithine (100 μg /ml) or arginine (100 μg /ml) as indicated or were left untreated. Note that in the absence of ornithine or arginine the growth rate of the mutant is similar to that of the wild type, whereas upon addition of ornithine or arginine wild type cells grow better than ΔspeF. Average of two biological experiments ± standard deviation.
(PDF)
Cultures were grown for 17 hours from a single colony in 5 ml (50 ml tubes) of E-Minimal supplemented with arginine (100 μg /ml). PLtetO and PLtetO-orf34 are P15A origin.
(DOCX)
(DOCX)
aPrimers used for fragment amplification. bPrimers used to generate mutations.
(DOCX)
(A) aPlus (+) and minus (-) strands are indicated. (B) aPlus (+) and minus (-) strands are indicated. (C) aPlus (+) and minus (-) strands are indicated. bAll mutant constructs are based on 616 nt fragment (1636–1637) consisting of PspeF-orf34-speF'-lacZ. cPositions of the mutations or deletions are relative to the transcription start site of speF operon transcript.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.