Abstract
Purpose
Primary congenital glaucoma (PCG) is a clinically and genetically heterogeneous disease. The present study was undertaken to find the genetic causes of PCG segregating in 36 large consanguineous Pakistani families.
Methods
Ophthalmic examination including fundoscopy, or slit-lamp microscopy was performed to clinically characterize the PCG phenotype. Genomic nucleotide sequences of the CYP1B1 and LTBP2 genes were analyzed with either Sanger or whole exome sequencing. In silico prediction programs were used to assess the pathogenicity of identified alleles. ClustalW alignments were performed to determine evolutionary conservation, and three-dimensional (3D) modeling was performed using HOPE and Phyre2 software.
Results
Among the known loci, mutations in CYP1B1 and LTBP2 are the common causes of PCG. Therefore, we analyzed the genomic nucleotide sequences of CYP1B1 and LTBP2, and detected probable pathogenic variants cosegregating with PCG in 14 families. These included the three novel (c.542T>A, c.1436A>G, and c.1325delC) and five known (c.868dupC, c.1168C>T, c.1169G>A, c.1209InsTCATGCCACC, and c.1310C>T) variants in CYP1B1. Two of the novel variants are missense substitutions [p.(Leu181Gln), p.(Gln479Arg)], which replaced evolutionary conserved amino acids, and are predicted to be pathogenic by various in silico programs, while the third variant (c.1325delC) is predicted to cause reading frameshift and premature truncation of the protein. A single mutation, p.(Arg390His), causes PCG in six (~43%) of the 14 CYP1B1 mutations harboring families, and thus, is the most common variant in this cohort. Surprisingly, we did not find any LTBP2 pathogenic variants in the families, which further supports the genetic heterogeneity of PCG in the Pakistani population.
Conclusions
In conclusion, results of the present study enhance our understanding of the genetic basis of PCG, support the notion of a genetic modifier of CYP1B1, and contribute to the development of genetic testing protocols and genetic counseling for PCG in Pakistani families.
Introduction
Glaucoma is characterized by progressive optic neuropathy leading to severe, often permanent, vision loss [1]. Glaucoma is considered one of the major causes of bilateral blindness worldwide, with an estimated prevalence of 64.3 million, of whom 8.4 million are bilaterally blind [2]. This prevalence is anticipated to rise with an alarming rate to 76.0 million in 2020 and 111.8 million in 2040 [3,4]. Among the glaucoma subtypes, primary congenital glaucoma (PCG; OMIM 231300), characterized by the presence of an underdeveloped trabecular meshwork, is an imperative cause of severe visual disability in children, and has a prevalence of 1:10,000 to 18,000 live births worldwide [2].
In PCG, the improperly developed aqueous outflow system results in increased intraocular pressure, enlargement (Buphthalmos) and opacification of cornea, edema, with an ultimate consequence of optic nerve damage and vision impairment [5,6]. PCG is a genetically heterogeneous disorder, and commonly segregates in a typical autosomal recessive fashion. However, linkage and positional cloning studies have also revealed autosomal dominant PCG loci or genes [5]. Among the known genes, biallelic variants of CYP1B1 (OMIM 601,771) and LTBP2 (OMIM 602,091) are the common cause of PCG worldwide [5,7,8]. As of September 2018, around 240 and 26 pathogenic variants of CYP1B1 and LTBP2, respectively, are listed in the Human Gene Mutation database. In patients with PCG, early diagnosis is important for effective treatment of the disease with surgical interventions (e.g., trabeculotomy, trabeculectomy, goniotomy, or deep sclerectomy), use of drainage implants, or correcting refractive errors and amblyopia. Therefore, identifying individuals at risk for inheriting pathogenic variant(s) as soon as possible after birth has considerable ramifications for diagnosis, management, and treatment of PCG and to prevent unnecessary health care costs [5].
To investigate the etiology of the disease at the molecular level and to improve genetic diagnosis, we ascertained a large cohort of consanguineous Pakistani families segregating PCG. We performed candidate genes screening with Sanger sequencing and whole exome sequence (WES) analysis to identify the underlying genetic cause of PCG in the cohort. In this study, we report identification of five known and three novel variants of CYP1B1 segregating in 14 families with PCG. We did not find a pathogenic allele in CYP1B1 and LTBP2 in 22 of the 36 families (61%), which suggests genetic heterogeneity of PCG in the Pakistani population.
Methods
Subjects and clinical evaluation
The present study was approved by IRB committees of the participating institutions in the United States and Pakistan. All methods used in the study followed the precepts of Declaration of Helsinki and were in compliance of the ARVO statement on human subjects. Informed written consent was obtained from all individuals before inclusion in the study. A total of 423 individuals including 227 males (of which 79 had glaucoma) and 196 females (71 affected) with the ages ranging from 1 year old to 65 years old were recruited from Sindh and Punjab Provinces of Pakistan. Detailed interviews were conducted with family members to gather information on pedigree structure, comorbidities, onset of disease, and initial symptoms. The clinical diagnosis was based on presenting symptoms, physical and ophthalmic examinations, fundoscopy, or slit-lamp microscopy. The DNA was extracted from whole blood by using the inorganic method as described previously Grimberg et al 1989 [9].
Known genes screening and exome sequencing
Before exome sequencing, we performed Sanger sequencing on one affected individual from 24 of the 36 families with glaucoma for known PCG genes, CYP1B1 and LTBP2. Primers for Sanger sequencing were designed using Primer3. PCR amplification was performed as follows: 95 °C: 2 min (1X), 95 °C: 30 sec; annealing temperature: 30 sec; 72 °C: 1 min (35X), 72 °C: 7 min (1X), 25 °C: 5 min (1X) by using EconoTaq Plus 2X Master mix. and DNA sequencing were performed as described previously [10]. WES was performed on the remaining 12 families and on the families negative for pathogenic variants in CYP1B1 and LTBP2. For WES, genomic libraries were recovered for exome enrichment using the Agilent SureSelect Human Expanded All Exon V5 (62 Mb) kit (Agilent corporation, Santa Clara, CA).. Libraries were sequenced on an Illumina HiSeq2500 (Agilent corporation) with average 100X coverage. Data analysis used the Broad Institute’s Genome Analysis Toolkit [11]. Reads were aligned with the Illumina Chastity Filter with the Burrows-Wheeler Aligner [12]. Variant sites were called using the GATK UnifiedGenotyper module. Single nucleotide variant calls were filtered using the variant quality score recalibration method [11]. Filtration of candidate variants was performed as described previously [13]. Sanger sequencing was used to confirm the segregation of the variants identified in the families.
In silico analysis
The following online pathogenicity prediction programs were used to evaluate the pathogenicity of missense variants: Polymorphism Phenotyping v2 (PolyPhen-2), Sorting Tolerant From Intolerant (SIFT), Mutation Taster, Mutation Assessor, and fathmm. Moreover, ClustalW alignment was used to assess the conservation of the affected amino acid residues.
Results
Clinical findings
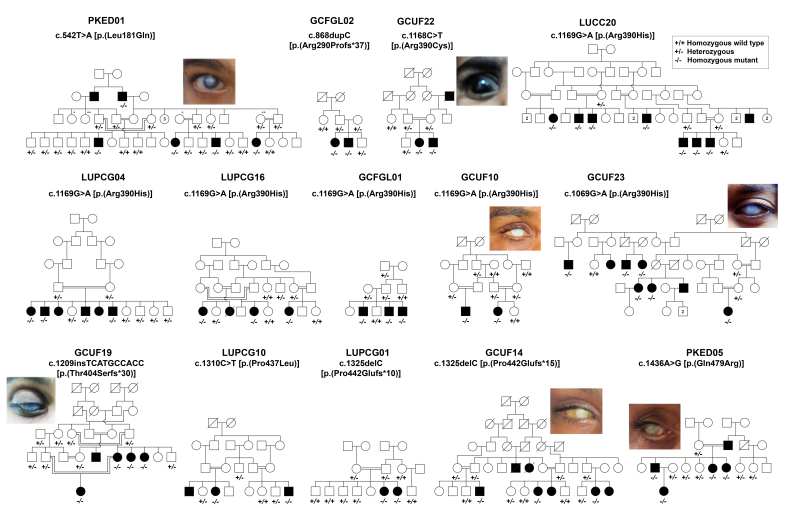
In this study, we enrolled 36 families segregating with primary congenital glaucoma from the Punjab and Sindh provinces of Pakistan (Figure 1). The neonatal clinical records of the affected individuals were not available at the time of enrollment. According to the family history, all the affected individuals were reported to have vision problems shortly after birth or in early childhood. The most common features consistent with PCG included high intraocular pressure, reduced visual acuity, increased corneal diameter, and corneal haze (Figure 1, Table 1). However, high inter- and intrafamilial variability was observed in the severity of PCG in these families (Table 1). For instance, one affected individual of family PKED05 had no apparent visual impairment or corneal haze, but ophthalmic evaluation revealed high intraocular pressure, while the other affected individuals of the same family had classical symptoms of PCG. Furthermore, a few affected individuals of the families with PCG had low vision and corneal haze only in one eye (Table 1). However, a few of the participating affected individuals had microphthalmia, and were completely blind at the time of enrollment.
Figure 1.
Pedigrees of Pakistani families with PCG with segregated variants in CYP1B1. Pedigrees of 14 multigenerational families who segregated recessive primary congenital glaucoma (PCG) due to disease-causing variants in CYP1B1. Filled and empty symbols represent affected and unaffected individuals, respectively. A double line connecting two individuals represents consanguineous marriage. Genotypes are written at the bottom of the enrolled individuals. Shown also are eye photographs of representative affected individuals. All the affected individuals have hazy corneas.
Table 1. Clinical details of the families segregating CYP1B1 variants.
| Family ID | Participant IDs | Age (Yrs) | Variant | IOP (mmHg) OS/OD | Visual Acuity OS/OD | Surgical procedure | Medicine used for IOP | Remarks |
|---|---|---|---|---|---|---|---|---|
| PKED01 |
IV 6 |
7 |
c.542T>A |
14/14 |
NA |
Trabeculectomy |
Yes |
Cornea clear, well-formed bleb |
| |
IV 12 |
6 |
p.(Leu181Gln) |
NA |
NA |
Trabeculectomy |
Yes |
OS/OD CD= 0.9 |
| |
IV 15 |
13 |
|
22/20 |
NA |
Trabeculectomy |
Yes |
NA |
| |
IV 18 |
1 |
|
17/17 |
NA |
Trabeculectomy |
Yes |
NA |
| GCFGL02 |
III:1 |
4 |
c.868dupC |
25/53 |
PL:NPL |
Corneal scraping |
Yes |
Bilateral corneal haze |
| |
|
|
p.(Arg290Profs*37) |
|
|
Trabeculectomy |
|
Enlarged anterior segment |
| |
III:2 |
7 |
|
18/29 |
PL:NPL |
Corneal scraping |
yes |
Bilateral corneal haze |
| |
|
|
|
|
|
Trabeculectomy |
|
Enlarged anterior segment |
| GCUF22 |
IV:2 |
5 |
c.1168C>T |
33/30 |
NPL: NPL |
No |
No |
Severe corneal haze |
| |
|
|
p.(Arg390Cys) |
|
|
|
|
Habb’s striae |
| |
IV:3 |
3 |
|
29/21 |
NPL: NPL |
OS: Trabeculectomy |
No |
Deep anterior segment bilateral |
| |
|
|
|
|
|
OD: No |
|
|
| LUCC20 |
IV:5 |
30 |
c.1169G>A |
NR/15 |
NPL: NPL |
Trabeculectomy |
No |
Bilateral corneal haze |
| |
IV:6 |
24 |
p.(Arg390His) |
32/28 |
NR: NR |
Trabeculectomy |
Yes |
Bilateral corneal haze |
| |
|
|
|
|
|
|
|
|
| LUPCG04 |
V:2 |
17 |
c.1169G>A |
30/NR |
NPL: NPL |
Trabeculectomy |
Yes |
Bilateral corneal haze |
| |
|
|
p.(Arg390His) |
|
|
|
|
|
| |
V:5 |
34 |
|
NR/NR |
NPL: NPL |
No |
No |
Bilateral corneal haze |
| LUPCG16 |
V:5 |
13 |
c.1169G>A |
40/35 |
NPL: NPL |
No |
No |
NA |
| |
|
|
p.(Arg390His) |
|
|
|
|
|
| GCFGL01 |
III:2 |
13 |
c.1169G>A |
18/20 |
NPL: NPL |
Trabeculectomy |
Yes |
OS/OD severe corneal haze |
| |
III:4 |
5 |
p.(Arg390His) |
32/29 |
NPL: NPL |
Trabeculectomy |
No |
OS: Habb’s Striae buphthalmol |
| |
|
|
|
|
|
|
|
OD: sever corneal haze |
| |
III:5 |
10 |
|
NA/26 |
NPL:20/50 |
No |
No |
OS: phthisis occurred |
| |
|
|
|
|
|
|
|
OD: severe corneal haze Enlarged anterior segment |
| GCUF10 |
IV:1 |
3 |
c.1169G>A |
28/24 |
20/100:20/70 |
Trabeculectomy |
Yes |
OS/OD Haze Cornea, AGV |
| |
IV:2 |
5 |
p.(Arg390His) |
29/26 |
20/70:20/70 |
Trabeculectomy |
Yes |
OS/OD Haze Cornea, AGV |
| GCUF23 |
III:1 |
22 |
c.1169G>A |
NA/16 |
NPL: NPL |
No |
No |
OS: phthisis occurred |
| |
|
|
p.(Arg390His) |
|
|
|
|
OD: severe haze cornea |
| |
III:4 |
43 |
|
|
NPL: NPL |
No |
No |
OS: severe corneal haze, deep cup |
| |
|
|
|
|
|
|
|
OD: phthisis occurred |
| |
III:5 |
40 |
|
26/NA |
NPL: NPL |
No |
No |
OS: severe corneal haze |
| |
|
|
|
|
|
|
|
OD: phthisis occurred |
| |
IV:2 |
30 |
|
20/NA |
PL: NPL |
No |
No |
OS: deep cup |
| |
|
|
|
|
|
|
|
OD: phthisis occurred |
| |
IV:3 |
27 |
|
21/16 |
PL: NPL |
No |
No |
OS: deep cup |
| |
|
|
|
|
|
|
|
OD: severe corneal haze |
| |
V:4 |
1.5 |
|
14062 |
PL:PL |
Lansectomy and Anterior Vetrectomy |
No |
OS: AGV done |
| |
|
|
|
|
|
|
|
OD: AGV done, Opaque cornea |
| GCUF19 |
IV:6 |
30 |
c.1209insTCATGCCACC |
NA/16 |
NPL:20/100 |
Goniotomy |
No |
OS: phthisis occurred |
| |
|
|
p.(Thr404Serfs*30) |
|
|
|
|
OD: deep cup |
| |
IV:7 |
25 |
|
43804 |
1/60: PL |
Trabeculectomy |
No |
OS/OD deep cup |
| |
IV:8 |
10 |
|
26/27 |
NPL: NPL |
Trabeculectomy |
Yes |
OS/OD deep cup |
| |
V:1 |
3 |
|
20/17 |
20/70:20/50 |
Trabeculectomy |
Yes |
severe corneal haze |
| LUPCG10 |
IV:8 |
11 |
c.1310C>T |
14/17 |
OS 6/24 |
Trabeculectomy |
Yes |
OS/OD CD= 0.8/0.9 |
| |
|
|
p.(Pro437Leu) |
|
OD 6/18 |
|
|
|
| LUPCG01 |
III:6 |
15 |
c.1325delC |
30/28 |
OS 6/20 |
Trabeculectomy |
Yes |
OS/OD CD= 0.6/0.5 |
| |
|
|
p.(Pro442Glufs*10) |
|
OD 6/10 |
|
|
|
| |
III:7 |
25 |
|
25/25 |
OS 6/24 |
Trabeculectomy |
Yes |
OS/OD CD= 0.7/0.6 |
| |
|
|
|
|
OD 6/24 |
|
|
|
| GCUF14 |
V:4 |
25 |
PCG |
35/33 |
NPL: NPL |
No |
No |
severe corneal haze |
| |
V:7 |
18 |
PCG |
30/32 |
NPL: NPL |
No |
No |
severe corneal haze |
| |
V:11 |
27 |
PCG |
NA |
NPL: NPL |
No |
No |
OS/OD phthisis occurred |
| GCFGL01 |
III:2 |
13 |
PCG |
18/20 |
NPL: NPL |
Trabeculectomy |
Yes |
OS/OD severe corneal haze |
| |
III:4 |
5 |
PCG |
32/29 |
NPL: NPL |
Trabeculectomy |
No |
OS: Habb’s Striae buphthalmol |
| |
|
|
|
|
|
|
|
OD: sever corneal haze |
| |
III:5 |
10 |
PCG |
NA/26 |
NPL:20/50 |
No |
No |
OS: phthisis occurred |
| |
|
|
|
|
|
|
|
OD: severe corneal haze enlarged anterior segment |
| PKED05 |
III 2 |
18 |
PCG |
NA |
NPL |
Trabeculectomy |
Yes |
OS: phthisis Bulbi |
| |
|
|
|
|
|
|
|
OD: phthisis Bulbi |
| |
III 6 |
13 |
PCG |
34/44 |
0.266666667 |
No |
Yes |
OS/OD CD= 1.0/0.8 |
| |
|
|
|
|
|
|
|
MG negative |
| |
III 7 |
11 |
PCG |
16/20 |
NA |
No |
Yes |
opaque cornea in both eyes |
| |
|
|
|
|
|
|
|
OD: leukocoria |
| IV 1 | 1 | PCG | NA | PL:PL | Trabeculectomy | Yes | photophobia, large eye balls, OS/OD severe corneal haze |
NPL: no light perception; PL: light perception; NA: not accessible due to phthisis; AGV: Ahmed’s glaucoma valve; TC: total cupping; NR: not recordable. IOP: Intraocular pressure; MG: myasthenia gravis; CD: cup-to-disc ratio
Genetic spectrum
We used candidate genes (CYP1B1 and LTBP2) screening and WES to identify the underlying genetic causes of PCG in the families. We identified eight variants of CYP1B1 cosegregating with PCG in 14 of the 36 families (Figure 1, Table 2). All of these 14 consanguineous families segregated PCG associated with CYP1B1 variants in a homozygous fashion. Five of these variants have been previously reported to cause PCG in humans, but we also identified three novel variants in CYP1B1 (Table 2). Almost 75% (six of eight) of the identified variants are present only once in the cohort (Table 2). However, a single variant p.(Arg390His) was found in six of the 14 families with CYP1B1-associated PCG (Table 2). Surprisingly, we did not find a disease-associated variant of LTBP2 in the cohort, which suggests genetic heterogeneity of PCG in Pakistani families.
Table 2. CYP1B1 alleles identified in this study.
| Family | Nucleotide change | Amino acid change | CADD | SIFT | Polyphen2 | MutationTaster | Mutation Assessor | Fathmm | gnomAD | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| PKED01 |
c.542T>A |
p.(Leu181Gln) |
23.6 |
Damaging |
Probably damaging |
Disease causing |
Medium |
Tolerated |
0 |
This study |
| PKED05 |
c.1436 A>G |
p.(Gln479Arg) |
11 |
Damaging |
Probably damaging |
Disease causing |
Medium |
|
0 |
This study |
| LUPCG01, GCUF14 |
c.1325delC |
p.(Pro442Glufs*15) |
- |
NA |
NA |
NA |
NA |
NA |
4.06E-06 |
This study |
| LUCC20, LUPCG04, LUPCG16, GCFGL01, GCUF10, GCUF23 |
c.1169G>A |
p.(Arg390His) |
35 |
Damaging |
Probably damaging |
Disease causing |
High |
Damaging |
0.000102 |
[27] |
| LUPCG10 |
c.1310C>T |
p.(Pro437Leu) |
33 |
Damaging |
Probably damaging |
Disease causing |
High |
Damaging |
2.16E-05 |
[27] |
| GCFGL02 |
c.868dupC |
p.(Arg290Profs*37) |
34 |
NA |
NA |
NA |
NA |
NA |
2.31E-05 |
[28] |
| GCUF19 |
c.1209InsTCATGCCACC |
p.(Thr404Serfs*30) |
25.5 |
NA |
NA |
NA |
NA |
NA |
0.000199 |
[29] |
| GCUF22 | c.1168C>T | p.(Arg390Cys) | 34 | Damaging | Probably damaging | Disease causing | NA | Damaging | 3.63E-05 | [30] |
NA: Not available; CADD: Combined annotation dependent depletion.
Causal variants
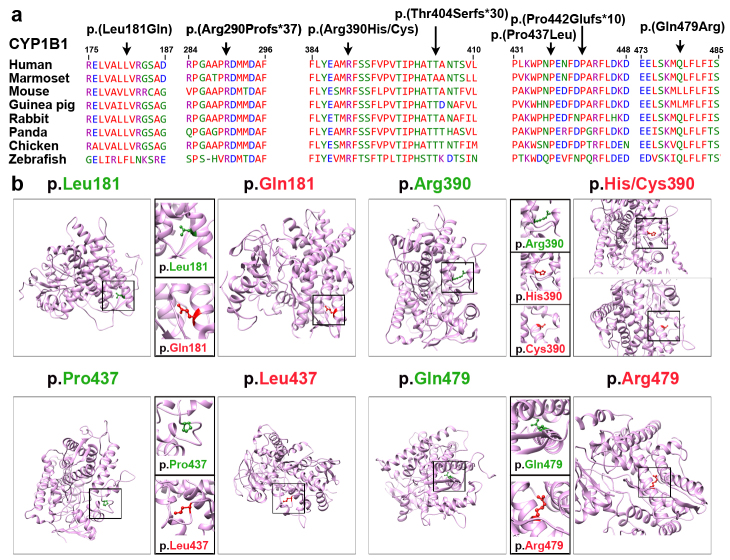
Of the causative variants identified in this study, two were novel missense variants [p.(Leu181Gln and p.(Gln479Arg)] of CYP1B1, cosegregating with PCG in families PKED01 and PKED05, respectively (Figure 1). These missense variants were not found in the Genome Aggregation (gnomAD), Human Gene Mutation (HGMD), and ClinVar databases, and were predicted to be deleterious by various in silico prediction algorithms (Table 2). Both missense variants affected amino-acid residues that are highly conserved among the CYP1B1 orthologs (Figure 2). We also used the HOPE [14] and Phyre2 [15] 3D modeling prediction programs to further assess the effects of these two missense variants on the secondary structure of the encoded protein. The p.Leu181 and p.Gln479 residues are located in the cytochrome p450 domain. Leucine at position 181 is part of an α-helix and is predicted to be buried in the protein core. Substitution of p.Leu181 with glutamine, which is smaller and less hydrophobic than leucine, does not prefer an α-helix as a secondary structure and leads to the loss of hydrophobic interactions in the protein core. Similarly, the substituting glutamine with arginine at position 479 is predicted to affect the protein stability by introducing protein-folding problems due to its positive charge, which, in turn, could affect the ligand contacts made by one of the neighboring residues.
Figure 2.
Multiple sequences alignment of CYP1B1 orthologs and molecular modeling of missense variants. A: ClustalW alignment of CYP1B1 proteins shows conservation of the residues at positions 181, 290, 390, 404, 437, 442, and 479 among eight species. B: Predicted three-dimensional (3D) structures of wild-type and mutant CYP1B1 proteins created using Phyre2. The positions of the wild-type and mutated forms of the amino acids in this cohort are shown in green and red, respectively. The p.Leu181 residue of CYP1B1 is located in the α-helix of the secondary structure, and replacing this residue with a smaller, less hydrophobic glutamine residue is predicted to slightly destabilize the local conformation and leads to the loss of hydrophobic interactions in the protein core. Modeling of the cytochrome domain of CYP1B1 revealed that the p.Arg390 residue forms a hydrogen bond and a salt bridge with glutamic acid at position 387, asparagine at position 428, and proline at position 437. Substitution with smaller residue histidine [p.(Arg390His)] or cysteine [p.(Arg390Cys)] is predicted to disrupt this hydrogen bond, and interrupt the signal transduction between the two domains of CYP1B1, cause an empty space in the core of the protein, and distort the correct protein folding. The p.Pro437 residue is located on the protein’s surface. Prolines are known to be rigid and therefore, induce a special backbone conformation. Replacing a proline with leucine might disturb this special conformation and distort the interactions with other molecules. Finally, the substituting glutamine with arginine at position 479 is predicted to affect the protein stability by introducing protein-folding problems due to its positive charge, which, in turn, could affect the ligand contacts made by one of the neighboring residues.
In two families with PCG (LUPCG01 and GCUF14), we found a novel single nucleotide deletion (c.1325delC), which is predicted to cause premature truncation [p.(Pro442Glufs*15); Table 2) of the encoded protein. This variant has low frequency in gnomAD (4.061 × 10−6), but is not registered in HGMD or ClinVar. In two other families (GCFGL02 and GCUF19), we identified two known frameshift variants [c.868dupC, p.(Arg290Profs*37); c.1209InsTCATGCCACC, p.(Thr404Serfs*30)], respectively. Both insertion variants are also predicted to result in premature stop codons and early truncation of CYP1B1.
In this study, the most commonly observed variant was c.1169G>A [p.(Arg390His)], which was found cosegregating with PCG in six families (Table 2). The c.1169G>A variant has relatively low frequency in the gnomAD database (1.017 × 10−4), which includes data of 123,136 exome and 15,496 whole-genome sequences. Therefore, we performed a haplotype analysis using eight closely linked single nucleotide polymorphisms (SNPs) in CYP1B1 to identify potential founder effects for this recurrent variant. Selection of these eight SNPs for haplotype analysis was based on the observance of high heterozygosity (>0.3) in 50 control samples randomly collected from the Pakistani population. The results were consistent with a common ancestor for the c.1169G>A recurrent allele in the Pakistani families in this study (data not shown).
The arginine at position 390 is highly conserved across CYP1B1 orthologs (Figure 2A), and is predicted to be protein damaging by several in silico bioinformatics tools (Table 2). The 3D protein modeling revealed that the p.Arg390 forms a hydrogen bond and a salt bridge with glutamic acid at position 387, asparagine at position 428, and proline at position 437 (Figure 2B). Substitution with a smaller residue histidine [p.(Arg390His)] is predicted to disrupt this hydrogen bond, and interrupt the signal transduction between the two domains of CYP1B1, and thus, is predicted to be pathogenic. In another family with PCG (GCUF22), arginine at position 390 is substituted with cysteine [p.(Arg390Cys); Table 2], which is also predicted to result in the loss of charge required to keep ionic interactions to make a salt bridge. The p.(Arg390Cys) substitution is also predicted to cause an empty space in the core of the protein and loss of hydrogen bonds and distort the correct protein folding.
Finally, through WES we identified a known missense variant [c.1310C>T, p.(Pro437Leu)] of the CYP1B1 gene in family LUPCG10 (Table 2). The proline residue at position 437 is located on the protein’s surface. As proline is rigid, p.Pro437 is predicted to induce a special protein backbone conformation at this position. The p.(Pro437Leu) substitution might disturb this special conformation and distort the interactions with other molecules.
Discussion
PCG is one of the most common genetic diseases worldwide. Pathogenic variants in CYP1B1 represent a broad spectrum for PCG at the global level. CYP1B1 is a constituent of cytochrome p450 family genes that are known to be involved in the detoxification of exogenous and endogenous molecules. CYP1B1 also plays a role in developmental processes of eye with an influence on metabolism. In ocular development, retinoic acid and estradiols are the two specific substrates for CYP1B1 [16]. CYP1B1 is known to regulate oxidative homeostasis, and ultrastructural and functional performance of trabecular meshwork tissue in the eye [17].
This study described the identification of eight allelic variants of CYP1B1 in 14 families with PCG ascertained from different regions of Pakistan. All of these variants were either substitution of the highly conserved residues or predicted to cause early truncation of the encoded protein with determined functional significance, and thus, considered to be disease causing. All of the identified variants segregated with the phenotype with an autosomal recessive pattern of inheritance. Seven of these variants were found only in one to two families, except the p.(Arg390His) allele, which was recurrently found in six families and likely originated from a single ancestral modification incidence. We also identified two novel homozygous missense variants [p.(Leu181Glu and p.(Gln479Arg)] in PKED01 and PKED05, respectively. Each variant results in a disturbed protein local secondary structure.
Ophthalmic evaluation of the affected individuals harboring homozygous variants in CYP1B1 revealed inter- and intrafamilial variability in the severity of PCG in these families. Variability in the disease manifestation and incomplete penetrance of CYP1B1 alleles have been previously found in consanguineous populations in Saudi Arabia and Pakistan [18,19]. It is possible that environmental or other epigenetic factors may be the cause of this incomplete penetrance. However, a possible dominant modifier locus has also been proposed [18], although no modifier gene has been reported yet. Genetically mapping a modifier variant of CYP1B1 in humans is challenging because of the usual limitations in the number of family subjects with incomplete penetrance [18,19]. Currently, we also cannot rule out the possibility of multiple modifier factors, including genetic and environmental, leading to incomplete penetrance. Modifiers that suppress a mutant phenotype of a Mendelian disorder provide insight into the mechanisms by which organisms can buffer biologic processes to accommodate the adverse effects of genetic mutations [20-24]. Thus, identification of the modifier of CYP1B1 may point toward possible therapies for individuals with PCG.
Considering the reported frequency of LTBP2 alleles in patients with PCG negative for variants in CYP1B1 [7,25], it is surprising that we did not find a pathogenic variant in the coding exons or in the splice junctions of LTBP2 in the PCG cohort. There are several possible reasons for our failure to detect variants of LTBP2 and CYP1B1 in the remaining 22 families with PCG. For instance, disease-causing variants may alter the sequence of cis-acting regulatory or splicing elements of LTBP2 or CYP1B1 that are necessary for their expression in ocular tissue. Presently, we do not know the location of the regulatory elements of CYP1B1 and LTBP2. Moreover, these results further support the genetic heterogeneity of PCG in the Pakistani population. Currently, we are investigating the exome data of the remaining 22 families with PCG for pathogenic variants at loci (GLC3B; OMIM 600975 and GLC3C; OMIM 613085) that were previously linked with PCG [26,27], as well as in genes that were not previously associated with glaucoma.
In summary, we identified three novel and five known variants of CYP1B1 that are associated with PCG in 14 Pakistani families. The data suggest that the variants in CYP1B1 might be responsible for extensive PCG cases in a highly consanguineous population. Detailed functional evaluation of pathogenic variants in CYP1B1 will add to better awareness of the PCG disease mechanism. This study extends the knowledge spectrum of PCG at the molecular level and could lead to the improvement of clinically relevant genetic diagnostic protocols.
Acknowledgments
We would like to thank the participating patients and their families, and the health care professionals involved in their care. We thank Dr. E. Richard and D. Gomes for critical review of the manuscript. Part of the study in Pakistan was supported by Pakistan Science Foundation Grant No: 101 and LUMHS Intramural Funds to A.M.W. Work at UMSOM was sponsored by the National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grant R01DC016295 to Z.M.A.
References
- 1.Pietrucha-Dutczak M, Smedowski A, Liu X, Matuszek I, Varjosalo M, Lewin-Kowalik J. Candidate proteins from predegenerated nerve exert time-specific protection of retinal ganglion cells in glaucoma. Sci Rep. 2017;7:14540. doi: 10.1038/s41598-017-14860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rulli E, Quaranta L, Riva I, Poli D, Hollander L, Galli F, Katsanos A, Oddone F, Torri V, Weinreb RN. Italian Study Group on Qo LiG. Visual field loss and vision-related quality of life in the Italian Primary Open Angle Glaucoma Study. Sci Rep. 2018;8:619. doi: 10.1038/s41598-017-19113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quigley HA. Glaucoma. Lancet. 2011;377:1367–77. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- 4.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–90. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Abu-Amero KK, Edward DP. Primary Congenital Glaucoma.Editors In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors.GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2019 [PubMed] [Google Scholar]
- 6.Sarfarazi M, Akarsu AN, Hossain A, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS. Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics. 1995;30:171–7. doi: 10.1006/geno.1995.9888. [DOI] [PubMed] [Google Scholar]
- 7.Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, Gilmour DF, Towns K, Murphy AL, Azmanov D, Tournev I, Cherninkova S, Jafri H, Raashid Y, Toomes C, Craig J, Mackey DA, Kalaydjieva L, Riazuddin S, Inglehearn CF. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84:664–71. doi: 10.1016/j.ajhg.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauf B, Irum B, Kabir F, Firasat S, Naeem MA, Khan SN, Husnain T, Riazuddin S, Akram J, Riazuddin SA. A spectrum of CYP1B1 mutations associated with primary congenital glaucoma in families of Pakistani descent. Hum Genome Var. 2016;3:16021. doi: 10.1038/hgv.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaworek TJ, Kausar T, Bell SM, Tariq N, Maqsood MI, Sohail A, Ali M, Iqbal F, Rasool S, Riazuddin S, Shaikh RS, Ahmed ZM. Molecular genetic studies and delineation of the oculocutaneous albinism phenotype in the Pakistani population. Orphanet J Rare Dis. 2012;7:44. doi: 10.1186/1750-1172-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, Song Y, van Beusekom E, Khan AA, Tomas-Roca L, Rashid M, Zahoor MY, Wissink-Lindhout WM, Basra MAR, Ansar M, Agha Z, van Heeswijk K, Rasheed F, Van de Vorst M, Veltman JA, Gilissen C, Akram J, Kleefstra T, Assir MZ. Uk10K, Grozeva D, Carss K, Raymond FL, O’Connor TD, Riazuddin SA, Khan SN, Ahmed ZM, de Brouwer APM, van Bokhoven H, Riazuddin S. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry. 2017;22:1604–14. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mookherjee S, Acharya M, Banerjee D, Bhattacharjee A, Ray K. Molecular basis for involvement of CYP1B1 in MYOC upregulation and its potential implication in glaucoma pathogenesis. PLoS One. 2012;7:e45077. doi: 10.1371/journal.pone.0045077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Wang S, Sorenson CM, Teixeira L, Dubielzig RR, Peters DM, Conway SJ, Jefcoate CR, Sheibani N. Cyp1b1 mediates periostin regulation of trabecular meshwork development by suppression of oxidative stress. Mol Cell Biol. 2013;33:4225–40. doi: 10.1128/MCB.00856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bejjani BA, Stockton DW, Lewis RA, Tomey KF, Dueker DK, Jabak M, Astle WF, Lupski JR. Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Hum Mol Genet. 2000;9:367–74. doi: 10.1093/hmg/9.3.367. [DOI] [PubMed] [Google Scholar]
- 19.Sheikh SA, Waryah AM, Narsani AK, Shaikh H, Gilal IA, Shah K, Qasim M, Memon AI, Kewalramani P, Shaikh N. Mutational spectrum of the CYP1B1 gene in Pakistani patients with primary congenital glaucoma: novel variants and genotype-phenotype correlations. Mol Vis. 2014;20:991–1001. [PMC free article] [PubMed] [Google Scholar]
- 20.Cazeneuve C, Ajrapetyan H, Papin S, Roudot-Thoraval F, Genevieve D, Mndjoyan E, Papazian M, Sarkisian A, Babloyan A, Boissier B, Duquesnoy P, Kouyoumdjian JC, Girodon-Boulandet E, Grateau G, Sarkisian T, Amselem S. Identification of MEFV-independent modifying genetic factors for familial Mediterranean fever. Am J Hum Genet. 2000;67:1136–43. doi: 10.1016/s0002-9297(07)62944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoblauch H, Schuster H, Luft FC, Reich J. A pathway model of lipid metabolism to predict the effect of genetic variability on lipid levels. J Mol Med (Berl) 2000;78:507–15. doi: 10.1007/s001090000156. [DOI] [PubMed] [Google Scholar]
- 22.Riazuddin S, Castelein CM, Ahmed ZM, Lalwani AK, Mastroianni MA, Naz S, Smith TN, Liburd NA, Friedman TB, Griffith AJ, Riazuddin S, Wilcox ER. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet. 2000;26:431–4. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- 23.Slavotinek A, Biesecker LG. Genetic modifiers in human development and malformation syndromes, including chaperone proteins. Hum Mol Genet. 2003;12:R45–50. doi: 10.1093/hmg/ddg099. [DOI] [PubMed] [Google Scholar]
- 24.Yousaf R, Ahmed ZM, Giese AP, Morell RJ, Lagziel A, Dabdoub A, Wilcox ER, Riazuddin S, Friedman TB, Riazuddin S. Modifier variant of METTL13 suppresses human GAB1-associated profound deafness. J Clin Invest. 2018;128:1509–22. doi: 10.1172/JCI97350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan AO. Genetics of primary glaucoma. Curr Opin Ophthalmol. 2011;22:347–55. doi: 10.1097/ICU.0b013e32834922d2. [DOI] [PubMed] [Google Scholar]
- 26.Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS, Sarfarazi M. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum Mol Genet. 1996;5:1199–203. doi: 10.1093/hmg/5.8.1199. [DOI] [PubMed] [Google Scholar]
- 27.Stoilov I, Akarsu AN, Alozie I, Child A, Barsoum-Homsy M, Turacli ME, Or M, Lewis RA, Ozdemir N, Brice G, Aktan SG, Chevrette L, Coca-Prados M, Sarfarazi M. Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am J Hum Genet. 1998;62:573–84. doi: 10.1086/301764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prokudin I, Simons C, Grigg JR, Storen R, Kumar V, Phua ZY, Smith J, Flaherty M, Davila S, Jamieson RV. Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet. 2014;22:907–15. doi: 10.1038/ejhg.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelberman D, Islam L, Jacques TS, Russell-Eggitt I, Bitner-Glindzicz M, Khaw PT, Nischal KK, Sowden JC. CYP1B1-related anterior segment developmental anomalies novel mutations for infantile glaucoma and von Hippel’s ulcer revisited. Ophthalmology. 2011;118:1865–73. doi: 10.1016/j.ophtha.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 30.Curry SM, Daou AG, Hermanns P, Molinari A, Lewis RA, Bejjani BA. Cytochrome P4501B1 mutations cause only part of primary congenital glaucoma in Ecuador. Ophthalmic Genet. 2004;25:3–9. doi: 10.1076/opge.25.1.3.28999. [DOI] [PubMed] [Google Scholar]