Mycoplasmas are small bacterial commensals or pathogens that commonly colonize host mucosal tissues and avoid rapid clearance, in part by stimulating inflammatory, immunopathogenic responses. We previously characterized a wide array of transcriptomic perturbations in avian host tracheal mucosae infected with virulent, immunopathologic Mycoplasma gallisepticum; however, mechanisms delineating these from protective responses, such as those induced upon vaccination, have not been thoroughly explored.
KEYWORDS: attenuated vaccine, cytokines, host response, inflammation, Mycoplasma gallisepticum, TLR, vaccine
ABSTRACT
Mycoplasmas are small bacterial commensals or pathogens that commonly colonize host mucosal tissues and avoid rapid clearance, in part by stimulating inflammatory, immunopathogenic responses. We previously characterized a wide array of transcriptomic perturbations in avian host tracheal mucosae infected with virulent, immunopathologic Mycoplasma gallisepticum; however, mechanisms delineating these from protective responses, such as those induced upon vaccination, have not been thoroughly explored. In this study, host transcriptomic responses to two experimental M. gallisepticum vaccines were assessed during the first 2 days of infection. Relative to virulent infection, host metabolic and immune gene responses to both vaccines were greatly decreased, including early innate immune responses critical to disease development and subsequent adaptive immunity. These data specify host genes and potential mechanisms contributing to maladaptive versus beneficial host responses—information critical for design of vaccines efficacious in both limiting inflammation and enabling pathogen clearance.
INTRODUCTION
Mycoplasma gallisepticum, a respiratory pathogen, contributes to the development of chronic respiratory disease (CRD) in poultry. CRD in chickens is frequently associated with clinical signs, including coughing, nasal discharge, and tracheal rales (1). Inflammatory lesions are most commonly found in the trachea, air sacs, and lungs and occasionally in the conjunctiva and oviduct (2). Despite efforts to control outbreaks, M. gallisepticum results in significant worldwide economic losses due to decreased feed conversion efficiency, downgrading of carcasses due to air sacculitis, and reduced egg production and hatchability (1, 3).
Virulent strains of M. gallisepticum, such as Rlow, avoid clearance by the host’s respiratory defense mechanisms by cytadhering to tracheal epithelium and colonizing the mucosal surface. Tight attachment is mediated by GapA and CrmA, cytadherence molecules required for colonization and pathogenesis (4). Disruption of genes encoding either of these cytadhesins results in limited colonization and a dramatically reduced host response and associated pathology (4–6).
The tracheal host response to Rlow is characterized by a marked influx of inflammatory cells into the lamina propria (7, 8) and is associated with robust signaling of Toll-like receptors (TLRs) in epithelial cells lining the tracheal lumen (9). This results in increased expression of proinflammatory chemokines and cytokines, including interleukin 6 (IL-6), IL-8, CCL20, CXCL-13 and -14, and RANTES, that are likely sustained due to increased signaling and activation of immune cells associated with Rlow colonization (8–10). Over time, the vigorous host response reduces the bacterial load; however, it is typically unable to clear the infection. The concomitant immunopathogenic sequelae predispose birds to coinfection and lead to the development of CRD (11–13).
Currently available vaccines, while highly effective, have limitations that render them unsuitable in certain situations (14–17). Efforts to develop more efficacious vaccines have resulted in laboratory-generated, live-attenuated vaccine (LAV) strains, including GT5 and Mg7. GT5 was created by complementing the avirulent strain Rhigh, a high-passage strain derived from Rlow, with wild-type gapA (5). GT5 induces lower levels of proinflammatory cytokines and chemokines than does Rlow and is capable of inducing the local recruitment of CD4+ and CD8+ T cells and B cells and ultimately the production of M. gallisepticum-specific IgG (IgY), IgA, and IgM antibodies (8, 18–20). Due to the number and magnitude of genomic changes identified in Rhigh (21), its limited ability to cytadhere (5), and the modest immune response induced by GT5 (18–20), this vaccine is considered to have a highly attenuated phenotype (22). Conversely, Mg7 is an isogenic mutant of Rlow, differing only by virtue of the transpositional inactivation of the dihydrolipoamide dehydrogenase (lpd) gene (23). As such, Mg7 is genetically identical to fully virulent Rlow but lacks a single metabolic gene, and hence, as reflected by data presented here, has a considerably less attenuated phenotype than GT5. Virulence assessments of Mg7 indicated that it has limited colonization capabilities and minimal lesions in the trachea and air sacs but still evokes levels of antibodies consistent with those of other vaccines (20, 23). Comparison of the efficacies of these two LAVs indicated that chickens could rapidly clear the challenge strain (Rlow) and had significantly reduced histopathologic lesion scores (20). Both GT5 and Mg7 outperformed the commercially available vaccines in laboratory-controlled experiments (20). Mg7 resulted in significantly lower levels of postchallenge recovery of Rlow suggestive of, and highly correlated with, prophylactic immunity (20). While GT5 and Mg7 have been shown to be protective, further understanding of the host response to these strains is needed to determine which elements of the host response are required to induce protective immunity versus those which are potentially maladaptive. As noted by others, there is attendant risk to using overly attenuated vaccines in that they may inadvertently favor the emergence of more pathogenic strains as weaker strains are eradicated from the population (22, 24).
Previous transcriptomic analysis conducted in our laboratory indicated a massive and dynamic host tracheal response associated with virulent Rlow infection over 7 days, highlighting the immunological, metabolic, and stress- and injury-related genes and pathways which may contribute to the exaggerated and dysregulated host response (10). To better understand differences between virulent and nonimmunopathologic host responses, we conducted a 2-day infection study followed by RNA sequencing (RNA-Seq) to assess the host transcriptional differences in genes and pathways leading to either a maladaptive or beneficial immune response to M. gallisepticum (25). Responses against intratracheally administered GT5 and Mg7 LAVs were compared to responses against Rlow at early time points postinfection. Previous studies utilizing similar methods (8, 18–20, 23) showed the importance of early host-pathogen interaction in establishing the trajectories for inflammation and pathogenic change versus the induction of prophylactic immunity. We anticipate these data to provide a framework to guide the rational development of protective vaccines—those which retain specific elements of protective immune activation absent perturbation of genes associated with immunopathogenic inflammation or cell recruitment/activation.
RESULTS
DEGs affected during virulent or LAV strain M. gallisepticum infection.
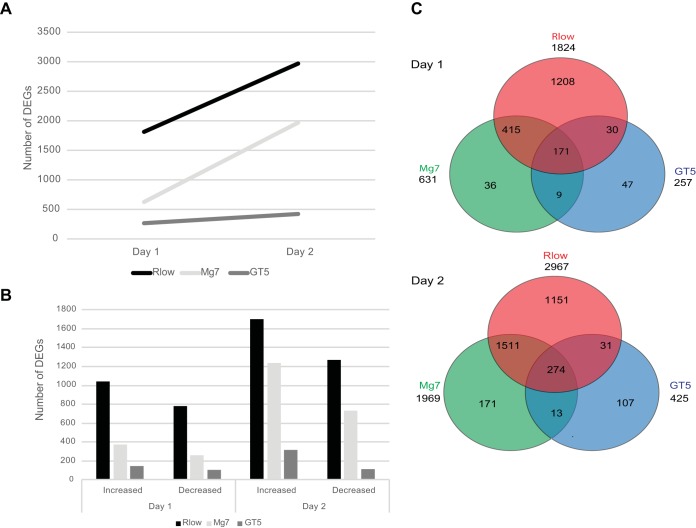
RNA collected from cells lining chicken tracheal lumens yielded transcripts that mapped to 17,935 genes within the Gallus gallus genome. On day 1 postinfection there were 1,824 differentially expressed genes (DEGs) observed in response to Rlow infection, 631 DEGs due to Mg7 exposure, and 257 due to GT5. Approximately 60% of the day 1 DEGs in each group showed increased expression compared to the control group. Day 2 postinfection resulted in greater numbers of DEGs for each infection group: 2,967 in Rlow with 57% increased, 1,969 in Mg7 with 63% increased, and 425 in GT5 with 74% increased (Fig. 1A and B). Of the total DEGs, only 171 were common to all three infection groups on day 1, and 274 were common on day 2 (Fig. 1C). As expected, on both days, the greatest numbers of unique DEGs were found in Rlow-infected groups.
FIG 1.
DEGs between control and Rlow or LAV strains. The number of DEGs was determined by comparing each infection group (Rlow, Mg7, and GT5) to the respective control group as identified by Cuffdiff on days 1 and 2 postinfection. (A) The overall number of DEGs identified in each group; (B) the number of DEGs displayed as having increased or decreased expression; (C) Venn diagram displaying the numbers of unique and common DEGs per infection group.
Host pathways affected during virulent or LAV strain M. gallisepticum infection.
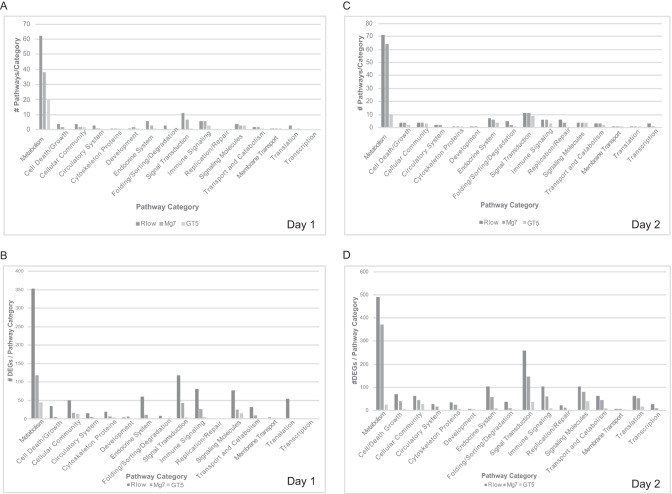
Cellular pathways affected during M. gallisepticum infection were predicted based on involvement of DEGs. Infection with virulent strain Rlow resulted in the greatest number of affected pathways on both days postinfection, with 110 pathways affected on day 1 and 134 on day 2. Exposure to Mg7 resulted in 69 pathways affected on day 1 and 115 on day 2, and GT5 exposure affected the fewest pathways, with 35 on day 1 and 41 on day 2 (Fig. 2A and C). The greatest number of pathways and DEGs across days and bacterial strains that were affected were classified in the metabolism category (Fig. 2). The pathways affected by Mg7 followed a trend similar to that of the Rlow-infected group; however, there were fewer pathways and DEGs observed within these pathway categories. The difference in the number of DEGs between Rlow and Mg7 infection was especially notable within metabolism-related pathways and those with immune-related functions, including signal transduction, immune signaling, and signaling molecules (Fig. 2B and D). GT5 exposure affected the lowest number of pathways in both metabolic and immune-related categories and showed the least variability in the number of DEGs between time points.
FIG 2.
Pathways activated due to Rlow or LAV strains. Pathways containing ≥2 DEGs were categorized based on function using DAVID bioinformatics software (24). (A and C) The number of identified pathways per category is indicated for each infected group on day 1 (A) and day 2 (C) postinfection. (B and D) The number of DEGs per pathway category is indicated for each infected group on day 1 (B) and day 2 (D) postinfection.
Functional GO of host DEGs affected during virulent or LAV strain M. gallisepticum infection.
Functional gene ontology (GO) assessment of the DEGs with a log2 fold change of ≥2.00 enabled comparison of enriched biological processes identified in each infection group (see Fig. S1 in the supplemental material). On day 1, Rlow, Mg7, and GT5 exposure resulted in 245, 79, and 24 DEGs, respectively. In keeping with our previous study (10), Rlow infection affected biological processes involving host immune and inflammatory processes, pathogen recognition, cytokine and chemokine production, and chemotaxis. Mg7 exposure affected fewer and thus functionally less broad host processes than did Rlow infection; however, the fewer processes affected by Mg7, such as chemotaxis and leukocyte migration, were similar to those affected by Rlow. GT5 exposure resulted in the fewest affected biological processes.
On day 2, Rlow, Mg7, and GT5 exposure resulted in 194, 123, and 125 DEGs, respectively. Many of the biological processes affected in the Rlow-infected group on day 1 were also affected on day 2, though 21% fewer were affected on day 2. One notable difference, however, was the presence of positive regulation of NF-κB import into the nucleus on day 2. Vaccine strains, unlike Rlow, affected far more DEGs on day 2 than day 1. On day 1, Mg7 exposure affected approximately two-thirds fewer processes than those affected by Rlow infection on day 1; however, by day 2 Mg7 affected only one-third fewer processes than Rlow. Notable differences between Mg7 and Rlow at day 2 included the absence of cellular response to interferon gamma affected by Mg7 and the presence of the following responses affected by Mg7: positive regulation of interleukin 6 production, positive regulation of interleukin 8 production, macrophage activation, and positive regulation of T-cell activation. The greatest within-group difference between days 1 and 2 was seen in the GT5-exposed group, with only 5 processes affected on day 1 and 64 processes affected on day 2. The leukocyte chemotaxis and neutrophil chemotaxis processes were not affected by GT5 exposure on day 2, nor were any processes involved in the recognition of a bacterial pathogen or the host immune response. The majority of unique processes that were identified in the GT5-exposed group included those involved in cell growth, repair, development, differentiation, and proliferation.
Host immune pathways affected during virulent or LAV strain M. gallisepticum infection.
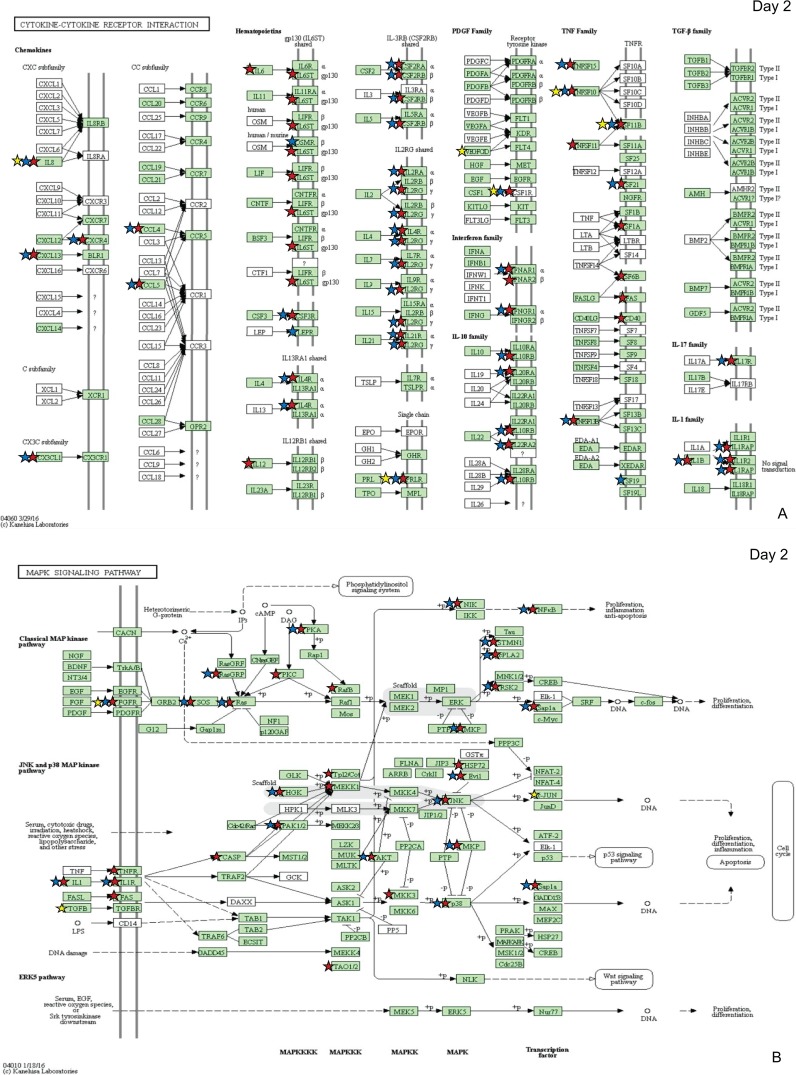
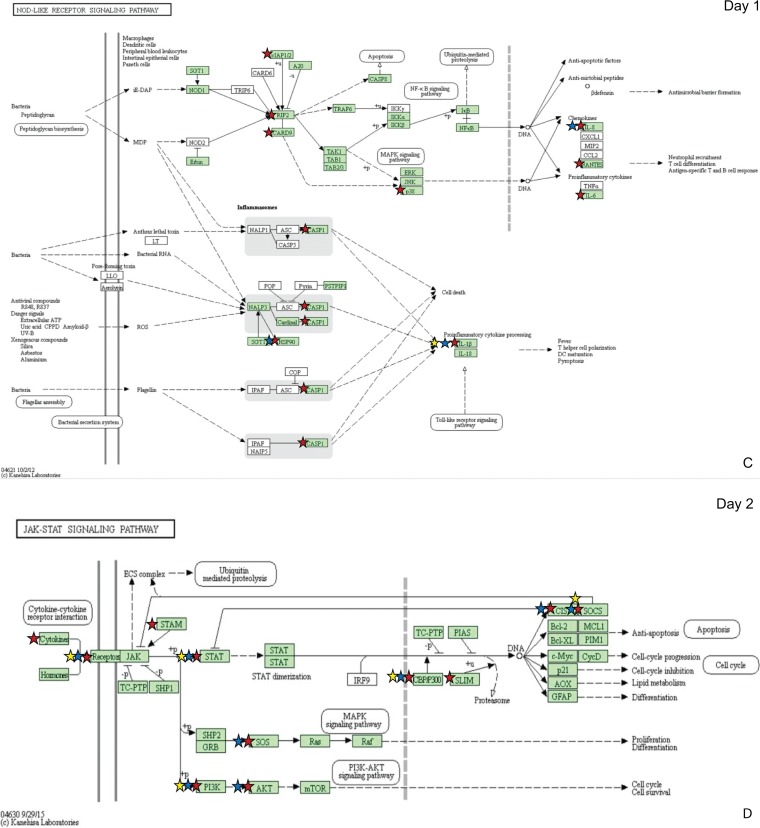
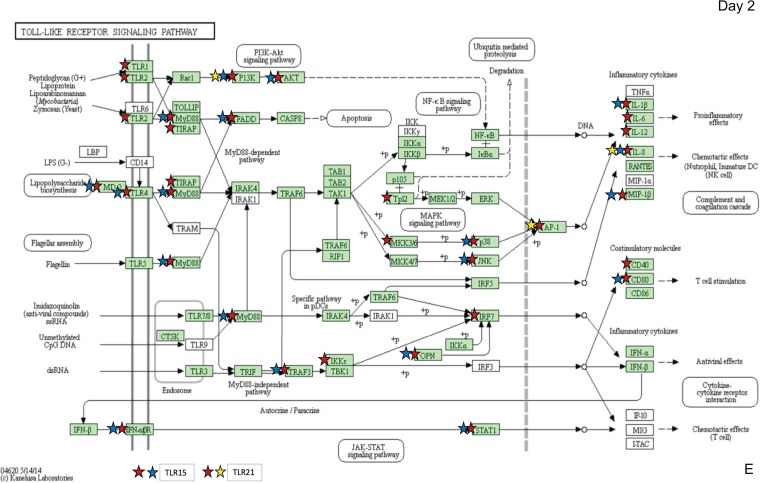
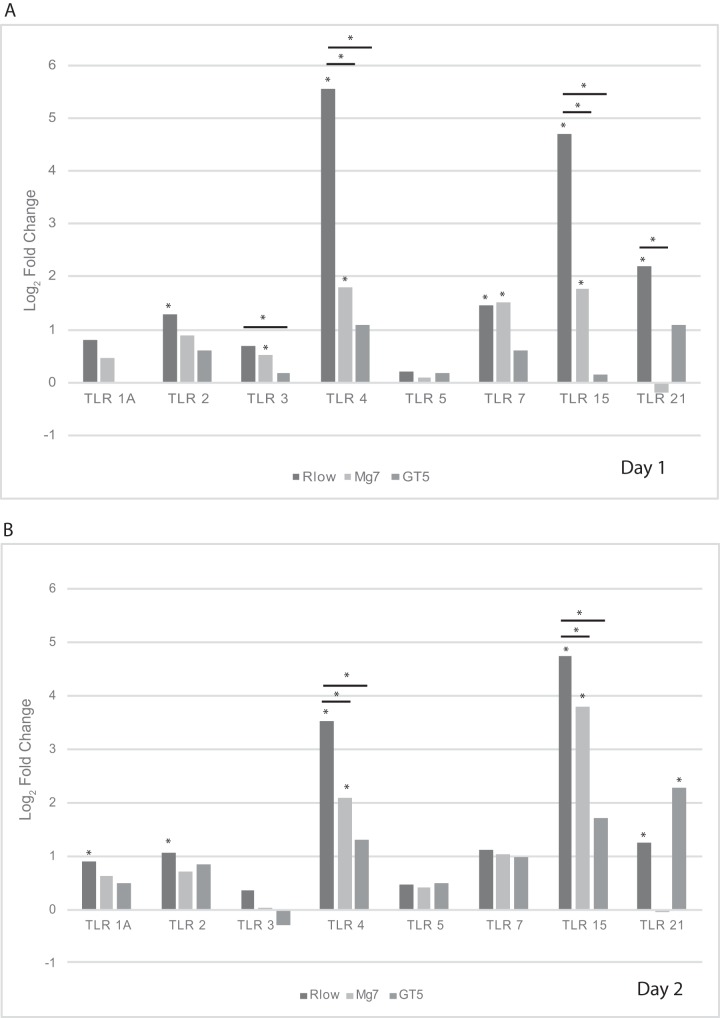
Pathways of interest were selected for specific analyses based on known differences in the host immune response to virulent and attenuated strains. These immune-related pathways included cytokine-cytokine receptor (CCR) interaction (Fig. 3A), the mitogen-activated protein kinase (MAPK) signaling pathway (Fig. 3B), the nucleotide oligomerization domain (NOD)-like receptor pathway (Fig. 3C), the Janus kinase signal transducer and activator of transcription (Jak-STAT) pathway (Fig. 3D), and the Toll-like receptor (TLR) pathway (Fig. 3E). As expected based on the total number of DEGs found in each group (Fig. 1), the Rlow-infected group contained the most DEGs in each of these pathways at both time points, and GT5 exposure resulted in only a limited number. On day 1, Rlow infection uniquely affected proinflammatory genes of the NOD-like receptor pathway, including IL-6, RANTES, and caspase 1 (CASP1), while GT5 exposure did not affect any genes in this pathway (Fig. 3C). On day 2, Rlow infection affected IL-6, IL-12, TNFSF11, and approximately 24% more receptors of the cytokine-cytokine receptor interaction pathway than did Mg7 exposure (Fig. 3A). DEGs in the MAPK pathway affected only by Rlow included protein kinase C (PKC), FAS, HSP72, and CASP, and those affected only by GT5 included transforming growth factor β (TGF-β) and c-JUN (Fig. 3B). Few differences were observed in the Jak-STAT pathway (Fig. 3D); one difference was that only Rlow infection affected the signal transduction adapter molecule (STAM). In the TLR pathway, the most prominent differences between Rlow infection and Mg7 exposure were seen in the Toll-like receptors and the downstream components, including both inflammatory cytokines and costimulatory molecules. These include IL-6, IL-12, and CD40, affected only in Rlow infection; IL-1β, RANTES, and CD80, affected in Rlow infection and Mg7 exposure; and IL-8, which was affected in all three groups (Fig. 3E). The greatest fold changes in the TLRs were seen in TLR-4 and -15 upon Rlow infection and were also significant in Mg7-exposed chickens (Fig. 4). On day 1, both Rlow infection and Mg7 exposure resulted in significant increased expression of several TLRs, while none of the TLRs were significant in response to GT5. However, by day 2, while the response to Rlow remained similar, only TLR-4 and -15 were significantly affected by Mg7 and only TLR-21 was significantly affected by GT5.
FIG 3.
Immune pathways highlighting differential expression of genes following exposure to Rlow and LAVs. Pathways include cytokine, cytokine receptor interaction (A), MAPK signaling (B), NOD-like receptor (C), Jak-STAT signaling (D), and Toll-like receptor (E). DEGs identified in pathways are indicated for each infection group by red stars (control versus Rlow), blue stars (control versus Mg7), and yellow stars (control versus GT5) as identified by DAVID bioinformatics software and the KEGG database (54–56). Green shading represents the completeness of the pathway identified within the Gallus gallus genome. TLR-15 and -21 were added to the original output, which is based on mammalian pathways. Copyright, Kanehisa Laboratories; reproduced with permission.
FIG 4.
Differential expression of Toll-like receptors. TLRs identified by RNA-Seq analysis are displayed. Differential expression of Rlow, Mg7, and GT5 relative to the uninfected control or between experimental groups is expressed as log2 fold change, with significant differential expression (Q ≤ 0.05) indicated with an asterisk above its respective bar.
Host genes with greatest differential expression during virulent and LAV strain M. gallisepticum infection.
The top 30 common DEGs were determined and ranked based on the log2 fold change values of the Rlow-infected group (Table 1). Many of the common DEGs encode proteins with immune-related functions, such as IL-13RA2, IL-1β, LYG2, IL-4I1, IL-20RA, IL-8L2, and BPI. However, a greater proportion of DEGs are related to functions including injury response, metabolism, cell adhesion and remodeling, extracellular matrix (ECM) degradation, and membrane transport. Assessing the top DEGs revealed that by day 2, the DEGs observed due to GT5 exposure involved cell growth/proliferation, wound repair, and maintenance of the ECM (Table S1). In a comparison of differences in the top 30 DEGs unique to Rlow infection and Mg7 exposure (i.e., not found in GT5) (Table S2), several DEGs potentially involved in the host’s proinflammatory response were indicated: TLR-4, TLR-15, IL-1β, IL-1R2, IL-8L1, IL-22RA2, CXCL1, CCLI10, and IRG1. On day 1, Rlow infection resulted in greater fold change values than did Mg7 exposure, whereas on day 2 differences in fold change between these two groups were less pronounced. Lastly, to deduce differences in the host response to Rlow, the DEGs unique to Rlow infection were assessed (Table S3). DEGs with immune-related functions included IL-6, IL-12B, IL-22, CCL4, CD80, CSF2RA, TNFSF11, and NCF2.
TABLE 1.
Top 30 DEGs common to chickens exposed or infected with Rlow or LAV strainsa
| Gene symbol | Log2 fold change | Q value |
|---|---|---|
| Day 1 | ||
| MMP7 | 10.7134 | 0.000757872 |
| NOXO1 | 6.65159 | 0.00811598 |
| IL13RA2 | 6.5396 | 0.000757872 |
| EX-FABP | 5.90677 | 0.000757872 |
| TLR-4 | 5.56618 | 0.000757872 |
| ENPP3 | 5.33232 | 0.000757872 |
| AVD | 5.33025 | 0.000757872 |
| IL-1β | 5.30879 | 0.000757872 |
| IL-22 | 5.27032 | 0.00441529 |
| CA4 | 5.26316 | 0.000757872 |
| CXCL1 | 5.14426 | 0.000757872 |
| ENSGALG00000004041 | 5.07205 | 0.000757872 |
| IRG1 | 4.98928 | 0.000757872 |
| MB | −4.74018 | 0.000757872 |
| IL8-L1 | 4.73203 | 0.000757872 |
| TLR-15 | 4.70932 | 0.000757872 |
| IL-22RA2 | 4.55899 | 0.000757872 |
| ENSGALG00000016556 | 4.54609 | 0.0231227 |
| SEMA3G | 4.47905 | 0.0102995 |
| CSF3R | 4.46757 | 0.000757872 |
| LYG2 | 4.4458 | 0.000757872 |
| MYL2 | −4.34357 | 0.00521464 |
| ENSGALG00000002343 | 4.25019 | 0.0437935 |
| CCL4 | 4.22887 | 0.00137722 |
| NOX1 | 4.20915 | 0.000757872 |
| SERPINE2 | 4.18123 | 0.000757872 |
| IL-4I1 | 4.14185 | 0.000757872 |
| IL-8-L2 | 4.09163 | 0.000757872 |
| GGT1 | 4.04279 | 0.000757872 |
| MYL3 | −4.08948 | 0.000757872 |
| Day 2 | ||
| MMP7 | 11.5514 | 0.000502174 |
| NOXO1 | 7.782 | 0.0180061 |
| AVD | 7.09226 | 0.000502174 |
| ENSGALG00000005099 | 6.57039 | 0.000502174 |
| ENSGALG00000004041 | 6.57034 | 0.000502174 |
| MMP7 | 11.5514 | 0.000502174 |
| OGCHI | 6.54007 | 0.000502174 |
| IL13RA2 | 6.36389 | 0.000502174 |
| ENSGALG00000024484 | 6.26863 | 0.000502174 |
| OLFM4 | 6.05065 | 0.000502174 |
| IL8-L1 | 5.94985 | 0.000502174 |
| CA4 | 5.80822 | 0.000502174 |
| EX-FABP | 5.55622 | 0.000502174 |
| ENSGALG00000006295 | −5.47359 | 0.000502174 |
| CSF3R | 5.45562 | 0.000502174 |
| CALCA | 5.45036 | 0.000502174 |
| ENPP3 | 5.43387 | 0.000502174 |
| MMP9 | 5.23091 | 0.000502174 |
| IL8-L2 | 4.99823 | 0.000502174 |
| ENSGALG00000023578 | −4.89811 | 0.000926417 |
| TLR15 | 4.75184 | 0.000502174 |
| PCDH1 | 4.74193 | 0.000502174 |
| IL1R2 | 4.63499 | 0.000502174 |
| ENSGALG00000021025 | 4.61934 | 0.000502174 |
| NOX1 | 4.58002 | 0.000502174 |
| IL-4I1 | 4.44538 | 0.000502174 |
| RARRES1 | 4.43871 | 0.000502174 |
| IRG1 | 4.41293 | 0.000502174 |
| SOCS3 | 4.39838 | 0.000502174 |
| SLC6A14 | 4.39808 | 0.000502174 |
| SEMA3G | 4.37535 | 0.000502174 |
DEGs were determined by Cuffdiff and ranked based on the log2 fold change values in the Rlow infection group.
DISCUSSION
The development of LAVs derived from pathogenic bacteria has traditionally involved methods which result in ill-defined and/or random attenuating mutations (e.g., serial passage, chemical/UV mutagenesis, or generation of transposon mutant libraries), followed by subsequent assessments consisting of immune response, histopathology, and protection against virulent challenge. Increasing knowledge regarding pathogenesis and host response raises hope that these time-honored yet inefficient and imprecise approaches to vaccine design may give way to rational vaccine design, in which host responses are specifically tailored to match correlates of immune protection and safety on a gene/pathway-specific basis.
As a first step, we must assess how current LAVs trigger beneficial host immune stimulation in contrast to maladaptive inflammatory or dysregulated responses evoked by virulent strains. In this regard, the current study examined the global transcriptional differences in the host response to virulent versus LAV strains of M. gallisepticum during early stages of infection. Previous studies have largely assessed the efficacy and adaptive host immune response to GT5 and Mg7 (19, 20), with only one study assessing the innate immune response to GT5 (8). Both LAVs provided protection against subsequent virulent Rlow challenge, and protection was highly correlated with interfollicular zones of CD4+ and CD8+ T cells (8, 20), IgG (IgY)- and IgA-secreting plasma cells in tracheal tissue, and anti-M. gallisepticum IgG (IgY) in serum and tracheal secretions (20). Mg7 induced greater tracheal thickness than did GT5, likely due to higher levels of immune stimulation and more robust cellular infiltration; however, Mg7 outperformed GT5 in protection, as indicated by lower rates of Rlow recovery 4 weeks after challenge (20). These differences in immune stimulation and efficacy are likely related to differences in vaccine attenuation. Thus, GT5 and Mg7 represent two distinct LAVs which were used to examine variations in nonpathogenic, protective host responses relative to those observed upon virulent Rlow infection.
To examine differential host response to M. gallisepticum vaccines, transcriptomic differential gene expression was examined in the chicken tracheal mucosae during early infection in vivo (1 and 2 days postinfection). A significant number of DEGs in metabolic pathways were observed upon Rlow infection and Mg7 exposure, ostensibly due to epithelial cell distress (26, 27), suggesting that the two strains induce nearly comparable levels of cell stress upon colonization of the mucosal surface (Fig. 2). Conversely, GT5 induced only very moderate perturbations in expression levels of these genes. Cell stress is known to trigger an immune response through activation of the inflammasome within the NOD-like receptor pathway (27, 28). Caspase 1 (CASP1), a component of the Nlrp3 inflammasome and regulator of the processing and secretion of IL-1β, -18, and -22, was significantly increased only in response to Rlow infection on day 1 (Fig. 3; Table S4). IL-1β was increased in response to Rlow infection and Mg7 exposure on days 1 and 2 (with the greatest increase observed in the Rlow group) but was only significantly increased on day 1 in GT5-exposed chickens (Table S1). The fact that GT5 exposure resulted in significantly fewer DEGs within metabolic pathways suggests that minimal cell stress had occurred. Therefore, the limited host response to the highly attenuated phenotype of GT5 may in part be due to the lower levels of cell stress and IL-1β expression.
Comparing functional gene ontologies between treatment groups further elucidated differences between virulent and LAV strains. While it is well established that the host response to Rlow is both vigorous and rapid (8, 9, 29, 30), its magnitude was further highlighted when comparing the day 1 response evoked by Rlow to the limited number of mitogenic and cytokine-related host response processes activated upon Mg7 and GT5 exposures. Mg7 exposure induced a number of proinflammatory host response processes not observed after exposure to GT5. On day 2, the processes affected by Mg7 exposure more closely resembled those seen in Rlow infection, indicating a more gradual and slightly blunted activation of the host response. The response to GT5 exposure on day 2 primarily involved repair and regeneration, with only 2 processes involved in proinflammatory host responses. These differences may in part be due to the diminished cytadhesion capability of GT5, thereby reducing its residence time in association with the host mucosal epithelium (5). Nonetheless, GT5 was still capable of inducing expression changes in host genes, including those involving leukocyte and neutrophil chemotactic processes.
Given that TLR signaling plays an important role in host response to Mycoplasma infections (9, 10, 30–32), it is important to consider the differences in expression of TLRs induced between virulent and LAV strains. Based on ex vivo analysis, TLR-2 signaling in tracheal epithelial cells likely contributes to the influx of inflammatory cells into the lamina propria during Rlow infection (10, 33–38). In the current study, TLR-2 expression was significantly increased only in the Rlow-infected chickens (Fig. 4). However, TLR-4 and -15 were significantly increased on both days in response to Rlow and Mg7, with the greatest expression observed in response to Rlow. GT5 exposure, however, resulted in a negligible increase in the expression of TLR-2, -4, and -15. As previously discussed (10), increased expression of TLR-4 may reflect an increased cellular response to extrinsic factors, including the presence of damage-associated molecular pattern molecules (DAMPs) such as heat shock proteins, heparin, fibrinogen, and fibronectin, as a result of damaged extracellular matrix (ECM) or cellular injury (33, 34, 39). Aside from cell stress-related injury, damage to cells and the ECM may be due to a disproportionate increase in the expression of matrix metalloprotease 7 (MMP7) and/or MMP9 (36). Our previous study also noted perturbations in MMP7 and/or -9 expression, with significant increases in response to Rlow infection at all time points (10). In the current study, the greatest increase in expression of MMP7 was observed in response to Rlow infection, with comparable levels observed in response to Mg7 exposure and lower expression values seen in response to GT5 (Table 1). TLR-15, which is unique to avian and reptilian lineages, is a broad-spectrum TLR that has been shown to recognize diacylated lipopeptide from Mycoplasma synoviae (32). It is possible that the cellular interaction with bacterial lipoproteins and the greater levels of cellular stress and damage due to Rlow infection collectively contribute to the increased expression of TLR-2, -4, and -15 genes, which conceivably play a role in immunopathogenesis through subsequent increases in proinflammatory cytokines and chemokines. Consistent with effects on other genes/pathways evaluated in this study, these effects on TLR and related pathways were slightly delayed and somewhat lower in magnitude in response to Mg7 and were considerably lower in response to GT5.
One of the most profound differences observed between M. gallisepticum strains was the number of DEGs in the cytokine and chemokine receptor (CCR) pathway (Fig. 3). As anticipated, Rlow exposure resulted in the greatest number of differentially expressed CCRs, with approximately 60% and 24% more than Mg7 on days 1 and 2, respectively. Most notable was the almost complete lack of CCR DEGs in response to GT5 exposure, with one DEG observed on day 1 and two on day 2. The limited expression of Toll-like, cytokine, and chemokine receptors suggests that GT5 exposure induces limited host cell sensitivity, which is consistent with a lack of immunopathology. In comparison, the widespread increase in expression of host cell receptors evoked by Rlow infection is likely a factor significantly contributing to its pathogenicity, perhaps in part by rendering epithelial cells more sensitive to ligands from M. gallisepticum and/or to other extrinsic factors (i.e., resident commensal bacteria).
Gene ontology analysis indicated that Mg7 induced genes associated with the macrophage activation processes 2 days postexposure (Fig. S1). Actual macrophage recruitment in response to Rlow has been established in previous in vivo and ex vivo studies; however, the nature of Rlow-induced macrophage activation has been obscure given conflicting expression data for IL-12p40 (IL-12β) and gamma interferon (IFN-γ), cytokines typically produced by activated macrophages and whose atypical expression may be yet another indicator of the dysregulated host response during Rlow infection (40, 41). In the current study, expression of IL-12β was significantly increased on both days in response to Rlow but was not significantly changed after Mg7 or GT5 exposure. Additionally, expression of IFN-γ was not significantly differential between groups. Whether the macrophage activation process indicated by gene ontology in this study would result in activated macrophages is unknown.
Certain genes encoding proteins with immune related functions were upregulated after Rlow infection but were unaffected after exposure to LAVs. These included IL-6 and IL-22, which were uniquely upregulated in Rlow-infected chickens. CCL4 (MIP-1β) upregulation was uniformly seen after Rlow infection but only at day 2 following exposure to one of the vaccines (Mg7). IL-6 has been shown to play a role in both host protection and disease exacerbation depending on the presence of other cytokines (40–42). IL-22, a proinflammatory cytokine, can contribute to maladaptive host immune responses and may also result in increased levels of IL-6 and IL-8 (43, 44). This may explain the unique expression of IL-6 and the greater expression levels of IL-8 in Rlow-infected chickens than with Mg7 and GT5 exposures. CCL4 is a chemoattractant for immune cells such as NK cells and monocytes (44) and may further contribute to immune cell infiltration and immunopathology.
Developing vaccines using LAVs for use at mucosal surfaces has traditionally faced a number of challenges, including the need to circumvent the host’s natural barriers. The vaccine strain must retain efficient binding and uptake by epithelial cells (such as M cells or their equivalent), processing and presentation by macrophage and dendritic cells, stimulation of antigen-specific T and B cells, and the production of durable anamnestic responses capable of providing long-term protection (45; reviewed in reference 46). While LAVs are often effective, they are precariously poised at the crossroads between overattenuation (hallmarked by moderate immune responses, poor durations of immunity and suboptimal vaccine “take” in genetically heterogeneous populations) and underattenuation, which is characterized by more robust immune responses often accompanied by moderate levels of inflammation/cell/tissue damage, the risk of reversion to a pathogenic form, and significant risks to immunocompromised individuals or more sensitive subpopulations (reviewed in reference 47). Here we have shown that LAV strains Mg7 and GT5 induce significantly different patterns of host gene expression at the transcriptional level. The moderately attenuated phenotype of the Mg7 isogenic mutant induced a response very similar to that observed during Rlow infection, whereas the highly attenuated phenotype of GT5, with limited cytadhesion capabilities, induced dramatically less perturbation in gene expression in a number of important pathways, including those affecting metabolism, chemokines/cytokines, inflammatory mediators, and TLRs.
While studies such as these can provide meaningful insights into the nature and degree of attenuation, they do little to suggest improved approaches to the bacterial attenuation process. In this regard, our group and others have observed that a relatively low number of bacterial gene products, such as the lipid-associated membrane proteins of Mycoplasma species, can trigger massive changes in gene regulation (9). An alternative approach to laborious and inefficient methods to attenuate bacteria could involve modulation of specific genes/pathways in the host using small interfering RNA (siRNA), clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, or other methods (20, 48, 49). This would provide a mechanism to “pick and choose” which genes and pathways to leave unchanged and which should be silenced. In the current study, the unique expression of IL-22, IL-12β, and IL-6 in Rlow-infected chickens suggests that they play a role in disease exacerbation. IL-8, IL-1β, and CCL4 are comparatively less expressed in response to Mg7 and/or GT5, suggesting that these cytokines may contribute to beneficial immune responses. Lastly, given the significant expression of TLR-4 and -15 as a result of Rlow infection, these genes may contribute to the subsequent increased expression of the aforementioned cytokines and chemokines and potentially to maladaptive responses. Therefore, enabling future vaccines to limit the expression of inflammatory modulators could prevent immune dysregulation and facilitate effective clearance of M. gallisepticum.
This study provides compelling evidence regarding the complexity of host responses to bacterial strains which were previously categorized as attenuated. Given the challenges presented in developing LAVs and achieving the desired host response, future attempts should consider alternative approaches, such as host-side immune response modulation in the production of safe and effective attenuated vaccines against M. gallisepticum and other bacterial pathogens.
MATERIALS AND METHODS
Animals.
Four-week-old female specific-pathogen-free White Leghorn chickens (SPAFAS, North Franklin, CT) were divided randomly into one of the three infection groups with 10 chickens per group or the control group with 9 chickens (due to one unrelated death). Chickens were placed in HEPA-filtered avian isolators and allowed to acclimate for 1 week. Nonmedicated feed and water were provided ad libitum throughout the experiment. All components of the study were performed in accordance with approved University of Connecticut IACUC protocol number A13-001.
Chicken infection.
Stocks of M. gallisepticum strain Rlow (passage 17) were grown in Hayflick’s complete medium overnight at 37°C until mid-log phase was reached, as indicated by a color shift from red to orange. Vaccine strains Mg7 (passage 6) and GT5 (passage 5) were grown in Hayflick’s complete medium with gentamicin selection (150 μg/ml) as previously described (20). Bacterial concentrations were estimated by the optical density at 620 nm (OD620) and verified by color-changing unit (CCU) serial dilutions. Bacteria were pelleted by centrifugation at 10,000 × g for 10 min and resuspended at 5 × 108 CFU/ml in Hayflick’s complete medium. Chickens were infected by pipetting 1 × 108 CFU/200 μl of Rlow (infected group) or Mg7 or GT5 (exposed groups) or 200 μl of Hayflick’s medium alone (control group) into the tracheal lumen as previously described (50).
Sample collection and RNA extraction.
Nineteen chickens (5 per infected and exposed groups and 4 control chickens) and twenty chickens (5 per infected and exposed groups and 5 control chickens) were humanely sacrificed at 24 and 48 h (day 1 and day 2) postinfection, respectively. Tracheas were excised and total RNA was immediately stabilized by pipetting 1 ml of TRIzol reagent four times through the lumen of each individual trachea (10, 51, 52) to minimize the changes in gene expression that may have occurred during the time between specimen collection and RNA purification (Invitrogen, Carlsbad, CA) and stored at −80°C. Total RNA was then purified using the Zymo-Direct-zol RNA miniprep kit according to the manufacturer’s instructions (Zymo Research Corporation, Irvine, CA). To confirm the presence of M. gallisepticum, an annular tissue section from the distal end of each trachea was placed in 1 ml of Hayflick’s medium for bacterial isolation. Samples were filtered through 0.45-μm Millipore filters to eliminate possible bacterial contaminants and kept at 37°C until a color shift from red to orange was observed.
Illumina sequencing.
cDNA libraries were created from RNA isolated from each individual chicken on days 1 and 2 postinfection using the Illumina TruSeq stranded mRNA library preparation kit (Illumina Inc., San Diego, CA) according to the Illumina TruSeq RNA Sample Preparation v2 (HT) protocol. Briefly, poly(A) selection was performed using RNA purification beads on 0.1 to 4 μg of total RNA per sample to obtain mRNA. Ten to 400 ng of purified mRNA was fragmented and used to synthesize first-strand cDNA using reverse transcriptase and random hexamer primers. Second-strand cDNA synthesis was performed using deoxynucleoside triphosphates (dNTPs), including dUTP, DNA polymerase, and RNase. End repair was performed by adding dATP to all free 3′ ends, and adapter ligation added unique index sequences to the fragments. The products were then amplified by PCR and purified.
The cDNA libraries were quantified using a Qubit 2.0 fluorometer (Invitrogen) and assessed for correct fragment size (∼260 bp) using the Agilent TapeStation 2200 (Agilent Technologies). Libraries were normalized to 2 nM, pooled, denatured, and sequenced on the NextSeq500 sequencing platform (Illumina Inc.) using a 75-bp paired-end approach targeting approximately 10 million reads per sample.
RNA-Seq analysis.
Fastq data from individual birds were mapped using the TopHat package (version 2.0.1) utilizing the Bowtie2 engine (version 2.2.5) to align reads to the Gallus gallus reference genome (WASHUC2), producing BAM alignment files (10). BAM files for each day were processed as experimental condition groups with Cufflinks (version 2.2.1) as previously discussed (52, 53). Within-condition normalization of data using Cuffdiff was achieved by calculating the fragments per kilobase of exon per million fragments mapped (FPKM) for each gene. The generated P value was then used to determine the significance of the differential expression based on the Benjamini-Hochberg correction with a false-discovery rate (FDR) of <5% to generate the Q value. Between-condition differences in expression values were considered significant when Q was ≤0.05, thus addressing within-condition variation between individual chickens (biological replicates) on each day. Fold change was determined from the log2 transformation of the ratio of FPKM between conditions (52). For this study, all references to differentially expressed genes (DEGs) imply statistical significance.
Pathway analysis.
Identification of biological pathways that ≥2 significant DEGs were involved in was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (54) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (55, 56).
Functional gene ontology.
DEGs with a log2 fold change of ≥2.00, as identified by Cuffdiff, were analyzed to identify enriched biological processes based on gene ontology (GO). This was performed using ClueGO v2.3.3, a plugin of the software platform Cytoscape v3.5.1 (28). “GO term fusion” was applied to reduce redundancy by comparing “parent-child” relations sharing similar genes and displaying the more representative parent or child terms. Processes that had a P value of ≤0.05 for each time point were considered significant and displayed.
Accession number(s).
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7675.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Szczepanek for helpful discussion during this study, Kirklyn Kerr for assistance at necropsy, and Gerald Kutish for assisting with the bioinformatic analysis pipeline.
We also thank the USDA National Institute of Food and Agriculture, Animal Health project numbers CONS00930 and CONS00874, and the Center of Excellence for Vaccine Research for support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00613-18.
REFERENCES
- 1.Ley DH. 2003. Mycoplasma gallisepticum infection, p 722–744. In Calnek B, Barnes H, Beard C, McDougald L (ed), Diseases of poultry, 11th ed. Iowa State University Press, Ames, IA. [Google Scholar]
- 2.Nunoya T, Kanai K, Yagihashi T, Hoshi S, Shibuya K, Tajima M. 1997. Natural case of salpingitis apparently caused by Mycoplasma gallisepticum in chickens. Avian Pathol 26:391–398. doi: 10.1080/03079459708419221. [DOI] [PubMed] [Google Scholar]
- 3.Evans JD, Leigh SA, Branton SL, Collier SD, Pharr GT, Bearson SMD. 2005. Mycoplasma gallisepticum: current and developing means to control the avian pathogen. J Appl Poult Res 14:757–763. doi: 10.1093/japr/14.4.757. [DOI] [Google Scholar]
- 4.Papazisi L, Frasca S, Gladd M, Liao X, Yogev D, Geary SJ. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect Immun 70:6839–6845. doi: 10.1128/IAI.70.12.6839-6845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papazisi L, Troy KE, Gorton TS, Liao X, Geary SJ. 2000. Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect Immun 68:6643–6649. doi: 10.1128/IAI.68.12.6643-6649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mudahi-Orenstein S, Levisohn S, Geary SJ, Yogev D. 2003. Cytadherence-deficient mutants of Mycoplasma gallisepticum generated by transposon mutagenesis. Infect Immun 71:3812–3820. doi: 10.1128/IAI.71.7.3812-3820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaunson JE, Philip CJ, Whithear KG, Browning GF. 2000. Lymphocytic infiltration in the chicken trachea in response to Mycoplasma gallisepticum infection. Microbiology 146:1223–1229. doi: 10.1099/00221287-146-5-1223. [DOI] [PubMed] [Google Scholar]
- 8.Javed MA, Frasca S, Rood D, Cecchini K, Gladd M, Geary SJ, Silbart LK. 2005. Correlates of immune protection in chickens vaccinated with Mycoplasma gallisepticum strain GT5 following challenge with pathogenic M. gallisepticum strain Rlow. Infect Immun 73:5410–5419. doi: 10.1128/IAI.73.9.5410-5419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumder S, Zappulla F, Silbart LK. 2014. Mycoplasma gallisepticum lipid associated membrane proteins up-regulate inflammatory genes in chicken tracheal epithelial cells via TLR-2 ligation through an NF-κB dependent pathway. PLoS One 9:e112796. doi: 10.1371/journal.pone.0112796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaudet J, Tulman ER, Pflaum K, Liao X, Kutish GF, Szczepanek SM, Silbart LK, Geary SJ. 2017. Transcriptional profiling of the chicken tracheal response to virulent Mycoplasma gallisepticum strain Rlow. Infect Immun 85:e00343-17. doi: 10.1128/IAI.00343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stipkovits L, Glavits R, Palfi V, Beres A, Egyed L, Denes B, Somogyi M, Szathmary S. 2012. Pathologic lesions caused by coinfection of Mycoplasma gallisepticum and H3N8 low pathogenic avian influenza virus in chickens. Vet Pathol 49:273–283. doi: 10.1177/0300985811415702. [DOI] [PubMed] [Google Scholar]
- 12.Chu H, Uppal P. 1975. Single and mixed infections of avian infectious bronchitis virus and Mycoplasma gallisepticum. Dev Biol Stand 28:101–114. [PubMed] [Google Scholar]
- 13.Sid H, Hartmann S, Petersen H, Ryll M, Rautenschlein S. 2016. Mycoplasma gallisepticum modifies the pathogenesis of influenza A virus in the avian tracheal epithelium. Int J Med Microbiol 306:174–186. doi: 10.1016/j.ijmm.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez R, Kleven SH. 1980. Pathogenicity of two strains of Mycoplasma gallisepticum in broilers. Avian Dis 24:800–807. doi: 10.2307/1589957. [DOI] [PubMed] [Google Scholar]
- 15.Abd-El-Motelib T, Kleven S. 1993. A comparative study of Mycoplasma gallisepticum vaccines in young chickens. Avian Dis 37:981–987. doi: 10.2307/1591903. [DOI] [PubMed] [Google Scholar]
- 16.Stallknecht DE, Luttrell MP, Fischer JR, Kleven SH. 1998. Potential for transmission of the finch strain of Mycoplasma gallisepticum between house finches and chickens. Avian Dis 42:352–358. doi: 10.2307/1592485. [DOI] [PubMed] [Google Scholar]
- 17.Kleven SH. 2008. Control of avian mycoplasma infections in commercial poultry. Avian Dis 52:367–374. doi: 10.1637/8323-041808-Review.1. [DOI] [PubMed] [Google Scholar]
- 18.Papazisi L, Silbart LK, Frasca S, Rood D, Liao X, Gladd M, Javed MA, Geary SJ. 2002. A modified live Mycoplasma gallisepticum vaccine to protect chickens from respiratory disease. Vaccine 20:3709–3719. doi: 10.1016/S0264-410X(02)00372-9. [DOI] [PubMed] [Google Scholar]
- 19.Mohammed J, Frasca S, Cecchini K, Rood D, Nyaoke AC, Geary SJ, Silbart LK. 2007. Chemokine and cytokine gene expression profiles in chickens inoculated with Mycoplasma gallisepticum strains Rlow or GT5. Vaccine 25:8611–8621. doi: 10.1016/j.vaccine.2007.09.057. [DOI] [PubMed] [Google Scholar]
- 20.Gates AE, Frasca S, Nyaoke A, Gorton TS, Silbart LK, Geary SJ. 2008. Comparative assessment of a metabolically attenuated Mycoplasma gallisepticum mutant as a live vaccine for the prevention of avian respiratory mycoplasmosis. Vaccine 26:2010–2019. doi: 10.1016/j.vaccine.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Szczepanek SM, Tulman ER, Gorton TS, Liao X, Lu Z, Zinski J, Aziz F, Frasca S, Kutish GF, Geary SJ. 2010. Comparative genomic analyses of attenuated strains of Mycoplasma gallisepticum. Infect Immun 78:1760–1771. doi: 10.1128/IAI.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinkler M, Leon AE, Kirkpatrick L, Dalloul RA, Hawley DM. 2018. Differing house finch cytokine expression responses to original and evolved isolates of Mycoplasma gallisepticum. Front Immunol 9:13. doi: 10.3389/fimmu.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson P, Gorton TS, Papazisi L, Cecchini K, Frasca S, Geary SJ. 2006. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect Immun 74:931–939. doi: 10.1128/IAI.74.2.931-939.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming-Davies AE, Williams PD, Dhondt AA, Dobson AP, Hochachka WM, Leon AE, Ley DH, Osnas EE, Hawley DM. 2018. Incomplete host immunity favors the evolution of virulence in an emergent pathogen. Science 359:1030–1033. doi: 10.1126/science.aao2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaudet J. 2017. Global transcriptional analysis of the chicken tracheal response to virulent and attenuated vaccine strains of Mycoplasma gallisepticum. PhD dissertation. University of Connecticut, Storrs, CT. [Google Scholar]
- 26.Ogura Y, Sutterwala FS, Flavell RA. 2006. The inflammasome: first line of the immune response to cell stress. Cell 126:659–662. doi: 10.1016/j.cell.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Lamkanfi M, Kanneganti TD. 2010. Nlrp3: an immune sensor of cellular stress and infection. Int J Biochem Cell Biol 42:792–795. doi: 10.1016/j.biocel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love W, Dobbs N, Tabor L, Simecka JW. 2010. Toll-like receptor 2 (TLR2) plays a major role in innate resistance in the lung against murine mycoplasma. PLoS One 5:e10739. doi: 10.1371/journal.pone.0010739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu T, Kida Y, Kuwano K. 2008. A triacylated lipoprotein from Mycoplasma genitalium activates NF-kB through Toll-like receptor 1 (TLR1) and TLR2. Infect Immun 76:3672–3678. doi: 10.1128/IAI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu T, Kida Y, Kuwano K. 2008. Mycoplasma pneumoniae-derived lipopeptides induce acute inflammatory responses in the lungs of mice. Infect Immun 76:270–277. doi: 10.1128/IAI.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oven I, Resman Rus K, Dušanić D, Benčina D, Keeler CL, Narat M. 2013. Diacylated lipopeptide from Mycoplasma synoviae mediates TLR15 induced innate immune responses. Vet Res 44:99. doi: 10.1186/1297-9716-44-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gharib SA, Altemeier WA, Van Winkle LS, Plopper CG, Schlesinger SY, Buell CA, Brauer R, Lee V, Parks WC, Chen P. 2013. Matrix metalloproteinase-7 coordinates airway epithelial injury response and differentiation of ciliated cells. Am J Respir Cell Mol Biol 48:390–396. doi: 10.1165/rcmb.2012-0083OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raulo SM, Sorsa T, Tervahartiala T, Pirilä E, Maisi P. 2001. MMP-9 as a marker of inflammation in tracheal epithelial lining fluid (TELF) and in bronchoalveolar fluid (BALF) of COPD horses. Equine Vet J 33:128–136. [DOI] [PubMed] [Google Scholar]
- 35.Elkington PTG. 2006. Matrix metalloproteinases in destructive pulmonary pathology. Thorax 61:259–266. doi: 10.1136/thx.2005.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visse R, Nagase H. 2003. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res 92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 37.Erridge C. 2010. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J Leukoc Biol 87:989–999. doi: 10.1189/jlb.1209775. [DOI] [PubMed] [Google Scholar]
- 38.Smiley ST, King JA, Hancock WW. 2001. Fibrinogen stimulates macrophage chemokine secretion through Toll-like receptor 4. J Immunol 167:2887–2894. doi: 10.4049/jimmunol.167.5.2887. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu T, Kimura Y, Kida Y, Kuwano K, Tachibana M, Hashino M, Watarai M. 2014. Cytadherence of Mycoplasma pneumoniae induces inflammatory responses through autophagy and Toll-like receptor 4. Infect Immun 82:3076–3086. doi: 10.1128/IAI.01961-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dann SM, Spehlmann ME, Hammond DC, Iimura M, Hase K, Choi LJ, Hanson E, Eckmann L. 2008. IL-6 dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J Immunol 180:6816–6826. doi: 10.4049/jimmunol.180.10.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asif M, Lowenthal JW, Ford ME, Schat KA, Kimpton WG, Bean AGD. 2007. Interleukin-6 expression after infectious bronchitis virus infection in chickens. Viral Immunol 20:479–486. doi: 10.1089/vim.2006.0109. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Q, Gu CQ, Hu XY, Wang DH, Li XM, Zhou SQ, Cheng GF. 2007. Role of interleukin-6 in the pathogenesis of an avian model of Staphylococcus aureus arthritis. Poult Sci 86:1245–1250. doi: 10.1093/ps/86.6.1245. [DOI] [PubMed] [Google Scholar]
- 43.Rutz S, Eidenschenk C, Ouyang W. 2013. IL-22, not simply a Th17 cytokine. Immunol Rev 252:116–132. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- 44.Menten P, Wuyts A, Van Damme J. 2002. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev 13:455–481. doi: 10.1016/S1359-6101(02)00045-X. [DOI] [PubMed] [Google Scholar]
- 45.Pavot V, Rochereau N, Genin C, Verrier B, Paul S. 2012. New insights in mucosal vaccine development. Vaccine 30:142–154. doi: 10.1016/j.vaccine.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Neutra MR, Kozlowski PA. 2006. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol 6:148–158. doi: 10.1038/nri1777. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava A, Gowda DV, Madhunapantula SV, Shinde CG, Iyer M. 2015. Mucosal vaccines: a paradigm shift in the development of mucosal adjuvants and delivery vehicles. APMIS 123:275–288. doi: 10.1111/apm.12351. [DOI] [PubMed] [Google Scholar]
- 48.Québatte M, Dehio C. 2017. Systems-level interference strategies to decipher host factors involved in bacterial pathogen interaction: from RNAi to CRISPRi. Curr Opin Microbiol 39:34–41. doi: 10.1016/j.mib.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Smith JL, Jeng S, McWeeney SK, Hirsch AJ. 2017. A microRNA screen identifies the Wnt signaling pathway as a regulator of the interferon response during flavivirus infection. J Virol 91:e02388-16. doi: 10.1128/JVI.02388-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin MY, Kleven SH. 1982. Cross-immunity and antigenic relationships among five strains of Mycoplasma gallisepticum in young Leghorn chickens. Avian Dis 26:496–507. doi: 10.2307/1589895. [DOI] [PubMed] [Google Scholar]
- 51.Pflaum K, Tulman ER, Beaudet J, Liao X, Geary SJ. 2016. Global changes in Mycoplasma gallisepticum phase-variable lipoprotein gene vlhA expression during in vivo infection of the natural chicken host. Infect Immun 84:351–355. doi: 10.1128/IAI.01092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das A, Chai JC, Kim SH, Park KS, Lee YS, Jung KH, Chai YG. 2015. Dual RNA sequencing reveals the expression of unique transcriptomic signatures in lipopolysaccharide-induced BV-2 microglial cells. PLoS One 10:e0121117. doi: 10.1371/journal.pone.0121117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang DW, Sherman BT, Lempicki R. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 55.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.