Abstract
This contribution provides a window into how strain differences impact interactions between host and symbiont during the first minutes to hours following initial partner engagement. Whereas biologists recognize that complex microbiomes, such as in the human gut, include the multiple strains of a resident species, the complexity of the microbial landscape and the inaccessibility of the colonization sites render these systems difficult to investigate. The intent of our study, using the squid-vibrio binary model of symbiont colonization of host epithelia, is to highlight possible evolutionarily conserved symbiont behaviors and their underlying genetic mechanisms. To this end, and using the model, confocal imaging and bacterial genetics, we characterize strain differences in the habitat transition between the free-living and symbiotic niche of the microbial partner. Taken together, the system offers a unique and unparalleled opportunity to document these critical events of symbiosis onset.
Summary
Among horizontally acquired symbioses, the mechanisms underlying microbial strain- and species-level specificity remain poorly understood. Here, confocal-microscopy analyses and genetic manipulation of the squid-vibrio association revealed quantitative differences in a symbiont’s capacity to interact with the host during initial engagement. Specifically, dominant strains of Vibrio fischeri, ‘D-type’, previously named for their dominant, single-strain colonization of the squid’s bioluminescent organ, were compared with ‘S-type’, or ‘sharing’, strains, which can co-colonize the organ. These D-type strains typically: (i) formed aggregations of 100s-1000s of cells on the light-organ surface, up to 3 orders of magnitude larger than those of S-type strains; (ii) showed dominance in co-aggregation experiments, independent of inoculum size or strain proportion; (iii) perturbed larger areas of the organ’s ciliated surface; and, (iv) appeared at the pore of the organ ~4Xs more quickly than S-type strains. At least in part, genes responsible for biofilm synthesis control the hyperaggregation phenotype of a D-type strain. Other marine vibrios produced relatively small aggregations, while an array of marine Gram-positive and -negative species outside of the Vibrionaceae did not attach to the organ’s surface. These studies provide insight into the impact of strain variation on early events leading to establishment of an environmentally acquired symbiosis.
Introduction
Many organisms acquire their symbiotic partners from the surrounding environment each generation, i.e., by horizontal transmission (McFall-Ngai, 1998; Bright and Bulgheresi, 2010; Salem et al., 2015, Cao and Goodrich-Blair, 2017). Unlike vertical transmission, in which provision of the symbiont cells in or on the host’s eggs assures specificity, symbiont selection in environmentally acquired partnerships usually occurs against an abundance of non-specific microbes in the host habitat. This mode of acquisition is best understood in terrestrial animals, such as insects (Aanen et al., 2002 Mueller et al., 2015; Powell et al., 2016; Takeshita and Kikuchi, 2017), where transfer of symbionts relies on direct interactions of the juveniles with infected conspecifics. In aquatic habitats, which can support a protracted free-living stage of the microbial partner, the mechanisms by which symbioses are established with fidelity each generation are largely undescribed. Typically, the potential symbiont cells represent only a fraction of the ambient environmental microbes, and efforts to observe the stochastic, initial interactions of these cells with host tissues over time and space face a formidable challenge.
Horizontal transmission also impacts the genetic features of the symbiont cells. For example, in model associations, such as the legume-rhizobia symbioses, specificity can be influenced by antagonistic interactions occurring between different combinations of host and symbiont genotypes, or between strains of the same symbiont species within a given host (Pellmyr and Huth, 1994; Frank, 1996; Kiers et al., 2003; Sachs et al., 2004, Poulsen and Boomsma, 2005; Bongrand et al., 2016). A recent study of the model symbiosis between Steinernema nematodes and Xenorhabdus bovienii (Murfin et al., 2015) showed that strain-level differences in the fitness benefits of microbial symbionts drive variation in the success of host-microbe associations, likely contributing to the maintenance of symbiotic associations over evolutionary time. In contrast, within the human gut microbiota, most bacterial species are represented by multiple co-colonizing strains (Caro-Quintero and Ochman, 2015, Greenblum et al., 2015; Noecker et al., 2017), although where they occur relative to one another, how such diversity is achieved and maintained, and whether strain diversity impacts metabolic function or host health, are questions that remain largely unanswered.
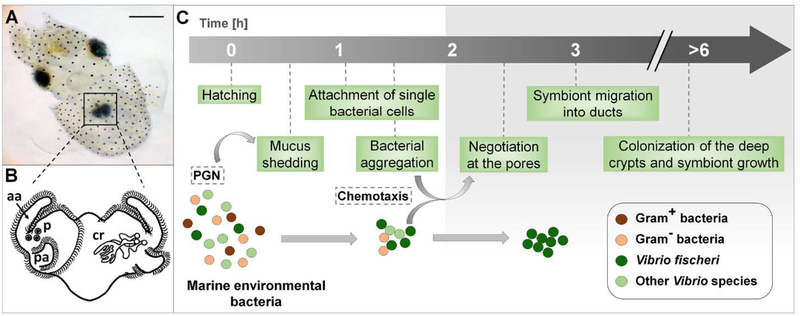
The association between the Hawaiian bobtail squid, Euprymna scolopes, and the luminous bacterium Vibrio fischeri has been studied for more than 25 years as an experimental model for the establishment, development and maintenance of horizontally transmitted symbioses (McFall-Ngai and Ruby, 1991, Nyholm and McFall-Ngai, 2004, McFall-Ngai, 2014). Females typically lay clutches of 100–400 eggs, a feature that enhances the statistical power of colonization experiments. During embryogenesis in the egg, the host develops a nascent organ poised to interact with symbionts upon hatching. The symbiosis, which is strictly specific for V. fischeri (McFall-Ngai and Ruby, 1991), is established within hours after the juvenile squid emerges from the egg. Species specificity is determined by both the biomechanical and biochemical landscape presented by the surface of the juvenile light organ. Briefly, two complex sets of ciliated epithelia, one on each lateral face of the hatchling organ, produce flow fields that entrain 2-μm particles (e.g., bacterial cells) into a region near pores (Nawroth et al., 2017), through which the symbionts enter the host tissues. Each of the superficial epithelial regions consists of cells with long, metachronally-beating cilia that populate the pairs of anterior and posterior ‘appendages’ on either side of the organ (Fig. 1), as well as a set of short cilia that surround and expand outward from the pores (Nawroth et al., 2017). The entrained particles associate with mucus, which contains antimicrobial compounds (e.g., NO, lysozyme, peptidoglycan-recognition proteins, and lipopolysaccharide-binding proteins; (McFall-Ngai, 2014)) that have been shed from the organ’s ciliated surface. The induction of mucus secretion is a nonspecific response to exposure to peptidoglycan released by environmental Gram-negative and Gram-positive bacteria (Nyholm et al., 2000; Nyholm et al., 2002).
Fig. 1.
The onset of the squid-vibrio symbiosis. (A) A newly hatched Hawaiian bobtail squid, Euprymna scolopes, with its bioluminescent light organ (black square) in the center of the body cavity; scale bar = 500 μm. (B) An illustration of the external (left side) and internal (right side) features of the nascent organ, as viewed ventrally: aa, anterior appendage; cr, crypts; p, pores; pa, posterior appendage. (C) Events in the recruitment of the specific symbiont, Vibrio fischeri, into the organ’s crypts, depicting the stages at which selectivity occurs. The gray shading (right) shows the internal portions of the organ, regions specific to V. fischeri. PGN, peptidoglycan released from bacteria in the ambient seawater.
Early studies of the process of symbiont initiation showed that several species of Gram-negative, but not Gram-positive, bacteria would aggregate within this mucus (Nyholm et al., 2000). The symbionts attach to host cilia (Altura et al., 2013), inducing a change in host gene expression that results in an increase in antimicrobials in the environment that are at higher concentrations in the host tissues into which the symbionts migrate (Kremer et al., 2013). This modification of the host microenvironment occurs during the first 1–3 h, when V. fischeri cells slowly aggregate and become the sole phylotype in the aggregations, suggesting that a biochemical environment is created that both favors association with the specific symbiont, and primes it for transit into antimicrobial-rich host tissues (Kremer et al., 2013). Once aggregated on the surface, the symbiont cells pass through the pores and proceed along a ~100-μm journey into the crypts (Fig. 1). On average, a single V. fischeri cell enters each of the 6 crypts (Dunn et al., 2006; Wollenberg and Ruby, 2009), where they grow to a total symbiont population of 105-106 cells (Ruby and Asato, 1993). Even in the absence of V. fischeri, no other bacterial phylotypes in the host’s environment are capable of entering host tissues beyond the pore region (Nyholm et al., 2000).
The majority of studies of this initiation process has been carried out using the V. fischeri symbiont strain ES114 (Boettcher and Ruby, 1990), in which genetic and genomic tools were first developed. Using this strain, the nascent symbiont has been shown to play a direct role in the aggregation process. For example, the regulator RscS is a critical factor for biofilm formation and subsequent colonization of host tissues by strain ES114 (Visick, 2009). RscS is a sensor kinase that responds to environmental conditions and enhances biofilm formation by inducing the transcription of a locus of symbiosis polysaccharide (syp) genes (Yip et al. 2005, Yip et al., 2006). A severe colonization defect results from the lack of either the rscS gene (Visick and Skoufos, 2001; Mandel et al., 2009), or the cluster of syp genes it controls (Yip et al., 2005, Shibata et al., 2012). Taken together, the data described above indicate that both the host and the symbiont contribute factors and activities that drive the initiation of a successful association.
Recent studies of the system have demonstrated that the symbiotic population in a naturally occurring E. scolopes is not clonal but, instead, contains several phenotypically and genomicly distinguishable strains of V. fischeri (Wollenberg and Ruby, 2009; Wollenberg and Ruby, 2012; Bongrand et al., 2016). In experiments where host-colonization potential was examined when strains were competed against one another, two phenotypes were noted: (i) a dominant (D-type) behavior, in which the host organ was usually colonized by a single strain; and, (ii) a sharing (S-type) behavior, in which more than one strain would co-colonize the same animal (Bongrand et al., 2016). The variation in the D- and S-type strains in their ability to dominate or co-colonize the crypts under these experimental conditions revealed a dominance hierarchy among these strains. Genomic analysis revealed that the D-strains are a recently diverged clade of V. fischeri (Bongrand et al., 2016). These phenotypic and genotypic differences among V. fischeri strains renders the squid-vibrio association an exceptional model system to study the impact of symbiont diversity on initiation, maintenance and persistence of host-microbe interactions.
In this study, we used the ability to view initial events of host-symbiont interaction directly and in real time to examine the impact of V. fischeri strain variation on initiation of the squid-vibrio symbiosis. The data provide evidence that strain variation is a major determinant of symbiosis initiation, affecting the very first interactions of the symbiont with host tissues. Genetic analyses suggested that strain variability in symbiosis initiation is controlled, at least in part, by differences in the relative level of a strain’s biofilm formation. In addition, we explored the capacity of environmental bacteria, including other Vibrio species, to interact with host tissues. The ability to form aggregates on the juvenile light organ surface was restricted to the Vibrionaceae tested. This study illustrates the wide range of intraspecies and interspecies variation that can occur in the initial interactions between a host and environmental bacteria during the minutes to hours surrounding the first encounter with the ambient environment, and underscores the impact of strain variation on symbiosis.
Results and discussion
V. fischeri strains exhibit differences in the extent of association with host cells during their initial recruitment into symbiosis.
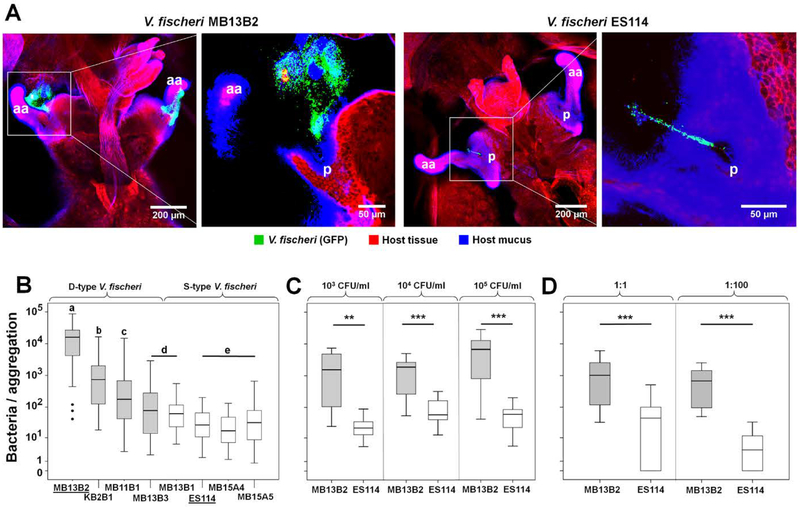
We examined the aggregation behavior of 8 V. fischeri strains, including 4 ‘niche-dominant’ (D) and 4 ‘niche-sharing’ (S) isolates (Table 1) from E. scolopes light organs. The D-type strains associated with the light organ (i.e., appeared at the pores, and aggregated) more quickly, in higher numbers, and across a larger area of the juvenile’s organ surface (Table 1). The D-type V. fischeri strains, MB13B2, KB2B1 and MB11B1, formed bacterial aggregations 1–3 orders of magnitude larger than all of the other strains, with MB13B2 having, on average, the largest aggregations (Fig. 2A & B). In addition, strains MB13B2 and KB2B1 formed aggregations as early as 10–15 min post-inoculation (Table 1), about 4-times faster than the well-studied S-type strain, V. fischeri ES114. The V. fischeri strains MB13B3 and MB13B1 showed no significant difference in size or time of aggregation, although they group with the D- and S-type strains, respectively. Similarly, the remaining S-type V. fischeri strains showed no significant difference in aggregation size or timing across strains (Fig. 2B). Based on these data, we established a ranking of these 8 strains in their ability to form bacterial aggregations on the surface of the host light organ as following: MB13B2 > KB2B1 > MB11B1 > MB13B3 = MB13B1 > ES114 = MB15A4 = MB15A5 (Fig. 2B).
Table 1.
Aggregation characteristics of D- and S-type V. fischeri symbiont strains
| Strain namea | Nb | Avg. number of cells cells in aggregate (range)c | Approx. time to form an aggregate (min)d |
|---|---|---|---|
| MB13B2 | 27 | 20,000 (41–85,000) | 10–15 |
| KB2B1 | 23 | 2,400 (18–16,000) | 10–15 |
| MB11B1 | 29 | 310 (3–3,900) | 30 |
| MB13B3 | 26 | 300 (2–2,900) | 60 |
| MB13B1 | 23 | 110 (6–550) | 60 |
| ES114 | 24 | 41 (1–200) | 60 |
| MB15A4 | 24 | 32 (1–130) | 60 |
| MB15A5 | 21 | 79 (1–750) | 60 |
Notes:
Strains above the dashed line are D-type
Number of animals analyzed; inoculated with 105 CFU/ml seawater for 3 h
Approximate number of cells in the aggregate (see Experimental procedures for details)
Time after inoculation at which the first evidence of aggregated cells was observed
Fig. 2.
Differences in aggregation behavior between strains of V. fischeri. (A) Two pairs of high and low magnification confocal-microscopy images illustrating the difference in the extent of bacterial aggregation on the surface of the squid’s light organ. Images captured at 3 h post-inoculation with GFP-expressing cells of either the V. fischeri D-type strain MB13B2 (left) or S-type strain ES114 (right); aa, anterior appendage; p, pores. Red, Cell Tracker Orange; blue, WGA (wheat-germ agglutinin) Alexa 633. (B) D-type V. fischeri strains generally had higher maxima as well as a large variation in average aggregation size; Kruskal-Wallis Test: Chi2=192, df=7, p<0.001 (for N values, see Table 1); the letters above the columns signify that those values are statistically significantly different from columns with another letter; bold horizontal lines represent medians; boxes comprise the interquartile ranges; bars indicate minimum and maximum values. (C) The size of aggregates produced by MB13B2 and ES114 cells was independent of the inoculum concentration; left panel, 103 CFU/ml, Mann-Whitney-U=7, Z=−3.13, p=0.002 (**); middle panel, 104 CFU/ml, Mann-Whitney-U=20, Z=−3.24, p=0.001 (***); right panel, 105 CFU/ml, Mann-Whitney-U=10, Z=−3.56, p<0.001 (***); N >5 for all treatments. (D) The relative aggregation effectiveness of MB13B2 and ES114 cells was independent of the co-inoculum ratio; left panel, ratio 1:1 = 103 CFU/ml each strain, Wilcoxon-signed-ranks test: Z=−4.11, n=22, p<0.001 (***); right panel, ratio 1:100 = 103 CFU/ml MB13B2 and 105 CFU/ml ES114: Wilcoxon-signed-ranks test: Z=−3.41, n=15, p=0.001 (***). Shaded boxes = D-type strains
To investigate the dynamics of aggregation in detail, we focused on one V. fischeri D-type strain, MB13B2, and one S-type strain, ES114. First, cohorts of animals were exposed to each strain at one of three concentrations: 103, 104, or 105 CFU/ml of seawater. The lower cell density (103 CFU/ml) approximates the reported abundance of V. fischeri cells that the squid host might encounter in its natural habitat (Jones et al., 2007) and, thus, provides insight into the aggregation behavior of these strains under typical environmental conditions. Similar to our results for initial strain comparisons (Fig. 2B), the D-type V. fischeri strain, MB13B2, formed significantly larger aggregations, under both naturally occurring and enriched bacterial concentrations, than the S-type strain, ES114 (Fig. 2C).
To determine the effect of co-inoculation on the patterns of aggregation, strains MB13B2 and ES114 were co-inoculated at ratios of 1:1 and 1:100. As expected, at a 1:1 inoculum (103 CFU of each strain per ml), strain MB13B2 outcompeted strain ES114. However, even when the concentration of ES114 was increased 100-fold in the inoculum (i.e., 105 CFU/ml), this strain comprised <1% of the bacteria appearing in the aggregates, which consisted principally of MB13B2 cells (Fig. 2D; Supporting information Fig. S1). Thus, the combined results indicate that the superiority of the D-type strain MB13B2 for aggregation on the organ surface was largely independent of both the initial bacterial density, and the relative abundance of D- and S-type strains that the host encounters during symbiosis initiation.
How might strains that form larger aggregations have a colonization advantage over those forming smaller ones (Bongrand et al., 2016)? One possible benefit would be the dilution of host-derived antimicrobials (McFall-Ngai, 2014) within the large, biofilm-rich (see below) aggregations; a second might be that these strains, unlike ES114, reach the pore sooner (Table 1) and are already ‘primed’ for successful navigation of the journey to the crypts (Wang et al., 2010; Kremer et al., 2013). In any case, the advantage, while uniform within the D-strains, is not directly proportional to their aggregate size (Fig. 2), or to the ratio of strains within the aggregate (Supplemental Fig. S1) (Bongrand et al., 2016).
Many studies have demonstrated that differences between strains of a pathogenic species are reflected in their various levels of persistence and host responses (for review, see (Palmer and Brayton, 2013). In addition, several studies have reported on the impact of strain variation in mutualistic symbioses, including the Steindernema-Xenorhabdus association (Murfin et al., 2015; McMullen et al., 2017) and in the plant-rhizobium interactions (Heath and Tiffin, 2007; Ezzakkioui et al., 2015; Batsone et al., 2017). In contrast, to the authors’ knowledge, few, if any, data are available on the differential behavior of a series of strains of a beneficial microbe during the initial minutes to hours of an animal-host association. For instance, while metagenomics have enabled descriptions of high levels of strain diversity within the mammalian gut microbiome (Greenblum et al., 2015; Ellegaard and Engel, 2016), the complexity of these communities has made understanding strain-level dynamics a challenging goal. Studies of binary associations, such as the squid-vibrio system and the nematode-Xenorhabdus association (Goodrich-Blair, 2007), as well as simple microbial consortia, such as those of the hydra (Deines et al., 2017), leech (Graf et al., 2006) and honeybee (Kwong and Moran, 2016), are offering opportunities for extensive experimental manipulation of strain variation. These invertebrate models promise to provide insight into the rules governing when, where and how strain-level variation is a significant factor in maintaining healthy, robust, symbiotic communities.
Aggregation of bacteria on the light organ surface perturbs the ultrastructure of the cilia, an effect whose extent reflects the aggregate size.
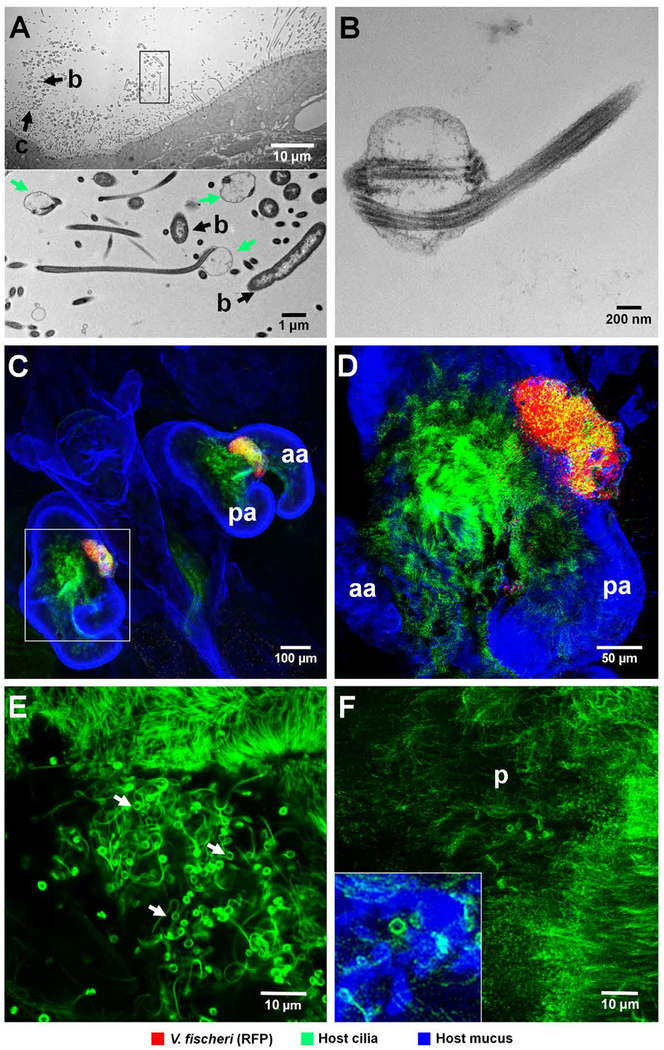
To determine how the host-cell surface responds to aggregating bacteria during initiation of symbiosis, we examined the interaction of different bacteria with the light organ’s ciliated field by both transmission electron microscopy (TEM) and confocal microscopy. A distinct structural modification of the cilia tips was observed when host cells were in association with V. fischeri cells (Fig. 3), independent of strain type. Such structurally altered cilia appeared exclusively in areas where bacteria were directly interacting with the ciliated surface; because the cells of a hyperaggregating strain are more abundant, the perturbations were more extensive. [NOTE TO EDITOR: This sentence was moved up from the end of the paragraph to clarify the point that this effect was not strain specific but rather due to degree of cell-cell contact between host and bacterial cells.] Aposymbiotic animals typically had only a few such modified cilia randomly distributed (Fig. 3E & F). Interestingly, this perturbation also occurred with V. campbellii strain KNH1, a bacterium that forms aggregates (Nyholm et al., 2000) but is unable to colonize the light organ (see below).
Fig. 3.
Association of V. fischeri cells with cilia on the surface of the light organ. (A, B) TEM images of cells of V. fischeri strain MB13B2, demonstrating structurally altered cilia in areas where bacteria directly interact with the ciliated surface of the light organ. (A) Upper panel: low magnification, showing the relationship of aggregating V. fischeri cells to the host’s ciliated epithelium; b, bacteria; c, structurally altered cilia. Lower panel: boxed area (rotated 90˚ clockwise) at higher magnification; altered cilia with swollen, paddle-shaped tips (green arrows) in association with V. fischeri cells. (B) High-magnification image, showing typical features of distorted cilia tip. (C-E) Confocal images at increasing magnification reveal host-cilia interactions with MB13B2 cells. (C) Low magnification, showing whole light organ. (D) Three-fold higher magnification of boxed area in C, showing aggregating bacteria. Yellow area indicates the co-localization of cilia (green) and V. fischeri cells (red). (E) Five-fold higher magnification than D, with only the green laser channel active to show the structurally altered cilia (green threads with terminal paddles) that form in areas of bacterial association (white arrows). The upper sector of this image shows structurally unaltered cilia for comparison (F) High-magnification confocal image of the ciliated surface of an aposymbiotic animal, where only an occasional cilium has an altered tip (inset, lower left); aa, anterior appendage; pa, posterior appendage; p, pore. Green,Tubulin Tracker; blue, WGA (wheat-germ agglutinin) Alexa 633.
The aposymbiotic data suggest that a few environmental cells capable of causing this perturbation were present in the ambient seawater, because the effect was not detected in animals exposed to bacteria-free filtered seawater (0.2 μ pore-size). Filtering does not remove soluble bacterial microbe-associated molecular patterns (MAMPs), such as lipopolysaccharide (LPS) and peptidoglycan derivatives that are known signaling molecules in the squid-vibrio system (McFall-Ngai et al., 2010), indicating that these bioactive soluble signals are not responsible for cilia-tip distortion. Because similar distortions can be an artifact of sample preparation (Short and Tamm, 1991), we confirmed this phenotype by live imaging both aposymbiotic and symbiotic animals, anesthetized either by cooling or with 2% ethanol.
The mechanisms mediating the perturbation of ciliary tips, as well as the functional significance of this ultrastructural modification, remain to be determined; however, similar modifications of cilia have been previously reported (i) in several marine invertebrate tissues (for review, see (Deiner et al., 1993)), and (ii) as a result of genetic defects in cilia biogenesis and maintenance (Siller et al., 2015). A possible cause of such distortion on the light organ’s ciliary surface could be a biomechanical consequence of the direct interaction of bacteria with the tip of the cilia (Nawroth et al., 2017). Specifically, the swelling and coiling of ciliary tips into paddle-like structures could result from changes in membrane tensile stress on the elastic axonemes of the cilia (Deiner et al., 1993), a force that may occur when motile V. fischeri cells attach to the cilia.
Studies of ES114 showed that, during first encounter of the partners, as few as 5 cells attaching to the short cilia on the light organ surface are sufficient to signal changes in host gene expression (Kremer et al., 2013; Nawroth et al., 2017), likely through the presentation of MAMPs like peptidoglycan and LPS (McFall-Ngai et al., 2010). The greater surface area covered by hyperaggregating strains, and their earlier appearance at the pores, predict that these strains may signal the host’s ciliated cells more strongly and quickly.
The hyperaggregating behavior of a V. fischeri D-type strain is regulated by genes required for biofilm formation in an S-type strain.
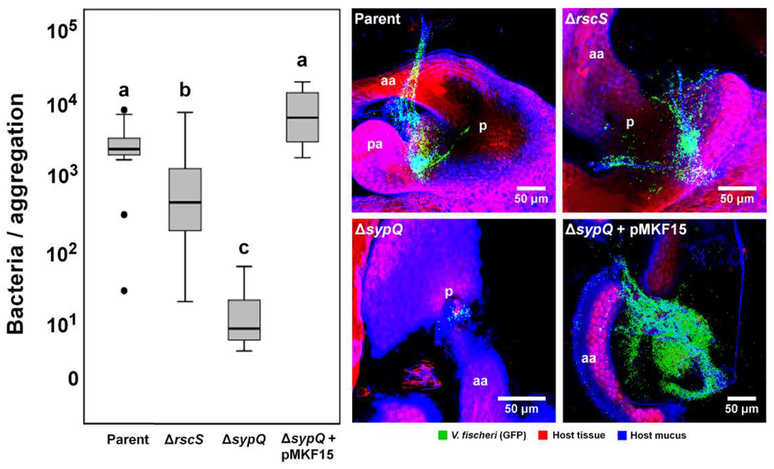
Studies of the S-type strain ES114 have demonstrated that genes for Syp-biofilm formation are essential for light-organ aggregation and colonization by V. fischeri (Yip et al., 2005; Yip et al., 2006, Shibata et al., 2012). To investigate whether these genes are required for the hyperaggregation of a D-type strain, we generated two mutations in V. fischeri strain MB13B2: (i) ΔrscS, defective for the Syp-biofilm positive regulator RscS, and (ii) ΔsypQ, missing a glycosyltransferase required for Syp-biofilm formation. Squid were inoculated with these strains at a concentration of 103 CFU/ml to mimic conditions that they might encounter in their natural environment. The ΔrscS mutation resulted in a statistically significant few-fold reduction in the number of cells amassed; however, the aggregate was still unexpectedly large (Fig. 4). In contrast, for most (>80%) squid inoculated with the strain with a sypQ deletion, no bacterial aggregations were observed at the light organ surface; similarly, those few aggregates that had formed were ~1000-fold reduced in the size, to a level characteristic of S-type strains (Table 1). Genetic complementation of the sypQ mutation restored the aggregation phenotype to that of the parental MB13B2 strain (Fig. 4).
Fig. 4.
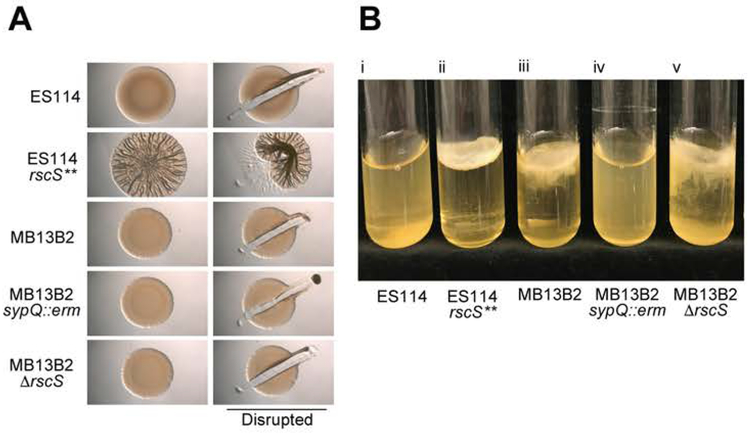
Role of bacterial biofilm-formation genes in a hyperaggregating strain. Left panel: Genetic manipulation of strain MB13B2 by deletion of the gene encoding RscS, a positive regulator of Syp-biofilm formation, only partially reduces the level of bacterial aggregation as compared to its wild-type parent. In contrast, a deletion of an essential Syp structural gene (sypQ) almost completely eliminated the aggregating phenotype of MB13B2 (Kruskal-Wallis Test: Chi2=20.6, df=3, p<0.001). The defect could be complemented by carriage of a wild-type copy of sypQ, in trans, on pMKF15 (Table S2), but not the vector (pKV282) alone. The letters above the columns signify that those values are statistically significantly different from columns with another letter; bold horizontal lines represent medians, boxes in this plot comprise the interquartile ranges (IQR); bars indicate 1.5 IQR of the data shown. The dots represent outliers. Right panel: confocal images showing the aggregation of mutants compared to the wild-type parent, V. fischeri strain MB13B2. aa, anterior appendage; pa, posterior appendage; p, pores. Red, Cell Tracker Orange; blue, WGA (wheat-germ agglutinin) Alexa 633.
The ability of most of the D-type strains to form symbiotic aggregates that were larger than those of ES114 suggested that these strains would also produce an extensive biofilm outside of the host. Thus, we assessed the ability of MB13B2, the D-type strain with the most extreme hyperaggregation phenotype, to form biofilms in two distinct assays, (i) wrinkled colony formation on solid agar media, and (ii) pellicle formation in static liquid cultures. The S-type strain ES114 is unable to form biofilms under either of those conditions, and so was used as a negative control, while KV4366 (rscS**), a previously described (Marsden et al., 2017), constitutively RscS-induced mutant of ES114 (Table S1), was used as a positive control.
On agar plates, the rscS**, biofilm-overproducing strain formed robust, cohesive, wrinkled colonies (Fig. 5A). In contrast, MB13B2 did not form colonies with either the surface architecture or cohesion of induced biofilms and, as expected, both the sypQ and rscS mutant derivatives of this strain also exhibited the smooth phenotype, similar to colonies of the negative control strain (data not shown). Consistent with its increased symbiotic aggregation phenotype, however, MB13B2 formed an extensive pellicle in static liquid culture following prolonged incubation (i.e., 10 days) at room temperature. While the ES114 culture formed a thin pellicle over that time period, it could be readily resuspended into a turbid culture (Fig. 5B–i). However, MB13B2 formed a thick pellicle at the air/liquid interface, similar to the biofilm-overproducing strain (Fig. 5B-ii & iii). These pellicles were robust, as manually shaking the culture dislodged the pellicle from the surface, but did not resuspend the cells as a turbid culture. Mutation of sypQ completely eliminated pellicle formation by strain MB13B2, indicating that this D-type strain forms syp-dependent biofilms under these conditions (Fig. 5B-iv). These data thus reveal that the D-type strain MB13B2 has an enhanced ability to form syp-dependent biofilms, relative to ES114, a result that correlates with its hyperaggregation during symbiosis initiation. Why MB13B2 responds differently in liquid culture and solid agar media by forming biofilms only in the former environment remains to be determined, but these data suggest that the liquid assay is more reflective of the fluid conditions of the symbiotic interaction.
Fig. 5.
In vitro biofilm phenotypes. (A) Development of a wrinkled-colony morphology over time. Ten microliters of a culture of each of three V. fischeri strains were spotted onto LBS agar plates. After 72 h of growth at 24 °C, colonies were disrupted with a toothpick to assess colony cohesiveness, which is an indicator of Syp-encoded polysaccharide production (Yip et al., 2005; Yip et al., 2006). (B) Pellicle formation in static liquid culture. Five strains of V. fischeri [(i) ES114, (ii) ES114 rscS** (KV4366), (iii) MB13B2, (iv) MB13B2 ΔsypQ (KV8195), and (v) MB13B2 ΔrscS (CBNR107)] were inoculated into 5 ml of LBS broth, and grown to log phase under shaking conditions. Cultures were then incubated statically at 24 °C for 10 days, and assessed for the presence of a surface pellicle.
In contrast to ΔsypQ, the rscS mutant retained the ability to form a robust pellicle that could be dislodged from the surface but not resuspended (Fig. 5B–v). These findings suggest that other regulatory inputs, in addition to RscS, control the hyperproduction of a syp-dependent biofilm by MB13B2. Indeed, in V. fischeri strain ES114, SypF is a required sensor kinase that activates biofilm formation and symbiotic colonization (Visick 2009, Norsworthy and Visick, 2015), as does HahK, which, like RscS, activates via SypF (Tischler et al. 2018). In contrast, BinK, which functions independently of RscS, directly inhibits syp-biofilm formation (Brooks and Mandel 2016). It is possible that SypF, HahK and/or BinK, all of which are encoded by MB13B2 (Bongrand et al., 2016) and share >99% amino-acid identity with their homologs in strain ES114, play a more important role in regulating biofilm formation in MB13B2. Another plausible explanation includes the upregulation of an additional biofilm component whose activity depends on the Syp polysaccharide; i.e., biofilms formed by other species involve outer membrane vesicles, extracellular DNA and proteins. Some of these factors are involved in, and/or upregulated coincident with, in vitro biofilm formation by strain ES114 (Shibata and Visick, 2012, Ray et al., 2015), and could contribute to hyperaggregation in a syp-dependent manner.
Should such differences in regulation exist between the normal and hyperaggregating strains, it is possible that they are mediated by genes present in the additional 250 Kb of genetic material found in the D-type strains (Bongrand et al., 2016); however, it should be noted that hyperaggregation can also result from overexpression of a single regulatory gene in a strain that does not carry the 250 Kb (Yip et al., 2006). Such questions are the subject of an ongoing comparative study focused on the different biofilm regulatory cascades in the two strains.
The formation of aggregates on the light organ’s surface is restricted to the Vibrionaceae.
Although V. fischeri is the only species that can colonize the light organ in nature, and V. fischeri ES114 can exclude other species from aggregating on the organ’s surface (Nyholm et al., 2000), when V. fischeri is absent, several marine bacteria will form a surface aggregation (Altura et al., 2013). We followed the development of such aggregates by confocal microscopy from 10 min to 6 h post-inoculation. Juvenile E. scolopes were inoculated with one of eight marine Gram-negative species (5 Vibrionaceae, and 3 non-Vibrionaceae) or three marine Gram-positive species, and their behavior compared to that of the squid symbiont V. fischeri ES114.
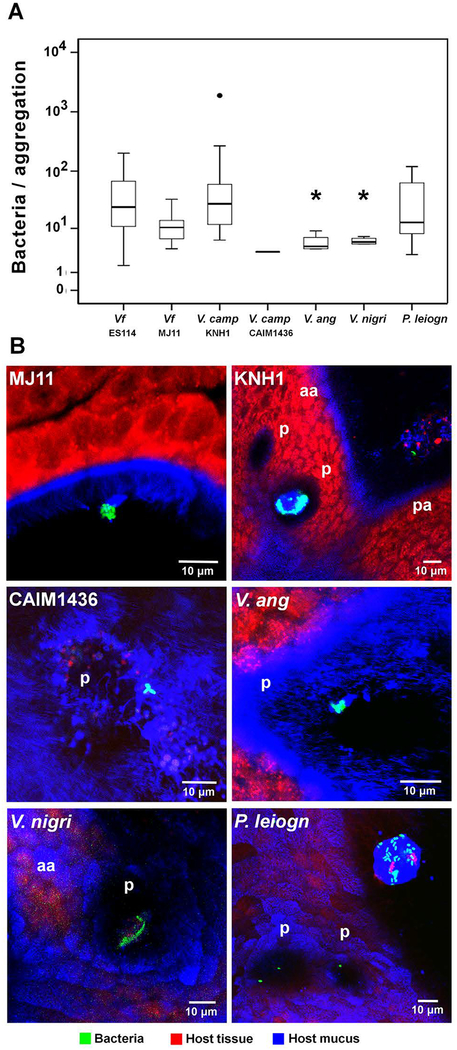
Consistent with previous reports (Nyholm and McFall-Ngai, 2003), even after 6 h, none of the Gram-positive strains had formed aggregations on the light organ (Table 2). This finding is surprising because biomechanical studies have shown that the light organ’s metachronally beating long cilia efficiently entrain bacteria-sized plastic beads into the region of short cilia above the pores, where aggregates form (Nawroth et al., 2017). It is here that V. fischeri cells attach to the tips of the cilia, and from which they colonize host tissues. The failure of Gram-positive bacteria to aggregate in this region may reflect their high sensitivity to antimicrobials such as peptidoglycan-recognition protein and lysozyme that are present there (Troll et al., 2010; Kremer et al., 2013). Like the Gram-positive strains, none of the three non-Vibrionaceae marine Gram-negative species were observed to aggregate, even though two of them (Pseudoalteromonas luteoviolacea HI1 and Leisingera sp. ANG1) associate with marine invertebrates (Shikuma et al., 2014; Gromek et al., 2016).
Table 2.
Aggregation characteristics of different bacterial phylotypes
| Speciesa (strain) | Phylum (Gram +/−) | Aggregates observedb | Avg. cells in an aggregate (range)c | Approx. time to form cells an aggregate (h)d |
|---|---|---|---|---|
| Vibrio fischeri (ES114) | γ-Proteo (−) | 24 / 24 | 41 (1–200) | 1 |
| V. fischeri (MJ11) | γ-Proteo (−) | 8 / 21 | 12 (4–33) | 4 |
| V. campbellii (KNH1) | γ-Proteo (−) | 16 / 37 | 161 (6–1,800) | 1.5 |
| V. campbellii (CAIM1436) | γ-Proteo (−) | 1 / 27 | 3 | 5.5 |
| V. anguillarum (CAIM8) | γ-Proteo (−) | 4 / 26 | 6 (4–9) | 5.5 |
| V. nigripulchritudo (CAIM323T) | γ-Proteo (−) | 4 / 22 | 6 (5–7) | 4.5 |
| Ph, leiognathi (KNH6) | γ-Proteo (−) | 18 / 29 | 35 (3–120) | 2 |
| Ps. luteoviolacea (HI1) | γ-Proteo (−) | 0 / 22 | -- | -- |
| Leisingera sp. (ANG1) | α-Proteo (−) | 0 / 21 | -- | -- |
| Cellulophaga lytica | Bacteroidetes (−) | 0 / 20 | -- | -- |
| Bacillus algicola | Firmicutes (+) | 0 / 18 | -- | -- |
| B. aquimaris | Firmicutes (+) | 0 / 22 | -- | -- |
| Exiguobacterium aestuarii | Firmicutes (+) | 0 / 13 | -- | -- |
Notes:
Species above the dashed line are Vibrionaceae; Ph.= Photobacterium; Ps.= Pseudoalteromonas
Number of animals with aggregates / total number observed; inoculated with 105 CFU/ml seawater
Approximate number of cells in the aggregate (see Experimental procedures for details)
Time after inoculation at which the first evidence of aggregated cells was observed
In contrast, two of the Vibrionaceae strains, V. campbellii KNH1 and Photobacterium leiognathi KNH6, produced aggregates indistinguishable from V. fischeri ES114 (i.e., on average 35–40 cells, in over half of the animals observed) within 2 h of inoculation. The other three Vibrionaceae strains (V. anguillarum CAIM8, V. nigripulchritudo CAIM323T, and V. campbellii CAIM1436) formed only small (~3–10 cells) aggregates on the organ that took 4–6 h to develop (Table 2; Fig. 6). Interestingly, just as different strains of V. fischeri displayed a range of aggregation behavior (Table 1), the two V. campbellii strains tested varied 100-fold in their level of aggregate formation. In summary, all these Vibrionaceae species were able to form surface associations; however, only V. fischeri could colonize the light organ. Thus, the ability to aggregate on the light-organ surface is a necessary, but not sufficient, step in initiating symbiosis. The finding that aggregation was restricted to the Vibrionaceae suggests a highly specific mechanism by which the host, after entraining bacteria-sized particles to the site of aggregation (Nawroth et al., 2017), must further discriminate among the Vibrionaceae to promote exclusive colonization by V. fischeri.
Fig. 6.
Aggregation behavior of environmental bacteria. (A) Sizes of aggregates of bacterial species that have the ability to form aggregations on the light organ surface at inocula of 105 CFU/ml (Kruskal-Wallis Test: Chi2=18.623, df=6, p=0.005); bold horizontal lines represent medians; boxes comprise the interquartile ranges; bars indicate the 1.5IQR of the data shown; dots represent outliers; asterisks indicate a significant difference in comparison to the control, S-type strain V. fischeri ES114 (V. anguillarum: Mann-Whitney-U=10.0, Wilcoxon W=20.0, Z=−2.324, p=0.018; V. nigripulchritudo: Mann-Whitney-U=9.0, Wilcoxon W=19.0, Z=−2.402, p=0.013). (B) Laser scanning-confocal micrographs show differences between the aggregation on the light-organ surface of V. fischeri and other environmental bacteria; aa, anterior appendage; pa, posterior appendage; p, pores. Environmental phylotypes (Table S1): V. camp, Vibrio campbellii; V. ang, Vibrio anguillarum; V. nigri, V. nigripulchritudo; P. leiogn, Photobacterium leiognathi. Red, Cell Tracker Orange; blue, WGA (wheat-germ agglutinin) Alexa 633.
Conclusions.
The present study characterizes the impact of species and strain differences on the first interactions of environmental bacteria with host tissues during the establishment of a natural symbiosis. Using the model squid-vibrio partnership, the data demonstrate that, while a small subset of environmental bacteria, i.e., members of the family Vibrionaceae, will aggregate on host mucociliary surfaces, only strains of V. fischeri are capable of colonizing host tissues. Within V. fischeri, different strains vary dramatically in both aggregation behavior and effect on host ciliated surfaces. Because these events are directly observable, and occur within minutes to hours, the data offer insight into how, under natural conditions, other mucociliary tissues might behave during their initial encounter with bacteria. While it will be difficult to apply this approach to the inaccessible mucociliary membranes of mammalian systems, such as the trachea and fallopian tubes, the results presented here promise to provide heuristic value for the design, modeling and experimentation of ciliated tissues in organ culture (Whitaker et al., 2017).
Experimental procedures
General
Adult Hawaiian bobtail squid, Euprymna scolopes, were caught in Maunalua Bay, Oahu, Hawaiʻi as previously described (Doino and McFall-Ngai, 1995) and maintained in a flow-through ocean seawater system at the Kewalo Marine Laboratory. Egg clutches laid in the system by female adults were transferred into smaller aquaria containing seawater free of V. fischeri, where they were incubated at 20°C (+/− 2°C). After hatching, the juvenile squid for all experiments were collected and transferred to UV-sterilized seawater prior to treatments and inoculations.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MI) or Thermo Fisher Scientific (Waltham, MA), unless otherwise stated. Restriction enzymes were purchased at New England BioLabs (Ipswich, MA).
Bacterial culture conditions
To initiate animal inoculations, all bacterial strains (Table 1) were recovered from glycerol stocks by spreading on a rich-medium (LBS) agar, and then grown overnight in LBS broth at 28 °C. LBS medium consists of 20 g NaCl, 50 ml of 1M Tris-HCl (pH 7.5), 10 g Bacto-Tryptone and 5 g yeast extract per liter (Graf et al., 1994; Stabb et al., 2001). For green fluorescent protein (GFP)- and red fluorescent protein (RFP)-labeled bacterial strains, 100 μg of kanamycin (Kn), or 2.5 μg of chloramphenicol (Cm), respectively, were added per ml of the overnight culture to ensure selection for cells carrying the plasmid-borne fluorescent protein (Table S2) (Dunn et al., 2006). Other antibiotics, at the indicated final concentrations: Cm, erythromycin (Em), or tetracycline (Tc) were added to growth media as necessary at 1, 2.5 or 5 μg ml−1, respectively.
E. coli DH5α was grown at 37 °C in LB medium (Davis et al., 1980) containing 10 g Bacto-Tryptone, 5 g yeast extract and 10 g NaCl per liter. As necessary, antibiotics were added to growth media at the indicated final concentrations: Kn, Cm or Tc, at 50, 12.5 or 15 μg per ml. Solid media were made using agar at a final concentration of 1.5%. Thymidine (0.3 mM) was added to media for growth of E. coli strain π3813.
For aggregation experiments, overnight cultures of marine bacteria were sub-cultured in a 1:100 dilution in seawater tryptone medium (SWT), containing 700 ml seawater, 300 ml deionized water, 5 g Bacto-Tryptone, 3 g yeast extract and 3 ml of glycerol per liter (Boettcher and Ruby 1990). Bacteria were grown to mid-exponential phase (OD600 ~ 0.3–0.6) with shaking before diluting them in seawater. Imaging of bacterial strains not genetically modified to carry GFP, was achieved by staining with 150 μl of ATTO495 fluorescent dye (0.5 mg/ml; AttoTec, Germany) in 3 ml of SWT medium, followed by ~2 h of bacterial growth in darkness, using the protocol described previously (Soto-Rodriguez et al., 2003).
Strain verification
16S amplicon sequencing was used to verify the identity of the environmental bacterial strains used in this study. Individual bacterial strains were grown in liquid culture and an aliquot was then centrifuged to pellet the bacteria. The supernatant was removed and the pellet used for nucleic acid extraction using the Epicentre MasterPure™ DNA extraction kit (Epicentre Technologies, Madison, WI). Finally, the DNA pellet was resuspended in 50 μl RNase-free low-TE buffer (1 mM Tris-HCl at pH 8.0, and 0.01 mM EDTA), and stored at −80 °C.
PCR with the general eubacterial primer pair fD1 and rP2 (Weisburg et al., 1991) amplified the 16S gene of each DNA sample. Amplification was performed on a BioRad C1000 Touch Thermocycler in a total reaction volume of 12.5 μl, containing 1 μl template DNA, 2.5 μl 5X GoTaq® Reaction Buffer (pH 8.5), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each primer and 0.25 U GoTaq® DNA-Polymerase (Promega, Madison, WI). Cycle parameters were as follows: an initial denaturation step at 95°C for 2 min, 35 cycles of 95 °C for 60 s, 65 °C for 60 s and 72 °C for 90 s, and a final extension step of 72 °C for 5 min. PCR success was verified by gel electrophoresis using a GelRed™ (Biotium, Fremont, CA) stained 1.5% TAE agarose gel (150 V, 20 min). The documentation of gel pictures was conducted using ChemiDoc-It2 Imager and the UVP VisionWorksLS software, version 8.6.15114.8618 (UVP®, Upland, CA). Positive bands were excised from the gel and purified using Wizard® SV Gel and PCR clean-up system (Promega, Madison, WI). The final DNA concentration of the purified PCR product was obtained from NanoDrop measurments (ND2000 photo-spectrometer). Sequencing was performed commercially at Genewiz® (South Plainfield, NJ).
Bacterial strains and mutant construction
The following E. coli strains were used for the purposes of cloning, plasmid maintenance, and conjugation: TAM1 (Active Motif, Carlsbad, CA, USA), DH5α and π3813 (Le Roux et al., 2007) (Table 1). Derivatives of V. fischeri were generated either by conjugation (DeLoney et al., 2002) or by natural transformation (Pollack-Berti et al., 2010, Brooks et al., 2014). Using the conjugation-based approach, an rscS deletion mutation was constructed in the D-type strain MB13B2 as described previously (Stabb and Ruby, 2002; Le Roux et al. 2007) with one modification. To build the recombinant plasmid, we first PCR amplified two fragments of V. fischeri MB13B2 DNA using primers 41 and 42, or primers 43 and 44 (Table S3). The two products were digested with SphI, and ligated together with T4 DNA ligase. The resulting sequence was amplified with primers 41 and 44. This PCR product, and the mobilizable suicide vector pKV363 (Shibata and Visick, 2012), were digested with SalI and XhoI and ligated together. The resulting plasmid was conjugated into MB13B2, and recombinants with a deletion of rscS were obtained.
For the transformation approach, PCR SOEing (splicing by overlap extension) (Ho et al. 1989) reactions were performed using EMD Millipore Novagen KOD high fidelity polymerase, and Promega Taq was used to confirm gene replacement events. Marked deletions of sypQ were generated by PCR SOEing using the primers listed in Table S3. Sequences (~500 bp) upstream (primers 1188 and 2174) and downstream (2175 and 443) of the target gene were amplified by PCR, then fused with an EmR antibiotic-resistance cassette (primers 2089 and 2090) in a PCR SOEing reaction. The final spliced PCR product was introduced into a tfoX-overexpressing ES114 strain carrying pLosTfoX-Kan (Brooks et al., 2014) by natural transformation (Pollack-Berti et al., 2010). Recombination of the PCR product into the chromosome resulted in the desired gene-replacement mutant, which was selected using the EmR marker. Chromosomal DNA was isolated from recombinants of V. fischeri strain ES114 using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) and used to introduce the desired mutation into tfoX-overexpressing MB13B2 cells.
Imaging of bacterial aggregations
To assess bacterial aggregation during symbiosis initiation, juvenile squid were incubated for between 10 min to 6 h in seawater containing 105 CFU/ml (unless stated otherwise) of either V. fischeri strains, or other Gram-negative and -positive marine bacteria (Tables 1 and 2). The animals were then stained with different fluorescent dyes such as Cell Tracker Orange (1:104) and wheat-germ agglutinin WGA Alexa Fluor 633 (1:40) (Thermo Fisher Scientific, Inc.) for 1–5 min, which labels host tissues and mucus, respectively. Bacterial strains were detected using either a GFP label (carried on pVSV102), or the ATTO495 fluorescent dye, which was used to label with V. nigripulchritudo CAIM323T as it was resistant to the triparental mating protocol for acquisition of the plasmid. Animals were anesthetized in 2% ethanol (EtOH), the mantle and funnel were removed to allow for visualization of the organ, and bacterial aggregation behavior on the organ surface was monitored at intermittent time points between 10 min and 6 h post-inoculation using a Zeiss LSM710 laser-scanning confocal microscope (Carl Zeiss Microscopy, Jena, Germany).
For the comparison of aggregation differences between the D- and S-type V. fischeri strains (Bongrand et al., 2016), squid were treated as described above with the following modification: animals were anesthetized in 2% EtOH after inoculation with bacteria, and subsequently fixed in 4% paraformaldehyde for 2 days. After fixation, the juvenile squid were rinsed 4 times, for 10–15 min, in 1X mPBS (marine phosphate buffered saline - 50 mM sodium phosphate, 0.45 M NaCl, pH 7.4) and were then fluorescently stained using the TOTO-3 fluorescent dye (Thermo Fisher Scientific, Inc.) at 1:650 in 1% Triton-X100 in mPBS for 1–2 h. After staining, samples were again rinsed two times in Triton X100-mPBS for 10–15 min, followed by two rinses of 10–15 min in 1X mPBS. These two different methods (the observation of live specimens, as well as samples that were fixed in 4% paraformaldehyde) allowed us to define the earliest time of bacterial aggregation, and to standardize the specific time point for aggregate-size measurement, respectively.
Estimation of aggregate size
The size of bacterial aggregates, as observed by laser-scanning confocal microscopy, was compared among strains to identify differences in the extent of bacterial aggregation. Two different methods were applied using the particle analysis tool of the FIJI-ImageJ imaging software (fiji.sc) to calculate spatial measurements based on fluorescence intensity. For fixed samples, a Z-stack was acquired using the LSM710 confocal microscope. Subsequently, multiple images in the Z-stack of one sample were projected onto one plane and the area (in μm2) of green (i.e., GFP) fluorescence was calculated, with 2 μm2 as the minimum threshold for bacterial size. Based on the acquired area, and the assumption of a bacterial cell cross-section being approximately 2 μm2, we estimated the number of cells present in a bacterial aggregation. All statistical analyses were performed using SPSS 25.0 Software (IBM, New York).
Interaction of D-type V. fischeri with light-organ cilia
To visualize the interaction of bacteria adhering to cilia on the organ surface, juvenile squid were exposed to 250 nM Tubulin Tracker in seawater after incubation with GFP- and RFP-labeled bacteria (Altura et al., 2013). Aposymbiotic animals were treated filtered seawater as a negative control. Subsequently, the animal tissues were fluorescently labeled as described above for observation by laser-scanning confocal microscope.
The density of interactions in aggregations of V. fischeri MB13B2 it made possible to locate and examine the ultrastructure of the paddle-like ciliary ends. For this transmission electron microscopy (TEM), we incubated juvenile squid with the V. fischeri MB13B2 for 30 min, anesthetized the animals with 2% EtOH, and then transferred them into fixative (2% paraformaldehyde and 2% glutaraldehyde in 1X mPBS) for 48 h at 4 °C. We then rinsed the samples 2 times for 15 min with 1X mPBS. For post-fixation, we incubated the samples with 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.4) for 1 h, followed by two rinses of 10 min each with 1X mPBS. The samples were dehydrated in a graded EtOH series and embedded in Spurr’s resin. Ultrathin (60–80 nm) sections were obtained on a Powertome ultramicrotome (RMC Boeckeler, Tucson, AZ), double stained with uranyl acetate (2% in distilled water) and lead citrate (Reynolds 1963), and viewed on a Hitachi HT7700 TEM at 100 kV.
Biofilm assays
To assess wrinkled-colony formation, V. fischeri strains were subcultured in 5 ml of LBS medium at 28 °C. Following growth to early-log phase, the cultures were standardized to an optical density at 600 nm (OD600) of 0.2. LBS agar plates were spotted with 10 μl aliquots, and incubated at 24 °C. Images of the spotted cultures were acquired over the course of wrinkled-colony formation at the indicated times using a Zeiss Stemi 2000-C dissecting microscope. At the end of the time course, the colonies were disrupted with a toothpick to assess colony cohesiveness, which is an indicator of the extent of Syp polysaccharide production (Ray et al., 2015).
Pellicle formation was visualized in V. fischeri strains grown overnight and subcultured in LBS with shaking. Following growth to mid-log phase, the cultures were allowed to stand statically at 24 °C for 10 days. Cultures were gently perturbed prior to imaging with an iPhone 7 camera.
Supplementary Material
Acknowledgements
We thank T Carvalho for technical assistance with transmission electron microscopy, M Flaherty for the construction of the sypQ complementation plasmid, and A Tischler for the construction of the ES114 ΔsypQ::EmR mutant. This study was made possible with funding from the National Institute of Health grants R37 AI150661 (to M McFall-Ngai and EG Ruby), R01 OD11024 (to EG Ruby and M McFall-Ngai), and GM114288 (to K Visick).
Footnotes
Conflict of interest
The authors declare no competing interests.
References
- Aanen DK, Eggleton P, Rouland-Lefevre C, Guldberg-Froslev T, Rosendahl S, and Boomsma JJ (2002) The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc Natl Acad Sci U S A 99:14887–14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, et al. (2013) The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol 15:2937–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastone RT, Dutton EM, Wang D, Yang M, and Frederickson ME (2017) The evolution of symbiont preference traits in the model legume Medicago truncatula. New Phytol 213:1850–1861. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, and Ruby EG (1990) Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, and McFall-Ngai M, et al. (2016) A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J 10:2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, and Bulgheresi S (2010) A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JF 2nd, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, et al. (2014) Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JF 2nd, and Mandel MJ (2016) The histidine kinase BinK Is a negative regulator of biofilm formation and squid colonization. J Bacteriol 198:2596–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, and Goodrich-Blair H (2017) Ready or not: Microbial adaptive responses in dynamic symbiosis environments. J Bacteriol 199: e00883–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, and Ochman H (2015) Assessing the unseen bacterial diversity in microbial communities. Genome Biol Evol 7:3416–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, and Roth JR (1980) Advanced bacterial genetics. Cold Spring Harbor, N.Y. [Google Scholar]
- Deiner M, Tamm SL, and Tamm S (1993) Mechanical properties of ciliary axonemes and membranes as shown by paddle cilia. J Cell Sci 104:1251–1262. [DOI] [PubMed] [Google Scholar]
- Deines P, Lachnit T, and Bosch TCG (2017) Competing forces maintain the Hydra metaorganism. Immunol Rev 279:123–136. [DOI] [PubMed] [Google Scholar]
- DeLoney CR, Bartley TM, and Visick KL (2002) Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol 184:5121–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doino JA, and McFall-Ngai MJ (1995) A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull 189:347–355. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, and Stabb EV (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, and Engel P (2016) Beyond 16S rRNA community profiling: Intra-species diversity in the gut microbiota. Front Microbiol 7:1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzakkioui F, El Mourabit N, Chahboune R, Castellano-Hinojosa A, Bedmar EJ, and Barrijal S Phenotypic and genetic characterization of rhizobia isolated from Hedysarum flexuosum in Northwest region of Morocco. J Basic Microbiol 55:830–837. [DOI] [PubMed] [Google Scholar]
- Frank SA (1996) Host-symbiont conflict over the mixing of symbiotic lineages. Proc Biol Sci 263:339–344. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair H (2007) They’ve got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr Opin Microbiol 10:225–230. [DOI] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, and Ruby EG (1994) Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Kikuchi Y, and Rio RV (2006) Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol 14:365–371. [DOI] [PubMed] [Google Scholar]
- Greenblum S, Carr R, and Borenstein E (2015) Extensive strain-level copy-number variation across human gut microbiome species. Cell 160:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromek SM, Suria AM, Fullmer MS, Garcia JL, Gogarten JP, et al. (2016) Leisingera sp. JC1, a bacterial isolate from Hawaiian bobtail squid eggs, produces indigoidine and differentially inhibits vibrios. Front Microbiol 7:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath KD, and Tiffin P (2007) Context dependence in the coevolution of plant and rhizobial mutualists. Proc Biol Sci 274:1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, and Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. [DOI] [PubMed] [Google Scholar]
- Jones BW, Maruyama A, Ouverney CC, and Nishiguchi MK (2007) Spatial and temporal distribution of the Vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb Ecol 54:314–323. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, and Denison RF (2003) Host sanctions and the legume-Rhizobium mutualism. Nature 425:78–81. [DOI] [PubMed] [Google Scholar]
- Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, et al. (2013) Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, and Moran NA (2016) Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F, Binesse J, Saulnier D, and Mazel D (2007) Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, and Ruby EG (2009) A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden AE, Grudzinski K, Ondrey JM, DeLoney-Marino CR, and Visick KL (2017) Impact of salt and nutrient content on biofilm formation by Vibrio fischeri. PLoS One 12:e0169521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Nyholm SV, and Castillo MG (2010) The role of the immune system in the initiation and persistence of the Euprymna scolopes--Vibrio fischeri symbiosis. Semin Immunol 22:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ (1998) The development of cooperative associations between animals and bacteria: Establishing detente among domains. Amer Zool 38:593–608. [Google Scholar]
- McFall-Ngai MJ (2014) The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, and Ruby EG (1991) Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1494. [DOI] [PubMed] [Google Scholar]
- McMullen JG 2nd, Peterson BF, Forst S, Blair HG, and Stock SP (2017) Fitness costs of symbiont switching using entomopathogenic nematodes as a model. BMC Evol Biol 17:100 oi: 10.1186/s12862-017-0939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NT, Bakacs E, Combellick J, Grigoryan Z, and Dominguez-Bello MG (2015) The infant microbiome development: mom matters. Trends Mol Med 21:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfin KE, Lee MM, Klassen JL, McDonald BR, Larget B, et al. (2015) Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio 6:e00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, et al. (2017) Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci U S A 114:9510–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker C, McNally CP, Eng A, and Borenstein E (2017) High-resolution characterization of the human microbiome. Transl Res 179:7–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norsworthy AN, and Visick KL (2015) Signaling between two interacting sensor kinases promotes biofilms and colonization by a bacterial symbiont. Mol Microbiol 96:233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, and McFall-Ngai MJ (2002) Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 68:5113–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, and McFall-Ngai MJ (2003) Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 69:3932–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, and McFall-Ngai MJ (2004) The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2:632–642. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, and McFall-Ngai MJ (2000) Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GH, and Brayton KA (2013) Antigenic variation and transmission fitness as drivers of bacterial strain structure. Cellul Microbiol 15:1969–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O, Huth CJ (1994) Evolutionary stability of mutualism between yuccas and yucca moths. Nature 372:257–260. [Google Scholar]
- Pollack-Berti A, Wollenberg MS, and Ruby EG (2010) Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M, and Boomsma JJ (2005) Mutualistic fungi control crop diversity in fungus-growing ants. Science 307:741–744. [DOI] [PubMed] [Google Scholar]
- Powell E, Ratnayeke N, and Moran NA (2016) Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol Ecol 25:4461–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray VA, Driks A, and Visick KL (2015) Identification of a novel matrix protein that promotes biofilm maturation in Vibrio fischeri. J Bacteriol 197:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, and Asato LM (1993) Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159:160–167. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Mueller UG, Wilcox TP, and Bull JJ (2004) The evolution of cooperation. Q Rev Biol 79:135–160. [DOI] [PubMed] [Google Scholar]
- Salem H, Florez L, Gerardo N, Kaltenpoth M (2015) An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc Biol Sci 282:20142957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, and Visick KL (2012) Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol 194:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Yip ES, Quirke KP, Ondrey JM, and Visick KL (2012) Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 194:6736–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, and Newman DK (2014) Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short G, and Tamm SL (1991) On the nature of paddle cilia and discocilia. Biol Bull 180:466–474. [DOI] [PubMed] [Google Scholar]
- Siller SS, Burke MC, Li FQ, and Takemaru K (2015) Chibby functions to preserve normal ciliary morphology through the regulation of intraflagellar transport in airway ciliated cells. Cell Cycle 14:3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rodriguez SA, Simoes N, Jones DA, Roque A, and Gomez-Gil B (2003) Assessment of fluorescent-labeled bacteria for evaluation of in vivo uptake of bacteria (Vibrio spp.) by crustacean larvae. J Microbiol Methods 52:101–114. [DOI] [PubMed] [Google Scholar]
- Stabb EV, Reich KA, and Ruby EG (2001) Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J Bacteriol 183:309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, and Ruby EG (2002) RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. [DOI] [PubMed] [Google Scholar]
- Takashita K, and Kikuchi Y (2017) Riptortus pedestria and Burkholderia symbiont: an ideal model system for insect-microbe symbiotic associations. Res Microbiol 168:175–187. [DOI] [PubMed] [Google Scholar]
- Tischler AH, Lie L, Thompson CM, and Visick KL (2018) Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol 200:e00016–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE,. et al. (2010) Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol 12: 2190–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL (2009) An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, and Skoufos LM (2001) Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, and Ruby EG (2010) Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 78:903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, and Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Shepherd ES, and Sonnenburg JL (2017) Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169:538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, and Ruby EG (2009) Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, and Ruby EG (2012) Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, and Visick KL (2006) The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, and Visick KL (2005) A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.