Significance
Animal–microbe associations are critical drivers of evolutionary innovation, yet the origin of specialized symbiotic organs remains largely unexplored. We analyzed the genome of Euprymna scolopes, a model cephalopod, and observed large-scale genomic reorganizations compared with the ancestral bilaterian genome. We report distinct evolutionary signatures within the two symbiotic organs of E. scolopes, the light organ (LO) and the accessory nidamental gland (ANG). The LO evolved through subfunctionalization of genes expressed in the eye, indicating a deep evolutionary link between these organs. Alternatively, the ANG was enriched in novel, species-specific orphan genes suggesting these two tissues originated via different evolutionary strategies. These analyses represent the first genomic insights into the evolution of multiple symbiotic organs within a single animal host.
Keywords: cephalopods, symbiosis, evolution, genomics, transcriptomics
Abstract
Microbes have been critical drivers of evolutionary innovation in animals. To understand the processes that influence the origin of specialized symbiotic organs, we report the sequencing and analysis of the genome of Euprymna scolopes, a model cephalopod with richly characterized host–microbe interactions. We identified large-scale genomic reorganization shared between E. scolopes and Octopus bimaculoides and posit that this reorganization has contributed to the evolution of cephalopod complexity. To reveal genomic signatures of host–symbiont interactions, we focused on two specialized organs of E. scolopes: the light organ, which harbors a monoculture of Vibrio fischeri, and the accessory nidamental gland (ANG), a reproductive organ containing a bacterial consortium. Our findings suggest that the two symbiotic organs within E. scolopes originated by different evolutionary mechanisms. Transcripts expressed in these microbe-associated tissues displayed their own unique signatures in both coding sequences and the surrounding regulatory regions. Compared with other tissues, the light organ showed an abundance of genes associated with immunity and mediating light, whereas the ANG was enriched in orphan genes known only from E. scolopes. Together, these analyses provide evidence for different patterns of genomic evolution of symbiotic organs within a single host.
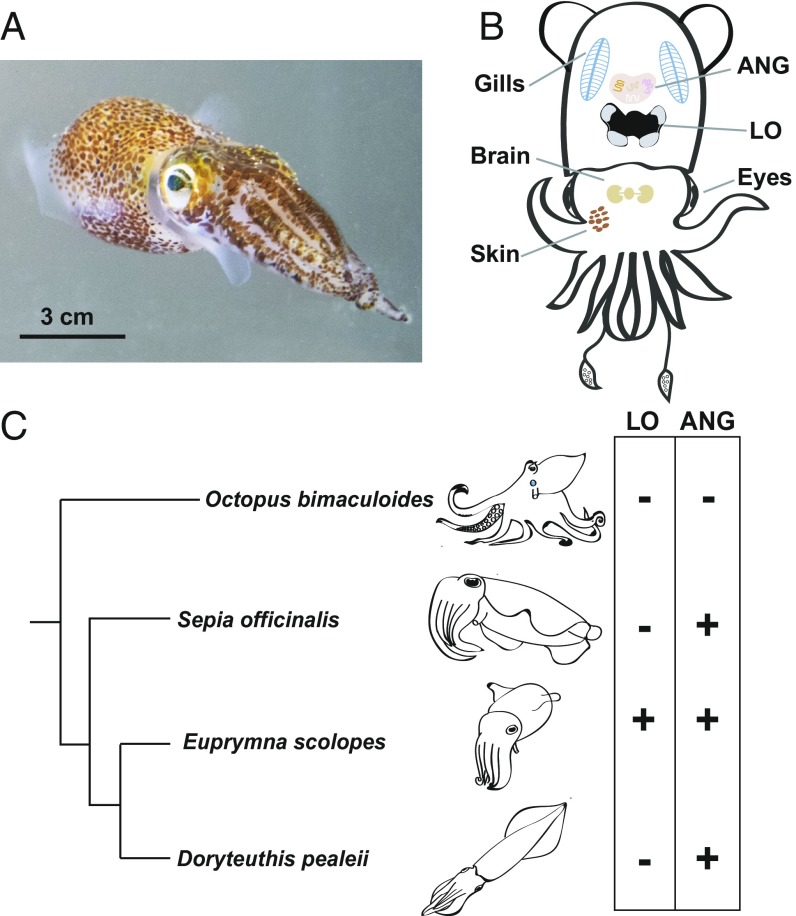
Numerous organisms have specialized organs to house their microbiota, yet the evolutionary processes underlying the origin of these tissues are not well understood (1–4). The Hawaiian bobtail squid, Euprymna scolopes, is a tractable animal model that has proven invaluable for interrogating symbiotic relationships and revealing common mechanisms by which successful colonization of animal epithelia by bacteria is established and maintained (Fig. 1A) (5). In the light organ (LO) of E. scolopes, a monospecific association with the bioluminescent bacterium Vibrio fischeri provides camouflage for the host and triggers a coordinated developmental remodeling of the surrounding epithelial tissues (6). Additionally, E. scolopes harbors a complex microbial consortium in the accessory nidamental gland (ANG), a component of the female reproductive system of many squids and cuttlefish that is hypothesized to play a role in egg defense (Fig. 1B) (7, 8). Octopus bimaculoides, the other cephalopod for which a genome has been reported to date, has neither a light organ nor an ANG making it an ideal organism for comparison (Fig. 1C). Therefore, to define elements of genome structure and function that are critical for symbiosis and the evolution of cephalopods, we sequenced and characterized the E. scolopes genome, the first example from the superorder Decapodiformes.
Fig. 1.
The Hawaiian bobtail squid, Euprymna scolopes, a model host for microbiome research and cephalopod innovations. (A) The animal, shown here in the water column, is a nocturnal predator that uses the luminescence of the LO symbiont Vibrio fischeri for camouflage. Scale bar, 3 cm. Image courtesy of Elizabeth Ellenwood (photographer). (B) Overview of key symbiotic and nonsymbiotic organs within E. scolopes. Whereas males only have an LO symbiosis, the females have the additional symbiosis of the ANG, a reproductive organ housing a consortium of bacteria from predominantly two phyla (8). In addition to these symbiotic tissues, gene expression was also analyzed from the gills, brain, eye, and skin. (C) Distribution of LOs and ANGs that occur only in coleoid cephalopods. Branch lengths are derived from Tanner et al. (35).
Results and Discussion
Our sequencing efforts have resulted in the most comprehensive cephalopod genome assembly to date, revealing the expansive and highly repetitive nature of the E. scolopes genome. Using a hybrid approach of shotgun and long-range linkage sequencing methods, an assembly estimated to be 5.1 Gigabases (Gb) was generated with a total half of the assembly in scaffolds of 3.7 Mb or longer (N50). The genome annotation was guided by 31 RNA-seq (Illumina) and ISO-seq (PacBio) transcriptomic libraries prepared from diverse tissues and developmental stages (SI Appendix, Table S1). The annotation resulted in 29,259 expressed protein-coding genes that yielded a BUSCO (Benchmarking Universal Single-Copy Orthologs) (9) score of 97% against the eukaryotic core set (Materials and Methods). A table summarizing the key statistics and a comparison with the O. bimaculoides genome is provided in SI Appendix, Table S2.
Genome analysis revealed differing histories of repeat element expansions between the bobtail squid and octopus, which may have contributed to the observed divergence in genome size (SI Appendix, Fig. S1). Although the estimated proportion of repetitive elements was >50% in both species, the abundance within individual repetitive element classes was strikingly different across the two species. Specifically, the abundance of long and short interspersed nuclear elements (LINEs and SINEs, respectively) differed; LINEs were the most abundant repeat class in E. scolopes, whereas SINEs dominated in O. bimaculoides (SI Appendix, Fig. S1B). Gene families in E. scolopes are similar to those reported in O. bimaculoides and also include tandem expansions in protocadherins and zinc finger transcription factors (SI Appendix, Fig. S2) (10). We infer that the last common coleoid cephalopod ancestor had 7,650 gene families, similar to the previously reported estimates for spiralian and bilaterian ancestors (10, 11). Because we do not find signatures of whole-genome duplication or evidence of extensive horizontal gene transfer in E. scolopes, the difference in the genome sizes between octopus and bobtail squid can be attributed to the increased LINE content in the E. scolopes genome. Future sequencing of other Decapodiformes genomes should enable a more exact estimate of this repeat element expansion.
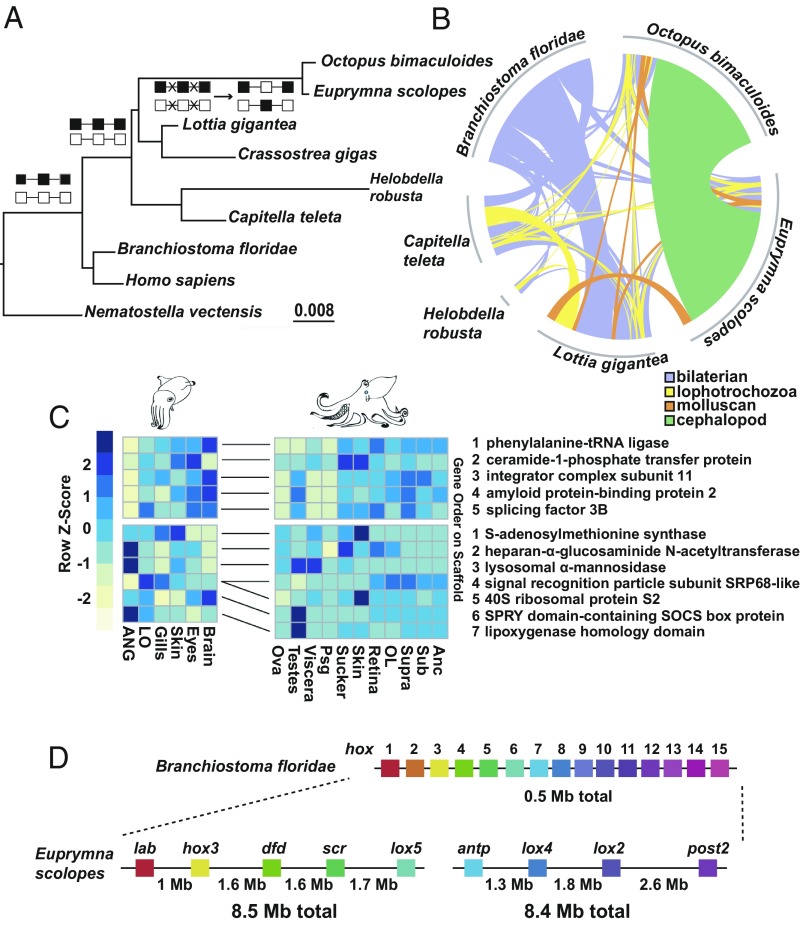
The E. scolopes genome revealed key genomic transitions toward cephalopod genomic architecture. We find that over half of the local gene linkages (i.e., microsynteny) conserved across many noncephalopod bilaterian genomes were disrupted in both O. bimaculoides and E. scolopes, indicating that a large genomic reorganization took place in the cephalopod ancestor (Fig. 2A). Additionally, we found numerous linkages shared between octopus and the bobtail squid that have not been previously identified in other animal genomes (Fig. 2 B and C). Those linkages contain genes expressed in highly developed organ systems (e.g., the central nervous system) as well as in symbiotic organs from E. scolopes and testes of O. bimacuolides (Fig. 2C). In total, this taxon-specific microsynteny constituted 67% (126 out of 189) of all conserved syntenic linkages in both cephalopod genomes (SI Appendix, Fig. S3 and Table S2).
Fig. 2.
Establishment of coleoid cephalopod genome architecture. (A) High rate of genome reorganization at the base of the coleoid cephalopods, as measured by the cumulative amount of microsyntenies lost and gained. Branch length estimation using MrBayes (SI Appendix) on a fixed tree topology using binary presence or absence matrix of shared orthologous microsyntenic blocks. Squares (black/white) connected by lines above the nodes indicate two hypothetical microsyntenic blocks illustrating a common scenario where conserved bilaterian and lophotrochozoan synteny is disrupted (crossed lines) followed by the emergence of a new order through rearrangement in the cephalopod stem lineage. (B) Prevalence of the unique cephalopod microsynteny (green shaded area) in O. bimaculoides and E. scolopes genomes. Total length of arches for individual species corresponds to the number of genes in microsyntenies. (C) Heat map of cephalopod unique microsyntenic clusters showing both neuronal (Upper) and broader gene expression (Lower) in E. scolopes and O. bimaculoides. Color bar indicates relative normalized gene expression level. Individual orthologous genes between E. scolopes and O. bimaculoides are connected by solid lines between heat maps with numbers indicating gene order along the scaffold. ANC, axial nerve cord; OL, optic lobe; Psg, posterior salivary gland; Supra/Sub, supraesophageal and subesophageal brain. (D) A large partial hox cluster on two separate scaffolds was recovered, totaling to a length of at least 16 Mb. Branchiostoma floridae hox cluster is shown for comparison with colors indicating orthologous genes.
The improved contiguity of the genome assembly also uncovered the presence of a partial hox cluster in E. scolopes with very large (1.5–2 Mb) intergenic distances that is in stark contrast to the previously reported atomized hox cluster in the octopus genome (10). These are to our knowledge the longest inter hox-gene separations (Fig. 2D) known in animals and are consistent with the long, gene-free regions surrounding hox genes in O. bimaculoides (10) and the identified expansion of repetitive elements. Such large intergenic distances may be responsible for the evolution of unique regulatory mechanisms. This hypothesis is further supported by unconventional hox gene expression in E. scolopes (12).
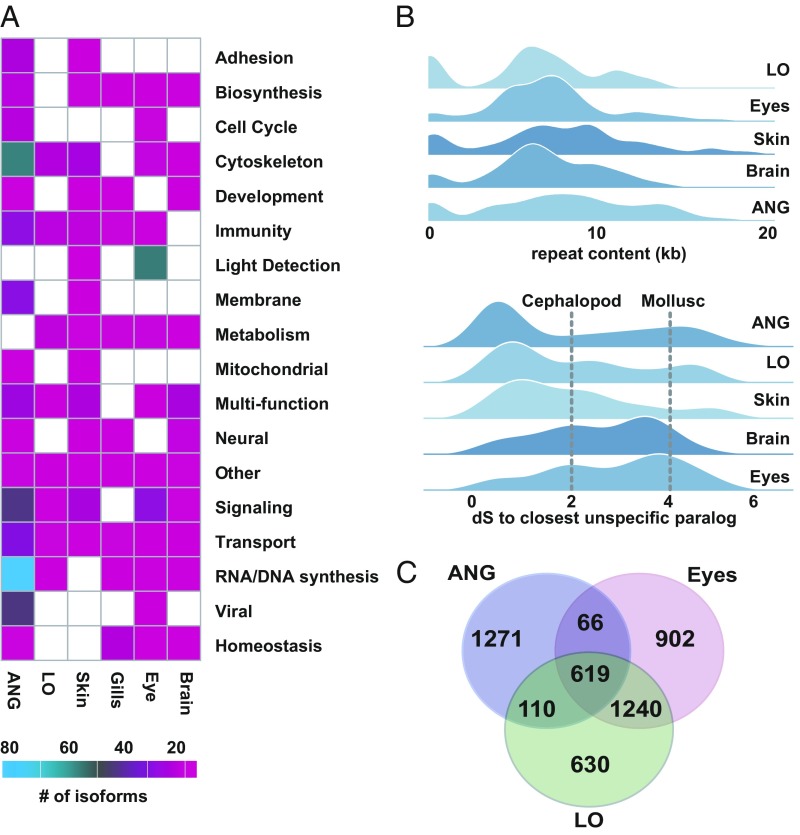
To understand how this genome reorganization drove the evolution of symbiotic organs in E. scolopes, we studied genomic signatures of genes with tissue-specific expression in symbiotic versus nonsymbiotic organs. To identify possible candidate genes that evolved in symbiotic organs after a duplication event, we searched the E. scolopes genome for all paralogous pairs of genes in which one gene had tissue-specific expression and the second did not. Using these genes, we then calculated the number of synonymous substitutions (dS) between each gene pair (Fig. 3). We found that for the LO and ANG, the dS distance between tissue-specific genes and the respective closest non-LO and non-ANG paralog was relatively small [dS < 1, ∼130 Mya, using a calibration point from Albertin et al. (10)]. These distances were in sharp contrast compared with genes specific to the eye and brain, which were much older (dS > 2, squid–octopus divergence, ∼270 Mya) (10, 13). These paralog ages are consistent with a scenario in which the ANG and LO, which harbor dense populations of bacteria, underwent relatively recent innovations within the bobtail squid lineage through gene duplication, compared with the more ancient duplications of genes that are involved in development and function of cephalopod eyes and the nervous system.
Fig. 3.
Characterization (i.e., functional categories) of key tissues revealing the high contribution of novel genes toward ANG evolution as well as strong similarity between LO and eye transcriptomes. (A) Total counts of unique isoforms across different functional categories in six adult tissues. (B) (Upper) Joy plot of the number of nucleotides within 20 kilobases (kb) windows located up and downstream of the tissue-specific genes from (A) that are attributed to repetitive elements. Regions around ANG genes show higher repeat content compared with other tissues (P < 0.1, Wilcoxon rank sum test). (Lower) Joy plot of the synonymous substitutions distances (dS) between the genes specifically expressed in a given tissue to their closest paralog expressed elsewhere. Distributions from tissues representing cephalopod synapomorphies (brain and eyes) show an older mean (P < 0.1, Wilcoxon rank sum test) compared with the distributions from ANG, LO, and skin tissues. (C) Venn diagram representing the number of shared transcripts among LO, ANG, and eye tissues identifying a significant overlap between LO and eye transcripts.
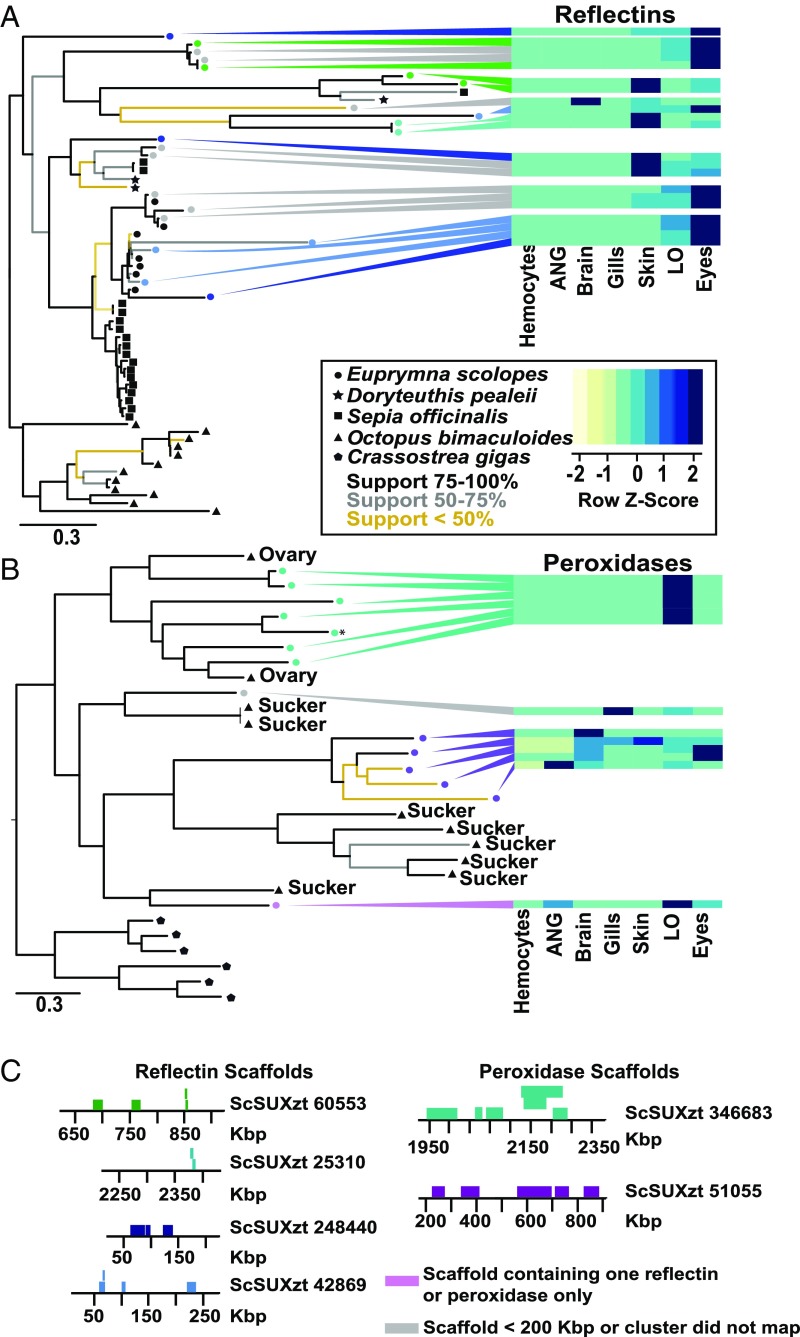
The main mechanism behind gene duplication in O. bimaculoides is through tandem or segmental duplication (10). Genes exclusively expressed in either LO, eyes, or ANG were typically found in tandem clusters of paralogs located on single scaffolds. In particular, we found two gene clusters, the first composed of reflectins and the second of peroxidases that were expressed in the LO (see below, Fig. 4, and SI Appendix, Figs. S4–S6). In both cases, the tandem gene duplications forming those clusters were E. scolopes-specific and occurred after the octopus–squid split, consistent with the emergence of ANG and LO in the subsequent squid lineage. A large 2-Mb cluster of S-crystallins, which are predominantly expressed in the eyes (SI Appendix, Fig. S7), was expanded in tandem in E. scolopes but was missing from the octopus genome. The omega-crystallins, which are typically found in the LO and the eyes of E. scolopes, on the other hand, did not show signs of any expansion (SI Appendix, Fig. S7). The characteristic cluster in the ANG (∼1 Mb) contained both novel genes and transposable elements. Overall, there were significantly more tandemly duplicated genes expressed in the LO and eyes compared with the number of the tandemly clustered genes specifically expressed in other tissues (Fisher’s exact test P < 0.001), which underscores the importance of gene duplication for those organ systems.
Fig. 4.
Independent tandem gene cluster formation in squid and octopus and the origin of light organ-specific gene expression. Phylogenetic trees highlight highly specific expansion patterns that correlate with general shared expression between the light organ and the eyes for reflectins (A) or the appearance of the light organ-specific expression pattern for heme peroxidases (B). Heat maps indicate relative normalized expression levels for each tissue (Z scores). Scale bar underneath phylogenetic trees indicates amino acid substitutions per site. Color of the nodes (A and B) identifies genomically colocalized genes (shown in C). Those genes are clustered in tandem on several scaffolds with positions and gene identifiers labeled. (C) Tandem clusters of reflectin and peroxidase genes in the E. scolopes genome. Scaffold ID and approximate location (kbp) is shown for each gene (represented by rectangle). Colors correspond to the sequences on the trees in A for reflectins and (B) for peroxidases.
Our data also reveal that despite the broad phylogenetic distribution among cephalopods (14), the ANG of E. scolopes is highly derived and shows an elevated evolutionary accumulation in both its coding (novel gene formation) and noncoding (turnover of the regulatory sequence) complements. The ANG is a secretory organ within the female reproductive system containing a bacterial consortium that is deposited into the egg capsule and is believed to play a role in defense from fouling and/or pathogens during embryogenesis (7, 8, 15, 16). Taxonomically restricted (i.e., orphan) genes have contributed to the evolution of unique tissues and organs in a number of animals (17–19). Within the two symbiotic organs of E. scolopes (LO and ANG), we profiled protein-coding transcripts that mapped to genome scaffolds from tissue-specific transcriptomes against the National Center for Biotechnology Information’s nonredundant database [using BLASTP (Basic Local Alignment Search Tool - Protein) e-value threshold of 1E-5; Fig. 3A] and classified the transcripts as either E. scolopes-specific, cephalopod, molluscan, bilaterian, metazoan, or premetazoan (SI Appendix, Fig. S8).
The ANG had the highest proportion of E. scolopes-specific transcripts (>35%) among all tissues. Additionally, regions 20 kb up- and down-stream of genes specifically expressed in the ANG also showed a higher proportion of repetitive element content compared with genes expressed in any other tissue (Fig. 3B), suggesting a high evolutionary turnover in regulatory regions. These higher rates of genomic innovation, specific to the E. scolopes ANG genes, are also supported by our analysis of previously reported ANG transcriptomes from both E. scolopes and the (loliginid) swordtip squid, Uroteuthis edulis, the latter of which had fewer unique genes specific to this organ (SI Appendix, Fig. S8) (20).
Within the LO of E. scolopes, there were several other genetic signatures contributing to the distinctive features of this symbiotic organ. First, the dominant transcripts within the light organ were reflectins (Fig. 4A), providing strong support for previous studies indicating that the anatomical features used to modulate light in the LO are similar to the eye, as are their physiology, biochemistry, molecular biology, and developmental induction (21–23). A comparison of adult LO and eye transcriptomes supports these previous studies (Figs. 3C and 4A). Identified reflectins (Fig. 4A) were almost exclusively (>99.99% of total expression) expressed in the LO, eyes, and skin. LO expression constituted around 19.5% of the total reflectin expression, whereas eyes and skin composed 72.8% and 7.6%, respectively. Reflectins usually shared two or more expression domains and were rarely expressed in a single tissue. The majority of genes showed a distinctive LO and eyes expression domain, and no reflectins were expressed exclusively in both LO and skin (Fig. 4 A and C). Evolutionary timing of the LO–eye reflectin clusters may thus hint at the origin of the LO. Molecular dating analysis using branch length and squid–octopus split as a calibration point (10, 13) revealed the age of those clusters at around 30 Mya or younger. Additionally, among the genomic expansion of 14 peroxidases unique to E. scolopes (Fig. 4 B and C and SI Appendix, Fig. S6), six genes [including a previously reported halide peroxidase (24, 25)] formed a single genomic cluster expressed exclusively in the light organ. Unlike reflectins, this expansion did not form a monophyletic group, suggesting high gain or loss of genes within this family, or gene conversion. The host is known to produce a halide peroxidase that is expressed in the LO and generates hypohalous acid, a potent antimicrobial compound thought to contribute to specificity in the squid–vibrio association (26, 27). This peroxidase, along with several others, is expressed in the tissues directly in contact with V. fischeri (28, 29). The expansion of the peroxidases and the unique expression of a subset of these genes exclusively in the LO may reflect the development of a unique microenvironment that helps to maintain specificity with V. fischeri (SI Appendix, Fig. S6).
Analysis of protein families also revealed a number of genes that had multiple immune-associated domains, many of which were expressed in the LO, ANG, and skin (SI Appendix, Fig. S9). All three of these tissues interact directly with environmental and/or symbiotic microorganisms. A number of cellular and biochemical components of the innate immune system are known to play critical roles in mediating specificity in the LO symbiosis with V. fischeri (30–32). Previous observations of host hemocytes infiltrating the ANG (7), along with expression of immune-related genes reported here, suggest that the host’s immune system plays a critical role in this symbiotic organ as well.
Although bacterial LOs are restricted to just two families of cephalopods (sepiolids and loliginids), the ANG is a more broadly distributed organ, present in these and a number of other squid and cuttlefish species (Fig. 1C). Their distinct evolutionary histories and origins are supported by the different evolutionary patterns detected. The prevalence of functional novelty through gene duplication in the LO compared with taxon-specific genes in the ANG suggests two different patterns of genomic innovation underlying symbiotic organ evolution in E. scolopes. Because these two associations are not nutritionally coupled, the mechanisms for the evolution of these symbiotic organs may be different from what has been proposed for insect bacteriocytes where metabolic complementation appears to be the major driver of innovation (33). Rapid gene evolution and/or gene loss in ancestral lineages may also have contributed to these unique signatures, especially in the ANG. Together, the presence of these two distinctive tissues within E. scolopes renders it a unique model organism to investigate the genetic mechanisms associated with the evolution of symbiotic organs (34).
Conclusions
Our analyses of the Hawaiian bobtail squid genome revealed large-scale genome reorganization, which preceded the coleoid cephalopod radiation, and provide evidence for two different patterns of genomic evolution that contributed to functional novelty. First, the extensive structural reorganization through the loss of ancient bilaterian microsynteny and increase in genome size through repetitive element expansions resulted in a unique genomic architecture that may have contributed to the innovations in the general cephalopod body plan. Second, the coding and surrounding regulatory regions of microbe-associated tissues suggest distinctive evolutionary patterns, such as the expansion of genes associated with immunity and light production in the LO. In the ANG, the prevalence of taxon-specific, or orphan, gene expression suggests a highly derived organ that evolved by a different means than the LO, perhaps due to unique selection pressures. Overall, our results set the stage for further functional analysis of genomic innovations that led to the evolution of symbiotic organs in a morphologically and behaviorally complex clade.
Materials and Methods
Data Access.
Genome and transcriptome sequencing reads have been deposited in the Sequence Read Archive as Bioproject PRJNA470951.
Genome Sequencing and Assembly.
Genomic DNA generated in this study was derived from a single adult male E. scolopes (SI Appendix). Illumina reads from several libraries (SI Appendix, Table S1) were assembled with Meraculous resulting in a preliminary assembly; scaffolds were generated using the in vitro chromatin conformation capture (Chicago) at Dovetail Genomics.
Gene Model Predictions.
Gene prediction models were generated using Augustus training with transcriptomic data. The prediction was filtered to exclude sequences that overlap with masked repeats over at least 50% of the lengths. From the remaining models, those with support from transcript evidence and significant homology to metazoan protein sequences were included in the nonredundant database. This gene modeling set was combined with transcriptome mapping (SI Appendix, Table S2) resulting in 29,089 protein sequences that passed the filtering stage.
Transcriptome Sequencing.
Thirty-one transcriptomes of E. scolopes were included in the reference transcriptome and sample metadata of the tissues and time points is provided in SI Appendix, Table S1. Different RNA extractions and sequencing platforms were used and are listed in SI Appendix, Table S1. For the tissue-specific transcriptomes, RNA was extracted from ANG, brain, eyes, gills, hemocytes, LO, and skin tissues, as well as juvenile head (white body, optic nerve, and brain), eyes, gills, and light organ. For the PacBio IsoSeq library, RNA was extracted, normalized, and pooled from adult brain, eyes, white body, optical lobe, gills, LO, and skin as well as whole juvenile hatchling, 24-h aposymbiotic, and 24-h symbiotic animals (SI Appendix, Table S1).
Synteny Analysis.
Using previously published methods of phylogeny-informed clustering, we have constructed the sets of orthologous gene families between the following species (11): Capitella teleta, Helobdella robusta, Lottia gigantea, Octopus bimaculoides, Euprymna scolopes, Crassostrea gigas, Nematostella vectensis, and Branchiostoma floridae. To account for differential gene loss that may impede our quantification of synteny loss/gain we focused only on 3,547 clusters that have an ortholog from each of the species. We have implemented microsynteny detection algorithm, as described in Simakov et al. (11), and found, in accordance with previous results (10, 34), ∼600 microsyntenic blocks (in 163 orthologous groups) that can be traced back to the bilaterian ancestor because they are shared between either both ingroups (protostome and deuterostome) or an ingroup and an outgroup species (Nematostella) (Dataset S1).
Supplementary Material
Acknowledgments
We thank Bo Reese from the Center for Genome Innovation, University of Connecticut (UCONN), for sequencing assistance and Jill Wegrzyn, Stephen King, and Ion Moraru of the Computational Biology Core, Institute for Systems Genomics, UCONN, for computational support. Computation was conducted at the Vienna Life Sciences Cluster (cube.univie.ac.at/) and the computing cluster at the Okinawa Institute of Science and Technology (OIST). This work was supported in part by NIH Grant R01-AI50661 (to M.M.-N. and E. G. Ruby, co-Principal Investigator); NIH Grant R01-OD11024 (to E. G. Ruby, co-Principal Investigator, and M.M.-N.); the University of Wisconsin (M.M.-N.); NASA Space Biology Grant NNX13AM44G (to J.S.F.); NSF Integrative Organismal Systems 1557914 and the Office of the Vice President for Research, UCONN (to S.V.N.); and the Molecular Genetics Unit of OIST Graduate University (to D.S.R.). O.S. and H.S. were supported by a grant from the Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung) P30686-B29.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome and transcriptome sequencing reads have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA470951).
See Commentary on page 2799.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817322116/-/DCSupplemental.
References
- 1.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 2.Markmann K, Parniske M. Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci. 2009;14:77–86. doi: 10.1016/j.tplants.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Matsuura Y, et al. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. 2012;6:397–409. doi: 10.1038/ismej.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griesmann M, et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361:eaat1743. doi: 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- 5.McFall-Ngai MJ. The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFall-Ngai MJ. Giving microbes their due—Animal life in a microbially dominant world. J Exp Biol. 2015;218:1968–1973. doi: 10.1242/jeb.115121. [DOI] [PubMed] [Google Scholar]
- 7.Collins AJ, et al. Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl Environ Microbiol. 2012;78:4200–4208. doi: 10.1128/AEM.07437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerwin AH, Nyholm SV. Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and developing eggs. Environ Microbiol. 2017;19:1463–1475. doi: 10.1111/1462-2920.13665. [DOI] [PubMed] [Google Scholar]
- 9.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 10.Albertin CB, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524:220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee PN, Callaerts P, De Couet HG, Martindale MQ. Cephalopod Hox genes and the origin of morphological novelties. Nature. 2003;424:1061–1065. doi: 10.1038/nature01872. [DOI] [PubMed] [Google Scholar]
- 13.Kröger B, Vinther J, Fuchs D. Cephalopod origin and evolution: A congruent picture emerging from fossils, development and molecules: Extant cephalopods are younger than previously realised and were under major selection to become agile, shell-less predators. BioEssays. 2011;33:602–613. doi: 10.1002/bies.201100001. [DOI] [PubMed] [Google Scholar]
- 14.Lindgren AR, Pankey MS, Hochberg FG, Oakley TH. A multi-gene phylogeny of Cephalopoda supports convergent morphological evolution in association with multiple habitat shifts in the marine environment. BMC Evol Biol. 2012;12:129. doi: 10.1186/1471-2148-12-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggs J, Epel D. Egg capsule sheath of Loligo opalescens Berry: Structure and association with bacteria. J Exp Zool. 1991;259:263–267. [Google Scholar]
- 16.Barbieri E, et al. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda:Loliginidae) Environ Microbiol. 2001;3:151–167. doi: 10.1046/j.1462-2920.2001.00172.x. [DOI] [PubMed] [Google Scholar]
- 17.Gould RM, et al. Myelin sheaths are formed with proteins that originated in vertebrate lineages. Neuron Glia Biol. 2008;4:137–152. doi: 10.1017/S1740925X09990238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 20.Pankey MS, Minin VN, Imholte GC, Suchard MA, Oakley TH. Predictable transcriptome evolution in the convergent and complex bioluminescent organs of squid. Proc Natl Acad Sci USA. 2014;111:E4736–E4742. doi: 10.1073/pnas.1416574111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong D, et al. Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crookes WJ, et al. Reflectins: The unusual proteins of squid reflective tissues. Science. 2004;303:235–238. doi: 10.1126/science.1091288. [DOI] [PubMed] [Google Scholar]
- 24.Tomarev SI, et al. Abundant mRNAs in the squid light organ encode proteins with a high similarity to mammalian peroxidases. Gene. 1993;132:219–226. doi: 10.1016/0378-1119(93)90199-d. [DOI] [PubMed] [Google Scholar]
- 25.Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- 28.Wier AM, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleicher TR, Nyholm SV. Characterizing the host and symbiont proteomes in the association between the bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS One. 2011;6:e25649. doi: 10.1371/journal.pone.0025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes–Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAnulty SJ, Nyholm SV. The role of hemocytes in the Hawaiian bobtail squid, Euprymna scolopes: A model organism for studying beneficial host-microbe interactions. Front Microbiol. 2017;7:2013. doi: 10.3389/fmicb.2016.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen F, et al. Bactericidal permeability-increasing proteins shape host-microbe interactions. MBio. 2017;8:e00040-17. doi: 10.1128/mBio.00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson AC, Duncan RP. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci USA. 2015;112:10255–10261. doi: 10.1073/pnas.1423305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simakov O, Kawashima T. Independent evolution of genomic characters during major metazoan transitions. Dev Biol. 2017;427:179–192. doi: 10.1016/j.ydbio.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Tanner AR, et al. Molecular clocks indicate turnover and diversification of modern coleoid cephalopods during the Mesozoic marine revolution. Proc Biol Sci. 2017;284:20162818. doi: 10.1098/rspb.2016.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.