Significance
Metabolism, one of the most conserved features across all domains of life, is interwoven with cellular signaling networks and the posttranslational modification state of proteins. Methionine is an essential amino acid in one-carbon metabolism that serves as the substrate for S-adenosylmethionine, the universal methyl donor for cellular methylation. Here we report that the use of methionine-depleted medium inhibits Wnt-induced endolysosomal formation and canonical Wnt/β-catenin signaling. Use of methotrexate, a folate cycle inhibitor used in cancer chemotherapy, prevented PRMT1 and GSK3 vesicular sequestration, decreased Wnt-driven endolysosomal activity, and reduced canonical Wnt β-catenin luciferase signaling. In sum, these results imply a role for one-carbon metabolism in the regulation of Wnt signaling and endolysosomal biology.
Keywords: Wnt signaling, arginine methylation, methotrexate, S-adenosylmethionine, nicotinamide
Abstract
The nutrient-sensing metabolite S-adenosylmethionine (SAM) controls one-carbon metabolism by donating methyl groups to biochemical building blocks, DNA, RNA, and protein. Our recent work uncovered a requirement for cytoplasmic arginine methylation during Wnt signaling through the activity of protein arginine methyltransferase 1 (PRMT1), which transfers one-carbon groups from SAM to many protein substrates. Here, we report that treatments that decrease levels of the universal methyl donor SAM were potent inhibitors of Wnt signaling and of Wnt-induced digestion of extracellular proteins in endolysosomes. Thus, arginine methylation provides the canonical Wnt pathway with metabolic sensing properties through SAM. The rapid accumulation of Wnt-induced endolysosomes within 30 minutes was inhibited by the depletion of methionine, an essential amino acid that serves as the direct substrate for SAM production. We also found that methionine is required for GSK3 sequestration into multivesicular bodies through microautophagy, an essential step in Wnt signaling activity. Methionine starvation greatly reduced Wnt-induced endolysosomal degradation of extracellular serum proteins. Similar results were observed by addition of nicotinamide (vitamin B3), which serves as a methyl group sink. Methotrexate, a pillar in the treatment of cancer since 1948, decreases SAM levels. We show here that methotrexate blocked Wnt-induced endocytic lysosomal activity and reduced canonical Wnt signaling. Importantly, the addition of SAM during methionine depletion or methotrexate treatment was sufficient to rescue endolysosomal function and Wnt signaling. Inhibiting the Wnt signaling pathway by decreasing one-carbon metabolism provides a platform for designing interventions in Wnt-driven disease.
A complex web of signal transduction events dictates cellular decisions by coordinating nutrient availability and intracellular signaling cascades. Canonical Wnt signaling is an evolutionarily conserved pathway that is intricately regulated (1) to govern a diverse set of processes through β-catenin activation of the T cell factor/lymphoid enhancer factor (TCF/LEF) family (2, 3). Recently, it has also been realized that Wnt signaling is an overall regulator of cellular protein catabolism (1, 4–6).
Glycogen synthase kinase (GSK3) is a key player in the Wnt pathway, targeting β-catenin for polyubiquitination and degradation in proteasomes (7, 8). Canonical Wnt stabilizes β-catenin through the inhibition of GSK3 (5, 9–11). Upon Wnt signaling, β-catenin becomes stabilized following the sequestration of active GSK3 from the cytosol inside late endosomes/multivesicular bodies (MVBs). This translocation is achieved through microautophagy of cytosolic components in small intraluminal vesicles via the Vacuolar protein sorting/endosomal sorting complexes required for transport (Vps/ESCRT) machinery (4, 5). Wnt signaling stabilizes many proteins in addition to β-catenin through a mechanism known as Wnt-dependent stabilization of proteins (Wnt/STOP) (4, 5, 12, 13), which facilitates processes such as cell growth during G2/M in preparation for division (4, 14).
GSK3 is a promiscuous kinase that processively phosphorylates consecutive consensus motifs (S/T-XXX-S/T) that are present in 20% of the human proteome (4, 5, 15, 16). In addition to GSK3, Wnt signaling also sequesters additional cytosolic proteins, many of which are GSK3 substrates, into endolysosomes (1, 12) to regenerate cellular building blocks (17, 18). We and others recently demonstrated that canonical Wnt signaling increases lysosomal activity and the endocytosis and digestion of extracellular BSA (1, 19). Intriguingly, pancreatic tumor cells adapt to surrounding microenvironments by acquiring nutrients from extracellular sources for metabolic reprogramming (20–23). Whether Wnt signaling through endolysosomes rewires metabolism is unknown.
A striking increase in endolysosomes/MVBs occurs within 5 min of Wnt stimulation (1). We recently discovered that protein arginine methyl transferase 1 (PRMT1) is required for canonical Wnt to signal and increase endocytosis of extracellular proteins (1). Of the nine PRMTs, PRMT1 is essential for development (24, 25) and accounts for 85% of cellular arginine methylation (26–28). Importantly, PRMT1 is expressed at high levels in the Wnt-dependent stem cells of the intestinal crypt where it is required for proliferation (29, 30) and intestinal remodeling (31–33).
PRMTs add methyl groups to arginine residues using the universal methyl-donor, S-adenosylmethionine (SAM, also known as AdoMet) as a substrate (28, 29, 34). Present in every living cell, SAM is known as a hub metabolite as it is used for myriad processes (35, 36). In contrast to phosphorylation where the kinase substrate ATP exists in vast excess, methylation is harmonized with the metabolic state of the cell as SAM levels are determined by nutrient availability (37–39). SAM is generated through one-carbon metabolism, a fundamental metabolic pathway that couples the folate and methionine cycles to coordinate nutritional status with physiological processes by transferring methyl groups to various substrates (40, 41). Within this cycle, SAM is generated directly from ATP and methionine, an essential amino acid whose levels are dictated by dietary factors (42–44). The rapid kinetics provided by posttranslational modifications are essential for regulating metabolic fluxes. Metabolic and transcriptional networks operate on different timescales where the turnover of a metabolite occurs within seconds (23), while Wnt/β-catenin luciferase assays usually involve periods of 12 h and MVB sequestration takes place within minutes after Wnt addition (1). Whether one-carbon metabolism influences PRMT1 or Wnt signaling is unknown.
In this study, we show that methionine availability regulates Wnt signaling by determining cellular SAM levels through one-carbon metabolism. Elucidating the timescales of cellular events in this study facilitates an intuitive understanding of these molecular mechanisms. The findings contribute to the relationship of Wnt and metabolism by focusing on early membrane trafficking events, supporting a model whereby Wnt signaling mediates rapid cellular responses to alterations in nutritional or environmental status. Importantly, inhibition of canonical Wnt with the antifolate methotrexate compromised endocytosis and lysosomal activity. This work reveals a role for the methionine cycle in fueling endocytosis and Wnt signaling, processes not previously known to be targeted by methotrexate.
Results
Methionine Is Required for Endocytosis and Wnt Signaling.
One-carbon metabolism is composed of the methionine cycle (Fig. 1A) and the folate cycle. The methylation cofactor SAM is generated through the ATP-driven adenylation of methionine by methionine adenosyltransferase (MAT) enzymes (45). SAM provides the methyl groups to substrates where PRMTs deposit methyl groups on the guanidino nitrogen atoms of arginines (28). Methyl transfer generates S-adenosylhomocysteine (SAH) as a byproduct, which is rapidly converted to homocysteine (41). Given that methionine represents an essential substrate upstream of SAM production and arginine methylation is required for canonical Wnt, we first set out to examine whether methionine levels affected Wnt signaling.
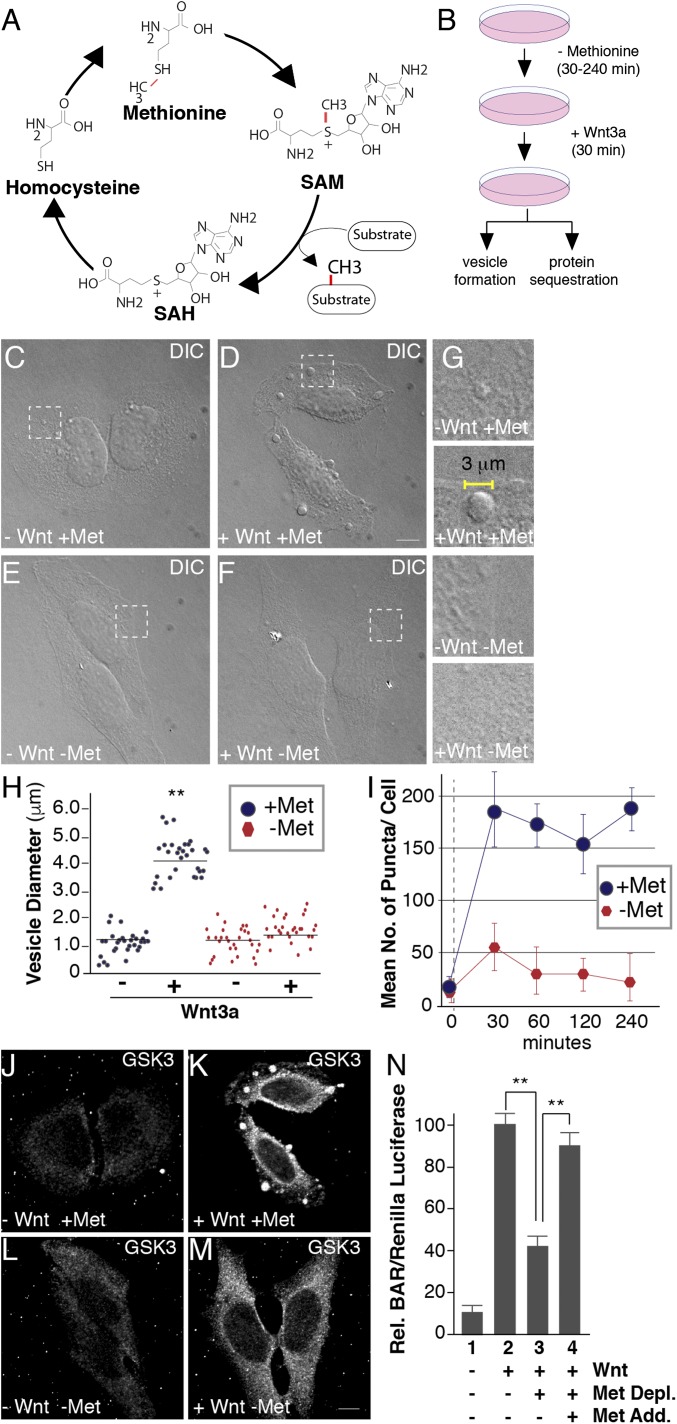
Fig. 1.
Methionine depletion inhibits Wnt signaling. (A) The universal cofactor for methylation SAM is generated through a biochemical mechanism known as the methionine cycle. Methionine is converted into SAM through an ATP-driven reaction. Methyltransferases use SAM as a cofactor to transfer methyl groups (in red) to a myriad of substrates, generating the byproduct SAH. SAH is rapidly converted to homocysteine, which can become remethylated to form methionine, completing the cycle. (B) Experimental design used for testing the effect of methionine deprivation on Wnt-induced endocytosis. Cultured cells were preincubated in methionine-depleted medium for 30 min and then treated with Wnt3a for 30 min (in the case of HeLa cell phase microscopy analyses or in situ protease protection assays) or 12 h (for HEK293T BAR/β-catenin luciferase assays). (C–F) Treatment with Wnt3a for 30 min triggered the formation of large puncta visualized by phase microscopy in HeLa cells; methionine depletion for 30 min strikingly inhibited vesicle formation in the presence of Wnt3a (compare D to F); the indicated stippled rectangles were enlarged. (G) Enlargements of Wnt3a-induced vesicles (2–5 μm in diameter) which were absent minus methionine. (H) Quantification of the diameters of Wnt-induced vesicles using ImageJ. (I) Time-course analysis of methionine depletion for 30, 60, 120, or 240 min followed by 30 min of Wnt3a signaling; note the significant decrease in vesicle formation after only 30 min (despite using nondialyzed serum in this case). (J–M) Wnt-induced GSK3 sequestration inside vesicles was inhibited by methionine starvation for 30 min, as assessed by in situ proteinase K protection assay (compare K to M). (N) Methionine depletion inhibited Wnt signaling in BAR luciferase reporter assays, while methionine readdition in column 4, rescued Wnt inhibition. Wnt luciferase reporter (BAR) was normalized using Renilla (49). (Scale bars: 10 μm.) **P < 0.01; n < 3.
Wnt3a treatment for 5–30 min generates large endolysosomal vesicles within the cell (1) that contain cytoplasmic proteins translocated by microautophagy (5, 12). To examine whether Wnt-induced vesicles were sensitive to methionine levels, HeLa cells were incubated in methionine-depleted RPMI culture medium with or without the addition of Wnt3a protein for 30 min and analyzed by phase contrast microscopy (Fig. 1B). Wnt3a protein generated the formation of large vesicles (Fig. 1 C and D). Vesicles also formed in the presence of cycloheximide, indicating that the Wnt-induced vesicles were not dependent on new protein synthesis (SI Appendix, Fig. S1 A–D). In methionine-depleted medium, vesicle formation was inhibited during Wnt activation, compared with cells incubated with methionine-containing medium (Fig. 1 E and F). Measurement of phase contrast images further showed that the Wnt3a-induced vesicles (2–5 μm in diameter) were greatly decreased or absent in methionine-depleted conditions (Fig. 1 G and H). Use of either dialyzed or nondialyzed (incomplete methionine depletion) FCS produced similar results; nondialyzed FCS was used in the experiments shown in Fig. 1. We next examined the temporal kinetics of vesicle formation and found that pretreatment for 30 min in low-methionine medium was sufficient to decrease Wnt-induced vesicles (Fig. 1I). This rapid timescale is consistent with a direct sensing mechanism rather than an indirect effect on de novo protein synthesis (Fig. 1I).
Canonical Wnt signaling requires the sequestration of GSK3 in late endosomes/MVBs (5, 46). Therefore, we next examined whether methionine starvation affected the vesicular translocation of specific Wnt targets using protease protection assays, the gold standard for analyzing membrane-bounded organelles (47). Cells were first treated with digitonin, which permeabilizes the plasma membrane while leaving intracellular vesicular membranes intact, and then with proteinase K that degrades cytosolic proteins not protected inside intracellular membrane-bounded organelles. Wnt3a-treated cells were incubated in medium with or without methionine, subjected to protease protection assays, fixed, and stained with anti-GSK3 antibodies. Following Wnt3a treatment, GSK3 was protected from proteinase K digestion inside vesicular endolysosomal membranes after digitonin permeabilization (Fig. 1 J and K). In the same conditions but in the presence of Triton-X 100, GSK3 was completely digested as shown previously (1). In contrast, methionine starvation dramatically reduced vesicular GSK3 relocalization by Wnt3a (Fig. 1M). These experiments indicate that suitable levels of methionine were required for GSK3 translocation from the cytosol into MVBs.
As GSK3 sequestration is required for canonical Wnt signaling and β-catenin stabilization (1, 5), we next examined whether methionine regulates transcriptional activity of the β-catenin–activated (BAR) Wnt luciferase reporter (48) using HEK293T cells stably transfected with BAR/Renilla (49). Wnt3a-treated cells were incubated in RPMI medium with or without methionine, and luciferase activity was measured. Consistent with the immunofluorescence results above, the loss of GSK3 sequestration in methionine-depleted medium caused a significant decrease of BAR luciferase activity (Fig. 1N). We next examined the nature of this response by reintroducing methionine and found that Wnt signaling was rescued, indicating that the effects of methionine depletion were reversible (Fig. 1N, compare lanes 3 and 4). Together, these results revealed that Wnt signaling and endolysosomal formation are sensitive to the availability of a particular amino acid, methionine.
The Methionine Cycle Regulates PRMT1 Localization and Endocytosis During Wnt Signaling.
The data above were compatible with a link between one-carbon metabolism and Wnt signaling. One-carbon metabolism is required for arginine methylation, a reversible posttranslational modification that occurs almost as frequently as phosphorylation (50). Work from the E.M.D.R. laboratory and others demonstrates that arginine methylation mediates growth factor signaling through PRMT1, a type I enzyme that catalyzes ω-NG,NG-asymmetric dimethylation (26, 28). Derynck and coworkers showed that PRMT1-mediated arginine methylation of cytoplasmic Smad proteins is essential for BMP and TGF-β signaling (51, 52). Similarly, we found that PRMT1 was required for Wnt signaling, catalyzing arginine methylation of cytosolic substrates that become sequestered in endolysosomes (1). These data challenge the view that methylation mostly operates in the nucleus. Despite the importance of PRMT1 in cell signaling, relatively little is known about the regulation of cytoplasmic arginine methylation.
Methionine availability dictates SAM levels, which in turn directly impacts PRMT activity (53). The methyltransferase inhibitor adenosine-2′,3′-dialdehyde (Adox), strongly inhibits PRMT1 relocalization into endolysosomes by Wnt (1). To investigate whether the loss of methionine similarly decreased the sequestration of PRMT1, cells were stained with PRMT1 and GSK3 antibodies and subjected to protease protection immunofluorescence analyses. Wnt3a led to the relocalization of PRMT1 into large endolysosomal vesicles that also contained GSK3 (Fig. 2 A and B) (1). Methionine depletion dramatically decreased Wnt-induced PRMT1 sequestration and colocalization with GSK3 in vesicles (Fig. 2 C and D). Quantification of this colocalization by Pearson’s coefficient underscored the significant effect shown in Fig. 2E, indicating that methionine is essential for PRMT1 endolysosomal translocation.
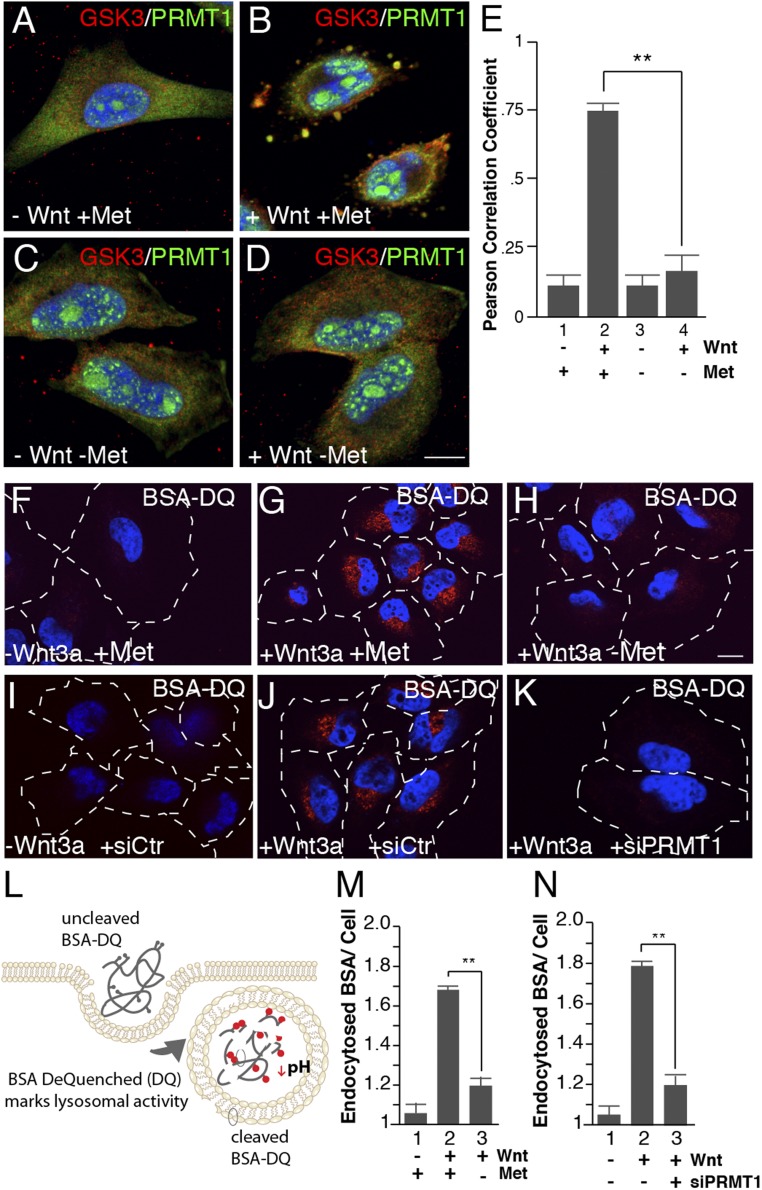
Fig. 2.
Wnt-induced endocytosis and lysosomal activity require methionine and PRMT1. (A–D) Methionine depletion inhibited PRMT1 sequestration into GSK3-containing vesicles by Wnt3a treatment in in situ protease protection analyses (compare B to D). (E) PRMT1 and GSK3 colocalization in vesicles quantified by Pearson’s correlation coefficient using ImageJ. (F–H) Methionine depletion decreased Wnt-induced endolysosomal activity, assessed by the endocytosis and digestion of BSA-DQ. (I–K) PRMT1 depletion with siRNA (siPRMT1) decreased Wnt3a-induced lysosomal activity compared with siScrambled control (siCtr) cells (compare J to K), as assessed by endocytosis of BSA-DQ. (L) Diagram of the BSA-DQ assay, which is endocytosed and fluoresces only after the protein is digested in lysosomes. (M) Lysosomal activity quantification of BSA-DQ fluorescence per cell after 30 min of methionine depletion. (N) Wnt-induced degradation of BSA-DQ in lysosomes requires the PRMT1 enzyme (ImageJ quantification per cell). (Scale bars: 10 μm.) **P < 0.01; n < 4.
Wnt-Induced Endocytosis Requires Methionine.
Canonical Wnt is emerging as a major regulator of the endocytic capacity of the cell (1, 19, 54, 55). Wnt signaling increased endocytosis and lysosomal digestion of BSA dequenched (BSA-DQ), a marker of nonreceptor-mediated endocytosis that fluoresces only after endocytosed BSA is digested in lysosomes (Fig. 2L) (1, 49). We next evaluated whether Wnt-induced BSA-DQ endocytosis and degradation after Wnt treatment was dependent on methionine levels. Wnt3a addition for 30 min increased BSA-DQ endocytosis into lysosomes in methionine-containing medium while methionine depletion significantly diminished BSA-DQ fluorescence (Fig. 2 F–H and M). To study the molecular requirements of Wnt-induced endocytosis, we next incubated cells with siRNA targeting PRMT1 and assessed endolysosomal activity during Wnt signaling. Comparison of cells treated with scrambled control siRNA (siCtr) or siPRMT1 showed greatly reduced endocytosis and lysosomal degradation of BSA (Fig. 2 I–K, quantitated in Fig. 2N), supporting the idea that PRMT1 is critical for Wnt endocytosis. Together, these data indicate that PRMT1 and methionine are required for Wnt-induced endolysosomal activity.
SAM Addition Rescues Endocytosis and Wnt Signaling.
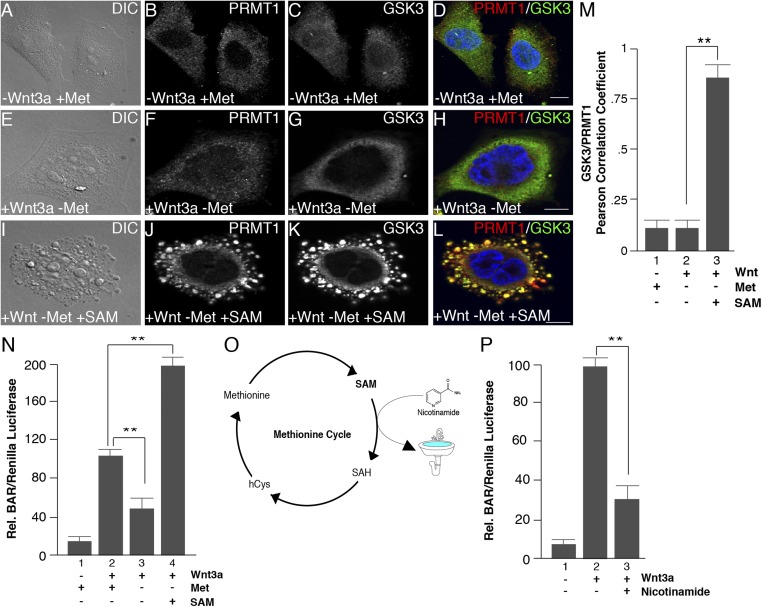
The response to methionine starvation underscores the plasticity of metabolism that allows cells to dynamically adapt to stresses such as nutrient depletion (53). To directly test whether methionine levels regulate Wnt signaling through SAM, the methyl cofactor for PRMT1, we first examined whether SAM was sufficient to rescue the effects of methionine depletion on Wnt signaling. SAM added to the culture medium at high concentrations has been shown to enter cultured cells (36, 56). We incubated HeLa cells in medium with or without methionine and examined the effects of SAM addition on vesicle formation and protein sequestration. The lack of vesicle formation observed during methionine depletion was completely rescued in the presence of 1 mM SAM following Wnt3a treatment (Fig. 3 A, E, and I). Further, the translocation of PRMT1 and GSK3 inside protease-protected vesicles during Wnt activation was also completely rescued when cells were incubated with SAM (Fig. 3 A–L; quantitated in Fig. 3M).
Fig. 3.
SAM addition to the culture medium rescues inhibition of Wnt signaling. (A–L) Methionine depletion decreased Wnt-induced vesicle formation and inhibited the Wnt-induced sequestration of PRMT1 and GSK3 after 30 min of Wnt3a treatment, and this effect was rescued by the addition of 1 mM SAM to the culture medium. (M) Quantification of the rescue of PRMT1 and GSK3 protease protection in vesicles by Pearson’s correlation coefficient. (N) Addition of 1 mM SAM increased Wnt-induced BAR luciferase activity when methionine was depleted. (O) Schematic depiction of the effect of addition of nicotinamide (vitamin B3) to the culture medium. Nicotinamide becomes readily methylated (generating 1-methylnicotinamide) and serves as a sink to store methyl groups and decrease SAM levels (59, 60). (P) Addition of 8 mM nicotinamide to the culture medium inhibited BAR/β-catenin luciferase activity. (Scale bars: 10 μm.) **P < 0.01; n < 3.
We next examined the effect of SAM addition on canonical Wnt signaling using the BAR/β-catenin luciferase reporter assay. Cells were incubated in medium with or without methionine, stimulated with Wnt3a, and supplemented with SAM. Wnt/β-catenin signaling in the absence of methionine was markedly increased by SAM addition to the culture medium (Fig. 3N). This indicated that the effects of methionine depletion on the Wnt pathway were mediated by a deficiency in SAM, the universal methyl donor. This also indicates that the inhibition of Wnt signaling by lowering methionine was not caused indirectly by a generalized defect in protein synthesis, since it could be replaced by a downstream metabolite. Intriguingly, SAM led to a twofold increase in Wnt signaling compared with cells without SAM even in the absence of methionine (Fig. 3N, lane 4). Perhaps the rapid endocytosis induced by Wnt may facilitate SAM incorporation into the cell (57).
Nicotinamide as a Methyl Sink.
Beyond the one-carbon metabolic pathway, additional metabolites can be exploited to alter global cellular methylation by directly affecting SAM concentrations (58). Nicotinamide (NAM, also known as vitamin B3) is the precursor to NAD+ and has been reported to act as a “methyl sink” to decrease SAM levels, draining methyl units from the cytoplasm to produce 1-methyl-NA (1MNA) (Fig. 3O) (59, 60). To examine whether decreasing SAM levels in this way would also affect Wnt signaling, we performed Wnt luciferase assays in permanently transfected BAR reporter HEK293T cells following treatment with nicotinamide. Compared with control cells, nicotinamide significantly decreased Wnt signaling activity (Fig. 3P). Taken together, the data with SAM rescue and the nicotinamide sink strongly support a model whereby SAM levels are intrinsically linked to the Wnt signaling pathway by promoting arginine methylation, sequestration of cytosolic PRMT1 and GSK3 in MVBs (1), and increasing endocytosis and lysosomal degradation of extracellular proteins.
Methotrexate Inhibits Wnt Signaling Through One-Carbon Metabolism.
One-carbon metabolism couples the methionine and folate cycles. Nutrients from both cycles determine the abundance of SAM (Fig. 4A). Folic acid is a vitamin found in green foliage required for mammalian physiology; folate deficiency leads to anemia and birth defects such as neural tube closure defects (61–63). Before entering the one-carbon pathway, folate must be reduced to dihydrofolate (DHF) and tetrahydrofolate (THF) by the NADPH-dependent activity of dihydrofolate reductase (DHFR) (Fig. 4A) (64, 65). Importantly, DHFR can be inhibited pharmacologically by methotrexate, a folic acid analog that represents one of the most effective chemotherapeutic agents and is used in the clinic to treat a variety of diseases including cancer and arthritis (66). We next investigated whether the disruption of the folate cycle could affect Wnt signaling.
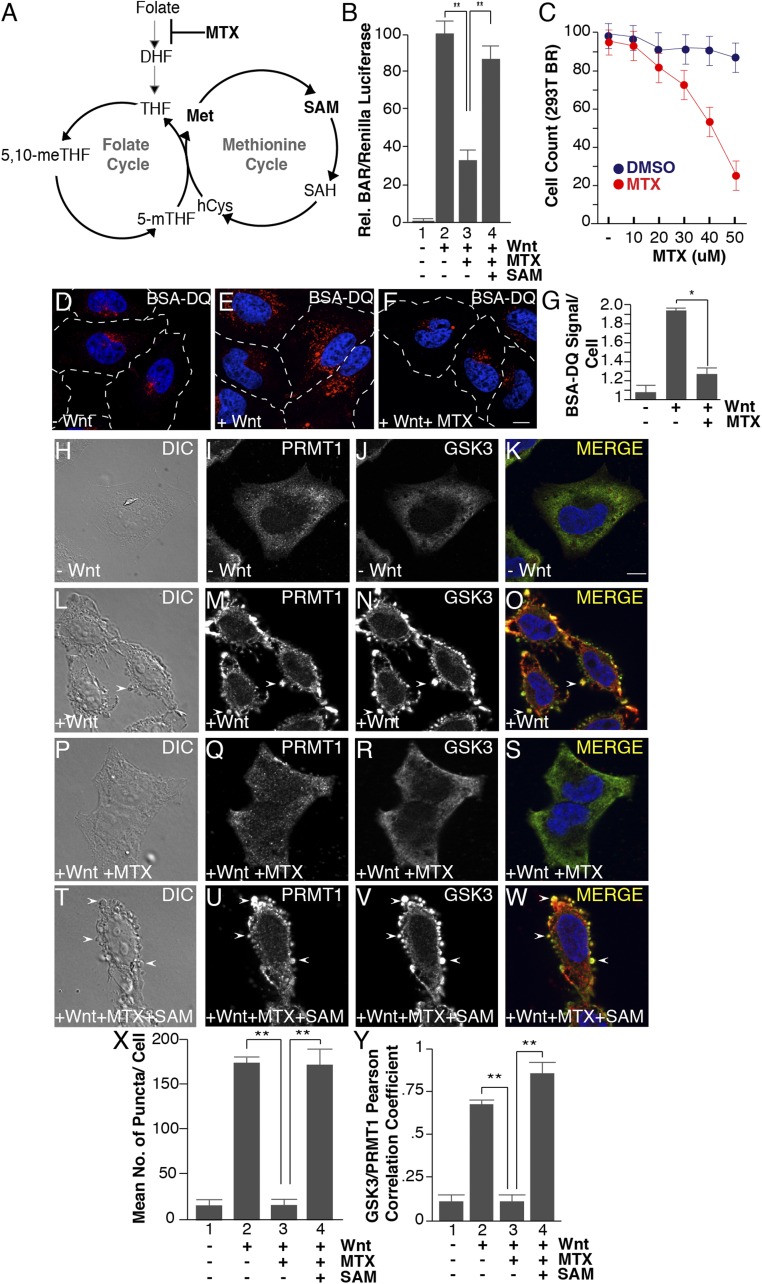
Fig. 4.
Methotrexate inhibits Wnt signaling, as well as Wnt-induced endocytosis, lysosomal activity, and GSK3 and PRMT1 sequestration through the folate cycle. (A) One-carbon metabolism is composed of the methionine and folate cycles; methotrexate (MTX) inhibits DHF from entering the cycle. (B) Methotrexate (20 μM) inhibited Wnt signaling (12 h), as assessed by BAR-Renilla luciferase assays. Note that Wnt inhibition was rescued by the addition of SAM (lane 4). (C) Methotrexate induced significant cellular toxicity only at concentrations above 30 μM in HEK293 cells stably expressing Renilla (12-h treatment with MTX). (D–F) Methotrexate treatment inhibited Wnt-induced lysosomal activity (assessed by the fluorescence of endocytosed BSA-DQ after 30 min of Wnt3a treatment). (G) Quantification of the inhibition of Wnt-induced endolysosomal activity by MTX using ImageJ. (H–W) MTX (20 μM) inhibited the formation of vesicles by Wnt3a (by phase contrast microscopy) and decreased PRMT1 and GSK3 sequestration (by in situ proteinase K protection). Note that the addition of 1 mM SAM rescued the effects of MTX (arrowheads). (X) Quantification of the inhibition by MTX, and rescue by SAM, of Wnt-induced endolysosomal vesicle formation assessed by phase contrast microscopy. (Y) Quantification of GSK3 and PRMT1 colocalization in endolysosomes/MVBs after 30 min of Wnt3a treatment by Pearson’s correlation coefficient. (Scale bars: 10 μm.) *P < 0.05; **P < 0.01; n < 3.
First, to assess canonical Wnt signaling, we performed BAR/β-catenin luciferase assays in cells treated with methotrexate (20 μM) in folate-free medium. Compared with DMSO-treated control cells, methotrexate significantly decreased Wnt-induced BAR activity (Fig. 4B, compare lanes 2 and 3). Importantly, the addition of SAM (1 mM) rescued Wnt signaling in the presence of methotrexate (Fig. 4B, lanes 3 and 4), supporting the hypothesis that SAM is the key metabolite that mediates Wnt signaling downstream of the folate pathway. To examine cell viability, the same HEK293T cells expressing Renilla luciferase from a constitutive promoter (49) were treated with methotrexate (5–60 μM) using the same time frame of the luciferase reporter assay (12 h). Cellular toxicity was significant only above 30 μM (Fig. 4C), consistent with previous findings (67). Second, endocytosis and BSA degradation was investigated in cells preincubated with 20 μM methotrexate (2 h) and subsequently treated with Wnt3a. Methotrexate inhibited Wnt-induced endocytosis and lysosomal degradation of BSA-DQ (Fig. 4 D–F, quantitated in Fig. 4G). Further, in situ protease protection assays indicated that both the formation of vesicles and the sequestration of GSK3 and PRMT1 were significantly inhibited following Wnt treatment in the presence of methotrexate (Fig. 4 H–W; quantified in Fig. 4X). Thus, folate cycle fluctuations have a similar effect as methionine depletion or nicotinamide addition described above. SAM addition to methotrexate-treated cells completely restored endolysosomal formation and vesicular localization of PRMT1 and GSK3 (Fig. 4 T–Y), indicating that the effects of methotrexate are through SAM. These data imply a role for the folate pathway in Wnt-regulated endolysosomal protein catabolism.
Discussion
One-Carbon Metabolism and Canonical Wnt.
We report that one-carbon metabolism can relay nutritional status of the cell to growth factor signaling through the levels of methionine and SAM. Methionine is required for Wnt-induced endolysosomal activity through the generation of SAM, which fosters the sequestration of PRMT1 and GSK3 inside late endosomes/MVBs. As Wnt signaling increases the endocytosis of extracellular proteins and the activity of lysosomes, two elements central for the regeneration of nutrients during cell growth, targeting this mechanism may provide a viable approach to decrease metabolic adaptations in Wnt-driven cancers. Methotrexate was introduced in 1948 by Farber and Diamond (64) and was one of the first chemotherapeutic agents targeting rapidly dividing cells. The regulation of Wnt signaling through one-carbon metabolism uncovered here identifies multiple attractive prospects for potential therapeutic intervention.
We propose that antagonizing Wnt signaling through the disruption of one-carbon metabolism by antifolate therapies like low-dose methotrexate, dietary alterations to reduce methionine levels, or oral administration of SAM-depleting metabolites (methyl sinks) such as nicotinamide (59, 68) could provide novel approaches to target Wnt-driven diseases such as familial adenomatous polyposis (FAP) and colon cancer (69–73). Mutations of the APC and Axin tumor suppressors may increase endocytosis (19, 72), as is the case with Wnt protein treatment (1). Other tumor suppressors that can activate the Wnt pathway are RNF43 or ZNRF3, which are potent transmembrane ubiquitin ligases that remove the Fzd and LRP6 coreceptors from the plasma membrane (while being themselves transcriptionally activated by nuclear β-catenin) (2). These transmembrane proteins are antagonized by the extracellular proteins R-spondins 1–4. The overexpression of R-spondin 2 or 3 through DNA translocations also cause cancer by inhibiting RNF43/ZNRF3 (2). Thus, progression of cancers caused by mutations in APC, Axin1, Axin2, RNF43, ZNRF3, and R-spondin 2 or 3 might be targeted by inhibiting endocytosis with methotrexate or methionine deprivation. The situation may be different for mutations stabilizing β-catenin (which interfere with GSK3 phosphorylation and degradation) that act at a transcriptional level, unless a β-catenin downstream transcriptional program that regulates endocytosis exists.
In intestinal tissue, Wnt signals drive a glycolytic gradient that corresponds with Wnt signaling, being highest at the base of the crypt and lowest at the top of the villi, respectively (2, 74, 75). Similarly, PRMT1 protein expression is also maximal in regions of high Wnt activity in the crypt where it is required for intestinal stem cell maintenance (30, 33, 76). There is evidence in the literature that one mechanism of action for methotrexate (Fig. 4A) is through the depletion of the methylation cofactor SAM (77–79). Thus, low-dose methotrexate could provide an already well-characterized pharmaceutical to target the PRMT1–Wnt axis in benign colon polyps and prevent colon cancer progression (72, 80, 81). Recent studies have identified key off-target effects contributing to cell death during methotrexate treatment such as histidine catabolism (82). As SAM metabolism exhibits flexibility in response to diet (42, 56, 83), dietary alterations to increase consumption of low-methionine foods (vegetable-based nutrients) (43, 84) or supplementation to increase histidine and nicotinamide (vitamin B3) (68) could aid in the efficacy of methotrexate treatment and reduce methylation in Wnt-driven disease. Folic acid, which in the United States is added as a supplement to many foods (such as bread) to prevent neural tube defects (85) may have a counterbalancing effect in the diet.
Arginine Methylation as a Metabolic Sensor.
Our study builds on the recent discovery that dietary methionine has the capacity to alter physiology through cellular methylation (56, 58, 59, 61, 62). We found that methionine starvation or nicotinamide treatment, which act as a cellular sink for methyl groups to decrease SAM (59), reduced Wnt signaling in cultured cells. As phospholipids also store methyl groups to decrease SAM (58, 84), future work to identify other methyl-sink metabolites could offer specialized therapeutic intervention of one-carbon metabolism. The possibility that endolysosomes could operate as a sink for methyl groups to decrease cytoplasmic SAM has never been investigated. This tantalizing concept should warrant future investigations based on two findings. First, we recently reported that endolysosomes dramatically accumulate methyl arginines in proteins sequestered by Wnt treatment (1). Second, SAM is present in lysosomes, which also contain unusually high concentrations of arginine (86–88). Future work evaluating if MVBs provide a repository for methylation could offer unexpected insights in the regulation of one-carbon metabolism.
Materials and Methods
Antibodies and Reagents.
Antibodies were obtained from the following sources: mouse monoclonal antibody against PRMT1 (sc-59648, 1:500; Santa Cruz Biotechnology) and antibody against GSK3 (610201, 1:500; BD Biosciences). BSA-DQ (D-12051, Thermo Fisher Scientific) was used to monitor lysosomes and fluid-phase endocytosis (49). Secondary antibodies Alexa 488 and 568 conjugated were purchased from Invitrogen. Cell culture reagents included DMEM, RPMI 1640 (Sigma-Aldrich, 11875093), RPMI 1640 methionine-free (Sigma-Aldrich, 1451701), RPMI 1640 folate-free (Sigma-Aldrich, 27016021), and dialyzed FBS (Invitrogen, 26400044). Folate and methionine were purchased from Sigma-Aldrich. The BioT transfection reagent (Bioland Scientific) was used for siRNA knockdown and overexpression in cultured cells. Reagents for protease protection assays were digitonin (Abcam), proteinase K (Invitrogen), and Triton X-100 (Sigma-Aldrich). Mounting medium for coverslips on slides was from Life Technologies, P36931. Chemical reagents used in this study included SAM (Cayman Chemical, 13956), methotrexate (Sigma-Aldrich, A6770), cycloheximide (Sigma-Aldrich, C7698), and nicotinamide (Sigma-Aldrich, N7559). Wnt3a was purchased from PeproTech (315-20).
Methionine Depletion and Nicotinamide Experiments.
Briefly, cells were grown in DMEM or RPMI 1640 supplemented with 10% FCS, glutamine, and 100 units/mL penicillin. For immunofluorescence methionine depletion experiments, HeLa cells were plated on coverslips at a density of 60% and incubated in either normal RPMI or methionine-free RPMI for the indicated times, with or without 100 ng/mL Wnt3a for 30 min. For luciferase assays, HEK293T cells stably transfected with BAR/Renilla DNA (49) were plated onto 12 wells and methionine depletion was performed over a 12-h time course, with or without Wnt3a (100 ng/mL) stimulation. Methionine depletion was performed with methionine-free RPMI 1640 supplemented with 10% of either dialyzed or nondialyzed FCS; both gave similar results. Nicotinamide has been previously reported to reduce SAM levels (59). Luciferase assays were performed in cells treated with nicotinamide (8 mM) with or without Wnt3a for 12 h.
Methotrexate Experiments.
Folate cycle inhibition with methotrexate (20 μM) was performed in HeLa cells for 1 h in folate-free RPMI medium 1640, before adding Wnt3a and assessing by immunofluorescence analyses. Cells displayed normal morphology. BAR-Renilla HEK293T cells were used to measure Wnt/β-catenin activity in the presence of 20 μM methotrexate after preincubation for 2 h in folate-free RPMI medium 1640 followed by overnight treatment with Wnt3a for 12 h. Cellular toxicity was assessed at methotrexate concentrations ranging from 5 μM to 60 μM.
Rescue Experiments.
Methionine depletion and methotrexate treatments were rescued by adding 1 mM SAM. For immunofluorescence assays, plated cells were incubated with methotrexate or in methionine-free medium for 2 h. The addition of 1 mM SAM to plated cells was performed for 30 min before Wnt3a stimulation for 30 min. For luciferase assays, cells were preincubated with methotrexate or methionine-free medium for 2 h before the addition of 1 mM SAM, and subsequently treated with Wnt3a protein (12 h). For rescue experiments with methionine, cells were incubated in methionine-depleted RPMI medium for 12 h and subsequently in standard RPMI 1640 for 12 h.
Cell Lines and Tissue Culture.
HeLa and HEK293T were cultured in DMEM or RPMI 1640 supplemented with 10% FCS (GIBCO), 1% penicillin, and 1% glutamine. Cells were maintained at 37 °C and 5% CO2.
Plasmids.
Plasmids used in this study were BAR-luciferase and Renilla (under the control of the elf1a promoter) reporters, a generous gift from Randall Moon, University of Washington, Seattle. Human PRMT1 siPRMT1 oligos (1) (sc-41069; Santa Cruz Biotechnology) targeted three regions with the following sequences: (i) 5′-gcaactccatgtttcataa-3′; (ii) 5′-gttccagtatctctgatta-3′; and (iii) 5′-cgacatgtcttgcatcaaa-3′. The negative control was Scrambled siRNA (D-001810, Thermo Fisher Scientific).
Wnt Treatments.
Recombinant murine Wnt3a protein (PeproTech, 315-20) was used for Wnt signaling studies (100 ng/mL). For protease protection studies, cells processed after 30 min of Wnt3a treatment at 37 °C. For luciferase assays, Wnt3a was added for 12 h.
Protease Protection and Endocytosis Assays.
In situ protease protection assays were performed to determine whether proteins were contained within membrane-bounded organelles as described previously (1). Briefly, cells were incubated with Wnt3a for 30 min, placed on ice, washed twice with PBS, incubated with 6.5 μg/mL digitonin in PBS for 15 min on ice, washed twice with ice-cold PBS to remove digitonin, and subsequently incubated with proteinase K (1 μg/mL) for 10 min (1). PFA fixation stopped the reaction and immunofluorescence preparation was performed as described below. To measure endocytosis and lysosomal activity following Wnt activation, we used BSA-DQ. Cells were plated onto 12-well dishes (60% confluency) and incubated with 5 μg/mL of BSA-DQ in the presence or absence of Wnt3a. Endocytosis was measured as the intensity of fluorescence from lysosome-digested BSA-DQ. For immunofluorescence analyses, cells plated on 12-well dishes were incubated with BSA-DQ for 30 min in the presence or absence of Wnt3a. Cells were subsequently washed with PBS and subjected to standard immunofluorescence processing which included fixation with 4% PFA for 15 min at room temperature in the dark, washing with PBS, and mounting with ProLong Gold antifade reagent with DAPI (Life Technologies).
Immunofluorescence Assays.
Immunofluorescence analyses were performed as described previously (1). In short, cells were grown in 10-cm dishes and replated onto 12-well dishes containing glass coverslips. Cells were allowed to attach to coverslips for 24 h before Wnt treatment. Cells were washed twice with PBS to remove medium, fixed in 4% (wt/vol) PFA (P6148, Sigma) in PBS for 20 min, and permeabilized with 0.2% (vol/vol) Triton X-100 in PBS for 10 min on ice. Next, coverslips were washed with PBS, blocked for 1 h in BSA blocking buffer composed of 5% (vol/vol) goat serum plus 0.5% (wt/vol) BSA at room temperature, and incubated with primary antibodies in blocking buffer overnight at 4 °C (1:100–1:1,000, diluted in blocking buffer). Finally, cells were washed three times with PBS, incubated with secondary antibodies produced in donkey for 45 min (1:1,000, diluted in blocking buffer), and mounted onto glass slides in ProLong Gold antifade reagent with DAPI (Life Technologies) to stain cell nuclei. Images were acquired on a Zeiss Imager Z.1 microscope with Apotome using a 63× oil immersion objective. Zeiss and ImageJ (NIH) imaging software were used for image analyses. The excitation filters used to capture the images were 488 nm, 568 nm, and 405 nm (DAPI) using Alexa 568- and Alexa 488-conjugated secondary antibodies.
Statistical Analyses and Image Quantification.
For the statistical analyses shown here, two-tailed t tests were used for two-sided comparisons, and *P < 0.05, **P < 0.01, and ***P < 0.001 were considered to be statistically significant. To quantitate the increased size in vesicles formed during Wnt signaling, light microscopy [differential interference contrast microscopy (DIC)] images were used to measure the size (diameter) of vesicles in different conditions using Zeiss (Zen) imaging software. The number of vesicles formed was quantified from the DIC images using computer-assisted particle analysis in ImageJ software, as described previously (1). Individual channels were first given thresholds using MaxEntropy, available in the FIJI ImageJ software package. Next, individual vesicles were separated using the binary watershed function and vesicles were counted using the analyze particles function at particle size 0.2–7 μm, circularity −1. More than 25 cells were counted per experiment, numbered, and outlined copied to a spreadsheet. Kinetics of vesicle formation were quantified in methionine-free medium (incubation times between 5 and 240 min) and in methotrexate-treated cells (1 h) using the DIC images from varying conditions. PRMT1 and GSK3 are cytoplasmic proteins that become colocalized following sequestration into vesicles during Wnt signaling (1). As such, Pearson’s correlation coefficients were calculated using ImageJ software to assess the degree of colocalization between two different channels (n > 20 cells per condition). For quantitatively analyzing BSA-DQ endocytosis, fluorescence was quantified in control and Wnt3a-treated cells using ImageJ software analyses (n > 20 cells per condition). Briefly, this includes normalizing fluorescence in images and measuring fluorescence in individual cells. Results from three or more independent experiments were presented as the mean ± SEM.
Detailed protocols for measuring endocytosis, lysosomal activity, methionine depletion, luciferase reporter assays, and media compositions are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Randy Moon (University of Washington) for reporter genes and Richard L. Watson (Cedar Sinai), Duane G. Albrecht (University of Texas, Austin), Douglas Geissert, Samantha Rundell, Alyssa Dsouza, Yi Ding, Gabriele Colozza, and Nydia Tejeda-Munoz [University of California, Los Angeles (UCLA)] for comments on the manuscript. L.V.A. is supported by an NIH postdoctoral fellowship (NIH F32 GM123622) and M.H.B. is funded by a UCLA Clinical and Translational Institute undergraduate fellowship (CTSI UL1TR000124). This work was supported by the Norman Sprague Endowment in Molecular Oncology and the Howard Hughes Medical Institute, of which E.M.D.R. is an investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820161116/-/DCSupplemental.
References
- 1.Albrecht LV, Ploper D, Tejeda-Muñoz N, De Robertis EM. Arginine methylation is required for canonical Wnt signaling and endolysosomal trafficking. Proc Natl Acad Sci USA. 2018;115:E5317–E5325. doi: 10.1073/pnas.1804091115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nusse R, Clevers H. Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Galluzzi L, Spranger S, Fuchs E, López-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2018;29:44–65. doi: 10.1016/j.tcb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acebron SP, Karaulanov E, Berger BS, Huang Y-L, Niehrs C. Mitotic Wnt signaling promotes protein stabilization and regulates cell size. Mol Cell. 2014;54:663–674. doi: 10.1016/j.molcel.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Taelman VF, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, et al. Bighead is a Wnt antagonist secreted by the Xenopus Spemann organizer that promotes Lrp6 endocytosis. Proc Natl Acad Sci USA. 2018;115:E9135–E9144. doi: 10.1073/pnas.1812117115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 9.Bilic J, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 10.Niehrs C, Acebron SP. Wnt signaling: Multivesicular bodies hold GSK3 captive. Cell. 2010;143:1044–1046. doi: 10.1016/j.cell.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Piao S, et al. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One. 2008;3:e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Vick P, Hedtke J, Ploper D, De Robertis EM. Wnt signaling translocates Lys48-linked polyubiquitinated proteins to the lysosomal pathway. Cell Rep. 2015;11:1151–1159. doi: 10.1016/j.celrep.2015.04.048. [DOI] [PubMed] [Google Scholar]
- 13.Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell. 2015;163:1225–1236. doi: 10.1016/j.cell.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Davidson G, et al. Cell cycle control of Wnt receptor activation. Dev Cell. 2009;17:788–799. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Kim N-G, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaidanovich-Beilin O, Woodgett JR. GSK-3: Functional insights from cell biology and animal models. Front Mol Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redelman-Sidi G, et al. The canonical Wnt pathway drives macropinocytosis in cancer. Cancer Res. 2018;78:4658–4670. doi: 10.1158/0008-5472.CAN-17-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palm W, Thompson CB. Nutrient acquisition strategies of mammalian cells. Nature. 2017;546:234–242. doi: 10.1038/nature22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson SM, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23:235–241. doi: 10.1038/nm.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamholz A, Phillips R, Milo R. The quantified cell. Mol Biol Cell. 2014;25:3497–3500. doi: 10.1091/mbc.E14-09-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Z, Chen T, Hébert J, Li E, Richard S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol. 2009;29:2982–2996. doi: 10.1128/MCB.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 27.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 28.Bedford MTM, Clarke SGS. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda H, Shi Y-B. An essential and evolutionarily conserved role of protein arginine methyltransferase 1 for adult intestinal stem cells during postembryonic development. Stem Cells. 2010;28:2073–2083. doi: 10.1002/stem.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda H, Paul BD, Choi CY, Hasebe T, Shi Y-B. Novel functions of protein arginine methyltransferase 1 in thyroid hormone receptor-mediated transcription and in the regulation of metamorphic rate in Xenopus laevis. Mol Cell Biol. 2009;29:745–757. doi: 10.1128/MCB.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto K, Matsuura K, Hu-Wang E, Lu R, Shi Y-B. Thyroid hormone activates protein arginine methyltransferase 1 expression by directly inducing c-Myc transcription during Xenopus intestinal stem cell development. J Biol Chem. 2012;287:10039–10050. doi: 10.1074/jbc.M111.335661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathioudaki K, et al. The PRMT1 gene expression pattern in colon cancer. Br J Cancer. 2008;99:2094–2099. doi: 10.1038/sj.bjc.6604807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu X, et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358:813–818. doi: 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai Z, Mentch SJ, Gao X, Nichenametla SN, Locasale JW. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat Commun. 2018;9:1955. doi: 10.1038/s41467-018-04426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locasale JW. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shlomi T, Fan J, Tang B, Kruger WD, Rabinowitz JD. Quantitation of cellular metabolic fluxes of methionine. Anal Chem. 2014;86:1583–1591. doi: 10.1021/ac4032093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mentch SJ, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: Understanding the specificity. Ann N Y Acad Sci. 2016;1363:91–98. doi: 10.1111/nyas.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finkelstein JD. Inborn errors of sulfur-containing amino acid metabolism. J Nutr. 2006;136(Suppl 6):1750S–1754S. doi: 10.1093/jn/136.6.1750S. [DOI] [PubMed] [Google Scholar]
- 46.Vinyoles M, et al. Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Mol Cell. 2014;53:444–457. doi: 10.1016/j.molcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
- 49.Ploper D, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larsen SC, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 51.Xu J, et al. Arginine methylation initiates BMP-induced Smad signaling. Mol Cell. 2013;51:5–19. doi: 10.1016/j.molcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsuno Y, et al. Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J Biol Chem. 2018;293:13059–13072. doi: 10.1074/jbc.RA118.002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mavrakis KJ, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 54.Pellón-Cárdenas O, Schweitzer J, D’Souza-Schorey C. Endocytic trafficking and Wnt/β-catenin signaling. Curr Drug Targets. 2011;12:1216–1222. doi: 10.2174/138945011795906552. [DOI] [PubMed] [Google Scholar]
- 55.Blitzer JT, Nusse R. A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clarke SG. 16 inhibition of mammalian protein methyltransferases by 5′-methylthioadenosine (MTA): A mechanism of action of dietary same? Enzymes. 2006;24:467–493. doi: 10.1016/S1874-6047(06)80018-1. [DOI] [PubMed] [Google Scholar]
- 58.Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. A metabolic function for phospholipid and histone methylation. Mol Cell. 2017;66:180–193.e8. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shlomi T, Rabinowitz JD. Metabolism: Cancer mistunes methylation. Nat Chem Biol. 2013;9:293–294. doi: 10.1038/nchembio.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia BA, Luka Z, Loukachevitch LV, Bhanu NV, Wagner C. Folate deficiency affects histone methylation. Med Hypotheses. 2016;88:63–67. doi: 10.1016/j.mehy.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadhu MJ, et al. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 2013;195:831–844. doi: 10.1534/genetics.113.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–257. doi: 10.1083/jcb.201604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 65.Hitchings GH, Burchall JJ. Inhibition of folate biosynthesis and function as a basis for chemotherapy. Adv Enzymol Relat Areas Mol Biol. 1965;27:417–468. doi: 10.1002/9780470122723.ch9. [DOI] [PubMed] [Google Scholar]
- 66.Howard SC, McCormick J, Pui C-H, Buddington RK, Harvey RD. Preventing and managing toxicities of high-dose methotrexate. Oncologist. 2016;21:1471–1482. doi: 10.1634/theoncologist.2015-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie L, et al. Methotrexate induces DNA damage and inhibits homologous recombination repair in choriocarcinoma cells. OncoTargets Ther. 2016;9:7115–7122. doi: 10.2147/OTT.S116387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pissios P. Nicotinamide N-methyltransferase: More than a vitamin B3 clearance enzyme. Trends Endocrinol Metab. 2017;28:340–353. doi: 10.1016/j.tem.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 70.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 71.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 72.Saito-Diaz K, et al. APC inhibits ligand-independent Wnt signaling by the clathrin endocytic pathway. Dev Cell. 2018;44:566–581.e8. doi: 10.1016/j.devcel.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pincus T. Guidelines for monitoring of methotrexate therapy: “Evidence-based medicine” outside of clinical trials. Arthritis Rheum. 2003;48:2706–2709. doi: 10.1002/art.11276. [DOI] [PubMed] [Google Scholar]
- 74.Pate KT, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33:1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Esen E, et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013;17:745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Papadokostopoulou A, et al. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer Res. 2009;29:1361–1366. [PubMed] [Google Scholar]
- 77.Clarke S, Vogel JP, Deschenes RJ, Stock J. Posttranslational modification of the Ha-ras oncogene protein: Evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci USA. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hilton MA, Hoffman JL, Sparks MK. Effect of methotrexate with 5-methyltetrahydrofolate rescue and dietary homocystine on survival of leukemic mice and on concentrations of liver adenosylamino acids. Cancer Res. 1983;43:5210–5216. [PubMed] [Google Scholar]
- 79.Kishi T, Tanaka Y, Ueda K. Evidence for hypomethylation in two children with acute lymphoblastic leukemia and leukoencephalopathy. Cancer. 2000;89:925–931. doi: 10.1002/1097-0142(20000815)89:4<925::aid-cncr28>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 80.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 81.Leibovitz A, et al. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 82.Kanarek N, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez-Chantar ML, et al. Importance of a deficiency in S-adenosyl-L-methionine synthesis in the pathogenesis of liver injury. Am J Clin Nutr. 2002;76:1177S–1182S. doi: 10.1093/ajcn/76/5.1177S. [DOI] [PubMed] [Google Scholar]
- 84.Stead LM, Brosnan JT, Brosnan ME, Vance DE, Jacobs RL. Is it time to reevaluate methyl balance in humans? Am J Clin Nutr. 2006;83:5–10. doi: 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- 85.Copp AJ, et al. Spina bifida. Nat Rev Dis Primers. 2015;1:15007. doi: 10.1038/nrdp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abu-Remaileh M, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536:229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.