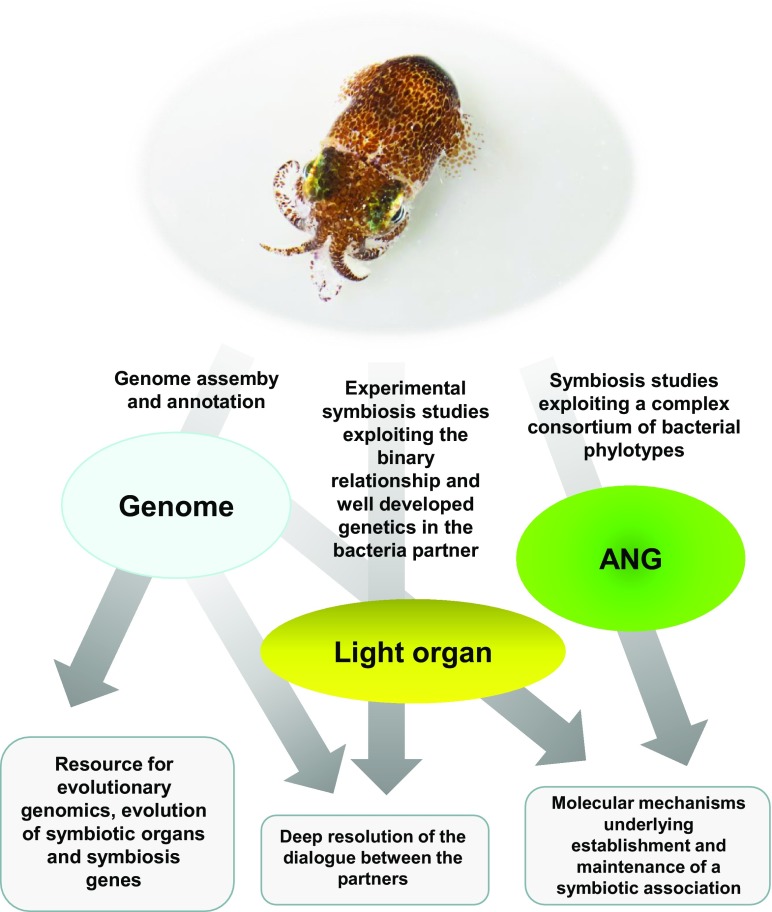
Animals are living in a microbial world. Bacteria, Archaea, and often eukaryotic microorganisms are ubiquitous and intimate associates of all multicellular organisms (1). Many of these microbes improve the fitness of their hosts by affecting multiple traits, including growth rates, immune function, nutrient allocation, and behavior (2). Squids are no exception in this regard. A particularly well-studied species is the bobtail squid Euprymna scolopes, which lives in a symbiotic relationship with light-producing Vibrio fischeri (3). Soon after hatching, the squid secretes mucous from a superficial epithelial field of cells. V. fischeri is selected in this mucus matrix as the sole symbiont, against the background of other environmental bacteria, before it moves into the light organ (LO) where it resides throughout the life of the host. The bacteria provide luminescence as a camouflage mechanism for their night-active hosts (4). The association between E. scolopes and V. fischeri offers a rich set of opportunities to study many aspects of symbiosis, from ecology and evolutionary biology to the molecular mechanisms underlying establishment and maintenance of a symbiotic association (5–7) (Fig. 1) to the role of symbionts in driving host circadian rhythms (8). In addition to the LO, the bobtail squid uses, like some other cephalopods, an accessory nidamental gland (ANG), a female-specific organ associated with reproduction, for hosting bacterial symbionts. In contrast to the LO, which is colonized by multiple strains of a single bacteria species (V. fischeri), a more complex consortium of bacterial phylotypes colonizes the ANG, presumably to protect the eggs (9, 10).
Fig. 1.
The Hawaiian bobtail squid E. scolopes provides insight into the evolutionary and genetic architecture of symbiosis. The genome of E. scolopes, an exceptionally well-studied model organism, is a useful resource to explore the genetic principles and evolutionary forces that shape symbiosis. Image courtesy of Elizabeth Ellenwood (photographer).
Nothing is known about the tools used by the bobtail squid to make up these symbiotic organs, which are present in certain species of cephalopods but absent in others such as the octopus. So, how do the organs, over the course of evolution, become a host for microbial residents? With the publication of the scientifically beautiful work of Belcaid et al. (11) in PNAS, that question became answerable.
Belcaid et al. (11) developed a straightforward method to annotate and analyze the genome of E. scolopes. A combination of shotgun and long-range linkage sequencing resulted in a genome assembly of 5.1 Gigabases (Gb) and 29,259 expressed protein-coding genes. Genome analysis was guided by the comparison with the genome of the California two-spot octopus, Octopus bimaculoides (12). The genome of E. scolopes is significantly larger than the 2.7-Gb O. bimaculoides genome. The difference appears to be due to an abundance of long interspersed nuclear elements in the E. scolopes genome. With regard to the overall genomic architecture and the presence of gene families, both genomes are remarkably similar. Notably, and in contrast to the reported Octopus genome (12), the E. scolopes genome contains a partial hox cluster with large inter hox-gene separations known from many other animals. The finding of numerous local gene linkages that are disrupted in both the E. scolopes and the O. bimaculoides genomes hints to a large genomic reorganization in the cephalopod ancestor.
Using elegant molecular techniques and biostatistics, Belcaid et al. (11) show some of the uses to which a genome can be put. To investigate the molecular architecture of E. scolopes’s symbiotic organs—the LO and the ANG—they used multiple tissue-specific transcriptomes and searched the E. scolopes genome for all paralogous pairs of genes in which one had tissue-specific expression and the second did not. By this approach, the authors could identify candidate genes that evolved in the symbiotic organs after a gene-duplication event. Calculating synonymous substitutions between each gene pair allowed them to propose (11) that the symbiotic organs in the bobtail squid are a relatively recent evolutionary innovation. Because genes exclusively expressed in the symbiotic organs were found in tandem clusters of paralogs on single scaffolds, tandem duplications appear to be the main driver of the bobtail squid’s body innovations.
A closer look at the data revealed that the genetic signature specific for the LO includes S- and omega-crystallin genes as well as a reflectin gene cluster. Since both crystallins and reflectins are typical features of the cephalopod (and the vertebrate) eye, the molecular architecture of the LO appears remarkably similar to the evolutionary conserved eye. The second symbiotic organ in E. scolopes, which is densely populated by bacteria, is the female-specific ANG. The genetic signature of the ANG differs greatly from the LO. In addition to a high content of repetitive elements, genes specifically expressed in the ANG include a high proportion (>35%) of taxonomically restricted (orphan)—and therefore E. scolopes-specific—genes, pointing to the ANG as the more evolutionary-derived symbiotic organ. Equally interesting is the finding that both symbiotic organs are characterized by an abundance of immune-related genes. Since both organs are densely colonized by beneficial microbes, the immune genes in E. scolopes’s symbiotic organs—similar to the components of innate immunity in other invertebrates (13)—may be critically involved in controlling the resident beneficial microbes, rather than in defending against invasive pathogens.
Now that Belcaid et al. (11) have given a first impression of how to interpret the bobtail squid’s genome, its further utilization can begin. At the top of the wish list is the development of methods to bring genetics into the squid host. The ability to produce transgenic Euprymna squids would allow researchers to exploit the newly available genomic resources for functional studies and to bring this already well-established symbiotic model to a new level. With a genome sequence available, there are ∼29,000 E. scolopes genes potential targets for disruption to understand their function. Zinc-finger, transcription activator-like effector nucleases, and/or CRISPR/Cas9 approaches for efficient gene disruption are waiting to be tested in Euprymna.
The availability of the E. scolopes genome sequence marks the beginning of a new era in the use of this model animal.
There is also a need for finding out the function of the many E. scolopes-specific genes expressed in the ANG. Detailed analysis of the genes differentially expressed in the ANG uncovered that >35% of the transcripts have no homologs in other animal taxa (11) and thus are taxon-restricted genes (TRGs). Such orphans are a widely underestimated class of genes. They have contributed to the evolution of unique tissues and organs in a number of animals (14). Uncovering their function is a tedious task because existing data banks will be of no use. Because the ANG is thought to protect female gametes, it seems obvious that a number of TRGs may be immune-related genes involved in the communication between the host and its microbial partners. Machine learning-based approaches (15) may help to shed light on the function of these puzzling gene sequences by, for example, predicting antimicrobial peptide-related properties such as membrane destabilizing activity. The identification of a large number of orphan genes in a cephalopod-specific organ supports the idea that they are key drivers of morphological specification (16). Understanding their role, therefore, seems essential for understanding the present ecology and way of living of Euprymna.
Another topic that deserves further consideration is the intriguing observation of the expression of reflectins and crystallins in the LO. Do the reflectin proteins in the LO work as they do in the eyes of other cephalopods (17)? Are the crystallins in the LO required for light transparency and refraction of the bioluminescence emitted by V. fisheri? Is the expression of crystallin genes in this evolutionarily novel organ controlled by evolutionary conserved transcription factors, including Pax6 and retinoic acid signaling (18), and how is this linked to the bioluminescent bacteria? Finding answers to these questions would be interesting because it would add to our understanding of the phylogenetic relationship between the eye and the LO.
In sum, the availability of the E. scolopes genome sequence marks the beginning of a new era in the use of this model animal. Analysis of the genome sequence has already led to a number of interesting findings. In the future, it is likely to hold the key to many more opportunities for understanding the evolutionary history, ecology, and developmental biology of Euprymna. As if to look through a strong magnifying glass, the Euprymna genome promises to show how, in the course of evolution, a cephalopod produces novel organs to accommodate a rich community of residential microbes. For developmental biologists, the Euprymna genome is another reminder that to understand the evolution of animal genomes, we have to bear in mind that genomes are evolving in a microbial world.
Acknowledgments
I thank the Wissenschaftskolleg (Institute of Advanced Studies) in Berlin for a sabbatical leave. Research in the T.C.G.B. laboratory is supported by the DFG’s Collaborative Research Centre (CRC1182; “Origin and Function of Metaorganisms”) and the Canadian Institute for Advanced Research.
Footnotes
The author declares no conflict of interest.
See companion article on page 3030.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas AE. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6:a016113. doi: 10.1101/cshperspect.a016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 4.McFall-Ngai M. Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bongrand C, Ruby EG. Achieving a multi-strain symbiosis: Strain behavior and infection dynamics. ISME J. October 23, 2018 doi: 10.1038/s41396-018-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandel MJ, Dunn AK. Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Front Microbiol. 2016;7:1982. doi: 10.3389/fmicb.2016.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehler S, et al. The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ Microbiol. August 22, 2018 doi: 10.1111/1462-2920.14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath-Heckman EA, et al. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. MBio. 2013;4:e00167–e13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins AJ, et al. Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl Environ Microbiol. 2012;78:4200–4208. doi: 10.1128/AEM.07437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerwin AH, Nyholm SV. Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and developing eggs. Environ Microbiol. 2017;19:1463–1475. doi: 10.1111/1462-2920.13665. [DOI] [PubMed] [Google Scholar]
- 11.Belcaid M, et al. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc Natl Acad Sci USA. 2019;116:3030–3035. doi: 10.1073/pnas.1817322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albertin CB, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524:220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch TCG. Rethinking the role of immunity: Lessons from Hydra. Trends Immunol. 2014;35:495–502. doi: 10.1016/j.it.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 15.Lee EY, Fulan BM, Wong GCL, Ferguson AL. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc Natl Acad Sci USA. 2016;113:13588–13593. doi: 10.1073/pnas.1609893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 17.DeMartini DG, Izumi M, Weaver AT, Pandolfi E, Morse DE. Structures, organization, and function of reflectin proteins in dynamically tunable reflective cells. J Biol Chem. 2015;290:15238–15249. doi: 10.1074/jbc.M115.638254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech Dev. 2014;131:111–126. doi: 10.1016/j.mod.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]