Significance
Mutations accumulate throughout life in every cell of the body. A long-standing question is to what extent these mutations contribute to aging. Age is the single greatest risk factor for cancer, a disease driven by random mutagenesis and selection for malignant phenotypes. We wondered whether cancer cells, being the most heavily mutagenized human cell, are short-lived due to mutagenesis. Single-cell measurements of replicative lifespan are not established for human cells. So, instead, we engineered diploid yeast to have cancer-associated defects that increase mutagenesis and then measured lifespan and mutation accumulation. Yeast cellular lifespan decreased as the total number of mutations increased to levels commonly found in human cancers, suggesting that the longevity of cancer cells may be inherently limited.
Keywords: aging, mutation accumulation, replicative lifespan, polymerase proofreading, mismatch repair
Abstract
Mutations accumulate within somatic cells and have been proposed to contribute to aging. It is unclear what level of mutation burden may be required to consistently reduce cellular lifespan. Human cancers driven by a mutator phenotype represent an intriguing model to test this hypothesis, since they carry the highest mutation burdens of any human cell. However, it remains technically challenging to measure the replicative lifespan of individual mammalian cells. Here, we modeled the consequences of cancer-related mutator phenotypes on lifespan using yeast defective for mismatch repair (MMR) and/or leading strand (Polε) or lagging strand (Polδ) DNA polymerase proofreading. Only haploid mutator cells with significant lifetime mutation accumulation (MA) exhibited shorter lifespans. Diploid strains, derived by mating haploids of various genotypes, carried variable numbers of fixed mutations and a range of mutator phenotypes. Some diploid strains with fewer than two mutations per megabase displayed a 25% decrease in lifespan, suggesting that moderate numbers of random heterozygous mutations can increase mortality rate. As mutation rates and burdens climbed, lifespan steadily eroded. Strong diploid mutator phenotypes produced a form of genetic anticipation with regard to aging, where the longer a lineage persisted, the shorter lived cells became. Using MA lines, we established a relationship between mutation burden and lifespan, as well as population doubling time. Our observations define a threshold of random mutation burden that consistently decreases cellular longevity in diploid yeast cells. Many human cancers carry comparable mutation burdens, suggesting that while cancers appear immortal, individual cancer cells may suffer diminished lifespan due to accrued mutation burden.
Accumulated somatic mutation burden, alongside a host of other factors, has long been proposed as a driver of aging (1–4). Early evidence that somatic mutations accumulate during aging came from observations of increased mutation frequencies in human lymphocytes and mouse models (reviewed in ref. 5). More recently, next-generation sequencing of hematopoietic stem cells (HPSCs), tumors, and organoids derived from human stem cells have confirmed that random age-dependent mutation accumulation (MA) occurs across the genome (6–17). For mutations to contribute to aging, they must impair cellular function individually or through negative epistatic interactions with other mutations, which increase dramatically with mutation burden. It is unclear what level of mutation burden may be required to consistently impact cellular aging and whether any human cells actually approach that value. The highest mutation burdens of any human cell occur in cancer (16). Conceivably, the proliferative nature of cancer may mask an accelerated “aging” phenotype affecting individual cells that compromises their replicative lifespan (RLS). The RLS of individual cancer cells has not been investigated primarily due to technical limitations of current human cell culture methods. Understanding how cellular lifespan is impacted by cancer-associated mutagenesis could lead to novel therapeutic strategies that capitalize on sensitivities resulting from elevated mutation burden.
Budding yeast are ideally suited to model the impact of mutation burden on cellular aging since they share many of the same DNA replication and repair pathways as human cells. Mutation rates can be monitored by fluctuation assays (18). Cost-effective next-generation sequencing methodologies allow whole-genome quantification of mutation burden (19), and simple yeast growth assays reveal the consequences of mutagenesis on overall health at the population level. Most importantly, yeast provide a means of assessing single-cell aging using RLS analysis (20, 21). RLS in budding yeast is defined as the number of daughter cells produced by a mother cell before irreversible cell cycle arrest and is typically quantified through manual or microfluidic dissection of daughter cells away from mother cells (22, 23). Common pathways influence aging in yeast and multicellular eukaryotes (24, 25), including caloric restriction (26), the mechanistic target of rapamycin (27), sirtuins (28), proteasomal function (29), and mRNA translation (30). Genome instability increases during replicative aging in yeast, especially around the rDNA locus, and may contribute to aging (31–34). Point MA, however, does not normally drive aging in WT yeast (35), in keeping with the observation that the descendants of old mother cells do not stably inherit a shorter lifespan (36). By inducing cancer-related mutagenesis in yeast, we can model the impact of accumulated mutation burden on RLS.
Among the most highly mutated human cancers are those that express a “mutator phenotype.” The mutator phenotype hypothesis holds that mutations that lead to genome instability, particularly those that decrease DNA replication and repair fidelity, arise in human cells and contribute to carcinogenesis and cancer evolution (37–40). Mutations that diminish DNA polymerase proofreading and/or mismatch repair (MMR) frequently occur in colorectal and endometrial cancers, as well as in a range of other tumor types (41–46). In model systems, simultaneous defects in these repair pathways elevate mutation rate synergistically to levels 1,000-fold or more above background mutation rates (47). Such extreme mutator phenotypes drive extinction of bacteria and haploid yeast through the inactivation of essential genes (47–50). Diploid yeast cells, which are buffered from this “error-induced extinction” by possessing two copies of every gene, nevertheless exhibit a catastrophic loss in fitness as mutation rates increase beyond a certain threshold (51). Interestingly, “ultramutator” tumors with combined defects in proofreading and MMR rarely surpass ∼300 mutations per megabase, suggesting that extreme mutation burden may also limit growth of human cancer cells (41).
Here, we model the consequences of cancer-related mutator phenotypes on RLS in yeast defective for MMR and/or leading strand (Polε) or lagging strand (Polδ) polymerase proofreading. We show that high rates of active mutagenesis reduce RLS in both haploid and diploid yeast and that combined defects in proofreading and MMR act synergistically to diminish lifespan. We then show that accumulated mutation burden in these strains explains their severely shortened lifespan. To understand how mutation burden impacts cellular aging and population doubling time, apart from an active mutator phenotype, we assessed transiently mutagenized diploid cells. We find that diploid yeast with mutation burdens past a certain threshold routinely exhibit diminished lifespan and an increased doubling time. Our results suggest that comparable levels of random mutation burden accrued in cells during cancer or aging may similarly diminish their longevity and fitness.
Materials and Methods
Media and Growth Conditions.
Yeast strains were cultured as previously described (52). Unless otherwise indicated, all reagents were purchased from Sigma–Aldrich or Fisher Scientific. For standard growth, YPD (1% wt/vol Bacto yeast extract, 2% wt/vol Bacto peptone, 2% wt/vol dextrose) or synthetic complete (SC) [1.7 g/L Difco yeast nitrogen base without amino acids and ammonium sulfate, 37.8 mM ammonium sulfate, 2% wt/vol dextrose, 2 g/L SC amino acid mix (Bufferad)] was used. Amino acid dropout medium was used for prototrophic selection and prepared as described (52). For haploid mutation rates, canavanine resistance (CanR) mutants were selected using SC medium lacking arginine and supplemented with 60 μg/mL l-canavanine. Diploid mutation rates used the above medium with the addition of 100 μg/mL nourseothricin (Research Products International) to maintain counterselection for the CAN1::natMX allele and 1 g/L monosodium glutamate instead of ammonium sulfate as the nitrogen source (51). Selection against Ura+ strains was performed using SC medium supplemented with 1 mg/mL fluoroorotic acid (FOA) (Zymo Research) (53). Sporulation in liquid medium [1% wt/vol potassium acetate, 0.1% wt/vol Bacto yeast extract, 0.05% wt/vol dextrose] was performed as previously described (54).
Yeast Strains.
A complete list of strains (SI Appendix, Table S1) and a list of all primers (SI Appendix, Table S2) used in their construction are provided. Strains AH0401 (CAN1::natMX/can1Δ::HIS3) and AH2801 (POL2/URA3::pol2-4 MSH6/msh6∆::LEU2 CAN1::natMX/can1Δ::HIS3) were previously described (19, 51) and are derived from the diploid BY4743 strain used to construct the systematic deletion collection (55). To construct AH2601, we first deleted one of the two copies of MSH6 in AH0401 to obtain AH0502, using a LEU2 transgene amplified from pRS415 with MSH6GU and MSH6GD primers (msh6Δ::LEU2) [Phusion Polymerase (New England Biolabs); 98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, 54 °C for 30 s, and 72 °C for 90 s]. We then integrated the pol3-01 allele by transformation with a URA3::pol3-01 DNA fragment amplified from pRS406-pol3-01 with POL3U and pldr6 primers, as described (51).
Haploid mutator spore clones (Fig. 1) were outgrown directly from dissected tetrads of AH2601 or AH2801 and genotyped via prototrophic growth and PCR genotyping assays. The pol3-01 genotyping assay has been previously described (50). For the pol2-4 assay, DNA was first PCR-amplified with pol2-4U and pol2-S5 primers (98 °C 1 min, followed by 30 cycles of 98 °C for 10 s, 48 °C for 30 s, and 72 °C for 1 min) and then digested with Alu I. A prominent 824-bp fragment observed in WT cells is cleaved into 447-bp and 377-bp fragments if the DNA carries the pol2-4 mutation.
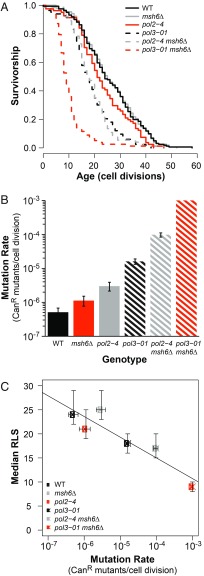
Fig. 1.
Mutation rate negatively correlates with RLS in haploid mutator yeast. (A) RLS of freshly dissected haploid spores. Mutator spores compared with WT using Wilcoxon rank sum tests with Bonferroni-corrected P values using p-adjust in R [WT (cells from both AH2601 and AH2801 backgrounds): median RLS = 24, n = 161; pol3-01: median RLS = 18, n = 74, P < 0.001; pol2-4: median RLS = 21, n = 81, P = 0.43; msh6∆ (cells pooled from both AH2601 and AH2801 backgrounds): median RLS = 25, n = 156, P = 1; pol2-4 msh6∆: n = 83, P < 0.001; pol3-01 msh6∆: n = 78, P < 0.001]. (B) Mutation rates for haploid spores. Colonies from independent spores served as replica cultures for fluctuation analysis using CanR [WT, n = 32 (combined from both genetic backgrounds); pol2-4, n = 14; msh6∆, (combined from both genetic backgrounds) n = 15; pol3-01, n = 22; pol2-4 msh6∆, n = 19]. The pol3-01 msh6∆ spores failed to form colonies; mutation rate was estimated from the synthetic relationship between proofreading and MMR in haploids (23) and confirmed by measurements of pol3-01/pol3-01 msh6Δ/msh6Δ diploids (SI Appendix, Fig. S1). Error bars represent 95% confidence intervals (CI). Mutation rates of viable mutator strains were compared with WT cells using a likelihood ratio test (all strains, P < 0.005). (C) Scatterplot of median RLS as a function of mutation rate (horizontal error bars, 95% CI; vertical error bars, SEM). Linear model adjusted R2 = 0.81, P = 0.009.
Diploid mutator strains described in Fig. 2 were obtained by mating freshly dissected AH2601 or AH2801 haploid spores. Mating occurred within two to four haploid cell divisions, and the resulting zygotes were moved by microdissection to a defined location on the agar plate to form a colony. We genotyped the pol2-4 and pol3-01 alleles, as described above, and assessed the status of MSH6 using a PCR assay with primers (msh6-upstream and msh6-downstream), which yielded distinct products for the MSH6 (3.9-kb) and msh6Δ::LEU2 (2.5-kb) alleles (98 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 2.5 min). Cells from genotyped colonies were subcloned to YPD plates, and these populations were directly harvested to make frozen stocks of each population.
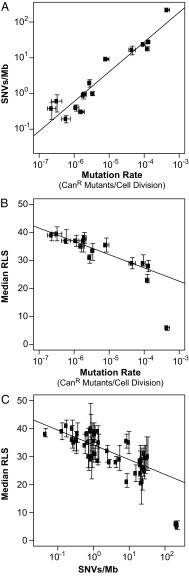
Fig. 2.
Decreased lifespan negatively correlates with increased mutagenesis in diploid yeast. (A) Correlation between mutation rate and mutation burden, expressed as SNVs per megabase (Mb). Horizontal error bars, 95% confidence intervals (CIs). Vertical error bars, SEM. Linear model adjusted R2 = 0.90, P < 0.001. (B) Median RLS as a function of mutation rate. Horizontal error bars, 95% CIs. Vertical error bars, SEM. Linear model adjusted R2 = 0.82, P < 0.001. (C) Median RLS as a function of mutation burden in diploid mutators. Instead of pooling on the basis of mutator genotype, individual mutator strains were plotted. Error bars, SEM. Linear model adjusted R2 = 0.55, P < 0.001. For B and C, linear regression modeling was performed after excluding the outlying datum (pol3-01/pol3-01 msh6Δ/msh6Δ).
To perform the genetic anticipation experiment in Fig. 3, we first dissected tetrads from AH2601 on SC plates lacking leucine and uracil (SC-LEU-URA) and identified URA3::pol3-01 msh6Δ::LEU2 haploid spores by their ability to divide on this selective medium. We mated the double-mutant haploid cells after two to four divisions and manually isolated the resulting pol3-01/pol3-01 msh6Δ/msh6Δ zygotes. To establish the initial lifespan cohort, we took the first diploid daughter cell from each zygote. We moved the initial daughter cells from each of these new mothers to a separate location on the plate to form a colony for genotyping and propagation. To test for a change in RLS with propagation, six of these colonies were patched onto new SC-LEU-URA plates and lifespan cohorts were isolated after an overnight incubation [day 3 (d3) cohorts, Fig. 3; LS_d3_7-10–LS_d3_14-19; Dataset S1]. At that time, cells were patched again to fresh SC-LEU-URA plates for a second set of lifespan measurements (d4 cohorts, Fig. 3; LS_d4_7-10–LS_d4_14-19; Dataset S1).
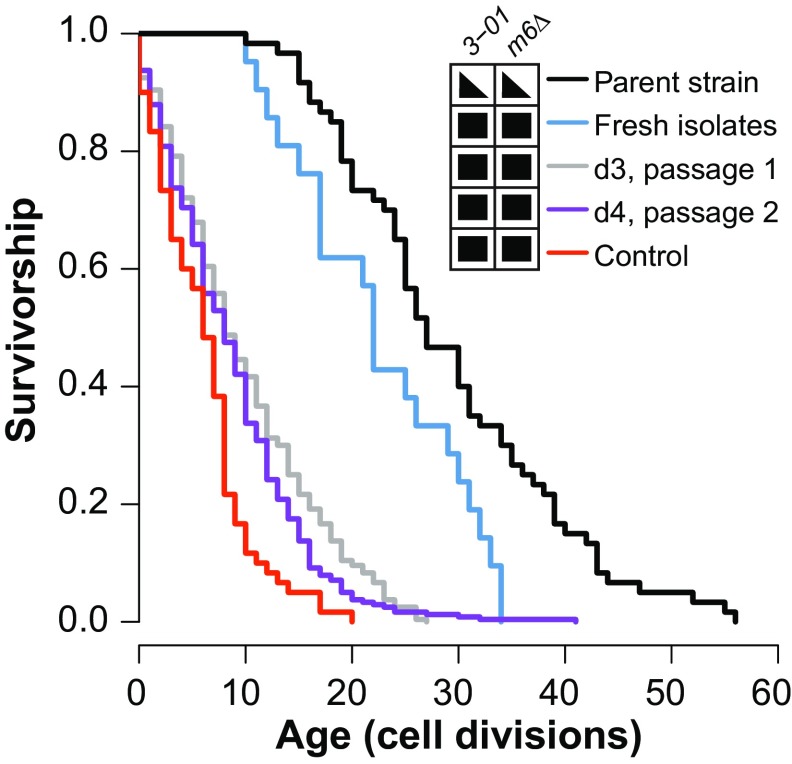
Fig. 3.
Genetic anticipation in the reduction of RLS of strong diploid mutators. The pol3-01/pol3-01 msh6Δ/msh6Δ zygotes were manually isolated from a cross of pol3-01 msh6Δ haploid cells that had undergone three or fewer cell divisions after tetrad dissection. RLSs of the first diploid daughter cells from these zygotes (fresh isolates, blue; median RLS = 22, n = 21) were compared concurrently with previously passaged pol3-01/pol3-01 msh6Δ/msh6Δ cells (control, red; median RLS = 6, n = 60, P < 0.001) and the POL3/pol3-01 MSH6/msh6Δ parent strain (parent strain, black; median RLS = 27, n = 60, P = 0.075). Daughter cells dissected from the fresh isolates were grown into colonies. Six colonies were subcloned twice to obtain two additional RLS cohorts (d3, gray; d4, purple) (combined median RLS = 8, n = 240, P < 0.001). Comparisons used the Wilcoxon rank sum test with a Bonferroni multiple testing correction. Genotype grid: 3-01, pol3-01; m6, msh6; filled triangles, heterozygous; filled squares, homozygous.
Mutagenically stable MA lines (Fig. 4) were derived from previously described POL3/URA3::pol3-01,L612M strains (51), which display an ultramutator phenotype due to the pol3-01,L612M allele. Ura− cells that had spontaneously lost the pol3-01,L612M allele through mitotic recombination were identified by plating frozen stocks onto FOA plates. We confirmed the absence of the mutator allele via pol3-01 PCR genotyping as well as by demonstrating WT mutation rates (Dataset S1).
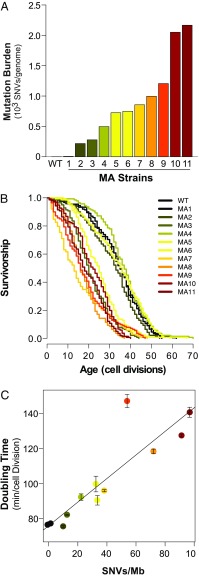
Fig. 4.
MA in the absence of active mutagenesis erodes RLS in diploid yeast. (A) Total mutation burdens [single nucleotide variants (SNVs)/genome] of diploid yeast MA lines transiently mutagenized using the pol3-01,L612M strong mutator allele. (B) Kaplan–Meier plots of survival of MA lines. (C) Peak doubling time (minutes per cell division) of MA lines as a function of mutation burden [SNVs per megabase (Mb)]. Error bars, SEM. Linear model adjusted R2 = 0.75, P < 0.001.
Mutation Rates via Fluctuation Analysis.
Mutation rates were determined via fluctuation analysis using CanR (56). For haploid mutators, individual spore colonies taken directly from the dissection plate were treated as replica colonies and suspended in 200 μL of water. Twenty microliters of cell suspension was used for 10-fold serial dilutions. The remaining undiluted suspensions, as well as 10 μL of each serial dilution, were plated onto canavanine selection plates to determine the number of CanR mutants in each colony. Dilutions were also plated onto SC plates to estimate the total number of cells per colony (Nt) and onto prototrophic selection plates for genotyping. All plates were grown for 2 d at 30 °C before determining colony counts. At least 12 independent spore clones of each genotype were used for mutation rate calculations. For diploid mutator strains, replica colonies were obtained by streaking frozen stocks onto YPD plates, followed by 2 d of incubation at 30 °C. Colonies were suspended in 100 μL of water, and 20 μL of cell suspension was used for serial dilutions. The remaining 80 μL of undiluted cell suspension, as well as 10 μL of all serial dilutions, was plated onto canavanine selection plates. Dilutions were also plated in SC to determine Nt values and on prototrophic selection plates to confirm genotypes. When possible, we counted CanR colonies arising from the undiluted cell suspensions. For stronger mutator strains, CanR mutants were quantified from dilutions. Between six and 12 colonies per genotype were analyzed in each experiment, and two to three experiments were performed for each strain. Only replica colonies with similar Nt values (1 – 5 × 106 cfu per colony) were used for the final mutation rate calculations. To pool mutation rate data by genotype (Fig. 2), we combined mutation counts from replicas with similar Nt values from multiple strains. The average number of mutational events occurring during the formation of the colonies (m) was estimated by maximum likelihood using newton.LD.plating in the R package rSalvador (57). To obtain CanR mutants per cell division, m was then divided by Nt. The 95% confidence intervals were calculated using confint.LD.plating in rSalvador, and mutation rates were compared by a likelihood ratio test using LRD.LD.plating, which yielded P values of the comparisons.
RLS.
A modified RLS protocol was developed based on previously described methods (58, 59), to minimize outgrowth in actively mutating and mutagenized strains. For haploid mutators, tetrads were dissected directly on lifespan plates and spores were used as mother cells. The first few daughter cells were transferred to another location on the lifespan plate to form colonies for genotyping. To determine genotypes, cells were suspended in 50 μL of water and 5 μL was plated on prototrophic selection plates. For diploid mutator and MA lines, frozen stocks were struck for single colonies onto YPD and outgrown for 2 d at 30 °C. Colonies were then patched directly onto lifespan plates, and virgin mother cells were selected from freshly divided cells. Survival curves were constructed, and Wilcoxon rank sum statistical analyses were performed using the RcmdrPlugin.survival package in R commander (60, 61).
Growth Analysis Using a Bioscreen C MBR System.
Doubling times of yeast were measured using a Bioscreen C MBR system (Growth Curves USA) as previously described (62). We obtained inoculums for these experiments in one of two ways. For mutator strains, we first isolated single colonies by dissecting haploid spores or streaking frozen diploid stocks onto YPD. We then suspended the fresh colonies in 50 μL of YPD without further propagation. For MA lines, single colonies were isolated from frozen stocks and then inoculated into 5 mL of YPD and incubated at 30 °C in a roller drum for 12–16 h. Two microliters of either cell suspension or outgrown culture was used as an inoculum into 148 μL of YPD for growth analysis. At least three colonies per strain were analyzed in triplicate for each experiment. Doubling times were calculated from the interval 0.2 ≤ x ≤ 0.5 OD420–580 nm using the online web tool Yeast Outgrowth Data Analyzer (YODA) (63). Two-tailed t tests with unequal variance were performed from the average doubling time of replicate cultures using R. Bonferroni-corrected P values were calculated using p-adjust.
Whole-Genome Sequencing.
To estimate the mutation burden of mother cells at the beginning of the RLS, we recapitulated how the aging cohorts were isolated. Frozen stocks of each strain were struck for single colonies on YPD plates and outgrown for 2 d at 30 °C. Three independent colonies were then patched directly onto fresh plates, and one individual virgin mother cell from each patch was moved to a defined location and allowed to form a colony. We inoculated 5 mL of YPD cultures from these colonies and purified genomic DNA using the ZR Fungal/Bacterial DNA Miniprep Kit (Zymo Research). Genomic DNA (200 ng) samples were sheared in a volume of 60 μL using a Covaris sonicator [peak incident power = 105 (for 96-well format), intensity = 3 (for single sample cuvettes), duty factor = 5%, cycles per burst = 200, 40 s]. After concentrating the DNA with a 0.8× Agencourt AMPure XP bead purification system (Beckman Coulter Life Sciences), we performed one-third volume 3′ end repair reactions using the NEBNext Ultra End Repair/dA-Tailing module (New England Biolabs), followed by ligation to annealed DNA adapter oligos (SI Appendix, Table S2) using the NEBNext Ultra Ligation Module (New England Biolabs). Following another 0.8× AMPure bead purification, the resulting libraries were single- or dual-indexed by quantitative PCR (primers are provided in SI Appendix, Table S2) (program: initial denature, 98 °C for 45 s; cycle denature, 98 °C for 15 s; annealing, 65 °C for 30 s; and elongation, 72 °C for 30 s) with Kapa HiFi Hotstart Ready Mix (KAPA Biosystems). The indexed DNA samples were purified using a double-bead purification procedure in which we first removed fragments larger than ∼1,000 bp by performing a 0.55× AMPure bead purification. We then precipitated fragments larger than ∼500 bp in the resulting supernatant by bringing the total volume of bead suspension added to 0.65×. We quantified the DNA using a Qubit dsDNA HS Assay Kit (Life Technologies) and pooled the samples by mass before sequencing the mixture on either a HiSeq 2500 [reagents: TruSeq Rapid SBS Kit v2 (500 cycles) (FC-402-4023; Illumina), TruSeq Rapid PE Cluster Kit v2 -HS (PE-402-4002; Illumina)] or NextSeq 500 [reagents: Mid-Output Kit V2 (FC-404-2003; Illumina)] system. An established computational pipeline was used to align sequencing reads and identify single nucleotide variants (SNVs) as well as insertion or deletion (indels) mutations within individual strains (19). We used a variant frequency cutoff of 0.22 for SNVs and 0.35 for indels. As part of this pipeline, polymorphisms present in the parental strain (AH0401), as well as recurring mapping artifacts, were identified and excluded from our analyses. We documented and followed the inheritance of strain-specific variants present in AH2601 and AH2801 (Dataset S2). The total number of mutations present in each strain was divided by the total number of bases scored and multiplied by 106 to obtain the number of mutations per megabase. Mutation burden was averaged among isolates of the same mutator genotypes, and SEM was used to express the variance in the data. Structural variants (SVs) were called with LUMPY Express (64), which successfully identified all of the known SVs engineered into these strains (Dataset S2).
Regression Analysis.
For correlation analyses, data from multiple strains of the same genotype were pooled after removing outliers. Outliers were identified in the RLS data by performing ANOVA. When a significant difference was detected by ANOVA (P < 0.05), pairwise t tests were used to identify the outlier. To identify outliers in the mutation rate data, pairwise likelihood ratio tests were performed in rSalvador (LRD.LD.plating). All correlation analyses were performed using R. Linear regression was performed using the lm function in R, and nonlinear regression was performed using the nls function in R. QQ-plots were used to assess normality of correlation analyses. Analysis of residuals and the Grubbs’ test were used to identify and confirm outliers. The R package “outliers” was used to perform the Grubbs’ test. Outliers were removed, and regression analyses were rerun to generate reported analyses.
Results
RLS Negatively Correlates with Increased Mutation Rate in Haploid Mutator Yeast.
To investigate how mutator phenotypes influence cellular aging, we first measured the RLS of haploid spores with mutations affecting base/base MMR (msh6Δ) and/or Polε proofreading (pol2-4) or Polδ proofreading (pol3-01). The spores were freshly dissected from sporulated diploid yeast strains that were heterozygous for msh6Δ and either pol3-01 (AH2601) or pol2-4 (AH2801). We reasoned that the low mutation rate of these diploid strains would ensure that the haploid spores began life with a modest mutation burden. Any background mutations with lifespan effects would be expected to segregate equally to each genotype, controlling for any adverse effects on lifespan. Spores defective for Polδ proofreading (pol3-01) exhibited a 30% decrease in median lifespan (median RLS = 18, n = 74) compared with WT (median RLS = 24, n = 161) (Fig. 1A; Wilcoxon rank sum test, P < 0.001). In the absence of Msh6, Polδ proofreading deficiency (pol3-01 msh6Δ) reduced median lifespan by 65% (median RLS = 9, n = 78) (Wilcoxon rank sum test, P < 0.001). Neither Msh6- nor Polε-proofreading–defective spores (pol2-4) were short-lived relative to WT (Fig. 1A). However, pol2-4 msh6Δ double-mutant spores were as short-lived as pol3-01 spores (median RLS = 17, n = 83) (Wilcoxon rank sum test, P < 0.001). Thus, mutator alleles negatively impact RLS, with the greatest deficits occurring when polymerase proofreading and MMR are simultaneously impaired.
Mutation rates in the strains used for the RLS analyses conformed to the reported values from previous studies (50) (Fig. 1B). Lack of Polε proofreading (pol2-4) or Msh6 elevated mutation rates two- to sixfold, while lack of Polδ proofreading (pol3-01) increased mutation rates 32-fold. Combined defects to Msh6 and Polε proofreading (pol2-4 msh6Δ) increased mutation rates synergistically to 200-fold greater than WT. A mutation rate for pol3-01 msh6Δ haploids could not be determined since they failed to form macroscopic colonies due to error-induced extinction. Mutation rates for these spores can be estimated at ∼1 × 10−3 CanR mutants per cell division based on our previous work on the haploid error threshold (50) and the measured mutation rates of pol3-01 msh2Δ diploid strains (51). A linear model of the relationship between mutation rate and median RLS reveals a strong negative correlation (Fig. 1C) (adjusted R2 = 0.81, df = 4, P = 0.009), suggesting that random mutagenesis drives reduced lifespan in these spores.
We previously found that pol2-4 msh6Δ cells accumulate an average of 2.6 mutations per division (19). Given a median RLS of 17 divisions, pol2-4 msh6Δ mother cells would have an average of ∼44 mutations at the time of death. In contrast, cells with either msh6Δ or pol2-4 alone, which display mutation rates ∼100-fold lower than pol2-4 msh6Δ haploids, would be expected to accumulate an average of less than one mutation over a lifetime (0.026 mutations per division × median RLS of 25). Assuming 0.5 mutations per division and an RLS of 18 divisions, pol3-01 cells would die with an average of nine mutations, while pol3-01 msh6Δ cells may have as many as 234 mutations (26 mutations per division × median RLS of 9). Thus, mutator spores with a reduced lifespan are those with the greatest opportunity to accumulate mutations within an average cellular lifetime.
Decreased RLS Strongly Correlates with Increased Mutation Rate and Mutation Burden in Diploid Mutator Yeast.
Since diploidy likely buffers human cells from the consequences of mutagenesis during cancer and aging, we sought to model the influence of mutation burden on diploid yeast cells. We created various diploid mutator strains by mating haploid spores from our mutator parent strains. To facilitate mutation rate measurements in diploid cells, the AH2601 and AH2801 parental strains had been engineered to be hemizygous for the CAN1 gene (CAN1::natMX/can1Δ::HIS3). We screened all diploid strains for this hemizygous configuration, which moderately extends lifespan (SI Appendix, section S1). Single colonies were isolated from frozen stocks of these diploid strains and patched onto fresh plates to isolate mother cells for 20-cell aging cohorts. Two to six independent cohorts were used for lifespan measurements of each strain (SI Appendix, Fig. S1A). We determined mutation rates by fluctuation analyses (SI Appendix, Fig. S1B) and mutation burden by whole-genome sequencing (SI Appendix, Fig. S1C). The mutator phenotypes elevated the rate of SNVs as well as indel mutations. We readily detected known SVs engineered into the strains, including both deletions and insertions. We also observed a known duplication in S288C (the genetic background of our strains), as well as a duplication of a nonessential gene specific to our strain background. We identified no other SVs. Thus, the lifespan phenotypes described below appear to be determined by point mutations. More information on compiled survival, mutation rate, and mutation burden datasets for diploid mutators is provided in Dataset S1. Detailed sequencing results and mutations observed in all strains, including SVs, are provided in Dataset S2.
We observed variability in RLS between diploid strains with low mutation rates and burdens that could be explained by strain-specific SNVs (SI Appendix, section S2 and Fig. S1). These observations suggest that relatively small numbers of random heterozygous mutations, acting alone or in combination, can reduce diploid yeast lifespan through specific mechanisms related to the affected genes. The effect of mutation burden on diploid longevity grew more consistent and pronounced in strains with stronger mutator phenotypes. The six pol2-4/pol2-4 msh6Δ/msh6Δ strains displayed a 24–34% reduction in median lifespan (P < 0.001, Wilcoxon rank sum test, Bonferroni-corrected). AH2601-derived strains with similar mutation rates and burdens as the pol2-4/pol2-4 msh6Δ/msh6Δ strains displayed similar reductions in lifespan (pol3-01/pol3-01, pol3-01/pol3-01 MSH6/msh6Δ, and POL3/pol3-01 msh6Δ/msh6Δ). A further 10-fold increase in mutation burden produced a dramatically shortened RLS in every pol3-01/pol3-01 msh6Δ/msh6Δ strain (combined median RLS = 6; n = 240; P < 0.001, Wilcoxon rank sum test), suggesting that lifespan declines precipitously when mutation burden exceeds a certain threshold.
We performed linear modeling to characterize the relationship between mutagenesis and reduced lifespan in diploid mutator strains. First, we compared the relationship between increased mutation rate and mutation burden after pooling the data on the basis of genotype. A linear relationship exists between increased mutation rate and the average burden of SNVs (Fig. 2A; adjusted R2 = 0.9, df = 13, P < 0.001) and indels (SI Appendix, Fig. S4A, adjusted R2 = 0.8, df = 13, P < 0.001). Next, we compared mutator median RLS and mutation rate (Fig. 2B; pooled lifespan curves are shown in SI Appendix, Figs. S2 and S3). In this and subsequent correlation studies, pol3-01/pol3-01 msh6Δ/msh6Δ strains were identified as outliers by analyzing residuals and were not included in the regression models. Median RLS decreased monotonically as log-transformed mutation rates increased across three orders of magnitude (adjusted R2 = 0.82, df = 12, P < 0.001). Finally, we compared median RLS with the burden of SNVs and indels (Fig. 2C and SI Appendix, Fig. S4C). To improve resolution, instead of pooling by genotype, we analyzed all individual strains where RLS and mutation burden had been quantified. Again, a negative relationship was observed between increased mutation burden and decreased RLS (SNVs: adjusted R2 = 0.55, df = 45, P < 0.001; indels: adjusted R2 = 0.43, df = 45, P < 0.001).
Genetic Anticipation in the Lifespan Reduction of Mutator Diploid Cells.
Most lifespan cohorts experience a period of low early-life mortality before the onset of age-associated death. In contrast, the pol3-01/pol3-01 msh6Δ/msh6Δ cells began to die at a constant rate from the very first division. Early death in pol3-01/pol3-01 msh6Δ/msh6Δ cells may simply derive from the combined effect of many deleterious mutations. Alternatively, the combination of pol3-01 and msh6Δ alleles could conceivably increase mortality independent of the mutator phenotype. Consistent with this second possibility, we observed that all POL3/pol3-01 MSH6/msh6Δ strains derived from mating lived shorter than other strains with similar mutation burdens (SI Appendix, Fig. S1). If reduced lifespan in pol3-01/pol3-01 msh6Δ/msh6Δ diploid strains indeed depends on mutation burden, then this phenotype should display a form of genetic anticipation: the longer a strong mutator lineage exists, the more mutations it will accumulate and the shorter lived it should become.
To test for genetic anticipation, we measured lifespans of pol3-01/pol3-01 msh6Δ/msh6Δ diploid cells freshly isolated from zygotes (Fig. 3). We dissected tetrads from the parental strain, AH2601, on media that selected for growth of double-mutant haploid spores. We randomly mated dividing cells to obtain zygotes and then used their first daughters to establish a cohort of mother cells (LS_d1; Dataset S1). The genomes in these cells underwent between four to six rounds of replication (haploid and diploid divisions, combined) with the full pol3-01 msh6Δ mutator phenotype before lifespan analysis. We compared the lifespan of this initial cohort with the lifespans of its propagated descendants (LS_d3 and LS_d4 series; Dataset S1). Since WT cells could not grow on this medium, as controls, we measured lifespans of cells from AH2601, as well as from one of the previously isolated pol3-01/pol3-01 msh6Δ/msh6Δ strains (yML421).
AH2601 displayed a similar lifespan (median RLS = 27, n = 60) as POL3/pol3-01 MSH6/msh6Δ strains derived from mating (median RLS = 30, n = 720), which suggests that there are mutagenesis-independent effects of the POL3/pol3-01 MSH6/msh6Δ genotype on cellular lifespan (Fig. 3 and SI Appendix, section S3). Freshly isolated pol3-01/pol3-01 msh6Δ/msh6Δ cells appear to be shorter lived (median RLS = 22, n = 21) than AH2601, especially over the last quartile, but this was not significant after a multiple-testing correction (P = 0.075, Wilcoxon rank sum test, Bonferroni-corrected). However, the freshly isolated pol3-01/pol3-01 msh6Δ/msh6Δ cells lived substantially longer than the propagated strains (d3 and d4: median RLS = 8, n = 240 for both lines), as well as the previously isolated pol3-01/pol3-01 msh6Δ/msh6Δ strain (SI Appendix, Fig. S1) (median RLS = 6; n = 60; P < 0.001, Wilcoxon rank sum test, Bonferroni-corrected). The mutation burden of clones derived from representative mother cells from the d3 and d4 cohorts averaged 176 SNVs per megabase and one indel per megabase, which were higher than the averages of 23 clonal SNVs per megabase and 0.2 indel per megabase present in the freshly isolated mother cells near the beginning of their lifespan, as estimated by sequencing DNA extracted from pooled cells from the d3 patches (Dataset S2). Thus, reduced RLS due to strong mutator phenotypes shows genetic anticipation, further establishing the ability of mutation burden to impact cellular lifespan.
Effect of Random Mutation Burden on RLS and Doubling Time in the Absence of an Active Mutator Phenotype.
To separate the effects of an active mutator phenotype from accumulated mutation burden, we utilized a transient hypermutation system to generate diploid yeast MA lines with varying numbers of fixed random mutations. To do this, we integrated a powerful mutator allele (pol3-01,L612M) into diploid yeast in the heterozygous state (51), and then identified mitotic recombinants that had spontaneously reverted back to a POL3/POL3 genotype. In all, we obtained 11 transiently mutagenized lines, each derived from an independently isolated POL3/pol3-01,L612M diploid strain. All strains exhibited WT mutation rates following reversion (Dataset S1), suggesting that the accumulated mutation burden had not introduced additional mutator alleles.
Whole-genome sequencing of the MA lines revealed a wide range of mutation burdens from 0.4 to 100 SNVs per megabase and from zero to 6.3 indels per megabase (Fig. 4A and SI Appendix, Fig. S4 and Dataset S2). Four of the five least mutagenized MA lines (MA1, MA2, MA4, and MA5) retained a normal lifespan and had mutation burdens ranging between 0.9 and 31.1 SNVs per megabase, respectively). The lone exception (MA3) had 12.4 SNVs per megabase (one indel per megabase) and a 43% reduction in median RLS (P < 0.001, Wilcoxon rank sum test, Bonferroni-corrected for multiple tests). All strains with a mutation burden greater than 31.6 SNVs per megabase showed at least a 29% decrease in lifespan (Fig. 4B and Dataset S1; P < 0.001 for all strains, Wilcoxon rank sum test, Bonferroni-corrected). The most severely affected strain (MA7), with a 63% decrease in median lifespan, had a mutation burden of 39.7 SNVs per megabase (2.4 indels per megabase). Thus, once mutation burdens exceeded a certain threshold, they consistently produced a diminished lifespan in the absence of an ongoing mutator phenotype.
In addition to survival, we sought to understand how MA impacted overall population health. One way to assess health in a microbial culture is to quantify peak doubling time (minutes per cell division) in an outgrowing population (65). We measured growth of the MA lines with a Bioscreen C MBR reader/shaker/incubator and calculated doubling time using YODA (63). Linear modeling revealed a strong negative correlation between mutation burden and population health as measured by doubling time (adjusted R2 = 0.77, df = 10, P < 0.001; Fig. 4C and Dataset S1).
The Role of Accumulated Mutation Burden in Driving Diminished Lifespan and Increased Doubling Time in Mutagenic Diploid Yeast.
We reexamined the relationship between accumulated mutation burden and diminished lifespan in all of our diploid strains (Fig. 5A) by combining data from the genetic anticipation and MA studies (Figs. 3 and 4) with our diploid mutator dataset (Fig. 2). At the lowest mutation burdens (0.04–0.4 mutation per megabase), median RLS was unchanged. Intermediate mutation burdens (0.4–35 mutations per megabase) were variable with regard to their impact on lifespan. This variability suggests that individual mutations or discrete epistatic interactions between mutations drive diminished survival in intermediately mutagenized populations. Above 35 mutations per megabase, lifespan is consistently and severely diminished (at least a 42% reduction in median RLS). Interestingly, this is true in both transiently mutagenized diploids and active mutator strains, suggesting that all strains carry sufficient numbers of these lifespan-limiting mutations.
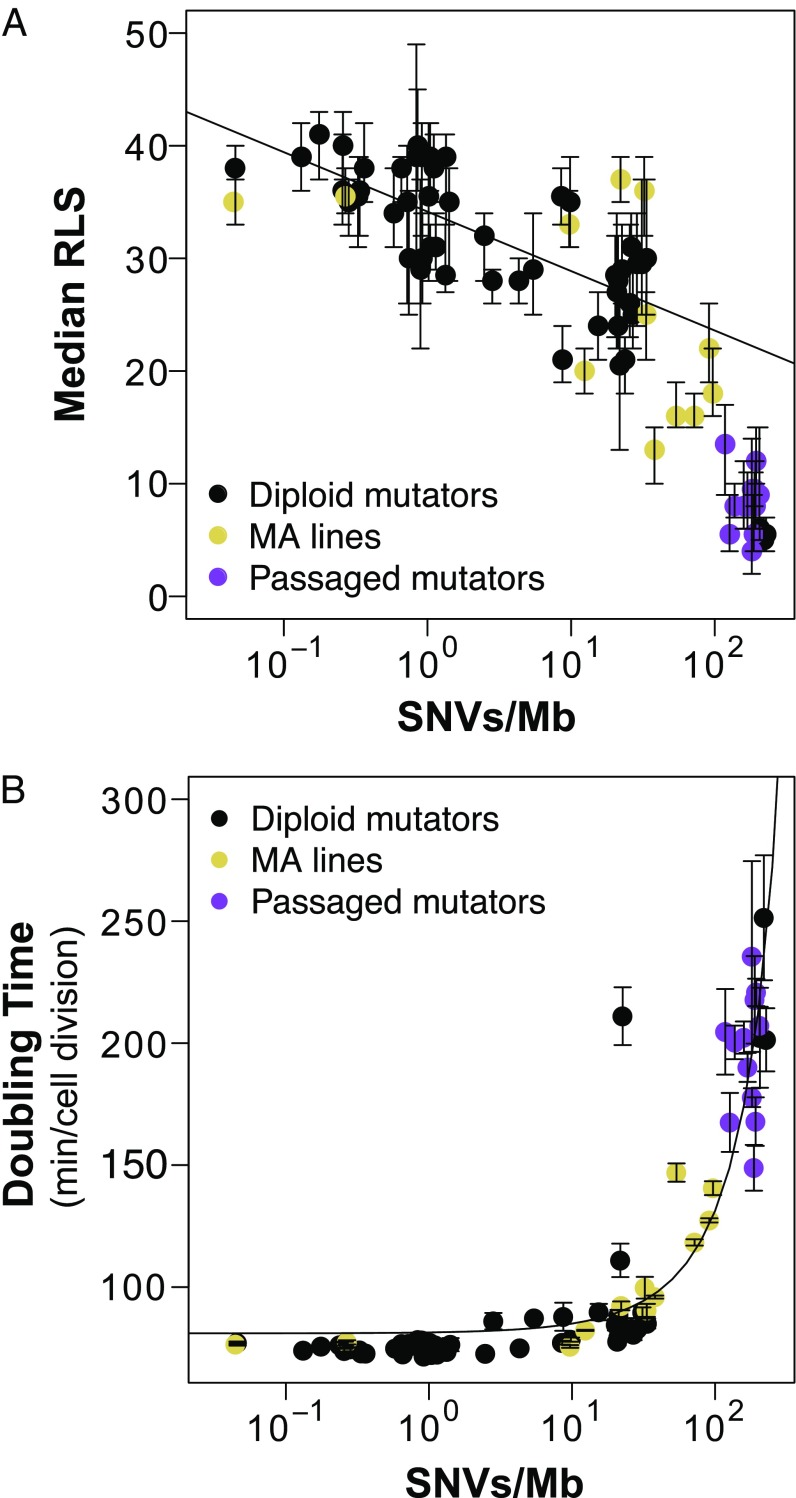
Fig. 5.
Role of accumulated mutation burden in driving diminished lifespan and increased doubling time in diploid yeast. (A) Scatterplot showing the relationship between mutation burden [SNVs per megabase (Mb)] and median RLS in diploid mutator yeast (black; Fig. 2), passaged pol3-01/pol3-01 msh6Δ/msh6Δ strong mutators (purple; Fig. 3), and transiently mutagenized MA lines (gold; Fig. 4). Error bars, 95% confidence intervals. (B) Correlation between log-transformed mutation burden and peak doubling time.
To better characterize how mutation burden impacted the health of our yeast populations, we expanded our analysis of peak doubling time to include all diploid strains Using untransformed data, linear modeling revealed a strong correlation between increased mutation burden and doubling time (adjusted R2 = 0.85, P < 0.001). Interpretation of this correlation is challenged, however, by the large number of data points from strains with very low mutation burdens. We log-transformed to normalize mutation burden data (Fig. 5B) and found that doubling time was consistent across mutation burdens for three orders of magnitude (0.04–30 mutations per megabase). Above 30 mutations per megabase, doubling time increased exponentially. As with median RLS, transiently mutagenized populations behaved similar to populations with an active mutator phenotype. Strikingly, the threshold for increased doubling time corresponds to the level at which mutation burden severely impacts RLS. This shows a clear link between decreased cellular lifespan and degraded population health resulting from elevated mutation burden.
Discussion
Historically, the contribution of MA to aging has been difficult to assess. Important progress is being made. Next-generation sequencing of clonally expanded cancers and HPSCs cells (6–17), aged single cells (66, 67), and complex cellular populations using highly accurate methodologies that dramatically reduce artifactual errors (68) now offers a window into how mutations accumulate in individual aged cells. However, the field lacks clear functional tests to distinguish the effects of mutation burden from those of other types of damage that accrue over a lifetime. To address this issue, we assessed how induced mutation burden changes the aging trajectory of young yeast cells. Our approach was inspired, in part, by the biology of mutator-driven cancer cells, which accumulate large numbers of unselected passenger mutations during tumorigenesis. If random MA accelerates cellular aging, then the highly proliferative nature of cancer may hide an underlying frailty of individual cancer cells. In what follows, we discuss the insights gained from our studies in yeast and their implications for aging and cancer.
Prior research showed that point mutations accumulate too slowly (less than one per RLS) to play a causative role in normal aging of haploid yeast (35). A mutator strain with a 10-fold elevation in mutation rate driven by oxidative damage also failed to decrease RLS. Our results confirm and expand upon this finding by showing that mutation rates must be three- to fivefold greater than those previously tested before consistent lifespan reduction occurs in haploid cells. This corresponds to the mutation rate at which appreciable MA occurs during the life of the mother cell. Single-gene deletions reveal that one-sixth of all genes are essential for macroscopic growth (69). Since an estimated 15–20% of all mutations within an average gene compromise function (51), it is not surprising that sufficient levels of mutagenesis during the lifespan of a haploid cell can diminish their longevity (Fig. 1).
The presence of two copies of every gene theoretically buffers diploid cells from the effects of MA. Nevertheless, we find evidence indicating that a wide range of mutation burdens can impact diploid longevity. First, we show that diploid strains with modest mutation rates and low mutation burdens exhibit variation in lifespan that can be attributed to random heterozygous mutations (SI Appendix, Fig. S1A). Second, we show that lifespan declines in a log-linear fashion as mutation burden increases (Fig. 2). Third, we show that the loss of longevity due to mutator phenotypes displays genetic anticipation, with later generations having shorter lifespans than earlier ones (Fig. 3). Finally, we find that MA lines that lack an active mutator phenotype nevertheless exhibit reductions in lifespan that correlate with mutation burden (Fig. 4). Combined analysis of all diploid strains tested shows the log-linear relationship between mutation burden and lifespan holds until a threshold of around 30 mutations per megabase, whereupon cellular lifespan diminishes more rapidly (Fig. 5).
In our experiments, the effect of MA on longevity likely depends on several factors, including the rate and severity of lifespan-shortening mutations, the power of purifying selection to remove these alleles from the population, and the exponential rise in epistatic interactions between genes. By one measure, lifespan-reducing mutations appear surprisingly common, as multiple lines with mutation burdens of fewer than two mutations per megabase displayed a 25% reduction in lifespan. While some mutations may accelerate a general process that contributes to normal aging, most mutations seem likely to increase mortality by specific mechanisms related to the affected genes. Strains with 10- to 30-fold the mutation burden show decreases in lifespan from 10–40%. If lifespan-reducing mutations are indeed common, why is there not a more dramatic effect of increasing mutation burden by an order of magnitude? There may be stochasticity in the number or severity of lifespan-limiting mutations between strains with similar mutation burdens. In addition, since many of the mutations in these strains arose during an outgrowth period before lifespan analysis, purifying selection may have partially sanitized the observed mutation burden by eliminating the most deleterious alleles. Consistent with purifying selection, few actively mutating strains show a dramatic increase in doubling time, except those with the highest mutation rates (SI Appendix, Fig. S1D). In strains with the highest mutation rates, scores of mutations become fixed in every cell in every division, which overwhelms the ability of purifying selection to remove deleterious alleles from the population. Epistatic interactions may also be influencing the effects of mutation burden in these lines. For instance, once cells carry one lifespan-reducing mutation in a pathway, they may absorb additional mutations in that pathway with limited phenotypic consequences due to positive epistasis. In addition, short-lived phenotypes may be offset by suppressor mutations, as well as by general lifespan-extending mutations in genes such as TOR1. However, once cells have one beneficial mutation in a pathway, due to “diminishing returns epistasis,” they gain little from additional mutations affecting the same pathway (70, 71). Eventually, the rise in deleterious biallelic mutations, dominant-negative mutations (Dataset S2), and negative epistatic interactions may overcome the beneficial effect of any lifespan-extending mutations. In the strongest diploid mutators, the high load of recessive lethal mutations may cause these cells to resemble haploids in their susceptibility to new inactivating mutations in essential genes.
In multicellular organisms, the phenotypic consequences of accumulated mutation burden are realized in a complex milieu of multiple lineages with variable mutation burdens. The level of mutation burden will depend on the proliferative nature of the tissue, as well as exposure to DNA damage. Recent work using data from the Cancer Genome Atlas project revealed a sixfold increase (0.37 vs. 2.21 mutations per megabase in samples from individuals aged under 20 y and over 80 y, respectively) in somatic mutation frequency with age (7). Our observations suggest that the mutation burdens of some human stem cells could plausibly restrict their longevity or the functionality of their terminally differentiated descendants. Phenotypic variability among different stem cell lineages is expected. The consequences of this variability for tissue function remain uncertain. Cellular dysfunction may lead to cell death, quiescence, or even stable senescent cells implicated in tissue aging (72). In the context of tissue homeostasis, reduced replicative capacity of one stem cell places a demand on other stem cell lineages to expand. Under these dynamic conditions, dividing cells with new mutations are subject to the evolutionary forces of selection and genetic drift. Thus, seemingly normal-looking tissues contain clonal expansions in which the phenotypic consequences of particular mutation burdens may predominate (8). In the context of cancer, sustained evolutionary pressure for adaptive mutations indirectly favors cells with higher mutation burdens. Exome sequencing from tumor populations shows that mutation burdens range from fewer than one to over 100 mutations per megabase, depending on cancer type (16, 73). Based on our results in yeast, many tumors may carry mutation loads that impact the longevity or replicative capacity of their cells. Our findings suggest that cells from the most extreme mutator-driven tumors, with mutation burdens greater than 200 mutations per megabase, may possess a profoundly shortened lifespan. Of course, cancer cells may adapt to high mutational stress in ways that preserve their replicative potential. However, if mutation burden does indeed restrict the lifespan of cancer cells, it would be an important refinement in our understanding of this disease. While cancer appears “immortal” at the population level, individual cancer cells may have diminished replicative potential.
The key challenge in developing cancer therapies is to identify targetable differences between cancer and normal cells. Tumor mutation burden is already being used as a prognostic factor for cancers most likely to respond to immunotherapy (74). It is important to understand what, if any, general sensitivities result from random deleterious mutation burden (75). Of course, treatments that further increase mutagenesis may synergize with the existing mutation burden to diminish lifespan, although this carries the potential for collateral damage to normal cells. For mutator-driven cancers, one solution may be to develop drugs that exploit the underlying DNA repair deficiency to preferentially mutagenize tumor cells (76). Another area to explore is whether mutagenized cells are sensitized to conditions that contribute to cellular aging. For instance, increased numbers of mutant proteins may tax proteostasis, and drugs that further exacerbate proteostatic stress may effectively discriminate between normal and mutagenized cells (75). Overall, understanding how accumulated mutation burden influences cell physiology may provide novel tools to be used in the fight against cancer.
Supplementary Material
Acknowledgments
We acknowledge Dr. Brad Preston for helpful conversations with A.J.H. at the inception of this work. This work was supported by the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging Invertebrate Longevity and Healthspan Core (Grant P30AG013280), and by Grants R01GM118854, R03AG037081, and P30 AG013280-18 (all to A.J.H.) from the National Institute of General Medical Sciences (NIGMS) and the National Institute on Aging (NIA). M.B.L. was supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Study, an NIH Cellular and Molecular Biology Training Grant (NIH T32GM007270), and the University of Washington Graduate Opportunities and Minority Achievement Program Bank of America Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIA, NIGMS, NIH, or HHMI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815966116/-/DCSupplemental.
References
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijg J, Montagna C. Genome instability and aging: Cause or effect? Transl Med Aging. 2017;1:5–11. [Google Scholar]
- 3.Szilard L. On the nature of the aging process. Proc Natl Acad Sci USA. 1959;45:30–45. doi: 10.1073/pnas.45.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Failla G. The aging process and cancerogenesis. Ann N Y Acad Sci. 1958;71:1124–1140. doi: 10.1111/j.1749-6632.1958.tb46828.x. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2011;133:118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milholland B, Auton A, Suh Y, Vijg J. Age-related somatic mutations in the cancer genome. Oncotarget. 2015;6:24627–24635. doi: 10.18632/oncotarget.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risques RA, Kennedy SR. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 2018;14:e1007108. doi: 10.1371/journal.pgen.1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podolskiy DI, Lobanov AV, Kryukov GV, Gladyshev VN. Analysis of cancer genomes reveals basic features of human aging and its role in cancer development. Nat Commun. 2016;7:12157. doi: 10.1038/ncomms12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy SR, et al. Volatility of mutator phenotypes at single cell resolution. PLoS Genet. 2015;11:e1005151. doi: 10.1371/journal.pgen.1005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 21.Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen KL, Crane MM, Kaeberlein M. Microfluidic technologies for yeast replicative lifespan studies. Mech Ageing Dev. 2017;161:262–269. doi: 10.1016/j.mad.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick MA, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith ED, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 27.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 28.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruegel U, et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffen KK, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 32.Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002015. doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinclair DA, Guarente L. Extrachromosomal rDNA circles–A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 34.Hu Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaya A, Lobanov AV, Gladyshev VN. Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae. Aging Cell. 2015;14:366–371. doi: 10.1111/acel.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 38.Loeb LA. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev. 2013;32:353–361. doi: 10.1007/s10555-013-9426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shlien A, et al. Biallelic Mismatch Repair Deficiency Consortium Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 42.Palles C, et al. CORGI Consortium; WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144, and erratum (2013) 45:713. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch HT, et al. Review of the Lynch syndrome: History, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayner E, et al. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 45.Tomkova M, McClellan M, Kriaucionis S, Schuster-Böckler B. DNA replication and associated repair pathways are involved in the mutagenesis of methylated cytosine. DNA Repair (Amst) 2018;62:1–7. doi: 10.1016/j.dnarep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Mertz TM, Baranovskiy AG, Wang J, Tahirov TH, Shcherbakova PV. Nucleotide selectivity defect and mutator phenotype conferred by a colon cancer-associated DNA polymerase δ mutation in human cells. Oncogene. 2017;36:4427–4433. doi: 10.1038/onc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: Defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giraud A, Radman M, Matic I, Taddei F. The rise and fall of mutator bacteria. Curr Opin Microbiol. 2001;4:582–585. doi: 10.1016/s1369-5274(00)00254-x. [DOI] [PubMed] [Google Scholar]
- 50.Herr AJ, et al. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7:e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herr AJ, Kennedy SR, Knowels GM, Schultz EM, Preston BD. DNA replication error-induced extinction of diploid yeast. Genetics. 2014;196:677–691. doi: 10.1534/genetics.113.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 53.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 54.Lundblad V, Struhl K. Yeast. Wiley Interscience; New York: 2010. [Google Scholar]
- 55.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Q. A new practical guide to the Luria-Delbrück protocol. Mutat Res. 2015;781:7–13. doi: 10.1016/j.mrfmmm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Steffen KK, Kennedy BK, Kaeberlein M. 2009. Measuring replicative life span in the budding yeast. J Vis Exp :e1209.
- 59.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fox J. The R commander: A basic-statistics graphical user interface to R. J Stat Softw. 2005;14:42. [Google Scholar]
- 61.Fox J, Carvalho MS. The RcmdrPlugin.survival package: Extending the R commander interface to survival analysis. J Stat Softw. 2012;49:1–32. [Google Scholar]
- 62.Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. J Gerontol A Biol Sci Med Sci. 2008;63:113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- 63.Olsen B, Murakami CJ, Kaeberlein M. YODA: Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MB, et al. A system to identify inhibitors of mTOR signaling using high-resolution growth analysis in Saccharomyces cerevisiae. Geroscience. 2017;39:419–428. doi: 10.1007/s11357-017-9988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lodato MA, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359:555–559. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong X, et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giaever G, Nislow C. The yeast deletion collection: A decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wünsche A, et al. Diminishing-returns epistasis decreases adaptability along an evolutionary trajectory. Nat Ecol Evol. 2017;1:61. doi: 10.1038/s41559-016-0061. [DOI] [PubMed] [Google Scholar]
- 71.Chou HH, Chiu HC, Delaney NF, Segrè D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–1192. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 74.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci USA. 2013;110:2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams LN, et al. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc Natl Acad Sci USA. 2015;112:E2457–E2466. doi: 10.1073/pnas.1422948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.